Abstract
The contribution of T cells to the host response to dengue virus (DENV) infection is not well understood. We previously demonstrated a protective role for CD8+ T cells during primary DENV infection using a mouse-passaged DENV strain and IFN-α/βR−/− C57BL/6 mice, which are susceptible to DENV infection. Here we examine the role of CD4+ T cells during primary DENV infection. Four I-Ab-restricted epitopes derived from three of the non-structural DENV proteins were identified. CD4+ T cells expanded and were activated after DENV infection, with peak activation occurring on day 7. The DENV-specific CD4+ T cells expressed intracellular IFN-γ, TNF, IL-2, and CD40L, and killed peptide-pulsed target cells in vivo. Surprisingly, depletion of CD4+ T cells before DENV infection had no effect on viral loads. Consistent with this observation, CD4+ T cell depletion did not affect the DENV-specific IgG or IgM Ab titers or their neutralizing activity, or the DENV-specific CD8+ T cell response. However, immunization with the CD4+ T cell epitopes before infection resulted in significantly lower viral loads. Thus, we conclude that whereas CD4+ T cells are not required for controlling primary DENV infection, their induction by immunization can contribute to viral clearance. These findings suggest inducing anti-DENV CD4+ T cell responses by vaccination may be beneficial.
Keywords: Dengue virus, T cells, vaccination, mouse, epitope
Introduction
Dengue virus (DENV) is a mosquito-borne RNA virus in the Flaviviridae family, which also includes West Nile Virus (WNV), Yellow Fever Virus (YFV), and Japanese Encephalitis Virus (JEV). The four serotypes of DENV (DENV1-4) share approximately 65–75% homology at the amino acid level (1). Infections with DENV can be asymptomatic, or cause disease ranging from dengue fever (DF) to dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (2). DF is a self-limiting illness with symptoms that include fever, headache, myalgia, retro-orbital pain, nausea, and vomiting. DHF and DSS are characterized by increased vascular permeability, thrombocytopenia, hemorrhagic manifestations, and in the case of DSS, shock, which can be fatal. The incidence of DENV infections has increased 30-fold in the past 50 years (2). DF and DHF/DSS are a significant cause of morbidity and mortality worldwide, and therefore a DENV vaccine is a global public health priority. However, vaccine development has been challenging, as a vaccine should protect against all four DENV serotypes (3).
Severe dengue disease (DHF/DSS) most often occurs in individuals experiencing a secondary infection with a heterologous DENV serotype, suggesting the immune response contributes to the pathogenesis (4, 5). One hypothesis is that serotype cross-reactive antibodies enhance infection of FcγR+ cells during a secondary infection resulting in higher viral loads and more severe disease via a phenomenon known as antibody-dependent enhancement (ADE) (6, 7). Recent studies have demonstrated DENV-specific Ab can enhance disease in mice (8, 9). It has also been proposed that serotype cross-reactive memory T cells may respond sub-optimally during secondary infection and contribute to the pathogenesis (10). Accordingly, studies have shown serotype cross-reactive T cells can exhibit an altered phenotype in terms of cytokine production and degranulation (11–13). However, another study found the breadth and magnitude of the T cell response during secondary DENV infection was not significantly associated with disease severity (14). Although many studies have investigated the role of T cells in DENV pathogenesis, few studies have examined the contribution of T cells to protection against DENV. Consequently, the role of T cells in protection versus pathogenesis during DENV infections is presently unknown. This is primarily due to the lack of an adequate animal model, as mice are resistant to infection with this human pathogen (15). We have previously shown that a mouse-passaged DENV2 strain, S221, does not replicate to detectable levels in wild-type C57BL/6 mice, but does replicate in IFN-α/βR−/− mice (16). Using S221 and IFN-α/βR−/− mice, we have previously demonstrated a protective role for CD8+ T cells in the response to primary DENV2 infection (16).
CD4+ T cells can contribute to the host response to pathogens in a variety of ways. They produce cytokines and can mediate cytotoxicity. They also help B cell responses by inducing immunoglobulin class switch recombination (CSR), and help prime the CD8+ T cell response. CD4+ T cells can help the CD8+ T cell response indirectly by activating APCs, for example via CD40L/CD40 (17). CD40L on CD4+ T cells is important in activating B cells as well (18). CD4+ T cells can also induce chemokine production that attracts CD8+ T cells to sites of infection (19). However, the requirement for CD4+ T cell help for Ab and CD8+ T cell responses is not absolute, and may be specific to the pathogen and/or experimental system. For instance, it has been shown that CSR can occur in the absence of CD4+ T cells (20), and the primary CD8+ T cell response is CD4-independent under inflammatory conditions (17).
Despite the known importance of CD4+ T cells in the host response to pathogens, to our knowledge no study has yet examined the role of CD4+ T cells during primary DENV infection, and no CD4+ T cell epitopes have been identified from DENV-infected mice. In this study we sought to define the contribution of CD4+ T cells to the host response to primary DENV2 infection using IFN-α/βR−/− mice. Infection with DENV2 resulted in CD4+ T cell expansion and activation. In order to study DENV-specific CD4+ T cells, a predictive algorithm was used to identify MHC class II (I-Ab)-binding peptides from the entire proteome of DENV2, which is approximately 3390 amino acids and encodes three structural (core (C), envelope (E), and membrane (M)), and seven non-structural (NS) (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) proteins. Four CD4+ T cell epitopes from the NS2B, NS3, and NS4B proteins were identified, one of which contains an immunodominant CD8+ T cell epitope that we identified previously (16).
DENV2-specific CD4+ T cells were of a Th1 phenotype, with intracellular expression of IFN-γ, TNF, IL-2, and CD40L, and could mediate in vivo cytotoxicity. However, depletion of CD4+ T cells did not have a significant effect on viral clearance, and CD4+ T cells were not required for the induction of the DENV2-specific Ab or CD8+ T cell responses. Immunization with dominant CD4+ T cell epitopes led to enhanced viral clearance, demonstrating that CD4+ T cells can contribute to the anti-DENV2 immune response, and supporting the development of a DENV vaccine that induces a robust CD4+ T cell response.
Materials and Methods
Mice and infections
C57BL/6 (H-2b) mice were obtained from The Jackson Laboratory and subsequently bred at the animal facility at La Jolla Institute for Allergy and Immunology (LIAI). IFN-α/βR−/− mice on the C57BL/6 background were obtained from Dr. Wayne Yokoyama (Washington University, St. Louis, MO) via Dr. Carl Ware (LIAI). B6.SJL mice were purchased from Taconic. Mice were used between 5 and 8 weeks of age. Mice were infected i.v. in the lateral tail vein or retro-orbitally (r.o) with 200 μl of the DENV2 strain, S221, in 5% FBS/PBS. Blood was obtained from anesthetized mice by r.o. puncture. All mouse experiments were approved by the Animal Care Committee at LIAI.
Cell culture and viral stocks
The hybridoma clones SFR3, GK1.5, and 2.43, which produce rat anti-human HLA-DR5, anti-mouse CD4, and anti-mouse CD8 IgG2b Ab, respectively, were from the American Type Culture Collection, and were grown in Protein-Free Hybridoma Medium supplemented with penicillin, streptomycin, HEPES, GlutaMAX, and 2-ME (all from Invitrogen) at 37°C, 5% CO2. C6/36, an A. albopictus mosquito cell line, was cultured in Leibovitz’s L-15 Medium (Invitrogen) supplemented with 10% FBS (Gemini Bio-Products), penicillin, streptomycin, and HEPES at 28°C in the absence of CO2. S221, a plaque-purified DENV2 strain, was derived from the clinical isolate, PL046 (21), as described previously (16). Viral stocks were amplified in C6/36 cells and purified over a sucrose gradient as previously described (22). Infectious doses were determined based on genomic equivalents (GE), which were quantified by real-time RT-PCR. There are approximately 5 × 104 GE/PFU for S221, based on plaque assay on baby hamster kidney cells.
Bioinformatic analyses
Candidate epitopes were predicted using a consensus approach described by Wang and colleagues (23). Briefly, all 15-mer peptides that are encoded in the DENV2 PL046 polyprotein were predicted for binding to H-2 I-Ab. Two independent algorithms (24) were used to rank the peptides by predicted binding affinity. The median of the two ranks was used to select the top 73 out of 3383 peptides, corresponding to the top 2% of all peptides.
Peptide synthesis
Peptides utilized in initial screening experiments were synthesized as crude material by A and A Labs. A total of 73 15-mer peptides were ordered and synthesized twice in different (alphabetical vs. predicted IC50) order. Positive peptides were re-synthesized by A and A Labs and purified to >90% homogeneity by reverse-phase HPLC. Purity of these peptides was determined using mass spectrometry. The HPLC-purified peptides were used for all subsequent experiments.
Flow cytometric analyses
For surface staining of germinal center B cells, splenocytes were stained with anti-B220-Alexa Fluor 647 (Biolegend), anti-CD4-PerCP (BD Biosciences), GL7-FITC (BD Biosciences), anti-IgD-eFluor 450 (eBioscience), and anti-Fas-PE (BD Biosciences). For intracellular cytokine staining of CD4+ T cells, 2 × 106 splenocytes were plated in 96-well U-bottom plates and stimulated with individual DENV2 peptides (3 μg/ml) for 2 h. Brefeldin A (GolgiPlug, BD Biosciences) was then added and cells were incubated for another 5 h. Cells were washed, incubated with supernatant from 2.4G2-producing hybridoma cells, and labeled with anti-CD4-eFluor 450 (eBioscience) and anti-CD8α-PerCP-eFluor 710 (eBioscience) or PE-Cy7 (BD Biosciences). The cells were then fixed and permeabilized using the BD Cytofix/Cytoperm Kit, and stained with various combinations of anti-IFN-γ-APC (eBioscience), anti-TNF-PE-Cy7 (BD Biosciences), anti-IL-2-Alexa Fluor 488 (BD Biosciences) or -PE (Biolegend), and anti-CD40L-PE (eBioscience). Foxp3 staining was done using the mouse regulatory T cell staining kit from eBioscience. The criteria for positivity in CD4+ T cell epitope identification were: 2x the percentage of IFN-γ produced by stimulated cells compared with unstimulated cells, positive in two independent crude peptide orders, and positive when ordered as HPLC-purified (>90% pure). For CD8+ T cell ICS, splenocytes (2 × 106) were stimulated in 96-well U-bottom plates for 5 h in the presence of 1 μg/ml H-2b-restricted epitopes that we identified previously: M60-67, NS2A8-15, and NS4B99-107 (16). Anti-CD107a-FITC (BD Biosciences) was added to the wells during the stimulation. Cells were then stained as described for CD4+ T cell ICS. Samples were read on an LSR II (BD Biosciences) and analyzed using FloJo software (Tree Star).
Immunohistochemistry
Tissues were embedded in O.C.T. compound (Sakura). Sections (6 μm) were cut and stored at −80° C. Frozen sections were thawed and fixed for 10 minutes in acetone at 25° C, followed by 8 minutes in 1% paraformaldehyde (EMS) in 100 mM dibasic sodium phosphate containing 60 mM lysine and 7 mM sodium periodate pH 7.4 at 4° C. Sections were blocked first using the Avidin/Biotin Blocking Kit (Vector Labs) followed by 5% normal goat serum (Invitrogen) and 1% BSA (Sigma) in PBS. Sections were stained overnight with anti-F4/80-biotin (clone BM8, Biolegend), anti-CD4-PE (clone RM4-5, eBioscience), anti-CD8β-Alexa Fluor 647 (clone YTS156.7.7, Biolegend), and anti-B220-FITC (clone RA3-6B2, BD Pharmingen). Sections were then washed and stained with streptavidin-Alexa Fluor 750 and rabbit anti-FITC-Alexa Fluor 488 (Invitrogen). Images were recorded using a Leica TCS SP5 confocal microscope, processed using Leica Microsystems software, stitched together using Adobe Illustrator, and adjusted using ImageJ.
T cell depletions
Hybridoma supernatants were clarified by centrifugation, dialyzed against PBS, sterile-filtered, and quantified by BCA Protein Assay Reagent (Thermo Scientific). IFN-α/βR−/− mice were injected i.p. with 250 μg of SFR3, or GK1.5, or 2.43 in PBS (250 μl total volume) 3 days and 1 day before or 1 day before and 1 day after infection, which resulted in depletion of ≥90% of CD8+ cells and ≥97% of CD4+ cells. In Fig. 4, one CD4-depleted mouse received GK1.5 only on day 1, which still resulted in ≥97% depletion.
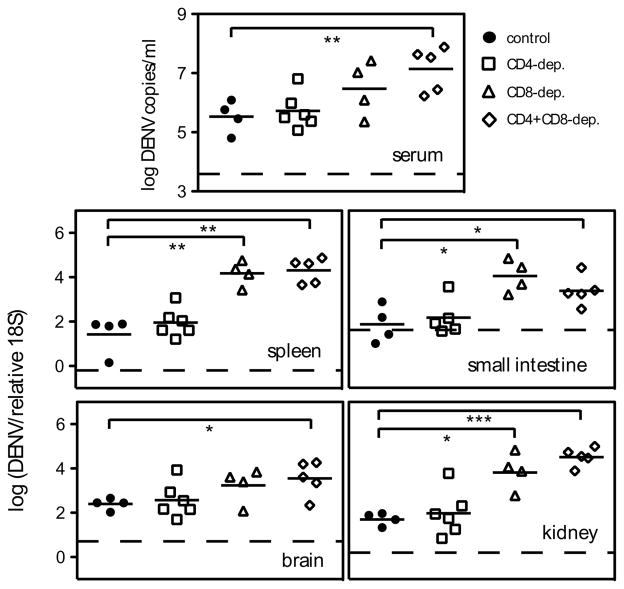
FIGURE 4. Depletion of CD4+ T cells prior to DENV2 infection does not affect viral RNA levels.
IFN-α/βR−/− mice were depleted of CD4+ or CD8+ cells, or both, by administration of GK1.5 or 2.43 Ab, respectively, (or given an isotype control Ab) 2 days before and 1 day after infection with 1010 GE of DENV2. Mice were sacrificed 5 days later, and DENV2 RNA levels in the serum, spleen, small intestine, brain, and kidney were quantified by real-time RT-PCR. Data are expressed as DENV2 copies per ml of sera, or DENV2 units normalized to 18S rRNA levels for the organs. Each symbol represents one mouse, the bar represents the geometric mean, and the dashed line is the limit of detection. * p < 0.05, ** p < 0.01, and *** p < 0.001 for viral RNA levels comparing T cell-depleted mice with control mice.
DENV2-specific antibody ELISA
Serum was harvested from control and CD4-depleted IFN-α/βR−/− mice 7 days after infection with 1010 GE of DENV2, or naïve mice. EIA/RIA 96-well plates (Costar) were coated with DENV2 (109 GE per well) in 50 μl 0.1M NaHCO3. The virus was UV-inactivated and plates left overnight at 4°C. The plates were then washed to remove unbound virus using 0.05% (v/v) Tween 20 (Sigma) in PBS. After blocking with Blocker Casein Blocking Buffer (Thermo Scientific) for 1 h at room temperature, 1:3 serial dilutions of serum in a total volume of 100 μl were added to the wells. After 1.5 h, wells were washed and bound antibody was detected using HRP-conjugated goat anti-mouse IgG Fc portion or HRP-conjugated donkey anti-mouse IgM μ chain (Jackson Immunoresearch) and TMB (eBioscience).
Antibody-virus neutralization assay
Serum was heat-inactivated at 56°C for 30 min. Three-fold serial dilutions of serum were then incubated with 5×108 GE of DENV2 for 1 h at room temperature in a total volume of 100 μl PBS. Next, approximately 6 × 105 C6/36 cells per well of a 24-well plate were infected with 100 μl of the virus-antibody mix for one hour at 28°C. Cells were washed twice with 500 μl of PBS, and then incubated at 28°C in 500 μl L-15 Medium containing 5% FBS, penicillin, and streptomycin for 24 h. For each antibody dilution, the percentage of infected cells was determined by flow cytometry as previously described (25) using 2H2-biotin (IgG2a anti-prM/M, DENV1-4 reactive) and streptavidin-APC (Biolegend). The percentage of infected cells was normalized to 100% (infection without serum).
CD8 in vivo cytotoxicity assay
IFN-α/βR−/− mice (recipients) were infected with 1010 GE of DENV2. Some mice were depleted of CD4+ T cells before infection. Splenocytes (targets) were harvested from donor B6.SJL congenic mice (CD45.1) 7 days later. RBC were lysed, and the target cells were pulsed with varying concentrations of a pool of 4 H-2b-restricted DENV2 peptides (M60-67, NS2A8-15, NS4B99-107, NS5237-245) or DMSO for 1 h at 37°C. The cells were then washed and labeled with CFSE (Invitrogen) in PBS/0.1% BSA for 10 min at 37°C. Cells were labeled with 1 μM CFSE (CFSEhigh) or 100 nM CFSE (CFSElow) or left unlabeled. After washing, the cell populations were mixed and 5 × 106 cells from each population were injected i.v. into naïve or infected recipient mice. After 4 h, the mice were sacrificed and splenocytes stained with anti-CD45.1-APC (eBioscience) and analyzed by flow cytometry, gating on CD45.1+ cells. The percentage killing was calculated as follows: 100 − ((percentage DENV peptide-pulsed in infected mice/percentage DMSO-pulsed in infected mice)/(percentage DENV peptide-pulsed in naïve mice/percentage DMSO-pulsed in naïve mice) × 100).
CD4 in vivo cytotoxicity assay
IFN-α/βR−/− mice (recipients) were infected with 1010 GE of DENV2. Some mice were depleted of CD4+ or CD8+ cells before infection. Splenocytes (targets) were harvested from donor B6.SJL congenic mice (CD45.1) 7 days later. RBC were lysed and the target cells were pulsed with 1.7 μg (approximately 1 μM) each of NS2B108-122, NS3198-212, and NS3237-251 (or DMSO) for 1 h at 37°C. The cells were then washed and labeled with CFSE in PBS/0.1% BSA for 10 min at 37°C. DENV2 peptide-pulsed cells were labeled with 1 μM CFSE (CFSEhigh) and DMSO-pulsed cells with 100 nM CFSE (CFSElow). After washing, the two cell populations were mixed and 5 × 106 cells from each population were injected i.v. into naïve or infected recipient mice. After 16 h, the mice were sacrificed and splenocytes stained and the percentage killing calculated as described for the CD8 in vivo cytotoxicity assay.
Quantitation of DENV burden in mice
Mice were euthanized by isoflurane inhalation and blood was collected via cardiac puncture. Serum was separated from whole blood by centrifugation in serum separator tubes (Starsted). Small intestines were put into PBS, flushed, filleted, chopped into small pieces, and put into RNAlater (Qiagen). Other organs were immediately placed into RNAlater and all organs were subsequently homogenized for 3 min in 1 ml tissue lysis buffer (Qiagen Buffer RLT) using a Mini-Beadbeater-8 (BioSpec Products) or Qiagen TissueLyser. Immediately after homogenization, all tissues were centrifuged (5 min, 4°C, 16,000 × g) to pellet debris, and RNA was isolated using the RNeasy Mini Kit (Qiagen). Serum RNA was isolated using the QIAamp Viral RNA Mini Kit (Qiagen). After elution, viral RNA was stored at −80°C. Quantitative RT-PCR was performed according to a published protocol (26), except a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) with One-Step qRT-PCR Kit (Quanta BioSciences) were used. The DENV2 standard curve was generated with serial dilutions of a known concentration of DENV2 genomic RNA which was in vitro transcribed (MAXIscript Kit, Ambion) from a plasmid containing the cDNA template of S221 3′UTR. After transcription, DNA was digested with DNase I, and RNA was purified using the RNeasy Mini Kit and quantified by spectrophotometry. To control for RNA quality and quantity when measuring DENV in tissues, the level of 18S rRNA was measured using 18S primers described previously (27) in parallel real-time RT-PCR reactions. A relative 18S standard curve was made from total splenic RNA.
Peptide immunizations
IFN-α/βR−/− mice were immunized s.c. with 50 μg each of NS2B108-122, NS3198-212, and NS3237-251 emulsified in CFA (Difco). After 11 days, mice were boosted with 50 μg peptide emulsified in IFA (Difco). Mock-immunized mice received PBS/DMSO emulsified in CFA or IFA. Mice were infected 13 days after the boost with 1011 GE of DENV2 (some mice were depleted of CD4+ or CD8+ T cells 3 days and 1 day before infection). Four days later, the mice were sacrificed and tissues harvested, RNA isolated, and DENV2 RNA levels measured as described above.
Statistical analyses
Data were analyzed with Prism software version 5.0 (GraphPad Software, Inc.). Statistical significance was determined using the unpaired t-test with Welch’s correction.
Results
CD4+ T cell activation and expansion following DENV2 infection
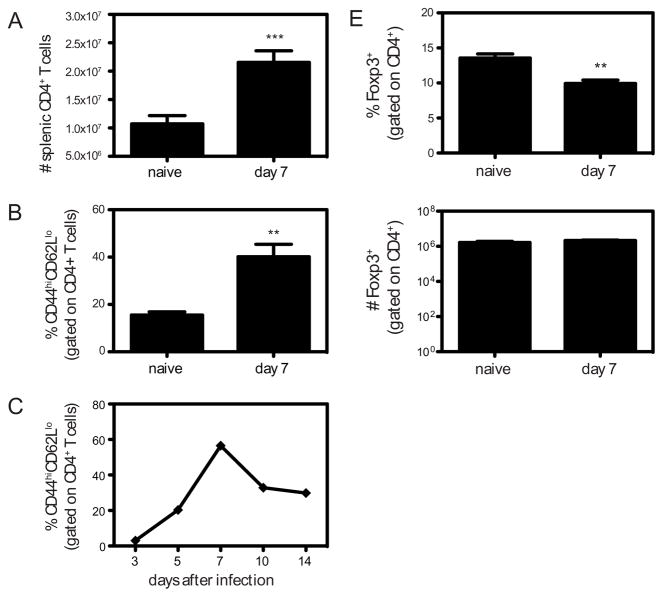
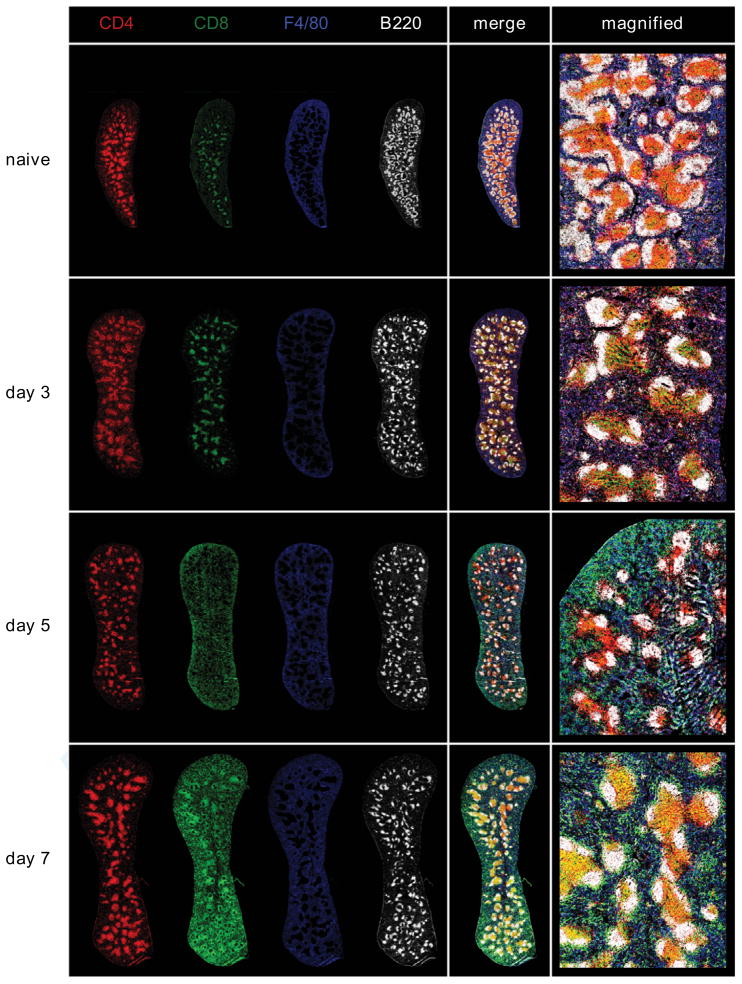
DENV2 (1010 GE of S221) infection of IFN-α/βR−/− mice results in an acute infection, with viral replication peaking between 2 and 4 days after infection (16). At this time the mice show signs of disease including hunched posture and ruffled fur, and the virus is subsequently cleared from the serum by day 6. To determine the role of CD4+ T cells during the course of this primary DENV2 infection, we first examined the expansion of CD4+ T cells in the spleens of IFN-α/βR−/− mice 7 days after infection with DENV2, and observed a 2-fold increase in CD4+ T cell numbers (Fig. 1A). The cells were activated, as measured by CD44 upregulation and CD62L downregulation on splenic CD4+ T cells (Fig. 1B) and on circulating blood CD4+ T cells, with the peak on day 7 after infection (Fig. 1C). To study the CD4+ T cell response in the spleen in more detail, we performed immunohistochemistry on spleen sections obtained from naïve mice and mice 3, 5, and 7 days after DENV2 infection. Sections were stained for CD4, CD8, B220 to highlight B cell follicles, and F4/80 to show red pulp macrophages. As expected, in naïve mice, we observed CD4+ and CD8+ T cells dispersed throughout the spleen, but preferentially in T cell areas, also known as the periarteriolar lymphoid sheath (PALS) (Fig. 1D). By day 3 after DENV2 infection, most of the CD4+ and CD8+ T cells had migrated to the PALS, with very few T cells observed in the red pulp. At day 5, the CD4+ cells were still concentrated in the PALS, at the border between the T cell area and B cell follicles, and also in the B cell follicles. At day 7 after infection, the spleen had increased in size dramatically, and CD4+ T cells were found primarily in the PALS and B cell follicles. The localization of CD8+ T cells differed from the CD4+ T cells mainly in that at day 5 after infection, many of the CD8+ T cells had left the T cell area and were found distributed throughout the red pulp and marginal zone (MZ). By day 7, the CD8+ T cells were observed in the PALS, MZ, and also the red pulp. These images illustrate the kinetics of the adaptive immune response to DENV2 in the spleen, and show CD4+ T cells in close proximity to both CD8+ T cells and B cells after DENV2 infection.
FIGURE 1. DENV2 infection results in CD4+ T cell activation and expansion in IFN-α/βR−/− mice.
A, The numbers of splenic CD4+ T cells in naïve IFN-α/βR−/− mice (n = 6) and IFN-α/βR−/− mice infected with 1010 GE of DENV2 (n = 11) are shown. *** p < 0.001 for naïve versus infected mice. B, The percentage of CD62LloCD44hi cells (gated on CD4+ cells) is shown for naïve (n = 4) and IFN-α/βR−/− mice infected with 1010 GE of DENV2 (n = 8). ** p < 0.01 for naïve versus infected mice. C, Blood lymphocytes were obtained from IFN-α/βR−/− mice on days 3, 5, 7, 10, and 14 after infection with 1010 GE of DENV2. The percentage of CD44hiCD62Llo cells (gated on CD4+ T cells) ± SEM (n = 6) is shown. D, Representative spleen sections from naïve mice, and mice 3, 5, and 7 days after DENV2 infection were analyzed by confocal microscopy. Red pulp macrophages (F4/80+) are shown in blue, B cells (B220+) in white, CD8+ T cells in green, and CD4+ T cells in red. E, The percentage and number of splenic Foxp3+ cells (gated on CD4+ cells) are shown for naïve (n = 4) and infected IFN-α/βR−/− mice (n = 4).
Regulatory T cells (Tregs) are a subset of CD4+ T cells that are characterized by the expression of the transcription factor, Foxp3 (28), and have been found to facilitate the early host response to HSV-2 (29) and help control WNV infection (30). To determine if DENV2 infection resulted in an expansion of Tregs, we examined the number of CD4+Foxp3+ cells in the spleen 7 days after infection, and found there was a decrease in the percentage of Tregs among total CD4+ cells, and no change in the number of Tregs, demonstrating that DENV2 infection does not lead to an expansion of Tregs in the spleen (Fig. 1E).
Identification of DENV2 CD4+ T cell epitopes
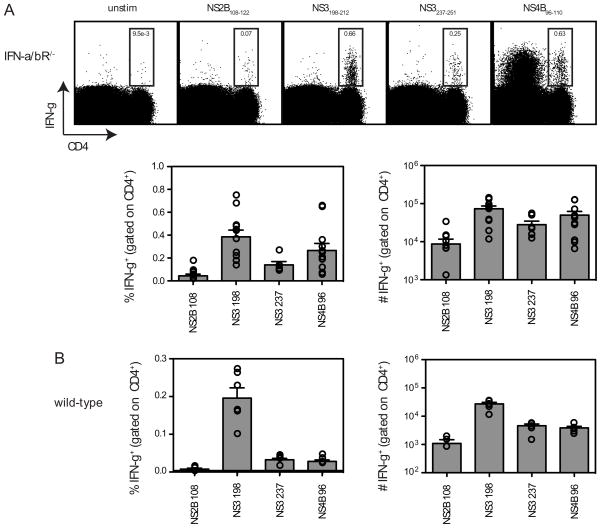
In order to study the DENV2-specific CD4+ T cell response, we sought to identify MHC class II (I-Ab)-restricted CD4+ T cell epitopes using a bioinformatics prediction method that has been previously used to map the CD4+ T cell response to mouse cytomegalovirus (31). Briefly, the proteome of DENV2 was screened and 73 15-mer peptides predicted to bind I-Ab were identified. The peptides were tested by IFN-γ ICS using splenocytes from DENV2-infected IFN-α/βR−/− mice. Positive peptides (2x background) were then re-ordered as HPLC-purified (>90%) and re-tested. Four positive peptides were identified: NS2B108-122, NS3198-212, NS3237-251, and NS4B96-110 (Fig. 2A and Table I). Similar to the DENV2-specific CD8+ T cell response (16), the epitopes identified in IFN-α/βR−/− mice were also recognized by CD4+ T cells from DENV2-infected wild-type mice (Fig. 2B), and the magnitude of the CD4+ T cell response was higher in IFN-α/βR−/− mice compared with wild-type mice, likely due to increased viral replication. Notably, NS3200-214 has been identified as a human HLA-DR15-restricted CD4+ T cell epitope (14, 32). It was also of interest that NS4B96-110 contains a CD8+ T cell epitope (NS4B99-107) that we had previously identified as the immunodominant epitope in both wild-type and IFN-α/βR−/− C57BL/6 mice infected with DENV2 (16).
FIGURE 2. Identification of DENV2-derived epitopes recognized by CD4+ T cells.
A, Splenocytes were obtained from IFN-α/βR−/− mice 7 days after infection with 1010 GE of DENV2 and re-stimulated in vitro with DENV2-derived 15-mer peptides predicted to bind I-Ab. Cells were then stained for surface CD4 and intracellular IFN-γ and analyzed by flow cytometry. The 4 positive peptides identified are shown. In the dot plots, the percentage of CD4+ T cells producing IFN-γ is indicated. The responses of individual mice as well as the mean and SEM are also shown (n = 7–11). The response of unstimulated cells was subtracted from the response to each DENV2 peptide, and the net percentage and number of splenic CD4+ T cells producing IFN-γ are indicated. B, Splenocytes were obtained from wild-type C57BL/6 mice 7 days after infection with 1010 GE of DENV2 and stimulated and stained as in A (n = 6).
Table I.
DENV2-derived CD4+ T cell epitopes
| Epitope | Sequence |
|---|---|
| NS2B108-122 | GLFPVSLPITAAAWY |
| NS3198-212 | GKTKRYLPAIVREAI |
| NS3237-51 | GLPIRYQTPAIRAEH |
| NS4B96-110 | IGCYSQVNPITLTAA |
Phenotype of DENV2-specific CD4+ T cells
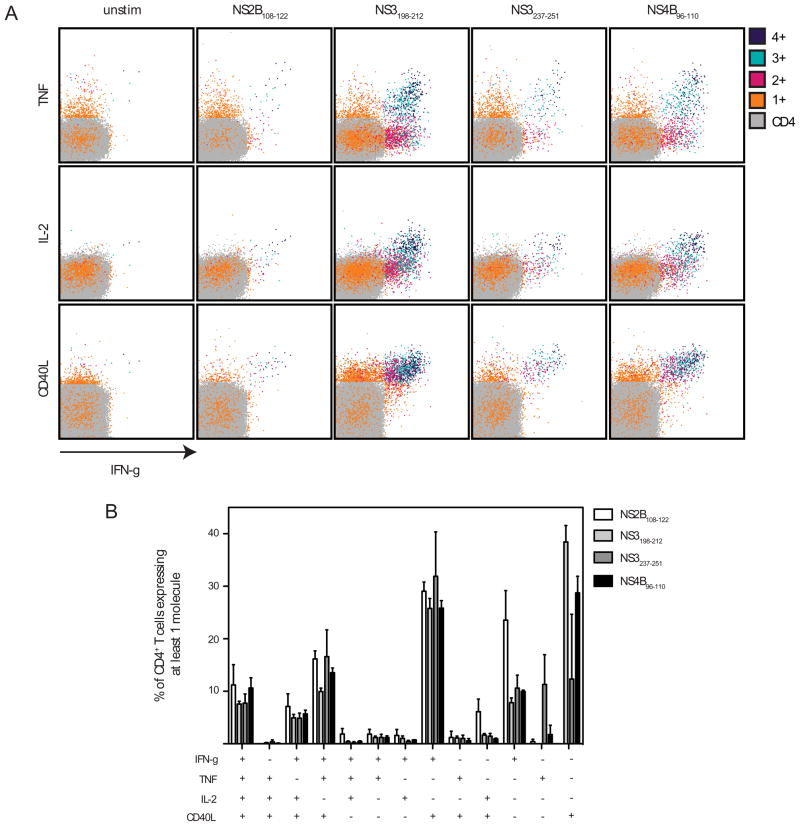
Multicolor flow cytometry was performed to study the phenotype of DENV2-specific CD4+ T cells. These cells produced IFN-γ, TNF, and IL-2 (Fig. 3). No intracellular IL-4, IL-5, IL-17, or IL-10 were detected (data not shown). The DENV2-specific CD4+ T cells also expressed CD40L, suggesting they are capable of activating CD40-expressing cells, which include DCs and B cells. The four DENV2-derived CD4+ T cell epitopes induced responses that differed in magnitude, but were similar in terms of phenotype. The most polyfunctional cells (those expressing IFN-γ, TNF, IL-2, and CD40L) also expressed the highest levels of the cytokines and CD40L. These results demonstrate that DENV2 infection elicits a virus-specific Th1 CD4+ T cell response in IFN-α/βR−/− mice.
FIGURE 3. DENV2-specific CD4+ T cells are polyfunctional.
Splenocytes were obtained from IFN-α/βR−/− mice 7 days after infection with 1010 GE of DENV2 and stimulated in vitro with individual peptides. Cells were then stained for surface CD4, and intracellular IFN-γ, TNF, IL-2, and CD40L, and analyzed by flow cytometry. A, Cells expressing 4, 3, 2, 1, or none of the molecules are color-coded. Representative dot plots are shown. B, The response of unstimulated cells was subtracted from the response to each DENV2 peptide, and the net percentages of the CD4+ T cells that are expressing at least one molecule are indicated. The mean and SEM of 3 mice is shown.
Effects of CD4+ and/or CD8+ T cell depletions on DENV2 viral RNA levels
To determine how CD4+ T cells contribute to controlling DENV2 infection, we depleted CD4+ T cells, CD8+ T cells, or both, from IFN-α/βR−/− mice and measured DENV2 RNA levels 5 days after infection with 1010 GE of DENV2. We found no difference in viral RNA levels between control undepleted mice and CD4-depleted mice in the serum, kidney, small intestine, spleen, or brain (Fig. 4). As we observed previously (16), CD8-depleted mice had significantly higher viral loads than control mice. Depletion of both CD4+ and CD8+ T cells resulted in viral RNA levels that were significantly higher than those in control mice in all tissues examined, and equivalent to the viral RNA levels in CD8-depleted mice. These data show that CD4+ T cells are not required to control primary DENV2 infection in IFN-α/βR−/− mice, and confirm an important role for CD8+ T cells in viral clearance.
CD4+ T cells are not required for the anti-DENV2 antibody response
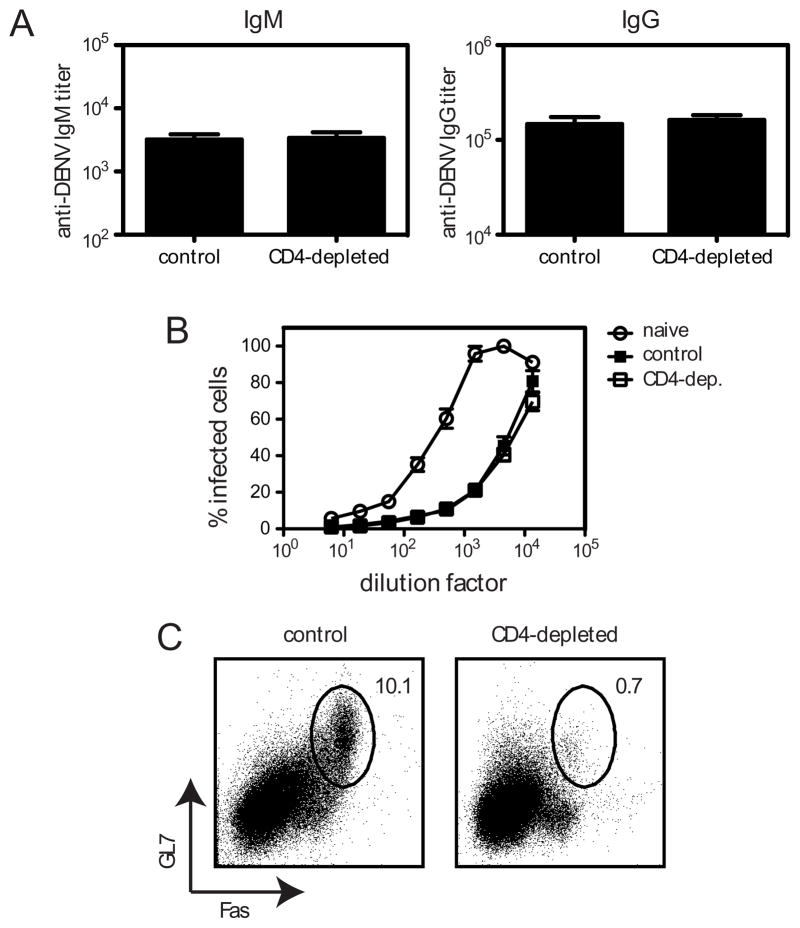
Although CD4+ T cells were not required for controlling DENV2 infection, we wondered whether they made any contribution to the anti-DENV immune response, for example by helping the B cell and/or CD8+ T cell responses. CSR, the process by which the immunoglobulin heavy chain constant region is switched so the B cell expresses a new isotype of Ab, can be induced when CD40L-expressing CD4+ T cells engage CD40 on B cells (20). However, CSR can also occur in the absence of CD4+ T cell help. To determine whether the anti-DENV2 Ab response depends on CD4+ T cells, we first measured DENV2-specific IgM and IgG titers in the sera of control and CD4-depleted mice 7 days after infection with 1010 GE of DENV2. As expected, we found no difference in IgM titers at day 7 between control and CD4-depleted mice (Fig 5A). There was also no difference in IgG titers between control and CD4-depleted mice. To measure the functionality of these DENV2-specific Ab, a flow cytometry-based neutralization assay was performed, in which C6/36 mosquito cells were infected with DENV2 in the presence of heat-inactivated sera obtained from control and CD4-depleted mice 7 days after infection. The sera from control and CD4-depleted mice could neutralize DENV2 equally well (Fig. 5B). As reported previously (8), naïve serum was able to prevent DENV infection of C6/36 cells, although not as efficiently as DENV-immune serum. We also looked for the presence of germinal center (GC) B cells, as the GC reaction is generally thought to be CD4+ T cell-dependent (33). As expected, we found GC B cells were absent in the CD4-depleted mice (Fig. 5C). Based on the lack of GC B cells in the DENV2-infected CD4-depleted mice, we conclude that the early anti-DENV2 Ab response is CD4- and GC-independent.
FIGURE 5. CD4+ T cells are not required for the anti-DENV2 antibody response.
IFN-α/βR−/− mice (control or CD4-depleted) were infected with 1010 GE of DENV2. A, IgM and IgG titers in the sera at day 7 were measured by ELISA (n = 5 control and 6 CD4-depleted mice). Data are combined from two independent experiments. B, Neutralizing activity of sera from naïve (n = 4) and control (n = 6) or CD4-depleted mice (n = 6) obtained 7 days after infection was determined by measuring the ability of the sera to reduce DENV2 infection of C6/36 cells. C, The percentage of germinal center B cells (GL7+Fas+, gated on B220+ cells) in the spleen 7 days after infection is shown. The plots are representative of 5 control and 5 CD4-depleted mice.
CD4+ T cells are not necessary for the primary DENV2-specific CD8+ T cell response
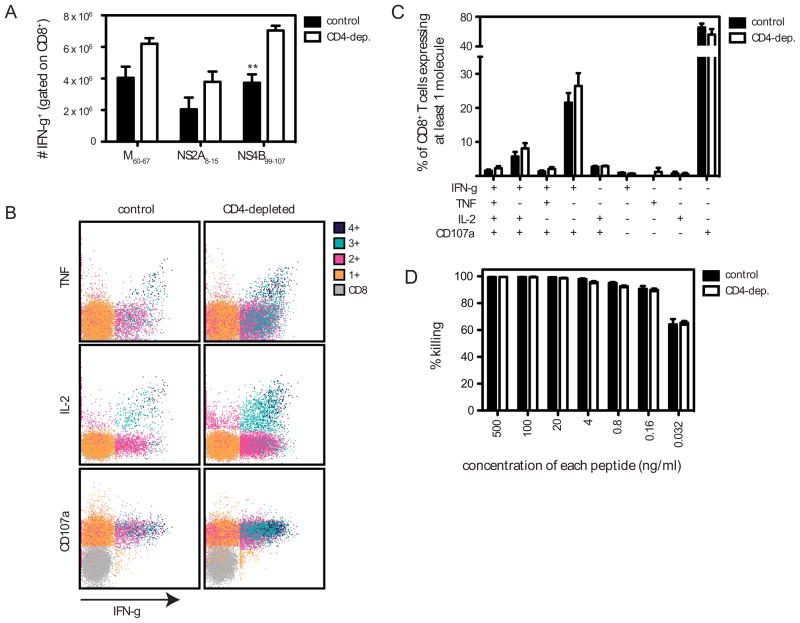
Next we assessed the role of CD4+ T cells in helping the CD8+ T cell response by examining the DENV2-specific CD8+ T cell response in control and CD4-depleted DENV2-infected mice. The numbers of splenic CD8+ T cells were equivalent in control and CD4-depleted mice (data not shown). IFN-γ ICS was performed using DENV2-derived H-2b-restricted immunodominant peptides that we identified previously (M60-67, NS2A8-15, and NS4B99-107) (16). Somewhat surprisingly, there was an increase in the number of DENV2-specific IFN-γ+CD8+ T cells in CD4-depleted mice compared with control mice (Fig. 6A). To further characterize the phenotype of the CD8+ T cells generated in the absence of CD4+ T cells, we examined expression of TNF, IL-2, and CD107a (a marker for degranulation) in cells stimulated with NS4B99-107 (Fig 6B and C). As also shown in Fig. 6A, the magnitude of the CD8+ T cell response was larger in the CD4-depleted mice, but the cytokine and CD107a expression profiles were comparable. Similar results were observed when cells were stimulated with M60-67 or NS2A8-15 (data not shown). Next we examined the functionality of the DENV2-specific CD8+ T cells using an in vivo cytotoxicity assay, in which splenocytes were pulsed with a pool of 4 H-2b-restricted immunodominant peptides and CFSE-labeled before injection into control or CD4-depleted DENV2-infected mice. CD8+ T cell-mediated-cytotoxicity was very efficient; almost 100% killing was observed at peptide concentrations of 500 ng/ml (Fig 6D). Therefore, the peptide concentrations were titrated down, and no difference in killing was observed between control and CD4-depleted mice at any concentration tested. These data reveal that the primary anti-DENV2 CD8+ T cell response, in terms of expansion, cytokine production, degranulation, and cytotoxicity, does not depend on CD4+ T cell help.
FIGURE 6. CD4+ T cells are not required for the primary DENV2-specific CD8+ T cell response.
A, Splenocytes were obtained from IFN-α/βR−/− mice (control or CD4-depleted) 7 days after infection with 1010 GE of DENV2, and stimulated in vitro with immunodominant DENV2-derived H-2b-restricted CD8+ T cell epitopes. Cells were then stained for CD8 and IFN-γ and analyzed by flow cytometry, and the number of CD8+ T cells producing IFN-γ is shown. Results are expressed as the mean ± SEM of 4 mice per group. ** p < 0 .01. B and C, Splenocytes were obtained as in A and stimulated with NS4B99-107 in the presence of an anti-CD107 Ab, and then stained for CD8, IFN-γ, TNF, and IL-2. B, Cells expressing 4, 3, 2, 1, or none of the molecules are color-coded. Representative dot plots are shown. C, The response of unstimulated cells was subtracted from the response to each DENV2 peptide, and the net percentages of the CD8+ T cells that are expressing at least one molecule are indicated. The mean and SEM of 3 mice is shown. D, CD8+ T cell-mediated killing. IFN-α/βR−/− mice (control or CD4-depleted) infected 7 days previously with 1010 GE of DENV2 were injected i.v. with CFSE-labeled target cells pulsed with a pool of DENV2-derived immunodominant H-2b-restricted peptides (C51-59, NS2A8-15, NS4B99-107, and NS5237-245) at the indicated concentrations (n = 3–6 mice per group). After 4 h, splenocytes were harvested, analyzed by flow cytometry, and the percentage killing was calculated.
In vivo killing of I-Ab-restricted peptide-pulsed target cells in DENV2-infected mice
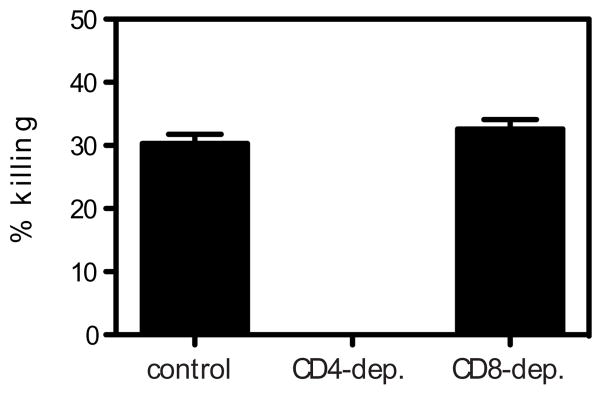
Although we found that the absence of CD4+ T cells had no effect on viral RNA levels on day 5 after infection, it was possible that CD4+ T cells could still be contributing to the anti-DENV2 host response by killing infected cells. We therefore performed an in vivo cytotoxicity assay using splenocytes pulsed with the three peptides that contain only CD4+ T cell epitopes (NS2B108-122, NS3198-212, and NS3237-251) and not NS4B96-110, as we wanted to measure only CD4+, not CD8+ T cell-mediated killing. Approximately 30% killing of target cells was observed (Fig. 7). No cytolytic activity was observed when CD4+ T cells were depleted, whereas depletion of CD8+ T cells had no effect on killing, demonstrating that the cytotoxicity was CD4+ T cell-mediated. Thus, DENV2-specific CD4+ T cells can exhibit in vivo cytolytic activity, although this effector function does not appear to significantly contribute to controlling primary DENV2 infection.
FIGURE 7. Cytotoxicity mediated by DENV2-specific CD4+ T cells.
In vivo killing of DENV2-derived I-Ab-restricted peptide-pulsed cells. IFN-α/βR−/− mice (control, CD4-depleted, or CD8-depleted) infected 7 days previously with 1010 GE of DENV2 were injected i.v. with CFSE-labeled target cells pulsed with the three epitopes that contain only CD4+ T cell epitopes (NS2B108-122, NS3198-212, and NS3237-51) (n = 6 control, 3 CD4-depleted, and 3 CD8-depleted mice). After 16 h, splenocytes were harvested, analyzed by flow cytometry, and the percentage killing was calculated.
Vaccination with DENV2 CD4+ T cell epitopes helps control viral load
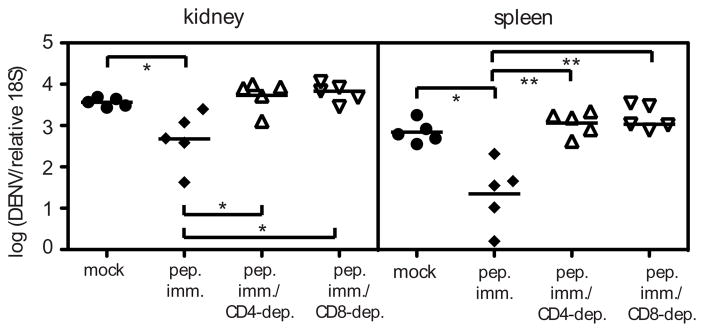
Having found that DENV2-specific CD4+ T cells can kill target cells, we next assessed whether immunization with CD4+ T cell epitopes could help control DENV2 infection. Mice were immunized with NS2B108-122, NS3198-212, and NS3237-251 before DENV2 infection, and we measured CD4+ T cell responses by ICS and viral RNA levels 4 days after infection. Peptide immunization resulted in enhanced CD4+ T cell cytokine responses (data not shown), and significantly lower viral loads in the kidney and spleen (Fig. 8). The protective effect was mediated by CD4+ T cells, as CD4-depletion before infection abrogated the protective effect. Similarly, CD8-depletion resulted in no protection, demonstrating that protection after CD4+ T cell peptide immunization requires both CD4+ and CD8+ T cells. These data suggest that CD4+ T cells elicited by immunization protect by helping the CD8+ T cell response. Thus, although CD4+ T cells are not required for the primary CD8+ T cell or Ab response, and the absence of CD4+ T cells had no effect on viral RNA levels, vaccination with CD4+ T cell epitopes can reduce viral loads.
FIGURE 8. Peptide immunization with CD4+ T cell epitopes results in enhanced DENV2 clearance.
IFN-α/βR−/− mice were immunized s.c. with 50 μg each of the three DENV peptides that contain only CD4+ T cell epitopes (NS2B108-122, NS3198-212, NS3237-51) in CFA, or mock-immunized with DMSO in CFA. Mice were boosted 11 days later with peptide in IFA, then challenged with 1011 GE of DENV2 13 days later, and sacrificed 4 days after infection. Separate groups of peptide-immunized mice were depleted of CD4+ or CD8+ T cells prior to infection. DENV2 RNA levels in the tissues were quantified by real-time RT-PCR and are expressed as DENV2 units normalized to 18S rRNA. Each symbol represents one mouse and the bar represents the geometric mean. * p < 0.05, ** p < 0 .01.
Discussion
Numerous studies have investigated the phenotype of DENV serotype cross-reactive T cells, which have been hypothesized to contribute to the pathogenesis of secondary heterologous infections, yet the actual contribution of T cells during DENV infection is unknown. The findings presented here, and in our previous study, reveal that CD8+ T cells play an important protective role in the response to primary DENV2 infection, whereas CD4+ T cells do not. CD4+ T cells expanded, were activated, and were located near CD8+ T cells and B cells in the spleen after DENV2 infection, yet they did not seem to affect the induction of the DENV2-specific CD8+ T cell or Ab responses. In fact, CD4+ T cell depletion had no effect on viral clearance. However, our data demonstrate that vaccination with CD4+ T cell epitopes prior to DENV infection can provide significant protection, supporting T cell peptide vaccination as a strategy for DENV immunization without the risk of ADE.
To the best of our knowledge, this is the first study to identify CD4+ T cell epitopes from DENV-infected mice. We found the DENV2-specific CD4+ T cells recognized epitopes from the NS2B, NS3, and NS4B proteins, and displayed a Th1 phenotype. CD4+ T cell epitopes have been identified in mice infected with other flaviviruses, including YFV, for which an I-Ab-restricted peptide from the E protein was identified (34), and WNV, for which six epitopes from the E and NS3 proteins were identified (35). DENV-derived epitopes recognized by human CD4+ T cells have been identified, primarily from NS proteins, including the highly conserved NS3 (10). One study identified numerous epitopes from the NS3200-324 region, and alignment of consensus sequences for DENV1-4 revealed that this region is more conserved (78%) than NS3 as a whole (68%) (14), suggesting that the region contains good candidates for a DENV T cell epitope-based vaccine. Interestingly, one of the NS3-derived epitopes we identified has also been described as a human CD4+ T cell epitope. This species cross-reactive NS3 peptide may bind human HLAs promiscuously, making it a good potential vaccine candidate. Another intriguing finding was that one of the CD4+ T cell epitopes identified in this study contained the most immunodominant of the CD8+ T cell epitopes we had identified previously. Overlapping epitopes have also been found in LCMV (36–38). The significance of overlapping epitopes is unknown, but is likely related to homology between MHC class I and MHC class II, and may be associated with proteasomal processing. Overlapping epitopes may turn out to be common once the complete CD4+ and CD8+ T cell responses to other pathogens are mapped.
CD4+ T cells are classically defined as helper cells, as they help B cell and CD8+ T cell responses. However, inflammatory stimuli can override the need for CD4+ T cell help, and therefore, the responses to many acute infections are CD4-independent (17). DENV2 replicates to high levels in IFN-α/βR−/− mice, the mice appear hunched and ruffled at the time of peak viremia, and they have intestinal inflammation (unpublished observations), suggesting that there is a significant inflammatory response to DENV2 in these mice. Accordingly, we found CD4+ T cells did not play a critical role in the immune response to primary DENV2 infection. The contribution of CD4+ T cells has been examined during infections with other flaviviruses. Adoptive transfer of primed CD4+ and CD8+ T cells, only in combination, protected mice from a lethal challenge with JEV when the cells and virus were administered intracerebrally (39). A protective role has been demonstrated for CD4+ T cells in response to WNV infection. One study showed CD4+ T cells contributed to protection by helping the Ab response and maintaining (but not priming) the CD8+ T cell response, and were important for clearance of virus from the CNS, but not the periphery (40). Another study found WNV-specific CD4+ T cells directly contributed to protection in the absence of CD8+ T cells and B cells, and produced IFN-γ, IL-2, and could kill peptide-pulsed cells (35). Thus, the contribution of CD4+ T cells to protection against flavivirus infection depends on the virus and experimental system.
Ab responses can be T cell-dependent or T cell-independent. In particular, the formation of GCs is thought to be CD4+ T cell-dependent, and is where high-affinity plasma cells and memory B cells are generated and where CSR can occur (20, 33). Classical CSR occurs in B cells after CD40L on activated CD4+ T cells binds CD40 on B cells. However, it is well established that CSR and Ab responses can occur in the absence of CD4+ T cells and organized secondary lymphoid structures (41). T-independent Ab responses to viruses have been demonstrated for vesicular stomatitis virus (42), rotavirus (43), and polyomavirus (44). In addition, EBV (via LMP1) can induce CD40-independent CSR (45), and mice deficient for CD40 or CD4+ T cells are able to mount an influenza-specific IgG response that is protective (46).
Similar to the studies mentioned above, our results demonstrate that the DENV2-specific IgG response at day 7 is CD4-independent. The lack of GC B cells in CD4-depleted mice shows that our CD4-depletions have a functional effect, and indicate anti-DENV IgG is being produced by extrafollicular B cells. It is possible that the absence of CD4+ T cells would have an effect on DENV2-specific Ab titers and/or neutralizing activity at later time points, however, the goal of our study was to determine how CD4+ T cells contribute to clearance of primary DENV2 infection, and we clearly show that the early anti-DENV2 Ab response is CD4-independent. The mechanisms by which T-independent Ab responses are induced are beginning to be elucidated. BLyS, also known as BAFF (B cell activating factor), and APRIL (a proliferation-inducing ligand) are induced on DCs after activation with IFN-α, IFN-γ, LPS, or CD40L, and mediate CD40-independent CSR in human B cells (47). BAFF and APRIL also induce CSR in mouse B cells in the absence of CD40 (48). T-independent CSR can also be induced via TLR signaling. For example, T-independent CSR to human papillomavirus virus-like particles occurs via TLR4 and MyD88 in mouse B cells (49), and TLR3 or TLR9 signaling can induce CSR in human B cells (50, 51). The pathways involved in mediating the CD4-independent CSR in DENV2-infected mice remain to be determined.
Like pathogen-specific Ab responses, the primary CD8+ T cell responses to many acute infections are also CD4-independent. CD4-independent CD8+ T cell responses have been demonstrated for Listeria monocytogenes (52, 53), LCMV (54), and influenza (55). Recently a mechanism for how DCs can activate CD8+ T cells in the absence of CD4+ T cell help has been described. DCs activated with TLR3 or TLR9 agonists, or by influenza infection, upregulated CD40L and activated naïve CD8+ T cells in the absence of CD4+ T cells (56). In accordance with the studies mentioned above, we found that the primary CD8+ T cell response to DENV2 did not depend on CD4+ T cells. In fact, we observed an enhanced DENV2-specific CD8+ T cell response in CD4-deficient mice compared with control mice at day 7, which has also been reported for influenza- (55) and WNV- (40) specific CD8+ T cell responses. In our study, this could be due to the depletion of Tregs, or an increased availability of cytokines (e.g. IL-2) in mice lacking CD4+ T cells. This enhanced CD8+ T cell response may explain why CD4-depleted mice have no differences in viral titers despite the fact that DENV2-specific CD4+ T cells demonstrate in vivo cytotoxicity.
Although CD4+ T cells did not play an important role in helping the Ab or CD8+ T cell responses, we found DENV2-specific CD4+ T cells could kill peptide-pulsed target cells in vivo. CD4+ T cells specific for other pathogens, including HIV (57) and influenza (58) demonstrate in vitro cytotoxicity. In vivo cytotoxicity assays have been used to show CD4+ T cell-mediated killing following infection with LCMV (59) and WNV (35). DENV-specific cytolytic human CD4+ T cell clones (60, 61) and a mouse (H-2d) CD4+ T cell clone (62) have been reported. Whether CD4+ T cells actually kill infected cells during DENV infection remains to be determined, but is possible, as MHC class II-expressing macrophages are targets of DENV infection (8). Based on the fact that CD4-depletion did not have a significant effect on viral clearance, it is unlikely that CD4+ T cell-mediated killing plays a major role in the anti-DENV2 response in this model.
An obvious caveat to using the IFN-α/βR−/− mice is that type I IFNs are known to help T cell and B cell responses through their actions on DCs, and can act directly on T cells (63). Type I IFNs were found to contribute to the expansion of CD4+ T cells following infection with LCMV, but not Listeria monocytogenes (64). Type I IFNs can induce the development of Th1 IFN-γ responses in human CD4+ T cells, but cannot substitute for IL-12 in promoting Th1 responses in mouse CD4+ T cells (65). Following Listeria infection, IL-12 synergized with type I IFN to induce IFN-γ production by CD4+ T cells (66). Although DENV does not replicate to detectable levels in wild-type mice, examining the CD4+ T cell response in these mice revealed that the same epitopes were recognized as in the IFN-α/βR−/− mice, but the magnitude of the epitope-specific response was greater in the IFN-α/βR−/− mice. This suggests that the high levels of viral replication in the IFN-α/βR−/− mice are sufficient to drive a DENV2-specific CD4+ IFN-γ response. While our results demonstrate a DENV2-specific CD4+ T cell response, including Th1-type cytokine production and cytotoxicity, in the absence of IFN-α/βR signaling, this response is not required for clearance of infection. It is, however, possible that CD4+ T cells contribute to protection during DENV infection of hosts with intact IFN responses.
We have previously shown that immunization with CD8+ T cells epitopes resulted in enhanced viral clearance (16), and the results presented here show that immunization with CD4+ T epitopes is also protective. Peptide immunization with CD4+ T cells epitopes has also been found to protect against WNV (35) and HSV-1 (67). Our results have significant implications for DENV vaccine development. Although a DENV vaccine is a global public health priority, designing a vaccine has been challenging, as it needs to induce protection against all four serotypes. The DENV vaccine candidates in development, some of which are in phase II trials, focus on eliciting an Ab response. The challenge is to induce and maintain a neutralizing Ab response against all four serotypes, as it is becoming increasingly clear that non-neutralizing Ab (or sub-neutralizing quantities of Ab) can actually enhance dengue disease (8, 9). A different approach would be a peptide vaccine that induces cell-mediated immunity, including both CD4+ and CD8+ T cell responses, which would not be able to prevent infection, but could reduce viral loads and disease severity, and would eliminate the risk of ADE. Such a vaccine should target highly conserved regions of the proteome, for example NS3, NS4B, and/or NS5, and ideally include epitopes conserved across all four serotypes. A vaccine containing only peptides from these particular NS proteins would also preclude the induction of any Ab against epitopes on the virion, which could enhance infection, or Ab against NS1, which could potentially contribute to pathogenesis (68). Our peptide vaccination was given along with CFA, which is commonly used in mice to induce Th1 responses (69), which was the type of response observed after natural DENV infection. CFA is not a vaccine adjuvant approved for human use, and thus, any peptide vaccine developed against DENV will have to be formulated with an adjuvant that is approved for human use and promotes a Th1 response. Numerous vaccine adjuvants are currently under development and can be tested in our experimental system in the future.
Although our results indicate CD4+ T cells do not make a significant contribution to controlling primary DENV2 infection, the characterization of the primary CD4+ T cell response and epitope identification will allow us to determine the role of CD4+ T cells during secondary homologous and heterologous infections. CD4+ T cells are often dispensable for the primary CD8+ T cell response to infection, but have been shown to be required for the maintenance of memory CD8+ T cell responses after acute infection (70). Finally, our findings support a DENV vaccine strategy that induces CD4+ T cell, in addition to CD8+ T cell, responses.
Acknowledgments
We thank Steven Lada for technical assistance, Dr. Carl Ware for mice, and Dr. Shane Crotty for reagents.
Footnotes
This study was supported by National Institutes of Health (NIH) grants AI077099 to S.S. and U54 A1057157 from the Southeast Regional Center of Excellence for Emerging Infections and Biodefense to P.F. Sparling, fellowships from The Center for Infectious Diseases and the Diabetes & Immune Disease National Research Institute to L.E.Y, and NIH contract HHSN272200900042C to A. S.
Disclosures
The authors have no conflicting financial interests.
References
- 1.Fu J, Tan BH, Yap EH, Chan YC, Tan YH. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90) Virology. 1992;188:953–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Dengue: Guidelines for diagnosis, treatment, prevention and control. World Health Organization (WHO) Press; Geneva, Switzerland: 2009. [PubMed] [Google Scholar]
- 3.Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. 2007;5:518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- 4.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 5.Guzman MG, Kouri G, Valdes L, Bravo J, Alvarez M, Vazques S, Delgado I, Halstead SB. Epidemiologic studies on Dengue in Santiago de Cuba, 1997. Am J Epidemiol. 2000;152:793–799. doi: 10.1093/aje/152.9.793. discussion 804. [DOI] [PubMed] [Google Scholar]
- 6.Morens DM. Antibody-dependent enhancement of infection and the pathogenesis of viral disease. Clin Infect Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- 7.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 8.Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 7:128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balsitis SJ, Williams KL, Lachica R, Flores D, Kyle JL, Mehlhop E, Johnson S, Diamond MS, Beatty PR, Harris E. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 6:e1000790. doi: 10.1371/journal.ppat.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev. 2008;225:300–313. doi: 10.1111/j.1600-065X.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 11.Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 12.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus PT, McMichael A, Malasit P, Screaton G. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 13.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, Khemnu N, Tangthawornchaikul N, Chotiyarnwong P, Sae-Jang K, Koch M, Jones Y, McMichael A, Xu X, Malasit P, Screaton G. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 14.Simmons CP, Dong T, Chau NV, Dung NT, Chau TN, Thaole TT, Hien TT, Rowland-Jones S, Farrar J. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J Virol. 2005;79:5665–5675. doi: 10.1128/JVI.79.9.5665-5675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yauch LE, Shresta S. Mouse models of dengue virus infection and disease. Antiviral Res. 2008;80:87–93. doi: 10.1016/j.antiviral.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol. 2009;182:4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bevan MJ. Helping the CD8(+) T-cell response. Nat Rev Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 18.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229:152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakanishi Y, Lu B, Gerard C, Iwasaki A. CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature. 2009;462:510–513. doi: 10.1038/nature08511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prestwood TR, Prigozhin DM, Sharar KL, Zellweger RM, Shresta S. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J Virol. 2008;82:8411–8421. doi: 10.1128/JVI.00611-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, Lund O, Lundegaard C, Nielsen M, Ponomarenko J, Sette A, Zhu Z, Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambeth CR, White LJ, Johnston RE, de Silva AM. Flow cytometry-based assay for titrating dengue virus. Journal of clinical microbiology. 2005;43:3267–3272. doi: 10.1128/JCM.43.7.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houng HH, Hritz D, Kanesa-thasan N. Quantitative detection of dengue 2 virus using fluorogenic RT-PCR based on 3′-noncoding sequence. J Virol Methods. 2000;86:1–11. doi: 10.1016/s0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- 27.Lacher MD, Tiirikainen MI, Saunier EF, Christian C, Anders M, Oft M, Balmain A, Akhurst RJ, Korn WM. Transforming growth factor-beta receptor inhibition enhances adenoviral infectability of carcinoma cells via up-regulation of Coxsackie and Adenovirus Receptor in conjunction with reversal of epithelial-mesenchymal transition. Cancer Res. 2006;66:1648–1657. doi: 10.1158/0008-5472.CAN-05-2328. [DOI] [PubMed] [Google Scholar]
- 28.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund JM, Hsing L, Pham TT, Rudensky AY. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanteri MC, O’Brien KM, Purtha WE, Cameron MJ, Lund JM, Owen RE, Heitman JW, Custer B, Hirschkorn DF, Tobler LH, Kiely N, Prince HE, Ndhlovu LC, Nixon DF, Kamel HT, Kelvin DJ, Busch MP, Rudensky AY, Diamond MS, Norris PJ. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest. 2009;119:3266–3277. doi: 10.1172/JCI39387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arens R, Wang P, Sidney J, Loewendorf A, Sette A, Schoenberger SP, Peters B, Benedict CA. Cutting edge: murine cytomegalovirus induces a polyfunctional CD4 T cell response. J Immunol. 2008;180:6472–6476. doi: 10.4049/jimmunol.180.10.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng L, Kurane I, Okamoto Y, Ennis FA, Brinton MA. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J Virol. 1996;70:3108–3117. doi: 10.1128/jvi.70.5.3108-3117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Most RG, Harrington LE, Giuggio V, Mahar PL, Ahmed R. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology. 2002;296:117–124. doi: 10.1006/viro.2002.1432. [DOI] [PubMed] [Google Scholar]
- 35.Brien JD, Uhrlaub JL, Nikolich-Zugich J. West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol. 2008;181:8568–8575. doi: 10.4049/jimmunol.181.12.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Homann D, Lewicki H, Brooks D, Eberlein J, Mallet-Designe V, Teyton L, Oldstone MB. Mapping and restriction of a dominant viral CD4+ T cell core epitope by both MHC class I and MHC class II. Virology. 2007;363:113–123. doi: 10.1016/j.virol.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mothe BR, Stewart BS, Oseroff C, Bui HH, Stogiera S, Garcia Z, Dow C, Rodriguez-Carreno MP, Kotturi M, Pasquetto V, Botten J, Crotty S, Janssen E, Buchmeier MJ, Sette A. Chronic lymphocytic choriomeningitis virus infection actively down-regulates CD4+ T cell responses directed against a broad range of epitopes. J Immunol. 2007;179:1058–1067. doi: 10.4049/jimmunol.179.2.1058. [DOI] [PubMed] [Google Scholar]
- 38.Dow C, Oseroff C, Peters B, Nance-Sotelo C, Sidney J, Buchmeier M, Sette A, Mothe BR. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. J Virol. 2008;82:11734–11741. doi: 10.1128/JVI.00435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murali-Krishna K, Ravi V, Manjunath R. Protection of adult but not newborn mice against lethal intracerebral challenge with Japanese encephalitis virus by adoptively transferred virus-specific cytotoxic T lymphocytes: requirement for L3T4+ T cells. J Gen Virol. 1996;77(Pt 4):705–714. doi: 10.1099/0022-1317-77-4-705. [DOI] [PubMed] [Google Scholar]
- 40.Sitati EM, Diamond MS. CD4+ T Cell Responses are Required for Clearance of West Nile Virus from the Central Nervous System. J Virol. 2006 doi: 10.1128/JVI.01650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagarasan S, Honjo T. T-Independent immune response: new aspects of B cell biology. Science. 2000;290:89–92. doi: 10.1126/science.290.5489.89. [DOI] [PubMed] [Google Scholar]
- 42.Freer G, Burkhart C, Ciernik I, Bachmann MF, Hengartner H, Zinkernagel RM. Vesicular stomatitis virus Indiana glycoprotein as a T-cell-dependent and -independent antigen. J Virol. 1994;68:3650–3655. doi: 10.1128/jvi.68.6.3650-3655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franco MA, Greenberg HB. Immunity to rotavirus in T cell deficient mice. Virology. 1997;238:169–179. doi: 10.1006/viro.1997.8843. [DOI] [PubMed] [Google Scholar]
- 44.Szomolanyi-Tsuda E, Brien JD, Dorgan JE, Garcea RL, Woodland RT, Welsh RM. Antiviral T-cell-independent type 2 antibody responses induced in vivo in the absence of T and NK cells. Virology. 2001;280:160–168. doi: 10.1006/viro.2000.0766. [DOI] [PubMed] [Google Scholar]
- 45.He B, Raab-Traub N, Casali P, Cerutti A. EBV-encoded latent membrane protein 1 cooperates with BAFF/BLyS and APRIL to induce T cell-independent Ig heavy chain class switching. J Immunol. 2003;171:5215–5224. doi: 10.4049/jimmunol.171.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee BO, Rangel-Moreno J, Moyron-Quiroz JE, Hartson L, Makris M, Sprague F, Lund FE, Randall TD. CD4 T cell-independent antibody response promotes resolution of primary influenza infection and helps to prevent reinfection. J Immunol. 2005;175:5827–5838. doi: 10.4049/jimmunol.175.9.5827. [DOI] [PubMed] [Google Scholar]
- 47.Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Castigli E, Wilson SA, Scott S, Dedeoglu F, Xu S, Lam KP, Bram RJ, Jabara H, Geha RS. TACI and BAFF-R mediate isotype switching in B cells. J Exp Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang R, Murillo FM, Delannoy MJ, Blosser RL, Yutzy WHt, Uematsu S, Takeda K, Akira S, Viscidi RP, Roden RB. B lymphocyte activation by human papillomavirus-like particles directly induces Ig class switch recombination via TLR4-MyD88. J Immunol. 2005;174:7912–7919. doi: 10.4049/jimmunol.174.12.7912. [DOI] [PubMed] [Google Scholar]
- 50.He B, Qiao X, Cerutti A. CpG DNA induces IgG class switch DNA recombination by activating human B cells through an innate pathway that requires TLR9 and cooperates with IL-10. J Immunol. 2004;173:4479–4491. doi: 10.4049/jimmunol.173.7.4479. [DOI] [PubMed] [Google Scholar]
- 51.Xu W, Santini PA, Matthews AJ, Chiu A, Plebani A, He B, Chen K, Cerutti A. Viral double-stranded RNA triggers Ig class switching by activating upper respiratory mucosa B cells through an innate TLR3 pathway involving BAFF. J Immunol. 2008;181:276–287. doi: 10.4049/jimmunol.181.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shedlock DJ, Whitmire JK, Tan J, MacDonald AS, Ahmed R, Shen H. Role of CD4 T cell help and costimulation in CD8 T cell responses during Listeria monocytogenes infection. J Immunol. 2003;170:2053–2063. doi: 10.4049/jimmunol.170.4.2053. [DOI] [PubMed] [Google Scholar]
- 54.Ahmed R, Butler LD, Bhatti L. T4+ T helper cell function in vivo: differential requirement for induction of antiviral cytotoxic T-cell and antibody responses. J Virol. 1988;62:2102–2106. doi: 10.1128/jvi.62.6.2102-2106.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol. 2002;76:12388–12393. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson S, Zhan Y, Sutherland RM, Mount AM, Bedoui S, Brady JL, Carrington EM, Brown LE, Belz GT, Heath WR, Lew AM. Selected Toll-like receptor ligands and viruses promote helper-independent cytotoxic T cell priming by upregulating CD40L on dendritic cells. Immunity. 2009;30:218–227. doi: 10.1016/j.immuni.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norris PJ, Moffett HF, Yang OO, Kaufmann DE, Clark MJ, Addo MM, Rosenberg ES. Beyond help: direct effector functions of human immunodeficiency virus type 1-specific CD4(+) T cells. J Virol. 2004;78:8844–8851. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor SF, Bender BS. Beta 2-microglobulin-deficient mice demonstrate class II MHC restricted anti-viral CD4+ but not CD8+ CTL against influenza-sensitized autologous splenocytes. Immunol Lett. 1995;46:67–73. doi: 10.1016/0165-2478(95)00024-y. [DOI] [PubMed] [Google Scholar]
- 59.Jellison ER, Kim SK, Welsh RM. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol. 2005;174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- 60.Gagnon SJ, Zeng W, Kurane I, Ennis FA. Identification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clones. J Virol. 1996;70:141–147. doi: 10.1128/jvi.70.1.141-147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurane I, Meager A, Ennis FA. Dengue virus-specific human T cell clones. Serotype crossreactive proliferation, interferon gamma production, and cytotoxic activity. J Exp Med. 1989;170:763–775. doi: 10.1084/jem.170.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rothman AL, Kurane I, Ennis FA. Multiple specificities in the murine CD4+ and CD8+ T-cell response to dengue virus. J Virol. 1996;70:6540–6546. doi: 10.1128/jvi.70.10.6540-6546.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 64.Havenar-Daughton C, Kolumam GA, Murali-Krishna K. Cutting Edge: The direct action of type I IFN on CD4 T cells is critical for sustaining clonal expansion in response to a viral but not a bacterial infection. J Immunol. 2006;176:3315–3319. doi: 10.4049/jimmunol.176.6.3315. [DOI] [PubMed] [Google Scholar]
- 65.Rogge L, D’Ambrosio D, Biffi M, Penna G, Minetti LJ, Presky DH, Adorini L, Sinigaglia F. The role of Stat4 in species-specific regulation of Th cell development by type I IFNs. J Immunol. 1998;161:6567–6574. [PubMed] [Google Scholar]
- 66.Way SS, Havenar-Daughton C, Kolumam GA, Orgun NN, Murali-Krishna K. IL-12 and type-I IFN synergize for IFN-gamma production by CD4 T cells, whereas neither are required for IFN-gamma production by CD8 T cells after Listeria monocytogenes infection. J Immunol. 2007;178:4498–4505. doi: 10.4049/jimmunol.178.7.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.BenMohamed L, Bertrand G, McNamara CD, Gras-Masse H, Hammer J, Wechsler SL, Nesburn AB. Identification of novel immunodominant CD4+ Th1-type T-cell peptide epitopes from herpes simplex virus glycoprotein D that confer protective immunity. J Virol. 2003;77:9463–9473. doi: 10.1128/JVI.77.17.9463-9473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin CF, Wan SW, Cheng HJ, Lei HY, Lin YS. Autoimmune pathogenesis in dengue virus infection. Viral Immunol. 2006;19:127–132. doi: 10.1089/vim.2006.19.127. [DOI] [PubMed] [Google Scholar]
- 69.Billiau A, Matthys P. Modes of action of Freund’s adjuvants in experimental models of autoimmune diseases. J Leukoc Biol. 2001;70:849–860. [PubMed] [Google Scholar]
- 70.Sun JC, Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]