SUMMARY
It was previously shown that the NF-κB pathway is downstream of oncogenic Notch1 in T cell acute lymphoblastic leukemia (T-ALL). Here we visualize Notch-induced NF-κB activation using both human T-ALL cell lines and animal models. We demonstrate that Hes1, a canonical Notch target and transcriptional repressor, is responsible for sustaining IKK activation in T-ALL. Hes1 exerts its effects by repressing the deubiquitinase CYLD, a negative IKK complex regulator. CYLD expression was found to be significantly suppressed in primary T-ALL. Finally, we demonstrate that IKK inhibition is a promising option for the targeted therapy of T-ALL as specific suppression of IKK expression and function affected both the survival of human T-ALL cells and the maintenance of the disease in vivo.
INTRODUCTION
NF-κB is an important regulator of cell survival, proliferation and differentiation and is frequently involved in malignant transformation (Karin, 2006). NF-κB is mainly activated in response to pro-inflammatory stimuli such as TNFα or IL1β, components of pathogenic bacteria and viruses or following TCR signaling in T-lymphocytes. Regardless of the activation of specific receptors that associate to different effectors, canonical NF-κB signaling invariably leads to the activation of IκB kinase (IKK) complex composed by two catalytic and one regulatory subunits called IKKα, IKKβ and NEMO (IKKγ), respectively. The main target of activated IKK complex is the NF-κB inhibitor, IκB, that it is rapidly phosphorylated in specific serine residues thus targeting the protein for ubiquitination, leading to proteasomal degradation. Degradation of IκB is sufficient to induce the nuclear translocation of the transcription factor that results in specific gene activation (Vallabhapurapu and Karin, 2009)). IκBα itself is one of the main transcriptional targets for NF-κB activity thus providing an efficient mechanism for signal termination (Hoffmann et al., 2002). However, other mechanisms are required to maintain the non-active state of NF-κB in the absence of stimulation but also to attenuate the signal following chronic stimulation of the pathway. They involve the deubiquitination enzymes A20 and CYLD, which remove the activator K63 ubiquitin chains from different elements of the NF-κB signalosome (Brummelkamp et al., 2003; Reiley et al., 2007; Trompouki et al., 2003; Wertz et al., 2004).
Despite the huge amount of gathered data on NF-κB activation, not much is known about the crosstalk of NF-κB with other signaling pathways, including pathways that control tissue differentiation like the Notch cascade. Notch receptors are crucial regulators of cell differentiation in multiple systems (Reviewed by (Egan et al., 1998)). Activation of Notch by Jagged or Delta ligands results in the cleavage of the intracellular fragment of the receptor by a γ-secretase activity (Mumm et al., 2000). Once released from the extracellular part of the molecule, the intracellular cleaved Notch translocates to the nucleus to bind the ubiquitous RBPjκ factor and to activate transcription of a number of target genes, including the Hes family of transcription factors (Iso et al., 2003; Jarriault et al., 1995). The classical function of Hes family proteins is the transcriptional repression of pro-differentiation genes by direct interaction with specific sequences of their promoters or by the association with other bHLH factors and combinatorial DNA binding (Kageyama and Ohtsuka, 1999; Kuroda et al., 1999). The Notch pathway was recently shown to be oncogenic in a wide variety of cancers, most notably in T cell acute lymphoblastic leukemia (T-ALL), a devastating pediatric tumor arising by the transformation of hematopoietic progenitor cells. In this disease, the majority of patients harbor either activating mutations of Notch1 or inactivating mutations affecting negative regulators of the Notch pathway (Aifantis et al., 2008)
Recent evidence has suggested an interaction between Notch and NF-κB, more specifically in the context of cell transformation. Cross-talk between the Notch and NF-κB pathways have been proposed in human T-ALL and in mouse models of T cell leukemia (Screpanti et al., 2003; Shin et al., 2006; Vilimas et al., 2007). More specifically, we have previously shown that Notch activation can induce the expression of a large fraction of classical NF-κB gene targets in T cell progenitors. We have also demonstrated that this Notch-induced NF-κB activation requires both the transcriptional activity of Notch and the function of the IKK complex (Vilimas et al., 2007). However, these studies failed to provide a definitive demonstration that this interaction can be translated into a new therapeutic tool. Moreover, neither the mechanism explaining the ability of Notch to impinge on IKK (and thus NF-κB) activation nor the importance of IKK activation for the maintenance of T-ALL in vivo was addressed. These key issues could introduce NF-κB inhibition as an effective therapy for the treatment of T-ALL and are all addressed here.
RESULTS
Oncogenic Notch induces NF-κB activation both in vitro and in vivo
We attempted here to visualize in vivo the Notch-induced NF-κB activation using an animal model of T-ALL. We combined an established transplantation model with a NF-κB-dependent in vivo luciferase reporter strain (κBLuc) to measure the activation of this pathway. We transplanted ICN1+κBLuc+ progenitors in irradiated hosts and monitored the progression of the disease and the activation of the reporter using both FACS analysis and in vivo bioluminescence. Disease was induced as evidenced by the appearance of leukemic CD4+CD8+ blast cells in the peripheral blood (not shown). Early during T-ALL, visual induction of NF-κB activation was undetectable, probably due to the low number of transformed cells at this stage of the disease. However, NF-κB activity increased rapidly and was readily detectable 3 wks post-cell transplantation with “hotspots” in tissues containing lymphoid cells, including the thymus and bone marrow areas (Figure 1A).
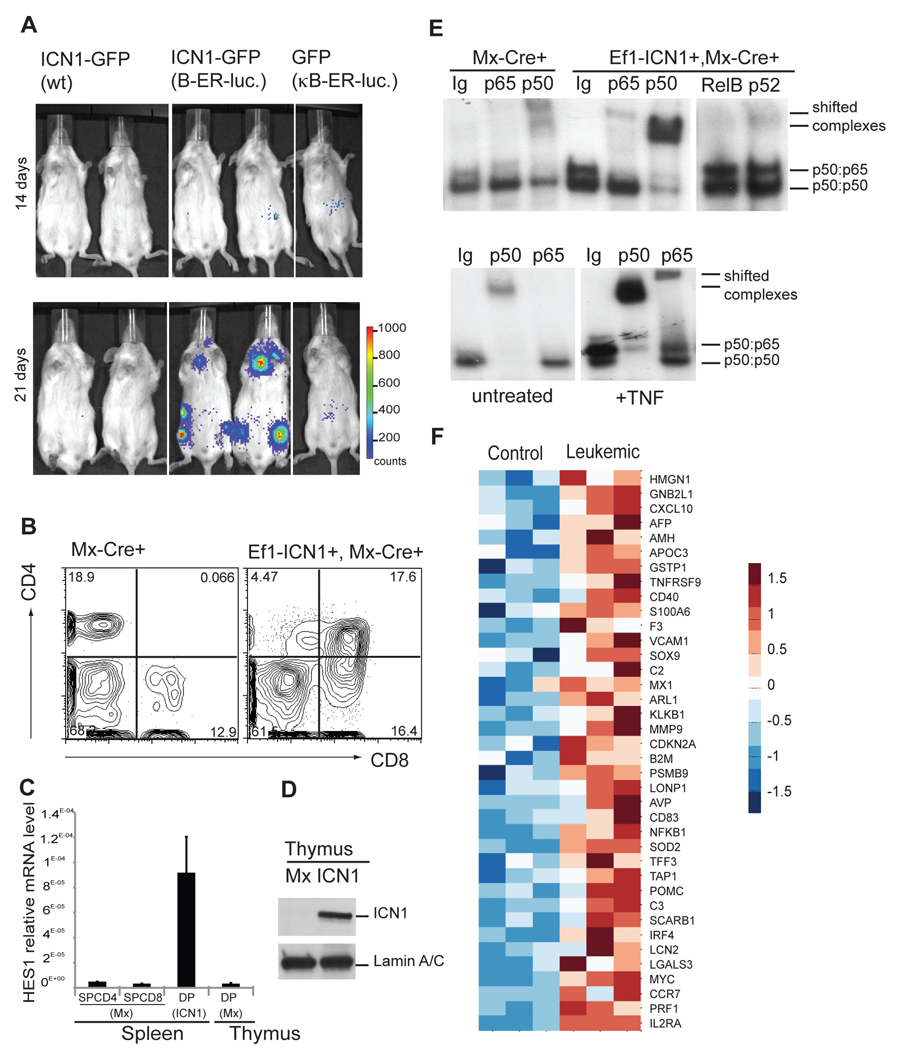
Figure 1. Oncogenic Notch induces NF-κB activation in T-ALL animal models.
(A) NF-κB luciferase reporter activity was detected in mice transplanted with: ICN1-GFP (WT), ICN1-GFP (NFκB-ER-luc) or GFP (NFκB-ER-luc). Bioluminescent imaging of mice 9 and 21 days after transplantation is shown. (B) Peripheral blood CD4/CD8 FACS analysis; (C) qPCR from sorted cells as indicated. Error bars denote +/− Standard Deviation. (D) Protein levels of Notch1-IC (ICN1) and Lamin A/C in nuclear extracts of thymocytes from Mx-Cre (Mx) and Ef1α-ICN1, Mx-Cre (ICN1) mice 4 weeks after pIpC injection. (E) NF-κB DNA binding activity was analyzed by EMSA using nuclear extracts of thymocytes from MxCre, Ef1α-ICN1 Mx-Cre mice 4 weeks after pIpC injection and wt thymocytes either untreated (Co) or treated with mTNFα. (F) Heat-map shows expression of NF-κB target genes selected from the literature (http://www.nf-kb.org) up-regulated in leukemic DP cells compared to control thymic DP cells by unpaired one-side t-test with α=0.05. See also related figure S1.
To further prove that Notch was responsible for induction of NF-κB activation, we used a distinct, recently generated animal model of the disease that does not require transplantation. In this model, the oncogenic ICN1 mutant is “knocked-in” the EF1a locus and induced by cre-recombinase expression (Buonamici et al., 2009). ICN1 expression rapidly induced T-ALL (Figure 1B) that overexpressed nuclear Notch1-IC and its target gene Hes1 (Figures 1C, D). To test whether NF-κB was activated in the leukemic cells, we sorted the CD4+CD8+ population and used a conventional EMSA approach. As shown in Figure 1E, this leukemic population showed significant NF-κB activation, as indicated by the increased DNA-binding activity. Antibody shift experiments demonstrated a predominance of p50-p50 (Nfκb1:Nfκb1) and p50:p65 (Nfκb1:RelA) dimers.
To support that Notch is responsible for promoting NF-κB activity in T-ALL cells, we measured the activation of a virally-driven NF-κB-dependent EGFP reporter in a representative human cell line (KOPT-K) that carries Notch mutations (O'Neil et al., 2007). We either kept the cells untreated or treated them with gamma-secretase inhibitors (GSI). Inhibition of the Notch pathway by GSI reduced the activation of the NF-κB-dependent EGFP reporter, the expression of the Notch-target genes Hes1, Deltex1, the NF-κB target Bcl2A1, as well as NF-κB DNA binding activity (Figure S1A–C).
These combined experiments support the hypothesis that NF-κB is targeted by Notch and demonstrate that this cross-talk between the two pathways takes place also in in vivo models of T-ALL. We were thus curious to see whether this in vivo Notch-induced NF-κB activation leads to the activation of a subset of genes that have been characterized as direct or indirect NF-κB targets. We have thus performed a meta-analysis of the transcriptional profiling comparisons between “control” and “leukemic” T cells performed by Li et al. (Li et al., 2008). As shown in Figure 1F, a large number of know NF-κB target genes, as proposed by multiple studies, were significantly induced in the CD4+8+Notch1-IC+ tumor samples. To further identify genes induced in leukemia that are direct NF-κB targets, we have combined our analysis to a recent report that combined whole-genome chromatin immunoprecipitation data (ChIP-seq) and RNA-sequencing data, identifying NF-κB p65 direct transcriptional targets (Kasowski et al., 2010). As shown in Figure S1D, we identified a significant number of NF-κB direct targets that are significantly upregulated in T-ALL cells. The combination of these data strongly suggest that a NF-κB gene expression “signature” is characterizing leukemic T cells in Notch-induced T-ALL.
Hes1 induces NF-κB activity and nuclear translocation of p65 by facilitating IκBα degradation
Next, we addressed the mechanism of Notch-induced NF-κB activation. To answer this question, we measured the activity of a NF-κB-dependent reporter in response to ectopic expression of ICN1 or its main target Hes1. We found that ectopic expression of doxycycline-inducible Hes1 (Figure 2A) or non-inducible flag-Hes1 resulted in a dose-dependent activation of NF-κB, comparable to that induced by ICN1 (Figure S2). Activation of NF-κB was concomitant with the nuclear accumulation of p65 in Hes1 expressing cells, as detected by immunofluorescence (Figure 2B).
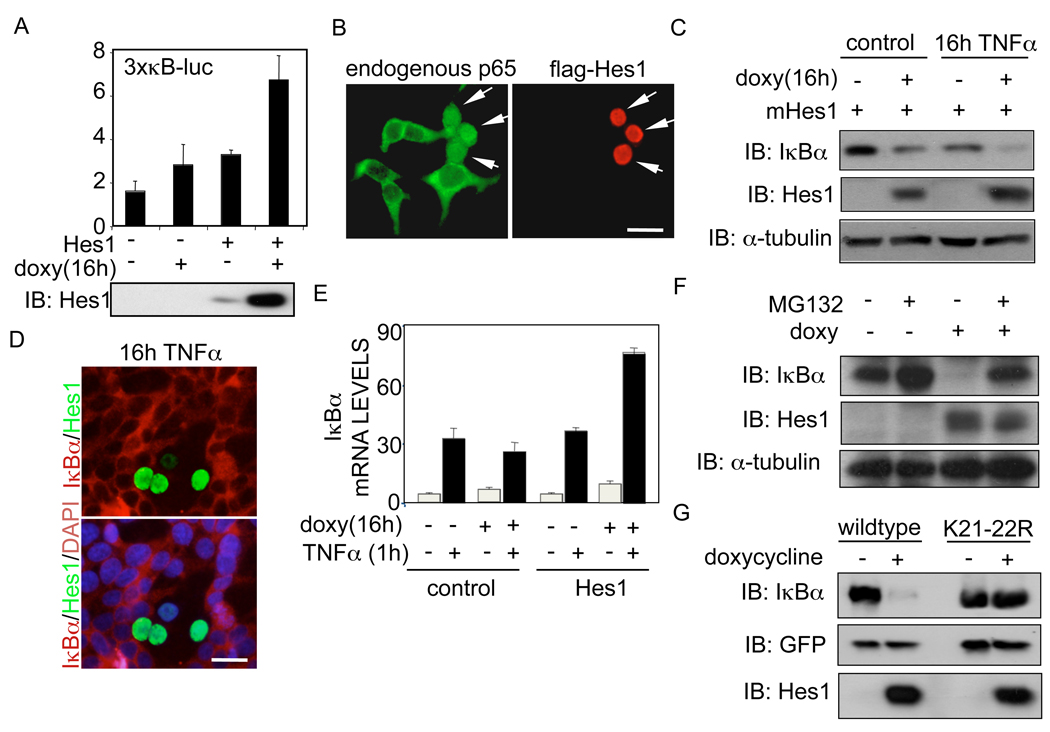
Figure 2. Hes1 enhances NF-κB-dependent activity by inducing IκBα degradation.
(A) Luciferase assay with the 3xκB-luc-reporter in HEK-293T cells transfected with a doxycycline-inducible Hes1 construct. Induction of Hes1 after doxycycline treatment is shown in the lower panel. (B) Immunofluorescence of HEK-293T transfected with flag-Hes1 showing the expression pattern of transfected Hes1 and endogenous p65. (C) Western blot of endogenous IκBα in HEK-293T cells transfected with inducible Hes1 in the presence or absence of TNFα. (D) Lack of IκBα immunostaining in cells transfected with ectopic Hes1. (E) Quantitative RT-PCR to determine the expression levels of IκBα mRNA in the presence or absence of Hes1. Error bars denote +/− Standard Deviation. (F) Western blot of IκBα levels in HEK-293T transfected with the doxycycline-inducible Hes1 construct in the presence or absence of the proteasome inhibitor MG132. (G) Western blot showing the levels of IκBαwt or IκBαK21-22R cotransfected with inducible Hes1. See also related figure S2.
We analyzed whether modification of the IκBα protein levels were responsible for the increased NF-κB-transcriptional activity induced by Hes1. We found that ectopic Hes1 expression correlated with a reduction in IκBα protein levels in the absence of stimulation that was further enhanced by chronic TNFα treatment (Figure 2C). Most importantly, IF staining of HEK-293T cells demonstrated that after TNFα treatment there was a complete absence of endogenous IκBα in cells expressing ectopic Hes1, whereas the rest of the cells have already re-synthesized IκBα (Figure 2D).
As Hes1 is a well-characterized transcriptional repressor (Iso et al., 2003), we speculated that Hes1 was regulating IκBα at the level of transcription. To test this possibility, we performed qRT-PCR from cells transfected with the control vector or the inducible Hes1 construct, untreated or treated with TNFα and/or doxycycline as indicated. We found that Hes1 expression did not exert any inhibitory effect on either basal or TNFα-induced IκBα transcription, but rather increased IκBα transcription as it is an NF-κB target (Figure 2E). These results suggested that Hes1 was modulating IκBα protein stability. In agreement with this, incubation with the proteasomal inhibitor MG132 or mutation of lysines 21 and 22 of IκBα into arginine, which are required for K48-linked ubiquitination (Rodriguez et al., 1996), completely abrogated the effects of Hes1 on IκBα levels (Figure 2F and 2G). These results demonstrated that Hes1 is able to induce IκBα degradation, which results in increased NF-κB activity.
Hes1 facilitates IκBα degradation by modifying IKK activity
As it is well established that IκBα degradation is regulated by the IKK complex, in response to the specific phosphorylation by IKKβ, we tested the effect of the IKKβ inhibitor BAY65-5811 on Hes1-induced degradation of IκBα. Inhibition of the IKK activity completely blocked the phopshorylation of IKK and the degradation of IκBα induced by ectopic expression of Hes1 in the presence of chronic TNFα treatment (as detected with the specific antibody recognizing phosphorylated serines 180 and 181 of IKKα and IKKβ respectively and IκBα) (Figure 3A). Next, we tested whether hes1 expression affected the phosphorylation/activation status of IKKα or β that are the catalytic subunits of the IKK complex. Figure 3B shows that doxycycline-inducible Hes1 expression significantly and specifically increased the amounts of active/phosphorylated HA-IKKβ but not HA-IKKα. As a control we show that this effect was not induced by doxycycline alone (Figure S3A). To further demonstrate the requirement of IKKβ in the Hes1-induced degradation of IκBα, we expressed HA-IκBα together with the inducible Hes1 construct in IKKβ-deficient MEFs. As shown in Figure 3C, Hes1 was not sufficient to induce IκBα degradation in cells lacking IKKβ, whereas reintroduction of IKKβ restored the ability of Hes1 to reduce IκBα levels. Finally, ectopic expression of the dominant negative form of TAK1 (TAK1K63W), the specific kinase that activates IKKβ, was sufficient to block the IκBα-degradation imposed by Hes1 (Figure 3D). These results indicate that Hes1 induces IκBα degradation by facilitating the activation of the IKK complex in both basal conditions and following TNFα treatment, and that this effect takes place upstream of IKKβ phosphorylation by TAK1.
Figure 3. Hes1 facilitates IκBα degradation by regulating IKKβ activity.
Western blots of cells transfected with an inducible Hes1 vector and treated for 16h with doxycycline and TNFα as indicated. (A) HEK-293T cells were treated with the IKK inhibitor BAY65-1185, (B) Cells were cotransfected with HA-IKKα or HA-IKKβ, (C) IKKβ-deficient cells were reconstituted with HA-IKKβ and (D) HEK-293T cells were transfected with the dominant negative TAK1 (K63W) construct. See also related figure S3.
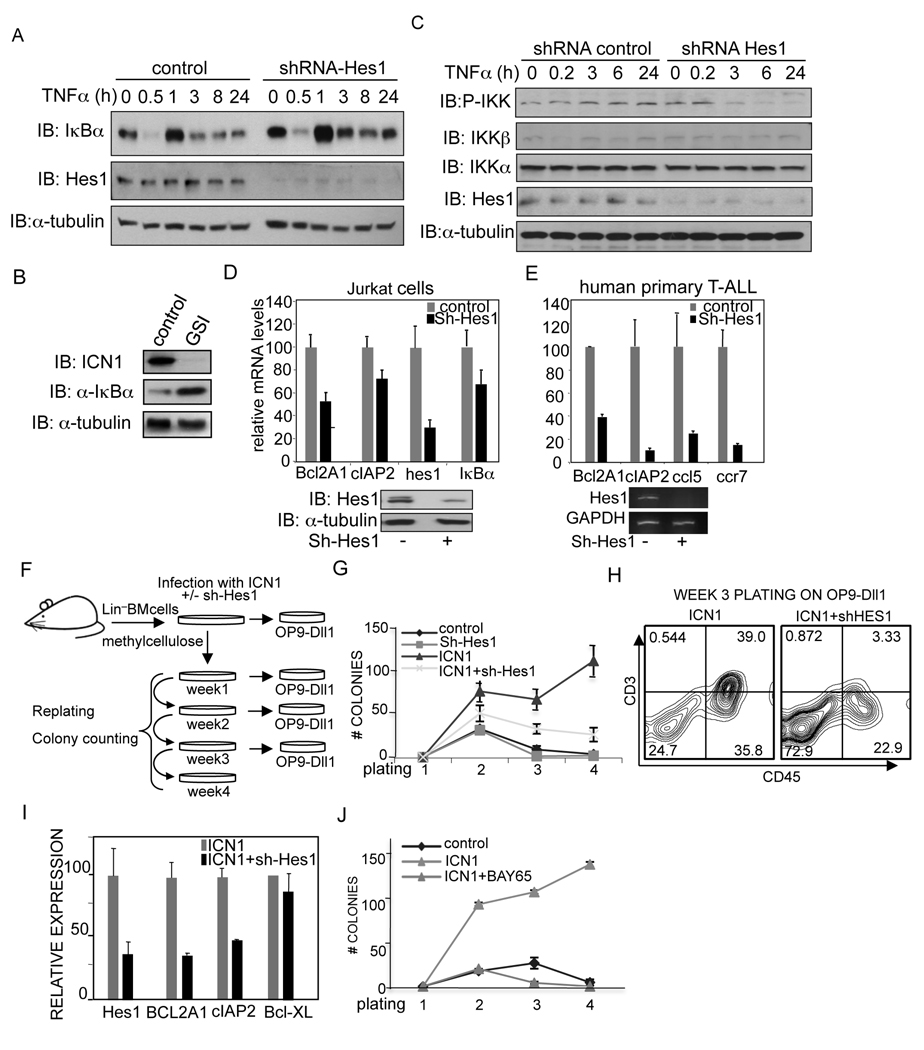
Hes1 is responsible for sustaining IKK/NF-κB activity in Notch-dependent leukemia
Based on these results, we speculated that the NF-κB activation that occurs in T-ALL with aberrant Notch activity might be mediated through Hes1. We confirmed that Notch1 activation (as detected by the Val1744 antibody) was mainly, although not exclusively, found in T-ALL lines, inversely correlating with the protein levels of IκBα (Figure S3B). To determine the contribution of Hes1 to the regulation of IκBα in T-ALL, we infected Jurkat cells with a lentiviral construct containing the shRNA against Hes1 or the control shRNA. Hes1 knockdown resulted in increased IκBα levels in non-induced cells and even more significant after chronic TNFα treatment (Figure 4A). Comparable results were obtained by inhibiting Notch activation in Jurkat cells with GSI (Figure 4B). Moreover, chronic activation of the endogenous IKK kinase complex following TNFα treatment was eliminated in cells with reduced Hes1 expression (Figure 4C). Next, we tested whether NF-κB activation was regulated by Hes1 in these cells and found a reduction in the levels of several NF-κB-target genes such as Bcl2A1 or cIAP2 by knocking down Hes1 in both Jurkat and most important, in primary human T-ALL cells (Figure 4D and 4E). Identical effects were obtained in Jurkat cells treated with GSI (Figure S4A). To study the functional relevance of Hes1 in cells containing active Notch ex vivo, we transduced lineage-depleted bone-marrow cells with a lentiviral vector expressing the oncogenic ICN1 alone or together with the Hes1-shRNA construct. Cells were serially plated every week and counted as an estimated measure of self-renewal capacity (Lavau et al., 1997) (Figure 4F). We found that ICN1-expressing cells exhibited a higher clonogenic capacity than the cells co-expressing the Hes1-shRNA as indicated by the increased number of colonies in these cultures (Figure 4G). Importantly, ICN1-transduced progenitors were able to produce increasing number of CD3+ cells when plated on OP9-Dll1 stroma compared to Hes1-shRNA-expressing cells (Figure 4H and Table S1). Moreover, in the absence of Hes1, cells expressing ICN1 showed an elevated number of apoptotic cells as measured by Annexin-V binding (Figure S4B) tconcomitant with a decrease in the expression of the pro-survival genes Bcl2A1 and cIAP2 (Figure 4I). To determine whether NF-κB provided pro-survival signals in ICN1-transduced cells, we plated these cells in the presence of the IKK inhibitor BAY65-5811. In these conditions, expression of oncogenic ICN1 was not sufficient to support colony and cell growth (Figure 4J).
Figure 4. Inhibition of Hes1 in cells with active Notch1 prevents NF-κB activity leading to increased apoptosis.
(A) Jurkat cells transduced with scrambled shRNA or Hes1-shRNA (80 to 90% reduction in Hes1 protein levels) were analyzed for the levels of IκBα protein during TNFα treatment, (B) IκBα is restored in the presence of GSI (L685,458) (C) levels of P-IKK in control and Hes1 knockdown cells by western blot and (D) expression of NF-κB-target genes by qRT-PCR in Jurkat or (E) primary human T-ALL leukemic cells transduced with the control or Hes1-shRNA lentivirus. (F) Experimental design to test the contribution of Hes1 to ICN1 transduced cells. (G–I) Primary mouse Lin− bone marrow progenitors transduced with the indicated lentivirus. (G) Colonies obtained from serial replatings on methylcellulose were counted. (H) T-cell differentiation was determined by CD3 expression after 7 days in co-culture on OP9-Dll1. One representative experiments out of three is shown. (I) Expression levels of the indicated genes were tested by qRT-PCR after 2 weeks in culture. (J) Number of colonies obtained in serial plating from ICN1 transduced cells in the presence or absence of BAY65-5811. Error bars denote +/− Standard Deviation. See also related figure S4.
The expression of the deubiquitinase CYLD is suppressed in human T-ALL
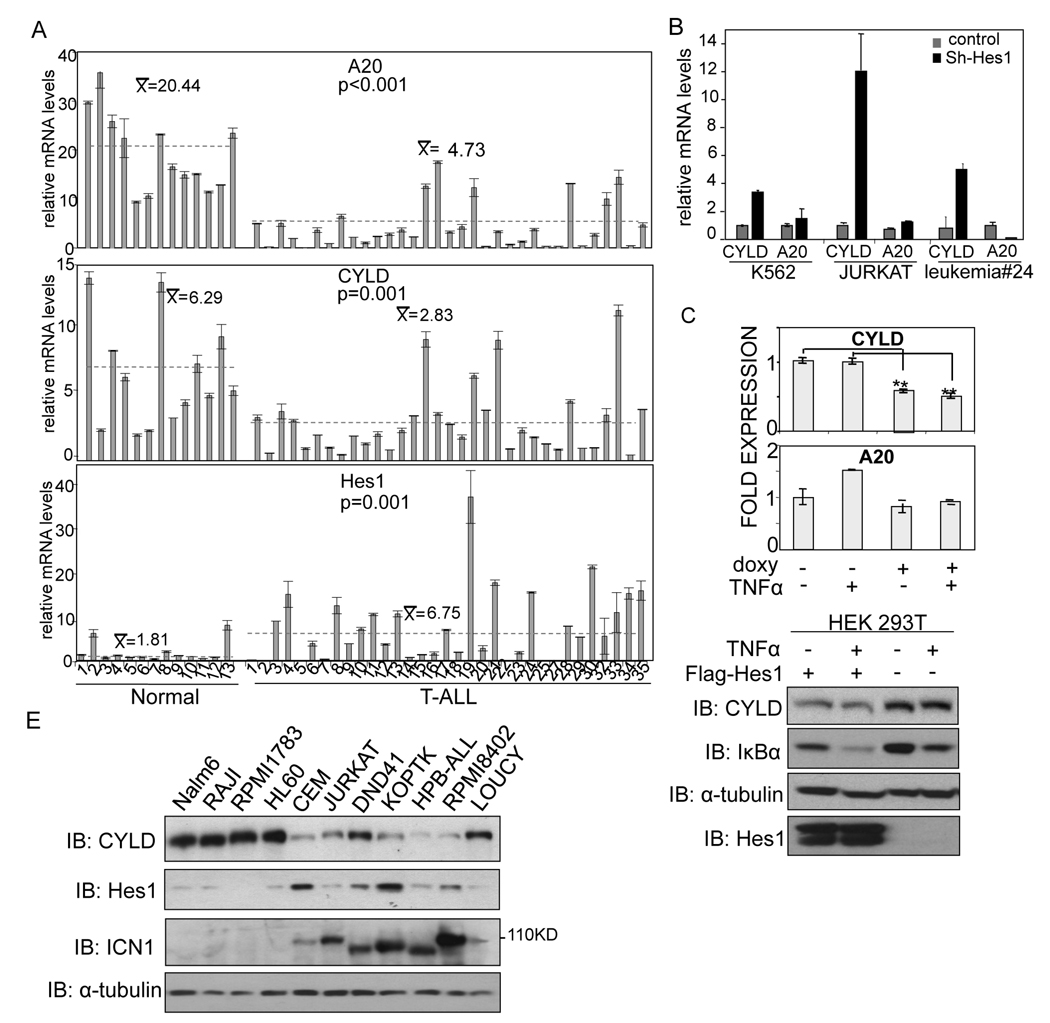
Since our results indicated that Hes1 was able to maintain IKK complex activation in T-ALL and we mapped this function upstream of TAK1, we tested whether the expression of the negative regulators of IKK activity, CYLD and A20, was decreased in human T-ALL. In this screen we included a group of 13 healthy control samples (9 whole peripheral blood (WPB) and 4 purified CD3+) and 33 WPB samples from T-ALL patients containing more than 60% of leukemic blasts, except sample number 18 with 44% leukemic cells (for details see Table S2). Using qRT-PCR we found that mRNA levels of both deubiquitinating enzymes were significantly decreased in the leukemic samples compared to the normal controls (p=0.001 for CYLD and p<0.001 for A20 using Student t-test) concomitant with a significant increase in the expression of Hes1 (p=0.001) (Figure 5A). Further meta-analysis of ALL gene expression databases showed that CYLD levels in T-ALL and significantly lower than in any B-ALL category (not shown). Next, we tested whether there is an inverse correlation between Hes1 and either CYLD or A20 genes in T-ALL by using Hes1-specific shRNA-mediated knock-down. Reduction of Hes1 levels resulted in a pronounced increase in the mRNA of CYLD whereas A20 levels remained unaffected (Figure 5B). In contrast, overexpression of Hes1 in HEK-293T cells resulted in a reduction in the levels of CYLD mRNA (Figure 5C) and protein (Figure 5D) associated with lower levels of IκBα, being this effect more dramatic after chronic treatment with TNFα (Figure 5D). We next tested whether Hes1 protein levels inversely correlated with CYLD protein levels in different T-ALL cell lines containing Notch1 mutations. As shown in Figure 5E, CYLD levels were reduced in T-ALL cells when compared to B-cell (Nalm6, RAJI and RPMI1783) or myeloid leukemia cell line (HL60) concomitant with variable, but higher levels of Hes1 protein. Importantly, inhibition of Notch1 by GSI resulted in an increase of both protein (Figure S5) and mRNA (not shown) levels of CYLD in these cells.
Figure 5. The expression of CYLD, a negative regulator of NF-kB, is suppressed in T-ALL.
(A) Levels of A20, CYLD and Hes1 mRNA in primary samples for normal WPB (1–9), purified normal CD3+ cells from PB (10–13) and PB from T-ALL patients (1–33) by qRT-PCR. (B) Analysis by qRT-PCR of CYLD or A20 levels in cells transduced with scrambled or Hes1-shRNA. For both A–B panels, error bars denote +/− Standard Deviation. (C–D) Expression of CYLD and A20 measured by qRT-PCR (C) or western blot (D) in HEK-293T cells expressing ectopic Hes1 untreated or treated with TNFα. (E) Western blot analysis of CYLD, active Notch1 and Hes1 protein levels in different leukemic cell lines. See also related figure S5.
The Notch pathway directly controls CYLD expression through Hes1
These data suggested that CYLD expression is kept suppressed in T-ALL, leading to an enhancement of IKK activity. To test whether the Notch pathway and specifically Hes1 was directly repressing CYLD transcription, we used the Genomatix software to identify putative Hes1 binding sites, and found three distinct N-box consensus sites in the promoter and 5’UTR of the CYLD transcriptional start site (Figure 6A, denoted as PRO1 and PRO2). Using a conventional ChIP assay in human Hes1+ T-ALL cells and a Hes1 antibody followed by PCR with specific primers flanking the identified putative N-box sites, we were able to show that endogenous Hes1 binds to the predicted PRO2 site in the CYLD 5’UTR (Figure 6B) and this association was lost following Hes1 knock-down (Figure 6C) as measured by qPCR. Ectopic expression of CYLD completely reverted the clonogenic growth of lineage negative BM cells transduced with ICN1 whereas downregulation of CYLD was sufficient to increase the number of colonies in this assay (Figure 6D). Consistent with these results, we found that ectopic expression of CYLD is sufficient to block Notch-induced activation of an NFκB-dependent reporter in HEK-293T (Figure S6A). Moreover, the capacity to produce CD3+ T cells by ICN1-tranduced progenitors in co-culture on OP9-Dll1 stroma is partially mimicked by CylD-shRNA-expressing cells and lost in cells co-expressing ICN1 and CYLD (Figure 6E and Figure S6) To further support the notion that CYLD expression in T-ALL is controlled by direct transcriptional repression by Hes1, we have quantified CpG methylation at the CYLD using the previously introduced human T-ALL samples. As shown in Figure 6F, there was no evidence of promoter hypermethylation suggesting that it is unlikely that DNA methylation can explain by itself the suppression of CYLD expression. These observations suggest that suppression of CYLD enhances the activity of the IKK kinase and the nuclear localization of NF-κB transcription factors. To directly test this hypothesis, we have totally eliminated CYLD expression in T-ALL using an animal model of CYLD deficiency (Reiley et al., 2006). As shown in Figure S6B–D, CYLD deletion is sufficient to significant enhance nuclear NF-κB activity in T-ALL cells as quantified by EMSA. All these observations strongly suggest that Hes1 sustains IKK complex activation and maintains the tumor phenotype in T-ALL by directly repressing CYLD expression and function.
Figure 6. Hes1 directly represses CYLD transcription in T-ALL.
(A) Scheme of the CYLD promoter indicating the putative Hes1 binding sites. (B) Detection of the CYLD PRO-2 Hes1 binding site by PCR using the indicated antibodies. (C) Relative enrichment of the CYLD promoter quantified by qPCR from ChIP with anti-Hes1 antibody in Jurkat T-ALL cells transduced with scrambled or Hes1-shRNA. Error bars denote +/− Standard Deviation. (D) Primary mouse Lineage-negative bone marrow progenitors were transduced with the indicated lentivirus and colonies obtained from serial replating on methylcellulose were counted every week. (E) Representative FACS analysis from week 3 progenitors after 7 days on OP9-Dll1 stroma. (F) EPITYPER methylation analysis showing no significant hyper-methylation of the CYLD promoter. Patient samples were analyzed utilizing the EpiTYPER Sequenom methylation assay. The heat map illustrates the range of methylation percentages, with red indicating 0% methylation and yellow indicating 100% methylation. Numbers along the top indicate CpG dinucleotide, patient sample numbers are listed along the vertical axis (p denotes the identity of the patient as presented in Figure 5A). Samples are run in duplicate. Two primers were run for each region. The positive control, IVD, has near 100% methylation, while the negative control, WGA, is unmethylated. See also related figure S6.
Targeted IKK in vitro silencing affects the growth of human T-ALL lines
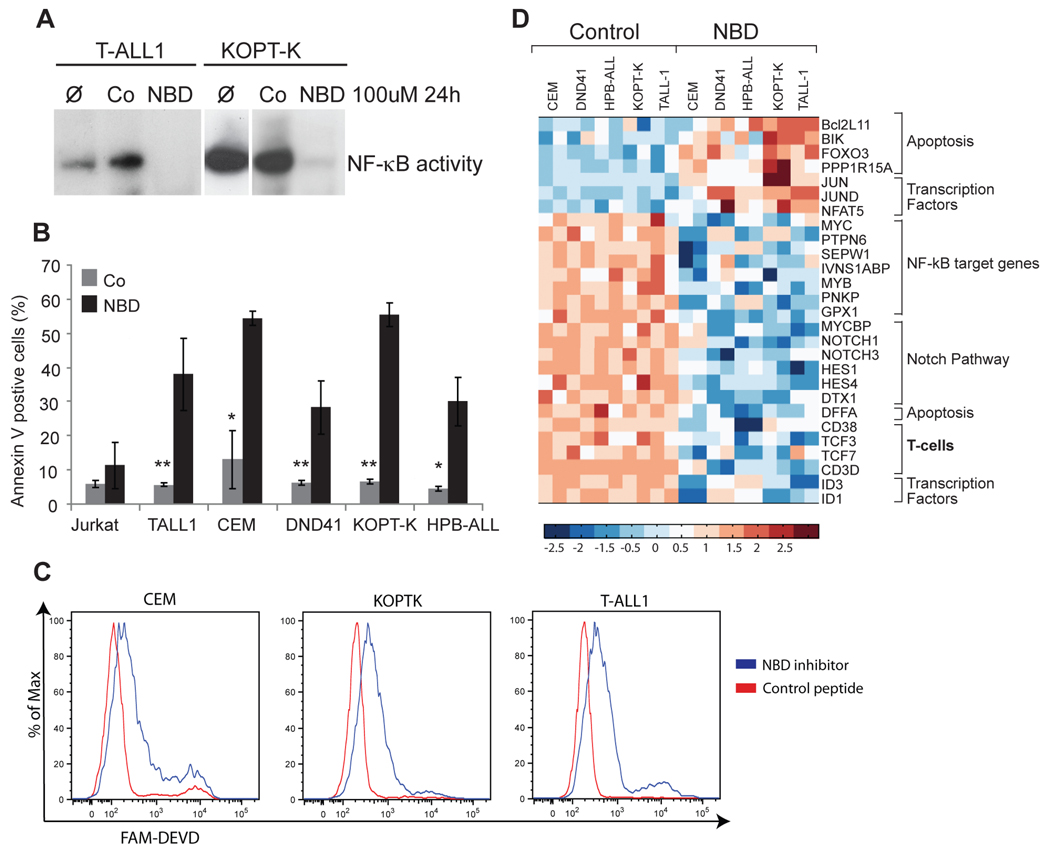
Our data so far suggest that targeting the IKK complex could be effective in the treatment of T-ALL. We have shown previously that the proteasome inhibitor Bortezomib could affect T-ALL cell line growth (Vilimas et al., 2007). As Bortezomib is not a specific NF-κB inhibitor (Hideshima et al., 2009), we have now tested the effects of a cell permeable peptide (NEMO BINDING DOMAIN) that specifically blocks IKK by disrupting the interaction of NEMO to IKKβ (May et al., 2000) on several (total of 9) T-ALL cell lines which have present an active Notch pathway and exhibit NF-κB activation. These human T-ALL cells lines harbour activating Notch1 mutations. As shown in Figure 7A, the NBD peptide was able to efficiently inhibit nuclear NFκB binding activity in T-ALL lines. Interestingly, silencing NF-κB led to specific biological effects, as shown by the rapid induction (24h) of apoptosis as determined by Annexin-V binding and caspase-3 activation (Figures 7B,C). No effects on cell cycle progression were noted suggesting that IKK inactivation is predominantly affecting cell survival (not shown).
Figure 7. IKK in vitro silencing affects the survival of human T-ALL cell lines.
Human T-ALL cell lines were treated with 100 µM of control (Co) or NBD inhibitory peptide for 24 hours. (A) Nuclear extract from treated and untreated cells were isolated and checked for NF-κB-DNA binding activity by EMSA. Untreated cells (Ø) are also shown. (B) Bar-graph indicating the proportion of Annexin V positive cells 24h after the NBD treatment. Error bars denote +/− Standard Deviation. (C) Caspase-3 activation as detected by FACS with fluorogenic cell permeable active caspase 3- specific probe (Fam-DEVD-fmk) in 3 representatives T-ALL lines 24h post treatment. All experiments shown in A–C were repeated at least three times. (mean ± SD of measurements: *, p<0.05; **, p<0.01 Student’s t test analysis). (D) Heat map showing up changes in expression of selected genes in response to NBD peptide treatment. See also related figure S7.
To better understand the molecular effects of IKK inhibition in vitro we used these five T-ALL lines to study transcriptome changes 16h after the addition of the NBD peptide. The initial conclusion from these experiments is that although the response to NBD treatment was uniform, gene expression changes were both general and cell-line specific (Figure 7D and Figure S7A). All five cell lines showed signs of induction of cell death characterized by the upregulation of Bcl2L11 (Bim), Bik and Foxo3, in agreement with their apoptotic phenotype, and an inhibition of NF-κB target genes (such as Myc, Ptpn6, Pnkp and Gpx1). Interestingly, another uniform target of IKK inhibition was the Notch pathway itself (including Hes1, Hes4, Notch1, Notch3, Dtx1) suggesting a mechanism of feedback inhibition. Using qRT-PCR we have verified the NBD-dependent regulation of several of these genes (Figure S7B). These findings suggested that IKK signalling was essential for disease maintenance and proposed that IKK inhibition could be a potential therapeutic tool in T-ALL.
IKK complex activity is essential for the in vivo maintenance of T-ALL
One of the key questions in cancer treatment is whether therapeutic targeting of a single signaling pathway is sufficient to affect tumor maintenance and progression. To address this key question in vivo we combined the Notch-dependent leukemia model with an inducible IKKγ (Nemo) deficiency (Mx1-cre+Nemof/f) model. Bone marrow stem and progenitor cells purified from Mx1-cre-inducible NEMO-deficient mice were transduced with ΔEN1-luciferase/ or EGFP-expressing retroviruses and transplanted into irradiated recipients. Two weeks post-transplantation, once the disease had been established as indicated the presence of double CD4+CD8+ lymphocytes in the peripheral blood (Figure 8A,B) and the expansion of the donor cells as detected by in vivo bioluminescence (Figure 8C, D), Nemo expression (and thus canonical NF-κB activation) was silenced by inducing the expression of cre-recombinase using polyI-polyC injections (Kuhn et al., 1995). As shown in Figures 8B–F, deletion of Nemo very efficiently reverted disease progression as judged by both the significant decrease in luminescence (Fig 8C, D) and the disappearance of transformed CD4+CD8+ T cells from the peripheral blood of the treated animals (Figure 8A,B) and T-cell infiltration in the spleen and liver (Figure 8F). In agreement to the effects of IKK inhibition on T-ALL cell lines, IKK/Nemo deletion lead to rapid induction of leukemic cell apoptosis (Figure 8E).
Figure 8. The NEMO/IKK complex activity is essential for the maintenance of T-ALL in vivo.
(A–B) Peripheral blood FACS analysis using CD4, CD8 and Ly5.2 antibodies at different time points post transplantation. (C) ΔEN1-luc Nemowt/wt, Mx-Cre and ΔEN1-luc Nemof/f, Mx-Cre chimeras were monitored before and after pI-pC injections using in vivo imaging system (IVIS) to quantify luciferase intensity. 2 weeks post-transplantation and before any deletion of NEMO, both chimeras harbored homogenous tumor load. Once this was verified mice were injected by pI-pC. (D) Quantification of luciferase intensity (dorsal and ventral) at different time-points. Error bars denote +/− Standard Deviation. (E) Determination of apoptotic cells gated on GFP+ cells 5 days after pIpC injections in ΔEN1-GFP Nemowt/wt, Mx-Cre and ΔEN1-GFP Nemof/f, Mx-Cre chimeras. (F) Micrographs of hematoxylin/eosin-stained liver and spleen sections prepared 40 days after transplantation. (G) Kaplan-Meyer survival curve of the different recipient animals (p<0.005). See also related figure S8.
Consistent with these phenotypic observations, pI-pC treated Nemowt/wt, MxCre+-expressing animals succumbed significantly faster than their pI-pC treated Nemof/f ,MxCre+ counterparts. However, if the treatment was not continuous, the disease was able to relapse and eventually kill the animals (Figure 8G). To determine whether the reappearance of the disease was due to the expansion of a small cell population in which Nemo was not deleted, we sorted leukemic cells, 61 days post transplant -approximately 40 days after the last pI-pC injection and tested them for the presence of Nemo. Further supporting the essential function of IKK signaling in T-ALL leukemias, the purified cells were bearing the “undeleted” NEMO allele (Figure S8A) and showed a significant increase of NF-κB DNA-binding activity in the nucleus (Figure S8B). These studies are the first to demonstrate that IKK activity is essential for the maintenance of T-ALL and open the way for inhibiting the IKK complex as a therapeutic target for patients with already established T-ALL.
DISCUSSION
Our studies demonstrate that the transcriptional repressor Hes1, a target of oncogenic Notch1, is able to induce the activation of the NF-κB pathway in human T-ALL lines and animal models of the disease. Moreover, they show that IKK/NF-κB signaling is essential for the maintenance of the disease, as T-ALL cells that are unable to activate the IKK kinase complex rapidly enter apoptosis. As Notch1 mutations or pathway hyper-activation are found in the vast majority of T-ALL patients, our findings are clinically important as they could be translated to either as single agent therapy targeting IKK (or NF-κB) activation or combinatorial drug treatments that could in parallel target additional elements of the Notch pathway.
The presented studies identify a previously unidentified mechanism of Notch-induced NF-κB activation in T-ALL. We show that Hes1 is a key mediator of Notch-induced transformation by targeting the deubiquitinase CYLD, a negative regulator of IKK activity. Our experiments demonstrate that Hes1 expression is sufficient to enhance NF-κB activity and this effect maps upstream of IκBα degradation, IKK phosphorylation, and TAK1 activity. A significant portion of these effects can be attributed to the suppression of CYLD expression by Hes1. In agreement with this notion, we have identified CYLD as a direct target for Hes1 repression. CYLD is a deubiquitinating enzyme that was originally identified as a tumor suppressor and is mutated in familial cylindromatosis (Bignell et al., 2000). More recently, CYLD inhibition has been associated with other tumors such as melanoma (Massoumi et al., 2009) and breast (Hutti et al., 2009). Although previous to this report, there was no information about an involvement of CYLD in leukemia, abnormal lymphopoiesis in CYLD-deficient animals had been reported (Reiley et al., 2006). This abnormal differentiation observed in the CYLD-deficient mice was initially associated with a defect in Lck kinase activation in response to TCR signaling, however it was later demonstrated that CYLD is also responsible for TAK1 deubiquitination resulting in the inactivation of the NF-κB pathway in T-lymphocytes (Reiley et al., 2007). Other well-characterized CYLD substrates that participate in the NF-κB signaling include TRAF2, Bcl3 and IKKγ/NEMO. We demonstrate here that CYLD expression is down-regulated in T-ALL cell lines and primary samples, and that knocking down Hes1 expression in these cells results in increased CYLD levels and attenuation of the NF-κB signaling. Interestingly, Hes1-mediated suppression appears to be the preferred mode of regulation as no inactivating mutations or CYLD promoter hyper-methylation were detected in human T-ALL (data not shown and Figure 6). We should also mention that CYLD suppression of expression is by itself unlikely to account for the initial NF-κB activation. Indeed, it is likely that basal NF-kB stimulation is required and is provided by either Notch itself (Vilimas et al., 2007) or/and the T cell receptor (TCR). We have previously shown that T-ALL cells express the pre-TCR that can activate constitutively the NF-kB pathway (Aifantis et al., 2001; Davis et al., 2001; Mandal et al., 2005). It is thus conceivable that the tumors have basal NF-kB activation and suppression of the negative regulator of the IKK complex (CYLD) further boosts pathway activation.
Another important point made by our studies is that Notch/Hes1 activation is targeting the canonical NF-κB pathway in T-ALL. NEMO is a regulatory subunit of IKK complex, which is responsible for activation of canonical NF-κB signaling pathway. The T lineage-specific deletion of NEMO prevents the generation of peripheral T cells due to apoptotic death mature thymocyte (Schmidt-Supprian et al., 2003). Consistent with this notion, silencing NEMO/IKKγ was sufficient to inhibit NF-κB activation in our animal model of ICN1-dependent T-ALL and affect tumor growth and animal survival. Explaining these effects, gene expression studies in IKK-inactivated T-ALL cells suggested that genes involved in regulating cell survival or apoptosis are targeted by canonical NF-κB signaling in T-ALL. Although several survival genes have been shown to be NF-κB targets, two of them, the anti-apoptotic Bcl2A1 and the pro-apoptotic Bcl2L11 (Bim), both members of the BCL2 family, showed contrasting modes of regulation in response to IKK silencing. Bcl2A1 appears to play a key role in Notch-induced T-ALL. We have previously shown that Bcl2A1 is over-expressed in T-ALL and that its silencing was able to affect the survival of both Notch1 and Notch3 over-expressing T cell leukemia lines (Mandal et al., 2005). We have also previously shown that Bim is expressed in low amounts in T cell leukemia and that it controls early T cell death induction (Mandal et al., 2008), making this molecule a putative therapy target.
Although the role of NF-κB in leukemogenesis has been previously reported (Davis et al., 2001), several recent studies in tumors as diverse as Diffuse Large B-cell lymphoma (DLBCL) (Compagno et al., 2009; Kato et al., 2009) or lung adenocarcinoma (Compagno et al., 2009; Kato et al., 2009; Meylan et al., 2009) suggest that the possibility to target this pathway in cancer therapy should be re-evaluated. In agreement with our findings, alterations in different elements of the NFkB pathway, including the enzymes A20 and CYLD, were found to be responsible for activating NF-κB in multiple myeloma and DLBCL, upstream of the IKK complex (Annunziata et al., 2007; Compagno et al., 2009; Kato et al., 2009; Keats et al., 2007; Lenz et al., 2008). In lung cancer cells with mutated K-ras, NF-κB is activated by the non-canonical TBK1/IKK interaction (Barbie et al., 2009) and blocking IKK activity reduces tumor growth in vivo (Meylan et al., 2009). We demonstrate here that in T-ALL, IKK can be activated by the Notch-Hes1-CYLD signaling cascade. However, whatever the trigger, the outcome is identical: activation of IKK, degradation of IκBα and nuclear translocation of the NF-κB factor, which provides the base to consider that pharmacological targeting of the IKK/NF-κB pathway could be a promising future treatment of a wide spectrum of tumors. The recent development of chemical inhibitors of the IKK complex, drugs that are currently in clinical trials for inflammatory disease, will enable us to test this hypothesis in future treatment protocols.
EXPERIMENTAL PROCEDURES
Mice and Animal procedures
C5/BL/6, or BALB/C mice were purchased from Jackson Laboratories and Taconic Farms. BALB/C-Tg(NF-κB-RE-luc [Oslo]) were purchased from Caliper. Nemof/f mice have been described previously (Schmidt-Supprian et al., 2000). Cyld knockout mice were generated as described previously (Reiley et al., 2007). For the polyI-polyC experiments, we injected 20µg pI-pC (Amersham) per gram of body weight. All mice were kept in specific pathogen-free animal facilities. All animal procedures were approved by and performed in accordance to the guidelines of the Institutional Animal Care and Use Committee of the NYU-SOM.
Human T-ALL samples
A total of 33 T-ALL samples and normal blood samples were obtained from the Hematology Department of Hospital de Sant Pau and the Department of Pathology of Hospital del Mar, Barcelona, Spain. All patients gave written consent to donate the blood speciment. The study was approved by the Ethics Committee of the Institution (Department of Pathology, Hospital del Mar).
Plasmids
3xκB-luciferase and HA-IκBαS32-36A (Espinosa et al., 2003), IκBαK21-22R (Arenzana-Seisdedos et al., 1997), HA-TAK1K63W (Ninomiya-Tsuji et al., 1999), ΔEN1-Luciferase, ΔEN1-GFP and ICN1-GFP retroviral constructs (Chiang et al., 2006; Vilimas et al., 2007) or, ICN1ΔOP (Bigas et al., 1998) have been previously described. Flag-Hes1 was obtained from M. Caudy (Castella et al., 1999) and used to clone Hes1 cDNA into the inducible vector pcDNA3.TO. pTHR1-NF-κB-dscGFP and pTHR1-mCMV-dscGFP lentiviral constructs were purchased from System Biosciences. CYLD retroviral and shRNA constructs were given by A. Pfeifer (Massoumi et al., 2009). Human and mouse-specific Hes1-shRNA lentiviral constructs was purchased from Sigma (TRCN0000018989 and TRCN0000028854 respectively).
Bioluminescence imaging
Imaging of κB-ER-luciferase and ΔEN1-luciferase chimeras were performed as previously described (Tseng et al., 2006).
qRT-PCR and Microarray
The primers used for RT-PCR analysis are listed in Supplemental Experimental Procedures. qRT-PCR was performed in LightCycler480 system using SYBR Green I Master or Probes Master kit (Roche). For microarray analysis, the dataset of the genome-wide RNA expression analyses of human T-ALL cells lines treated by control peptide versus NBD peptide has been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/); the accession numbers is GSE20667. RNA was labeled on hybridized to the Human U133 Plus 2.0 microarray (Affymetrix). The Affimetrix gene expression profiling data was first normalized using previously published Robust Multi-array Average (RMA) algorithm implemented in RNAExpress software version 1.0.4 (http://rmaexpress.bmbolstad.com/) and log2 transformed. Then genes that are differentially expressed between NBD peptide treated and control peptide treated cells were selected using a paired two-side t-test and requiring fold change ≥1.2 or ≤0.833. The multiple comparison correction was performed using the approach of (Reiner et al., 2003) with FDR threshold equal to 0.10. The application of the above procedure resulted in 387 genes (531 oligonucleotide probes).
Quantitative DNA methylation analysis
DNA methylation analysis was carried out using the Epityper system from Sequenom (San Diego, CA). The EpiTYPER assay is a tool for the detection and quantitative analysis of DNA methylation using base-specific cleavage of bisulfite-treated DNA and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS). Specific PCR primers for bisulfite-converted DNA were designed using the EpiDesigner software (www.epidesigner.com), for the entire CpG island of the genes of interest. T7-promoter tags are added to the reverse primer to obtain a product that can be in vitro transcribed, and a 10-mere tag is added to the forward primer to balance the PCR conditions. Primer sequences, target chromosomal sequence, and Epityper specific tags, are available upon request. A detailed description of the protocol used can be found at the Supplemental Experimental Procedures.
Statistical analysis
Student’s T-test was performed to assess the significance of expression levels. These data are presented as mean ± SD.
SIGNIFICANCE.
Although aberrant activation of the Notch pathway is a recurring event in T-ALL, the downstream signaling effects of this activation are poorly defined. Our work demonstrates that the Notch-Hes1-CYLD-IKK axis plays a crucial role in this type of leukemia, and that blocking NF-κB (or the IKK kinase) is sufficient to eliminate leukemic cells carrying activating Notch mutations. These findings suggest that NF-kB targeting could be a key component of successful future T-ALL clinical trials.
HIGHLIGHTS.
NF-κB activity is active and essential in established Notch-dependent T-ALL leukemias. Notch through Hes1 sustains NF-κB activity by repressing the deubiquitinase CYLD. Inhibition of IKK/NFκB activity reduces leukemic load and increases survival in mice.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the NYU Flow Facility for expert cell sorting, the NYU Cancer Institute Genomics Facility for help with micro-array processing, and the Meruelo Lab (NYU) for animal imaging. Adriana Heguy and the Geoffrey Beene Translational Oncology Core for assistance with sequencing and methylation analysis. Drs. T. Look and H. von Boehmer for gene expression database sharing. Drs. M. Schmidt-Supprian and A. Klinakis for valuable animal model advice. M. Caudy (Cornell University), A. Pfeifer (University of Bonn), M. Karin (UCSD), A. Cerutti (IMIM), R. Kageyama (Kyoto University) and R. Gimeno (Research Institute Hospital Vall d’Hebron) for valuable reagents and BAYER for providing the BAY65-1185. We thank C. Recasens, J. González (IMIM), Philmo Oh and Camille Lobry (NYU) for experimental advice, F. Torres (UAB) for statistical analysis. JG, TD and VR are recipients of FPI (BES-2008-005708), FPU (AP2008-01883) and FIMIM predoctoral fellowship respectively. LE is an investigator at the Carlos III program and, SC is supported by the Hope Street Kids Foundation. This work was supported by Instituto de Salud Carlos III Grant PI041890 and Fundación Mutua Madrileña to LE, by MEC2007-60080 and RTICCS/FEDER (RD06/0020/0098) to AB, by the National Institutes of Health grants R01CA133379, R01CA105129, R01CE149655, R21CA141399 (to I.A.), R56LM007948, 1UL1RR029893 (to A.S), the American Cancer Society (RSG0806801 to IA), the Irma T. Hirschl Trust, The Dana Foundation, The Mallinckrodt Foundation, the Alex’s Lemonade Stand Foundation, The Leukemia and Lymphoma Society (TRP grant to IA) and the Gabrielle’s Angels Foundation (R.L.L and I.A.). IA is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aifantis I, Gounari F, Scorrano L, Borowski C, von Boehmer H. Constitutive pre-TCR signaling promotes differentiation through Ca2+ mobilization and activation of NF-kappaB and NFAT. Nat Immunol. 2001;2:403–409. doi: 10.1038/87704. [DOI] [PubMed] [Google Scholar]
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay RT, Virelizier JL, Dargemont C. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigas A, Martin DI, Milner LA. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. MolCell Biol. 1998;18:2324–2333. doi: 10.1128/mcb.18.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Buonamici S, Trimarchi T, Ruocco MG, Reavie L, Cathelin S, Mar BG, Klinakis A, Lukyanov Y, Tseng JC, Sen F, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castella P, Wagner JA, Caudy M. Regulation of hippocampal neuronal differentiation by the basic helix-loop-helix transcription factors HES-1 and MASH-1. J Neurosci Res. 1999;56:229–240. doi: 10.1002/(SICI)1097-4547(19990501)56:3<229::AID-JNR2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Xu ML, Histen G, Shestova O, Roy M, Nam Y, Blacklow SC, Sacks DB, Pear WS, Aster JC. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol Cell Biol. 2006;26:6261–6271. doi: 10.1128/MCB.02478-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan SE, St Pierre B, Leow CC. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- Espinosa L, Ingles-Esteve J, Robert-Moreno A, Bigas A. IkappaBalpha and p65 Regulate the Cytoplasmic Shuttling of Nuclear Corepressors: Cross-talk between Notch and NFkappaB Pathways. Mol Biol Cell. 2003;14:491–502. doi: 10.1091/mbc.E02-07-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K, Mitsiades C, Munshi NC, Richardson PG, Carrasco RD, Anderson KC. Bortezomib induces canonical nuclear factor-kappaB activation in multiple myeloma cells. Blood. 2009;114:1046–1052. doi: 10.1182/blood-2009-01-199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkB-NF-kB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, Hahn WC, Cantley LC. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol Cell. 2009;34:461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205:2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Borowski C, Palomero T, Ferrando AA, Oberdoerffer P, Meng F, Ruiz-Vela A, Ciofani M, Zuniga-Pflucker JC, Screpanti I, et al. The BCL2A1 gene as a pre-T cell receptor-induced regulator of thymocyte survival. J Exp Med. 2005;201:603–614. doi: 10.1084/jem.20041924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Crusio KM, Meng F, Liu S, Kinsella M, Clark MR, Takeuchi O, Aifantis I. Regulation of lymphocyte progenitor survival by the proapoptotic activities of Bim and Bid. Proc Natl Acad Sci U S A. 2008;105:20840–20845. doi: 10.1073/pnas.0807557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fassler R, Bosserhoff AK. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med. 2009;206:221–232. doi: 10.1084/jem.20082044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May MJ, D'Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, Leonard TO, Norbury CC, Fitzpatrick L, Zhang M, Sun SC. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–1485. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, Sun SC. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7:411–417. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Wright J, Thompson J, Thomas D, Baleux F, Virelizier JL, Hay RT, Arenzana-Seisdedos F. Identification of lysine residues required for signal-induced ubiquitination and degradation of I kappa B-alpha in vivo. Oncogene. 1996;12:2425–2435. [PubMed] [Google Scholar]
- Screpanti I, Bellavia D, Campese AF, Frati L, Gulino A. Notch, a unifying target in T-cell acute lymphoblastic leukemia? Trends Mol Med. 2003;9:30–35. doi: 10.1016/s1471-4914(02)00003-5. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israel A, Rajewsky K, Pasparakis M. Mature T cells depend on signaling through the IKK complex. Immunity. 2003;19:377–389. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. Embo J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Tseng JC, Zanzonico PB, Levin B, Finn R, Larson SM, Meruelo D. Tumor-specific in vivo transfection with HSV-1 thymidine kinase gene using a Sindbis viral vector as a basis for prodrug ganciclovir activation and PET. J Nucl Med. 2006;47:1136–1143. [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, Thompson B, Spaulding C, Macaroun S, Alegre ML, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13:70–77. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.