Abstract
Cell diversity in the central nervous system (CNS) is achieved by a highly regulated process of differentiation from multipotential neural stem cells. The spatial specificity and timing control of neural differentiation is achieved by the interplay between various genetic and epigenetic regulators. Oligodendrocytes, the myelinating cell in the CNS, play an important role in brain development and neuronal function. At present, multiple signaling pathways have been implicated in regulating in oligodendrocyte differentiation, however, the integration of these pathways with transcriptional and posttranscriptional regulatory networks are not fully understood. This review will focus on exploiting epigenetic mechanisms underlying oligodendrocyte development including chromatin remodeling by histone deacetylases and gene silencing by non-coding RNAs (e.g., microRNA), and attempts to summarize the recent advance as to the genetic and epigenetic interaction in controlling oligodendroglial differentiation and myelination.
Keywords: HDAC, HAT, miRNA, non-coding RNA, oligodendrocyte, myelination, chromatin, transcriptional repressors
Introduction
The formation and maintenance of electrically active neural circuitry depends on functional integration between neurons and supporting glial cells. Oligodendrocytes are the major glial cell in the CNS. By producing a multilayered myelin sheath wrapping neighboring axons, they provide nerve insulation and facilitate saltatory conduction of nerve impulse by invoking the myelin-dependent ion channels distribution.1 The failure of oligodendrocyte remyelination leads to a variety of acquired and inherited neurological disorders such as multiple sclerosis (MS) and leukodystrophies,2,3 and even a major psychiatric disease like schizophrenia.4 The current prognosis for individuals with MS or any other demyelinating disease is gloomy, as an effective treatment allowing for remyelination to occur has not been discovered. Understanding the mechanisms underlying oligodendrocyte development and myelination would provide effective therapeutic avenues for myelin repair after injuries.5
What is the source of oligodendrocytes? Oligodendrocyte precursor cells (OPCs) are specified from distinct regions of the embryonic ventricular zone under the influence of diverse extracellular signaling molecules such as ventral expressing sonic hedgehog and dorsal expressing bone morphogenetic proteins (BMP) in developing neural tube.6,7 Although the fate of regionally distinct oligodendrocytes is not identical, at present, no functional discrepancy is observed among oligodendrocytes derived from different regions. Oligodendrocyte differentiation derived from different CNS regions could be achieved using distinct combinatorial arrangement of transcriptional regulators. In developing brain, ventrally-derived early oligodendrocyte precursors appear to compete with their dorsal counterparts.8 The ventral-most lineage is eliminated during postnatal life,8 however, dorsal precursors derived from cortical progenitors in forebrain prevail at late developmental stages.8,9 In contrast, a different pattern is found in the spinal cord where competition is won by the primary wave derived from the ventral region during early embryogenesis. Only a subset of oligodendrocytes is generated from progenitor cells in the dorsal spinal cord at later embryonic stages.7,10,11 Collectively, oligodendrocytes can be derived from several distinct regional origins in a temporally controlled manner. Oligodendrocyte specification and differentiation/maturation in different CNS regions are likely controlled by distinct genetic and epigenetic programs.
Oligodendrocyte maturation is characterized by successive stages of lineage progression from OPCs to immature premyelinating oligodendrocytes and finally to mature myelinating oligodendrocytes.12 The precise mechanisms underlying the progressive stages of oligodendrocyte differentiation are not fully understood at present. Oligodendrocyte development is controlled by dynamic interaction of a series of intrinsic and extrinsic regulators in a spatiotemporally specific manner including negative regulatory pathways such as Notch/Hes, Wnt/β-catenin/TCF and BMP/ID pathways, and positive regulators that enhance myelin gene expression such as bHLH transcription factors (e.g., Olig1/2 and Ascl1/Mash1), HMG-domain transcription factors (e.g., Sox10), Zinc-finger transcription factors (e.g., YY1, Zfp488 and Myt1), NDT80 domain transcription factor (e.g., MRF), homeodomain transcription factors: (e.g., Nkx2.2 and Nkx6.2) and nuclear hormone receptors [e.g., thyroid hormone receptors (THRs) and retinoid X receptors (RXRs)].13–15 It is believed that the balance of these positive and negative factors controls the timing of oligodendrocyte lineage progression.
Recent evidence indicates that epigenetic factors, which regulate gene expression without changing underlying primary DNA sequence such as modulation of the chromatin structure, play a critical regulatory role for neural development including oligodendrocyte formation. Major epigenetic mechanisms includes covalent modifications of nucleosomal histones by histone deacetylases (HDACs) and histone acetyltransferases (HATs), ATP-dependent SWI/SNF mediated chromatin remodeling, DNA methylation and non-coding RNAs such as microRNA (miRNA) mediated silencing.
This review summarizes recent findings of epigenetic mechanisms by HDACs and miRNAs in regulating oligodendrocyte differentiation and myelination, with a focus on how genetic and epigenetic regulators integrate to control the progression of oligodendrocyte differentiation.
Genetic and Epigenetic Control of Oligodendrocyte Development by Histone Modification Enzymes
One of the best-characterized chromatin remodeling is nucleosomal histone modification by acetylation on lysine residues, which is dynamically mediated by HATs and HDACs. HATs add acetyl groups and favor a transcriptionally competent chromatin structure while HDACs remove acetyl groups from lysine tails, resulting in chromatin compaction. HDACs have been classified into four broad classes: ubiquitously expressed Class I (HDAC-1, -2, -3 and -8), tissue-specific Class II (HDAC-4, -5, -6, -7, -9 and -10), NAD-dependent Class III (SIRT1-7) and Class IV (HDAC11), which comprise a highly conserved enzyme family.16–19 HATs or HDACs can be recruited as components of multiprotein transcriptional complexes on the genome and serve as epigenetic co-activators or co-repressors to either facilitate or limit gene expression.
Although the role of HATs in oligodendrocyte development remains elusive, a series of experiments have demonstrated that HDAC activity is essential for OPC differentiation and myelin gene expression. Oligodendrocyte differentiation is inhibited by pharmacological HDAC inhibitors (e.g., valproic acid and trichostatin A) in OPC culture, during animal development20,21 and remyelination.22 Effects of histone deacetylation appear to be temporally specific. HDAC activity is critical for early stage of oligodendrocyte precursor differentiation, but is not required for myelin gene expression after myelination onset.21
RNAi-mediated gene silencing in cultured primary OPCs indicated that HDAC1 and HDAC2 but not other class I HDAC members are critical for their differentiation.22 These molecules were also detected on the promoter of differentiation inhibitors during the remyelination phase of cuprizone-induced demyelination in young mice that efficiently repair, but not in old mice that are unable to remyelinate,22 thereby suggesting an important role for OPC differentiation also in the adult brain.
Recently, our studies with oligodendrocyte lineage-specific HDAC knockouts demonstrated the importance of histone deacetylation mediated by HDAC1 and HDAC2 in vivo.23 HDAC1:HDAC2 double mutant mice developed severe deficits in oligodendrocyte differentiation and myelination deficits leading to tremor and postnatal lethality,23 although HDAC1 or HDAC2 single knockouts did not yield obvious developmental defects, pointing to the crucial redundant roles of HDAC1 and HDAC2 during oligodendrocyte development. HDAC1/2 regulate oligodendrocyte differentiation, at least in part, by functionally counteracting the activity of β-catenin-TCF complexes and therefore inhibiting Wnt signaling.23
Unlike class I HDACs, the role of cell type-specific class II HDACs in oligodendrocyte development remains unclear. They are predominantly in the cytosol of the cells where they can exert distinctive functions. Another family of cytosolic enzymes with deacetylase activity is the class III HDAC (NAD+)-dependent deacetylases, also called “sirtuins.” Sirtuin 2 (SIRT2) is highly enriched in oligodendrocytes and localized to the outer and juxtanodal loops in the myelin sheath.24,25 Interestingly, tetraspan proteolipid protein PLP is required for SIRT2 transport into the myelin compartment.25 The main substrate for the SIRT2 deacetylase is identified as acetylated α-tubulins, but not histones H3 or H4.24 RNAi-mediated knockdown study reveals that SIRT2 is required for oligodendrocyte process arborization.24 Finally, recent studies have reported an important role for the class IV family member HDAC11, in myelination. HDAC11 expression is increased in maturing oligodendrocytes26 and HDAC11 is critical for the removal of acetyl groups from lysine K9 and K14 during oligodendrocyte maturation. HDAC11 knockdown in an oligodendrocyte cell line reduced myelin gene expression and arrested oligodendrocyte maturation.26
HDACs can modulate gene expression in several ways. First, they initiate repression by modulating the acetylation state of nucleosomal histones or transcriptional regulators. Second, they can physically bind to transcriptional regulators and function as transcriptional co-repressors.27 The function of HDAC-mediated repressive complexes in oligodendrocyte development has been related to inhibit expression of differentiation inhibitors or block the activation of neuronal differentiation genes. For instance, in the mitogen-withdrawal model of OPC differentiation, HDAC1 binds to YY1 and is recruited to promoters of several inhibitors of differentiation, including Sox11, ID4 and TCF7l2 (also known as TCF4), to repress their transcription.21,28
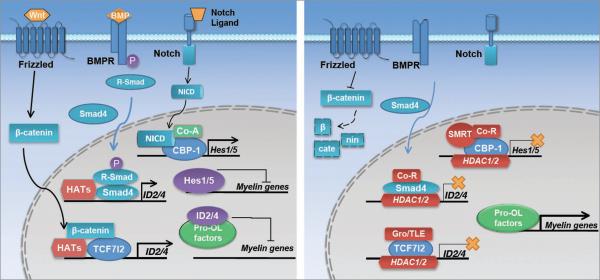
In progenitor cells, with active Notch signaling, Hes5 is likely maintained at high levels and the formation of an activating complex containing the intracellular domain of Notch (NICD) and Cp-binding factor 1 (CBF-1) [also known as recombination signal sequence-binding protein-Jκ (RBP-JΚ)].29,30 HDACs can form inhibitory complexes with Groucho-related co-repressors (GRO/TLE/GRG) and SMRT (Silencing Mediator of Retinoid and Thyroid Hormone Receptor), which in turn displaces the association of NICD and transcription co-activators with CBF-1 on the Hes5 promoter30 (Fig. 1).
Figure 1.
Interplay of genetic and epigenetic factors in controlling oligodendrocyte differentiation. (Left) Upon Wnt, BMP or Notch signaling activation, β-catenin, Smads or NICD induce the transcription of differentiation inhibitors such as ID2, ID4 and Hes1/5 during oligodendrocyte differentiation. ID2/4 sequestrate pro-oligodendrocyte factors, such as Olig1/2, away form myelin gene promoters, whereas Hes1/5 bind to regulatory elements of genes required for oligodendrocyte maturation. (Right) HDAC1/2, together co-repressors such as Gro/TLE and SMRT, compete or displace Wnt, BMP and Notch signaling components, β-catenin, Smads and NICD, respectively, to inhibit expression of differentiation inhibitors, which de-repressing or activating the transcription of myelin genes.
In addition to decreasing Notch signaling, HDACs can inhibit the Wnt signaling pathway. Canonical Wnt signaling was shown to negatively regulate OPC differentiation.31 In progenitor cells, expression of high levels of β-catenin in canonical Wnt signaling allows the formation of a transcriptional activating complex containing β-catenin and TCF7l2, which upregulates oligodendrocyte differentiation inhibitors such as ID2/4. However, repressive complexes with HDAC1/2 and Groucho/TLE/GRG can compete with β-catenin for binding to TCF7l2, thereby interfering with the negative effect of Wnt signaling by decreasing ID2/4 expression.23,32 Thus, HDAC1/2 binding converts TCF7l2 from a transcriptional activator to a repressor, which prevents transcription of Wnt target genes ID2/4, inhibitors of oligodendrocyte differentiation. Similarly, HDAC1/Sin3A co-repressor complex can interact with BMP-dependent Smads to negatively regulate expression of ID2 and ID4,33,34 (Fig. 1). This would explain why HDAC1/2-mediated repressive complexes might counteract Wnt or BMP signaling and allow timely oligodendrocyte myelination and remyelination after injury.32
In the developing mouse spinal cord, TCF7l2 is expressed mainly in premyelinating oligodendrocytes beginning at late embryonic stage; it is later down-regulated in mature myelinating oligodendrocytes.23,35 This is consistent with the possibility that TCF7l2 modulates myelin gene expression and is downregulated due to recruitment of YY1/HDAC1 complexes on its promoter.28 However, if TCF7l2 functions merely as a repressor for oligodendrocyte differentiation, one would intuitively expect that loss of function of TCF7l2 will result in an increase or precocious expression of myelin genes in developing CNS. Gene targeting studies in mice indicated that TCF7l2 is essential for oligodendrocyte differentiation but not for OPC formation. One possible explanation for this paradox is that TCF7l2 plays distinct roles in cells that differ in β-catenin expression. TCF7l2 might be required for early differentiation of β-catenin expressing OPCs during early lineage progression, but not at the later myelination stages, TCF7l2 might have an inhibitory function that is β-catenin independent in oligodendrocyte terminal differentiation. This is consistent with the fact that mutant TCF7l2 lacking the β-catenin binding site retains the ability to inhibit myelin gene expression (He Y and Casaccia P, unpublished data).
HDACs might function to increase chromatin compaction around genes that encode differentiation repressors or activators for other neural lineages such as neuronal differentiation and block their transcription, while providing permissive conditions for oligodendroglial gene expression. How this selectivity on HDAC-mediated gene expression is achieved is currently not known. HDACs might recruit co-repressors such as the non-neuronal expressing repressor REST/NRSF to repress neuronal gene expression, while facilitating oligodendroglial gene expression. In addition, timing control of oligodendrocyte linage progression can be achieved by a sequence of repression events that result in silencing of stage-specific differentiation inhibitors and de-repression of myelin-specific transcription. The stage-specific transcriptional repression is probably attributed to interaction of HDACs with distinct transcription factors and/or regulatory complexes (Fig. 1).
Can HDACs interact directly with other oligodendrocyte lineage specific transcription factors such as Olig1/2 and Sox10 during oligodendrocytes development? How might HDAC activity or expression be regulated during oligodendrocyte development? These questions remain to be addressed in this field. Nonetheless, by modulating the outcome of multiple signaling pathways, HDACs undoubtedly function as convergent points that control oligodendrocyte differentiation and myelination.36
miRNA-Modulated Transcriptional Network in Neural/Glial Cell Differentiation
The discovery of functional noncoding RNAs reveals a novel epigenetic mechanism that occurs at posttranscriptional level. Noncoding RNAs including small and long forms control or fine-tune the output of gene transcription in the genome of complex organisms. Mature miRNAs are a class of small non-coding RNAs with a length of approximately 22 nucleotides. They inhibit gene expression by perfectly or imperfectly base-pairing with 3' untranslated region (3'UTR) of target mRNAs. A short “seed” sequence in 5' miRNA is an important determinant of miRNA targeting.37 To date, several hundred of miRNAs have been identified and each miRNA is predicted to regulate thousands of targeted genes.38,39 In addition, identification of many long non-coding RNAs (ncRNAs) in the brain adds another level of control of transcriptional networks.40
miRNAs are shown to regulate neuro-genesis, neural patterning, axonal path-finding, synaptic plasticity and neuronal degenerative diseases.41 For instance, miR-124 mediates neuronal differentiation and adult neurogenesis.42,43 miRNA-134 regulates the growth of dendritic spines and modulates neuronal plasticity.44 Recently, miRNA-9* and miR-124a are shown to repress the expression of a transcriptional regulator BAF53a, a subunit of SWI/SNF chromatin remodeling complexes, which in turn inhibits neural progenitor proliferation.45 Conversely, expression of miRNA-9* and miR-124b can be negatively regulated by the silencing transcription repressor REST/NRSF, an inhibitor of neuronal differentiation.45,46
miRNA-transcription factor feedback circuitry plays an important role in fine-tuning of neural differentiation and function. miR-133b was found specifically expressed in midbrain dopaminergic neurons. A paired-like homeodomain transcription factor Pitx3 specifically induces transcription of miR-133b, which in turn, downregulates Pitx3 post-transcriptionally, suggesting that a feedback control between miRNA and transcription factors fine-tunes maturation and activities of dopaminergic neurons.47
Recently, miRNA expression profiling analysis of OPCs (A2B5+/GalC−) and oligodendrocytes (A2B5+/GalC+) from neonatal rat brains revealed an alteration of a cohort of 43 miRNAs during the transition from OPCs to post-mitotic oligodendrocytes.48 An OPC-enriched miR-9 was identified to target 3'UTR of mRNA encoding peripheral myelin protein (PMP) 22. PMP22 protein is only found in Schwann cells, but its mRNA transcript is present in oligodendrocytes. This study suggests that miRNAs may play a role in maintaining oligodendroglial property.
miR-23 was identified as a negative regulator of lamin B1 during oligodendrocyte development and myelin formation. Excessive lamin B1 has been shown to cause severe CNS myelin loss in adult-onset autosomal dominant leukodystrophy patients. Thus, the negative regulatory role in lamin B1 expression suggests that miR-23 plays an important role in myelin maintenance.49
A majority of regulatory miRNAs appears to have a negative regulatory role, however, there are cases that microRNAs execute activating functions to upregulate translation.50–52 Thus, translation regulation by miRNAs may oscillate between repression and activation depending on their association of different transcriptional or chromatin-modifying complexes. Although many miRNAs have yet been characterized in oligodendrocyte development at present, modulation of transcriptional networks by miRNAs undoubtedly contributes to the control or maintenance of oligodendrocyte identity and differentiation. It would be crucial to determine how extensively and precisely miRNAs regulate the networks for oligodendrocyte differentiation and, perhaps, myelin regeneration after injuries.
Conclusions and Perspectives
It has become increasingly clear that epigenetic mechanisms play an essential role in regulating oligodendrocyte development. Recent studies indicate that histone modification regulates multiple stages of oligodendrocyte lineage progression. Particularly, HDACs appear to serve as a hub integrating several signaling pathways and may set the threshold for remyelination in pathological demyelinating conditions. However, our current research concerning epigenetic function in oligodendrocyte development still in its infancy, the function of many epigenetic regulators such as ATP-dependent SWI/SNF chromatin remodeling complexes and miRNAs in oligodendrocyte differentiation remain not clear, and knowledge about functional interactions between epigenetic factors and transcriptional regulators remains scant. Although curative treatments for demyelinating diseases currently eludes our grasp, understanding genetic and epigenetic mechanisms of oligodendrocyte development will facilitate to dissect the disease etiology and eventually provide the framework for the treatment of demyelinating diseases such as MS.
Acknowledgements
We thank Ms. Ning Xie for assistance and discussions. Q.R.L. is supported by the US National Multiple Sclerosis Society (RG3978) and the US National Institutes of Health (NS050389). P.C. acknowledges the support of NIH-NINDS, grants RO1-NS42925 and RO1-NS52738.
References
- 1.Lai HC, Jan LY. The distribution and targeting of neuronal voltage-gated ion channels. Nat Rev Neurosci. 2006;7:548–62. doi: 10.1038/nrn1938. [DOI] [PubMed] [Google Scholar]
- 2.Trapp BD, Peterson J, Ransohoff RM, Rudick R, Mork S, Bo L. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338:278–85. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 3.Berger J, Moser HW, Forss-Petter S. Leukodystrophies: recent developments in genetics, molecular biology, pathogenesis and treatment. Curr Opin Neurol. 2001;14:305–12. doi: 10.1097/00019052-200106000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Karoutzou G, Emrich HM, Dietrich DE. The myelin-pathogenesis puzzle in schizophrenia: a literature review. Mol Psychiatry. 2008;13:245–60. doi: 10.1038/sj.mp.4002096. [DOI] [PubMed] [Google Scholar]
- 5.Franklin RJ. Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci. 2002;3:705–14. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 6.Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–67. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- 7.Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, et al. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of nkx6 regulation and shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 8.Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–9. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue T, Xian K, Hurlock E, Xin M, Kernie SG, Parada LF, Lu QR. A critical role for dorsal progenitors in cortical myelination. J Neurosci. 2006;26:1275–80. doi: 10.1523/JNEUROSCI.4717-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogarty M, Richardson WD, Kessaris N. A subset of oligodendrocytes generated from radial glia in the dorsal spinal cord. Development. 2005;132:1951–9. doi: 10.1242/dev.01777. [DOI] [PubMed] [Google Scholar]
- 11.Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer SE, Warrington AE, Bansal R. The oligodendrocyte and its many cellular processes. Trends in Cell Biology. 1993;3:191–7. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- 13.Peru RL, Mandrycky N, Nait-Oumesmar B, Lu QR. Paving the axonal highway: from stem cells to myelin repair. Stem Cell Rev. 2008;4:304–18. doi: 10.1007/s12015-008-9043-z. [DOI] [PubMed] [Google Scholar]
- 14.Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19:479–85. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- 16.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 17.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grozinger CM, Schreiber SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 19.Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–55. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 20.Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–45. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–89. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008 doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–38. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27:2606–16. doi: 10.1523/JNEUROSCI.4181-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, et al. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27:7717–30. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Hu Q, D'Ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57:1–12. doi: 10.1002/glia.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–30. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–89. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol. 2005;282:397–410. doi: 10.1016/j.ydbio.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 32.Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, Kaing S, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–85. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DW, Lassar AB. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol Cell Biol. 2003;23:8704–17. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–42. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 35.Fu H, Cai J, Clevers H, Fast E, Gray S, Greenberg R, et al. A genome-wide screen for spatially restricted expression patterns identifies transcription factors that regulate glial development. J Neurosci. 2009;29:11399–408. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Richardson WD. Genetics meets epigenetics: HDACs and Wnt signaling in myelin development and regeneration. Nat Neurosci. 2009;12:815–7. doi: 10.1038/nn0709-815. [DOI] [PubMed] [Google Scholar]
- 37.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 39.Schaefer A, Jung M, Kristiansen G, Lein M, Schrader M, Miller K, et al. MicroRNAs and cancer: Current state and future perspectives in urologic oncology. Urol Oncol. 2010;28:4–13. doi: 10.1016/j.urolonc.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 42.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 43.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 45.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–6. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–30. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2:178–88. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–23. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 52.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]