Summary
Alternative splicing is generally regulated by trans-acting factors that specifically bind pre-mRNA to activate or inhibit the splicing reaction. This regulation is critical for normal gene expression, and dysregulation of splicing is closely associated with human diseases. Here we engineer artificial splicing factors by combining sequence-specific RNA-binding domains of human Pumilio1 with functional domains that regulate splicing. We applied these factors to modulate different types of alternative splicing in selected targets, examine the activity of effector domains from natural splicing factors, and modulate splicing of an endogenous gene, Bcl-x, an anti-cancer target. The designer factor targeted to Bcl-x increased the pro-apoptotic Bcl-xS splicing isoform, thus promoting apoptosis and increasing chemosensitivity of cancer cells to common anti-tumor drugs. Our approach permits the creation of artificial factors to target virtually any pre-mRNA, providing a new strategy to study splicing regulation and manipulate disease-associated splicing events.
As a key regulatory step in gene expression, alternative splicing is widespread in humans with most genes producing multiple splicing isoforms with distinct and sometimes opposing functions 1. The choice of splicing isoforms is tightly regulated in different tissues and developmental processes, and disruption of such regulation is a common cause of various human diseases 2. Therefore novel approaches to specifically modulate alternative splicing will improve our understanding of splicing regulation and have therapeutic potential.
Generally, the splicing process is regulated through cis-elements that recruit trans-acting splicing factors to affect use of nearby splice sites. Many splicing factors have modular organization, with separate sequence-specific RNA binding modules and splicing effector domains. For example, serine/arginine-rich (SR) proteins contain N-terminal RNA recognition motifs (RRMs) that bind to exonic splicing enhancers (ESEs) in pre-mRNAs and C-terminal arginine-serine (RS) rich domains that promote exon inclusion 3. Analogously, the hnRNP A1 binds to exonic splicing silencers (ESSs) through its RRM domains and inhibits exon inclusion through a C-terminal Glycine-rich domain 4. Following this configuration we envisioned engineering unique factors by combining an RNA recognition module to recognize targets with a functional module to affect splicing. Ideally, such an engineered splicing factor (ESF) should be able to recognize any target and modulate splicing in desired ways.
Here we report the first attempt to engineer splicing factors with designed sequence specificity and activities. These ESFs can promote or suppress splicing, and can specifically recognize target genes undergoing different types of alternative splicing. We used such ESFs to study the functional domains of natural splicing factors, and designed a new ESF to specifically modulate splicing of endogenous gene. The ‘designer’ ESF can shift splicing of Bcl-x to increase the pro-apoptotic isoform in multiple cultured cancer cells, therefore sensitizing these cells to common anti-tumor drugs.
Results
Design principles of ESFs
To generate an RNA-binding module with predictable specificity, we used the unique RNA recognition mode of PUF proteins (named for Drosophila Pumilio and C. elegans fem-3 binding factor) that are involved in mediating mRNA stability and translation 5. Most splicing factors recognize their targets through RRM or K homology domains that bind to short RNA elements with moderate affinities. However, it is impractical to engineer an RNA recognition module using these domains due to their weak binding affinity and the absence of a predictive RNA recognition code. The PUF domain of human Pumilio1 binds tightly to cognate RNA sequences and its specificity can be modified. It contains eight PUF repeats that recognize eight consecutive RNA bases with each repeat recognizing a single base 6 (Fig. 1a). Two amino acid side chains in each repeat recognize the Watson-Crick edge of the corresponding base and determine the specificity of that repeat, thus a PUF domain can be designed to specifically bind most 8-nt RNA 6, 7. To generate a sequence-specific ESF, we fused the modified PUF with a splicing regulatory domain (e.g. RS domain for a splicing activator or Gly-rich domain for a splicing repressor). We also included a nuclear localization sequence (NLS) to direct the ESF to the nucleus where splicing occurs and a FLAG tag to facilitate detection (Fig. 1b).
Fig. 1. Design ESFs to modulate exon skipping.
(a) Interaction of the human Pumilio1 PUF domain with RNA. The PUF domain has eight repeats to recognize an 8-nt RNA, with repeat R1 to R8 recognizing nucleotides N8 to N1, respectively. (b) Modular domain organization of ESFs. (c) Binding affinities of three PUF domains to their 8-mer targets. The dissociation constants (Kd) of RNA-protein binding were plotted. Wild-type PUF specifically binds Nanos Response Element (NRE) RNA (UGUAUAUA), whereas the mutated PUF3-2 (C935S/Q939E) recognizes A6G sequence and the mutated PUF6-2/7-2 (N1043S/Q1047E and S1079N/E1083Q) recognizes GU/UG sequence 6, 7. Asterisks indicate the cognate pairs of RNA-PUF. (d) Inhibition of exon inclusion by Gly-PUF type ESFs in exon-skipping reporters. Inclusion of cassette exon (yellow) containing the 8-nt targets (light blue) is inhibited by Gly-PUF type ESFs. All combinations of splicing reporters and ESF expression vectors listed in panel c were co-transfected into 293T cells. The inclusion of cassette exon was analyzed 24 hours after transfection. All transfections were repeated at least twice, and the results for representative experiments are shown. The extra band (asterisk) in lane 2 (Gly-PUFwt) is caused by an experimental artifact. (e) Fold change of cassette exon inclusion relative to the reporters alone (none ESF). The means of replicated experiments are plotted with error bars indicating the range of replicated experiments. (f) ESF expressions were detected using western blot with anti-FLAG antibody, with anti-actin antibody as loading controls. Samples are in the same order as panel d. (g-i) Promotion of exon inclusion by RS-PUF type ESFs using exon-skipping reporters containing different 8-nt target sequences. All experiments and quantification were carried out similarly to panel d-f.
Specifically modulating exon inclusion with ESFs
To prove design concept, we created six ESFs by fusing either the Gly-rich domain of hnRNP A1 or the RS domain of ASF/SF2 with three different PUF domains (wild type human Pumilio1 and two modified PUF domains) with distinct RNA binding specificity (Fig. 1c). We generated a panel of exon-skipping reporters containing the corresponding 8-nt target sequences of the ESFs in the alternatively spliced cassette exon. These reporters were co-transfected into 293T cells with the ESF expression constructs, and changes in splicing were analyzed using body labeled RT-PCR.
Consistent with our prediction, the Gly-PUF type ESFs repressed inclusion of the cassette exon containing a cognate target sequence (Fig. 1d-f). Such inhibition is sequence specific, with the maximal inhibition of exon inclusion occurring between cognate ESFs and reporters (lanes 2, 7 and 12, Fig. 1e). The splicing repressor activities of Gly-PUF type ESFs correlated roughly with the binding affinities between PUF motifs and their targets (Figs. S1 and 1d), suggesting that the binding affinity to its target contributes to splicing factor strength.
In line with the above observations, RS-PUF type ESFs had opposite activity on splicing and promoted inclusion of cassette exons containing cognate targets (Fig. 1g-i). The activities of these ESFs again correlated roughly with the binding affinities between PUF motifs and their targets (Fig. S1), supporting the modular configuration of splicing factors 8, 9. In addition, both types of ESFs are effective in the different cell types tested, suggesting that their activities are not cell line specific (Fig. S2).
Despite a clear trend, the correlation between ESF activities and their binding affinities to targets is not completely linear (Fig. S1), as considerable levels of exon skipping or, less prominently, inclusion, were observed between some non-cognate pairs of ESFs and targets (e.g. Fig. 1d, NRE and A6G sequences). This nonlinearity is likely due to a combination of sequence specific □and non-specific effects of splicing factors. We used a 1:5 ratio of Gly-PUF expression plasmid to splicing reporter plasmid, which represents the lower end of the range that gave robust activity and is substantially lower than the ratios typically used in splicing factor/splicing reporter co-transfection experiments 10. This ratio was optimized by titrating the amount of ESF expression plasmid to a fixed amount (0.2 μg) of splicing reporter plasmid (Fig. S3). Higher amounts of splicing factor expression vector will cause exon skipping for all reporters, including those with pseudo-specific target sequences (Fig. S3). We previously observed similar effects of hnRNP A1 and ASF/SF2 in tethering experiments with MS2-coat protein 11. Therefore, like other process involving protein-RNA binding, the sequence specificity of splicing factors can only be considered in the context of certain concentration ranges.
ESFs can specifically modulate the use of alternative splice sites
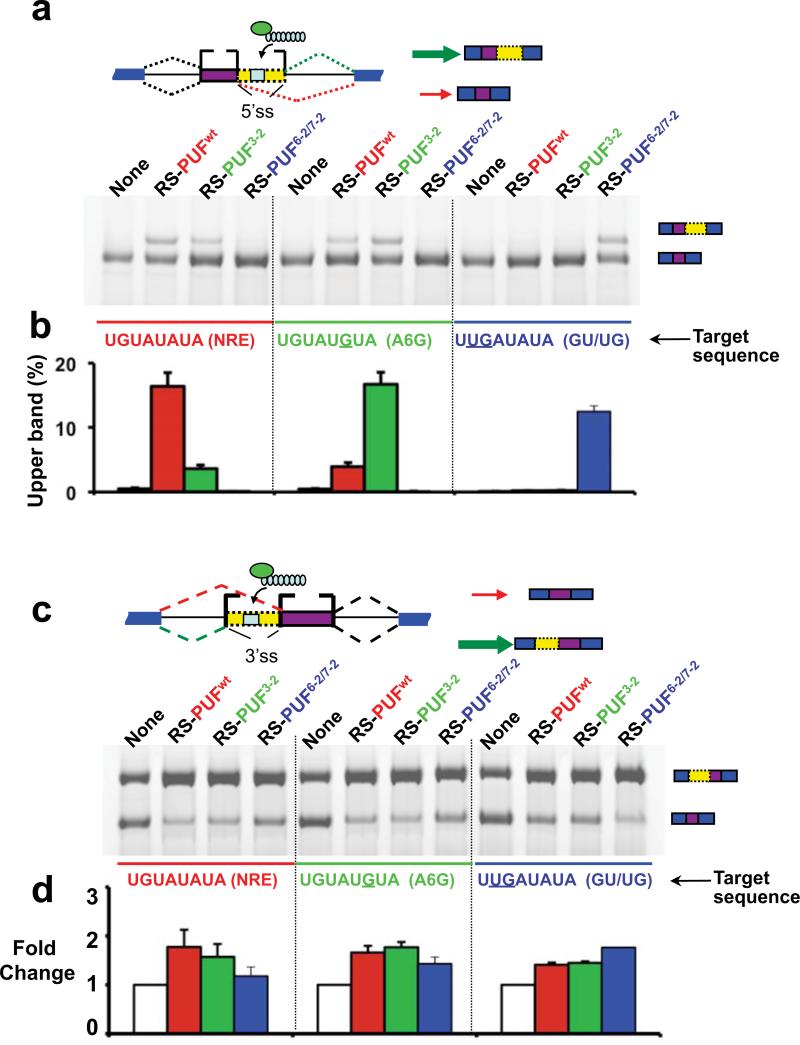
In addition to regulating exon skipping/inclusion, some splicing factors regulate alternative use of splice sites (ss) by recognizing regulatory sequences between two alternative sites. ASF/SF2 can recognize ESEs and promote the use of intron proximal sites, whereas hnRNP A1 can bind ESSs and shift splicing towards the use of intron distal sites 12-14. Therefore we next examined whether ESFs can predictably regulate alternative 5' or 3' ss usage with ESFs containing same panel of PUF domains (Fig. 1c). We designed reporters containing cognate target sequences between tandem 5' ss 11, and co-transfected them with the RS-PUF type ESFs. As expected, RS-PUFs increased the use of the downstream intron proximal 5' ss, with the strongest effect on reporters bearing their cognate sequence (Fig. 2a-b). Similarly, the RS-PUFs modulated alternative 3' ss usage (by promoting upstream 3' ss) in a sequence-specific fashion (Fig. 2c-d), indicating that these ESFs are capable of modulating different alternative splicing events. Due to the combination of sequence specific □and non-specific effects (see above), we also found that ESFs can affect splicing of pseudo-specific targets in alternative 3' ss reporter.
Fig. 2. Effect of ESFs on alternative splice site usage.
(a) Regulation of alternative 5' ss usage by RS-PUF type ESFs. Splicing reporters containing tandem 5' ss and different PUF target sequences (light blue) were co-transfected with RS-PUFs, which are expected to increase the use of the downstream site (indicated with the heavy green arrow). All combinations of splicing reporters and RS-ESF expression vectors listed in panel 1c were co-transfected into 293T cells. All experiments and quantification were carried out in duplicate as described in Fig. 1, and quantification of the results is plotted in (b). Instead of relative changes compared to no ESF controls, we plotted percentage of proximal 5' ss usage in panel b to avoid exaggerating the relative change because there is little or undetectable upper band in the absence of ESF (lanes 1, 5 and 9). (c) Regulation of alternative 3' ss usage by RS-PUF type ESFs. Splicing reporters containing tandem 3' ss and different PUF target sequences (light blue) were co-transfected with RS-PUFs, which are expected to increase the use of upstream 3' ss (indicated with the heavy green arrow). All experiments and quantification were carried out in duplicate as described in Fig. 1, and quantification of the results is plotted in (d) as fold changes of upstream 3' ss usage relative to no ESF controls.
Examining various functional domains in ESFs
The above experiments established the concept that artificial factors can be engineered with diverse activities and designed specificities. These ESFs provide a new approach to study the activities of natural splicing factors by specifically recruiting different proteins/domains to certain pre-mRNA regions. Compared to conventional tethering experiments using the MS2 coat protein or lambda N-B box systems 15, ESFs can recognize the pre-mRNA in a natural context without introducing foreign RNA, thus are more advantageous for in vivo applications.
Previous studies using in vitro splicing systems had shown that RS domains from various proteins (or even a short peptide of RS repeats) can function as splicing activators 3, 16. To test whether other RS domains can function as the effector module of ESFs, we generated new ESFs by fusing a PUF domain (PUF3-2) with RS domains from other SR proteins (9G8, SC35, SRP40) or a short peptide of RS repeats, and examined if these new ESFs promote exon inclusion when co-transfected with splicing reporters containing their targets (Fig. 3a, right panel). Compared to non-ESF control (Fig. 3b, lane 5), all RS-PUFs promoted inclusion of the cassette exon containing their cognate sequences (Fig. 3b, lane 6-10). The short RS repeat had similar activity to the other RS domains, with the only exception of the RS domain from 9G8, which has slightly higher active than the rests (Fig. 3c).
Fig. 3. Various RS domains or Gly-rich domains can function as the effector module of ESFs.
(a) Exon-skipping reporters containing the 8-nt cognate sequence of PUF3-2 ESFs in the cassette exon were used to examine the splicing suppressor activities of Gly-rich domains/fragment (left panel) or the splicing activator activities of RS domains/fragment (right panel). The experiments were similar to Fig. 1d and 1g except that different functional domains were fused to the same PUF3-2. (b) Inhibition (lanes 1-4) or promotion (lanes 6-10) of exon inclusion by ESFs with different splicing effector domains. The experiment conditions were same as Fig. 1d (for Gly-rich domains/fragment) or Fig. 1g (for RS domains/fragment), and all transfections were repeated at least twice. The functional domains of ESFs are lane 1 to 3, the Gly-rich domains of hnRNP A1 family proteins; lane 4, a short peptide of Gly-rich sequence (GYGGGGPGYGNQGGGYGGG); lane 6-9, the RS domains of SR proteins; lane 10, a short RS repeat (RS)6. (c) Fold change of cassette exon inclusion relative to the reporter alone (lane 5, no ESF). The means of replicated experiments are plotted with error bars indicating the range of replicated experiments.
Gly-rich domains are also found in many known splicing factors, especially members of hnRNP A1 family. However, it is unclear whether these Gly-rich domains can function as general splicing inhibitor, as only the domain from hnRNP A1 was shown to repress splicing when tethered to the exonic region 4. To test whether Gly-rich domains have intrinsic splicing inhibitory activity, we generated new ESFs by fusing PUF3-2 with the Gly-rich domains from additional hnRNP A1 family members (hnRNP A2/B1, hnRNP A3) or with a Gly-rich short peptide (19 aa in length), and examined whether they can inhibit exon inclusion when co-transfected with a splicing reporter (Fig. 3a, left panel). We found that all Gly-PUFs indeed suppressed the inclusion of the cassette exon containing their cognate sequence (Fig. 3b, lanes 1-4), indicating that the Gly-rich domains in members of hnRNP A1 family are likely responsible for their splicing suppression activity. More importantly, a 19-aa Gly-rich sequence was sufficient for splicing inhibitory activity (Fig. 3b-c, lane 4), suggesting that other splicing factors with a Gly-rich domain may function as splicing suppressors when binding to exons. PUF domains alone did not affect splicing of the same reporters (Fig. S4), suggesting that Gly-rich domains are responsible for splicing inhibition. We used the well-studied Gly-rich domain from hnRNP A1 for the remaining experiments.
Designing an ESF to modulate endogenous gene splicing
An important application for ESFs is to modulate alternative splicing of endogenous genes, particularly disease-associated splicing events. To demonstrate this, we chose to target the Bcl-x pre-mRNA that produces two splicing isoforms of opposite function using alternative 5' ss 17 (Fig. 4a). The long splicing isoform Bcl-xL encodes a potent apoptosis inhibitor in long-lived postmitotic cells and is up-regulated in many cancer cells to protect them against apoptotic signals. The short isoform Bcl-xS is pro-apoptotic and expressed predominantly in cells with a high turnover rate (e.g. developing lymphocytes) 18. Several labs have used antisense oligonucleotide-based methods to inhibit use of the intron proximal 5' ss and increase pro-apoptotic Bcl-xS 19-22.
Fig. 4. Design of an ESF to modulate splicing of endogenous Bcl-x pre-mRNA.
(a) Alternative splicing of Bcl-x pre-mRNA. The sequence UGUGCGUG between the two 5' ss was chosen as ESF target, and mutations (asterisks) were introduced in PUF repeats 1, 3 and 5 (Q867E/Q939E/C935S/Q1011E/C1007S) to recognize this sequence. The resulting PUF531 was fused with the Gly-rich domain of hnRNP A1 to inhibit downstream 5' ss (indicated with the heavy red arrow). (b) HeLa cells were transfected with different amounts of expression constructs of Gly-PUF531 and Gly-PUFwt (as control). Two Bcl-x isoforms were detected with RT-PCR using primers at exons 1 and 3 of the Bcl-x, and the Bcl-xS % was quantified. (c) Protein levels of Bcl-xL and Bcl-xS in the presence of ESFs. Samples are in the same order as panel b. The expression of ESFs is detected with anti-Flag antibody, and the tubulin level is used as control. The blot was exposed for a longer time for Bcl-xS because the available Bcl-x antibody detects Bcl-xL with much higher sensitivity. (d) Cleavage of PARP and caspase 3 in HeLa cells transfected with ESF expression constructs. Samples are detected by western blot at 24 hours after transfection. (e) Localization of ESFs in transfected HeLa cells detected with anti-FLAG antibody. The cells were co-stained with DAPI to show nuclei. Some nuclei, especially in cells transfected with Gly-PUF531, are fragmented due to apoptosis. Bar equals to 5μm. (f) Percentage of apoptotic cells (i.e. with fragmented nuclear DNA). Two independent experiments were conducted and > 200 cells were counted from pictures of randomly chosen fields.
The ratio of the two Bcl-x splicing isoforms is regulated by multiple cis-elements that are located near the two alternative 5' ss 23. To shift the splicing of Bcl-x, we designed a Gly-PUF type ESF to recognize the 8-nt sequence (UGUGCGUG) in the exon extension region, a sequence that otherwise does not affect splicing (not shown). This designer ESF should be able to “reprogram” the splicing regulation of Bcl-x to increase Bcl-xS isoform, which in turn can promote apoptosis (Fig. 4a). We mutated the first, third and fifth repeats of the wild-type PUF and verified that this modified domain (named PUF531) can recognize its new target (UGUGCGUG) with very high affinity (Kd ≈ 4 pM, Fig. S5) compared to the wild type PUF affinity for this sequence (Kd= 660± 17 nM). When transfected into HeLa cells where Bcl-xL is the predominant form, Gly-PUF531 increased splicing of the Bcl-xS isoform in a dose-dependent manner, whereas the control ESF (Gly-PUFwt) or PUF531 alone did not affect Bcl-xS level (Figs. 4b and S4b). The increase of Bcl-xS protein level was also observed using western blots (Fig. 4c), suggesting that the change of steady-state Bcl-xS/xL ratio is probably due to splicing shift rather than destabilization of Bcl-xL by PUF531 binding. The Bcl-xL protein levels showed no notable difference under our experimental conditions, probably due to a higher sensitivity of the Bcl-x antibody to detect Bcl-xL than Bcl-xS (Fig. S6) 19, 20.
Designer ESF induces apoptosis
Induction of the pro-apoptotic Bcl-xS isoform by Gly-PUF531 led to cleavage of caspase 3 and poly (ADP-ribose) polymerase (PARP), two known molecular markers in apoptosis pathway (Fig. 4d). Using immunofluorescence microscopy, we observed that many cells expressing Gly-PUF531 have fragmented nuclear DNA, indicating that they are undergoing apoptosis (Fig. 4e). Examination of >200 cells from randomly chosen fields in two independent experiments indicated that ~10% of cells transfected with Gly-PUF531 have fragmented nuclear DNA vs. only ~3% in control cells (Fig. 4f). We also noted that, as designed, the ESFs are localized predominantly in the nuclei of transfected cells (Fig. 4e), suggesting ESFs dissociate from their targets when the fully spliced mRNAs are transported into the cytoplasm.
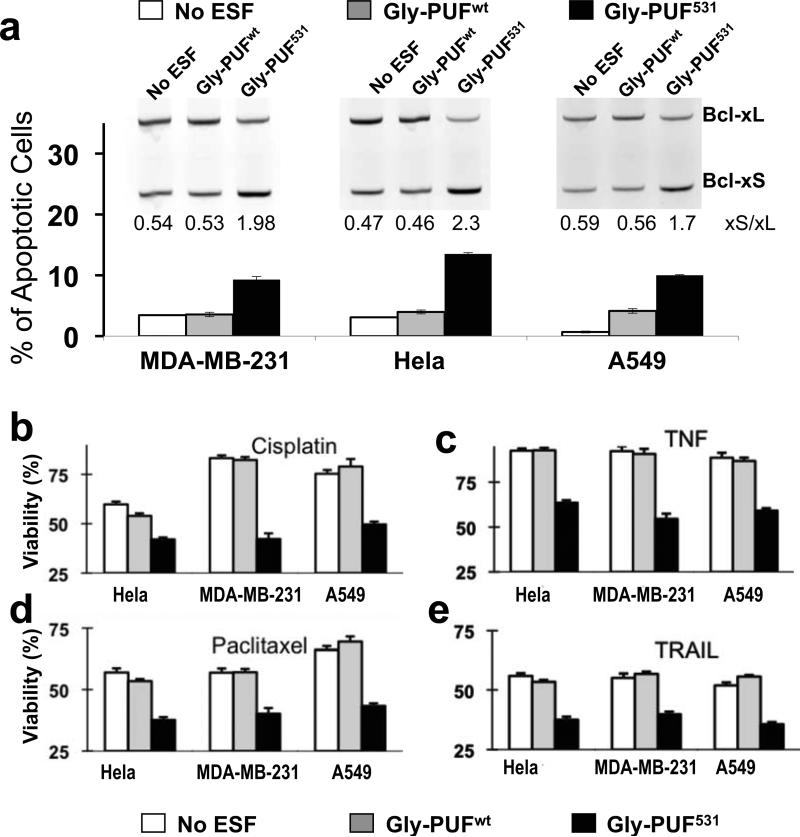
In addition to HeLa cells, we tested Gly-PUF531 in other cells including a breast cancer cell line (MDA-MB-231) and a lung cancer cell line (A549). We infected the cells with lentivirus expressing Gly-PUF531 or control ESF, and found that the designer ESF caused a significant shift of splicing to produce more Bcl-xS isoform in all cell types tested (Fig. 5a, inset). Such splicing shift increased apoptosis as determined by flow cytometry of propidium iodide-stained cells (Fig. 5a). In the absence of an exogenous stimulus that induces apoptosis, the increases of apoptotic cells are modest (~3 fold) but statistically significant in all cell types tested (p < 0.05, paired T-test), consistent with Bcl-xL being an important apoptosis inhibitor for most cancers 18.
Fig. 5. ESFs affect Bcl-x splicing in multiple cancer cells.
(a) Splicing modulation of Bcl-x in three cancer cell lines infected with lentivirus expressing Gly-PUF531, control ESF (Gly-PUFwt) or GFP (as mock infection). Bcl-x splicing isoforms were detected by RT-PCR, with Bcl-xS% being quantified and listed (inset). For reasons we are not completely clear, the lentivirus infected Hela cells have elevated basal level of Bcl-xS. Propidium iodide stained cells were analyzed with flow cytometry to determine the fraction of dead cells. (b) Effect of ESFs on cisplatin sensitivity of different cancer cells. Cells were infected with lentivirus to express the same designer ESF and controls, and cisplatin was added to a final concentration of 5 μM at 72 hours after infection. Cell viability was measured with the WST-1 assay 24 hours after drug treatment. All treatments were repeated at least twice, and the means with error bars representing the standard deviation (n=3) from representative experiments are plotted. White bars represent cells of mock infection, grey bars represent control Gly-PUFwt infection and black bars represent Gly-PUF531 infection. (c, d and e) Effect of ESFs on the sensitivities to paclitaxel, TNF-alpha and TRAIL in different cancer cell lines. Experimental conditions are the same as described for panel b except final concentrations of 10 nM paclitaxel (c), 20 ng/ml TNF-alpha (d), or 100 ng/ml TRAIL (e) were used. The significant differences (P<0.05, judged by paired T-test) of cell viabilities were observed for all drug treatments between the Gly-PUF531 and Gly-PUFwt infected cells.
Designer ESF sensitizes cancer cells to anti-tumor drugs
Since the increase of Bcl-xS releases inhibition of apoptosis, we expect a more apparent effect on cell survival during induction of apoptosis by chemotherapeutic drugs. We treated the different cancer cells with either Gly-PUF531 or control Gly-PUFwt in combination with anti-tumor drugs (cisplatin and paclitaxel) or cytokines (TNF-α and TRAIL, TNF-related apoptosis inducing ligand), which are commonly used in cancer treatments. To achieve robust expression during entire period of drug treatment, we infected the MDA-MB-231, A549 and HeLa cells with lentivirus expressing Gly-PUF531 or control ESF, and then treated the infected cells with low doses of drugs for 24 hours, conditions under which most mock-infected cancer cells are viable (Fig. 5b-e).
In all cell types tested, expression of Gly-PUF531 sensitized cells to the anti-cancer drugs tested, leading to significant decreases of cell viability compared to controls (p<0.05) as judged by WST-1 assay. Different cell lines responded to the combinations of anti-cancer drugs and ESFs with varying sensitivities (Figs. 5b-5e), consistent with the diverse mechanisms by which these drugs kill cancer cells. The enhancements of drug sensitivity in ESF-treated cancer cells were in the same range as cells treated with small molecule inhibitors of Bcl-xL 24, 25.
Discussion
Because ~90% of all human genes undergo alternative splicing 1, engineering novel factors to specifically modulate splicing offers tremendous promise for both basic and applied research. However, the development of such factors has been limited in the past mainly by the inability to design a sequence-specific RNA-binding module. By manipulating specificity of PUF domains, we are able to generate ESFs using an RNA-binding motif that is not found in natural splicing factors. This approach allows us to direct a novel splicing factor to almost any location in an endogenous pre-mRNA to either activate or repress splicing. A similar design could be adopted to study other RNA processing steps (e.g. polyadenylation or RNA degradation) that are regulated through interactions between RNA cis-elements and protein trans-factors.
The development of ESFs also provides a novel strategy to manipulate disease-associated splicing events, potentially leading to new treatment for diseases associated with aberrant splicing. Previous methods to modulate disease-related splicing have used antisense oligonucleotides to mask the splicing signals 26 or to recruit additional factors through an extended unpaired tail 27, 28, which require relatively high doses of oligonucleotides to effectively change splicing. The design of ESFs uses a fundamentally different strategy: to directly recognize targets through protein-RNA interaction and thus to reprogram the splicing regulation code. Compared to antisense methods, ESFs are more flexible because they can positively or negatively affect multiple types of alternative splicing. By optimizing various combinations of ESF modules, this approach will allow fine-tuned adjustment of alternative splicing. In addition, ESFs could be stably expressed in vivo using the current arsenal of gene therapy and expression tools, whereas the delivery and in vivo stability of antisense oligonucleotides are still difficult to control. Finally, in addition to manipulating disease associated splicing, this new approach has immediate impact as a new system to study factors that regulate splicing or other RNA processing pathways.
The RNA-binding module of our ESFs, PUF domain, specifically recognizes an 8-nt target sequence, giving us target discriminatory power similar to that of microRNAs which recognize targets mainly by a 7-nt seed match. Improving ESF specificity will minimize off-target effects, which may be achieved by recognizing longer target sequences using two tandem PUFs or creating a PUF with more repeats. A possible concern with the PUF domain is that repeats can be designed to recognize A, G or U bases, but amino acid residues that specify recognition of C have not been determined. This might present a limitation if a target sequence must be chosen in a small region. However, a C can be tolerated at position 5 in the target sequence, providing us with the ability to target a sequence with one C. Future work to determine the recognition code for a C base by a PUF repeat could address this issue.
In addition to manipulating disease associated splicing events, ESFs may be designed to create animal models of splicing diseases or to examine the physiological effects of particular alternative splicing choices. Another important application of ESFs is to study the self-regulation of splicing factors. Many splicing factors regulate their own splicing through a feedback loop 29, 30, but it is difficult to dissect such a regulatory network because these factors usually also regulate the splicing of essential genes for cell survival. Designing ESFs to direct the splicing of their own pre-mRNAs provides a unique opportunity to construct new splicing regulatory feedback loops or even new regulatory networks, thus could be used to model how splicing regulation is achieved on a systematic level.
Method
Methods and any associated references are available in the online version of the paper.
Supplementary Material
List of supplementary items
Supplementary Figure 1 Correlation of ESF activities with the binding affinity of their PUF domains to the target sequences
Supplementary Figure 2 Activities of ESFs in HeLa cells
Supplementary Figure 3 Increased amount of ESF expression can modulate splicing in sequence non-specific fashion
Supplementary Figure 4 PUF domain alone does not affect splicing
Supplementary Figure 5 Binding affinity between PUF531 and Bcl-x RNA sequence
Supplementary Figure 6 Detection of Bcl-xL and Bcl-xS protein in same Western blot after the ESF transfection
Supplementary Table 1 Primers used in constructing ESF expression vectors
Acknowledgments
We thank Dr. Chris Burge and our colleagues at UNC and NIEHS for critical reading of our manuscript. This work was supported by a grant from the Beckman Foundation (Z.W.) and the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (T.M.T.H).
Online Method
ESF expression constructs
To express ESFs in cultured cells, we generated expression constructs using the pCI-neo vector (Promega). We started with an expression construct that encodes from N- to C-terminal, FLAG epitope, Gly-rich domain of hnRNP A1 (residues 195-320 of NP_002127), and the MS2 coat protein (gift of Dr. R. Breathnach form Institut de Biologie-CHR 1). The fragment encoding the MS2 coat protein fragment was removed using BamHI/SalI digestion and replaced with a fragment encoding a NLS (PPKKKRKV) and the PUF domain of human Pumilio1, which was amplified using primers Pum-F1 and Pum-R1 (Supplementary Table 1). The resulting construct expresses a Gly-PUF type ESF under the control of a CMV promoter (Fig. 1b). To make an expression construct for an RS-PUF type ESF, we removed the fragment encoding the FLAG/Gly-rich domain with NheI/BamHI digestion, and replaced it with a fragment that encodes the RS domain of ASF/SF2 protein with an N-terminal FLAG epitope, which was amplified using primers ASF-RS-F and ASF-RS-R (Supplementary Table 1). To generate constructs for different ESFs with mutated PUF domains, we introduced the point mutations in consecutive steps using a QuikChange Site-Directed Mutagenesis kit (Stratagene) following manufacturer's instructions 2. The additional functional domains of ESF (RS domains/fragment or Gly rich domains/fragment) were amplified by PCR (or synthesized oligonucleotides for short fragments), and cloned between XhoI/BamHI. The RS domains are residues 123-238 of 9G8 (NP001026854), residues 180-272 of SRp40 (NP008856), residues 117-221 of SC35 (NP003007) and an (RS)6 sequence. The Gly-rich domains are residues 203-353 of hnRNP A2 (NP112533), residues 211-378 of hnRNP A3 (NP919223), and the short peptide of GYGGGGPGYGNQGGGYGGG.
Splicing reporter constructs
To assess the effects of ESFs on exon skipping, we used a modular reporter system that allowed us to change an inserted splicing regulatory sequence near a test exon (Exon 12 of the human IGF-II mRNA-binding protein 1, IGF2BP1, Ensembl ID ENSG00000159217) and assay for the inclusion of this test exon. The test exon, together with its flanking introns, was inserted between two GFP exons in pEGFP-C1 vectors as described previously 3. To insert target sequences of PUF domains into this reporter vector, we synthesized and annealed oligonucleotides containing the candidate sequences (designated by N8) flanked by XhoI and ApaI sites. The resulting DNA fragments were digested and ligated into the XhoI/ApaI digested vector (inside the test exon). The inclusion of the test exon is assayed by body-labeled RT-PCR, as described previously, with primers corresponding to the first and third exon (two GFP exons) of the reporter minigene 4.
To assess the effect of ESFs on the alternative use of 5' and 3' ss, we used reporters with competing 5' and 3' ss 4. The same target sequences of PUF domains were inserted into these reporters using either XhoI/ApaI sites (for the competing 3' ss reporter) or XhoI/EcoRI sites (for the competing 5' ss reporter) as described above.
Electrophoretic mobility shift assays
Recombinant proteins were expressed and purified and electrophoretic mobility shift assays were performed as described previously (Cheong and Hall, 2006). RNA oligonucleotides were obtained from Dharmacon, Inc. (Lafayette, CO) and radiolabeled at the 5' end by using [γ-32P]-ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England BioLabs) following manufacturer directions. The radiolabeled RNAs were purified on 20% polyacrylamide gels (Invitrogen, Carlsbad, CA) run with 1X Tris-Borate-EDTA buffer (89 mM Tris, 89 mM boric acid, 2.5 mM EDTA, pH 8.3) at room temperature.
Binding reactions included radiolabeled RNA (100 pM for A6G and GU/UG RNAs and 1-2 pM for Bcl-x RNA) and varying concentrations of protein incubated in binding buffer (10 mM Hepes, pH 7.4, 50 mM KCl, 1 mM EDTA, 0.01% (v/v) Tween-20, 0.1 mg/ml BSA, 1 mM DTT). Binding reactions were incubated for 1–2 h at room temperature and immediately analyzed by electrophoresis on 6% nondenaturing polyacrylamide gels (Invitrogen). Gels were dried and exposed to storage phosphor screens (GE Healthcare, Piscataway, NJ), scanned with a Typhoon 8600 Imager (GE Healthcare), and analyzed with Image Quant 5.2 software (Molecular Dynamics/GE Healthcare). The data were analyzed using Origin 7.5 software (OriginLab). All binding assays were performed at least in triplicate. The binding of PUF531 to its target RNA sequence was too tight to allow an accurate determination of its Kd. Thus, a value of ~4 pM is reported (Fig. S5).
Cell culture, transfection, RNA purification and semi-quantitative RT-PCR
The human embryonic kidney cell line 293T, lung cancer cell line A549, breast cancer cell line MDA-MB-231, and human cervical cancer cell line HeLa were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Breast cancer cell line BT474 was cultured in F12/DMEM supplemented with 10% fetal bovine serum.
For cell transfections, cells were seeded onto 24-well plates 1 day prior to transfection. For each well, in separate tubes 0.2 μg of splicing mini-gene reporter constructs were mixed with appropriate amounts of ESF expression vector (0.04 μg for Gly-PUF and 0.4 μg for RS-PUF) and 2 μl of Lipofectamine 2000 (Invitrogen) were added to 50 μl of Opti-MEM I (Invitrogen). The solutions were then combined and mixed gently, allowed to sit for 20 min at room temperature, and added to the plates for 24 h at 37 °C and then harvested for RNA or protein isolation.
Total RNA was isolated from transfected cells with TRIzol reagent (Invitrogen) according to the manufacturer's instructions, followed by 1 h DNase I (Invitrogen) treatment at 37 °C and then heat inactivation of DNase I. Total RNA (2 μg) was then reverse-transcribed with SuperScript III (Invitrogen) with poly T primer (for Bcl-x) or gene specific primer (for splicing reporter), and one-tenth of the RT product was used as the template for PCR amplification (25 cycles of amplification, with trace amount of Cy5-dCTP in addition to non-fluorescent dNTPs). The primers used to detect splicing reporter (GFP exons) were AGTGCTTCAGCCGCTACCC (forward) and GTTGTACTCCAGCTTGTGCC (reverse) and for Bcl-x were CATGGCAGCAGTAAAGCAAG (forward) and GCATTGTTCCCATAGAGTTCC (reverse). RT-PCR products were separated on 10% PAGE gels run with 1X TBE buffer, and scanned with a Typhoon 9400 scanner (Amersham Biosciences). The amount of each splicing isoform was measured with ImageQuant 5.2. All experiments were repeated at least three times.
Immunofluorescence
HeLa cells were seeded onto poly-lysine coated glass coverslips in a 6-well plate, and then transfected with Gly-PUFwt or Gly-PUF531 constructs using Lipofectamine 2000. At 24 h after transfection, the cells were fixed on the coverslips with 4% formaldehyde in 1X PBS for 20 min at room temperature and washed with 1X PBS three times. Cells were then permeabilized with 0.2% Triton X-100 in 1X PBS for 10 min and washed with 1X PBS three times. Cells were blocked with 3% BSA in 1X PBS for 10 min, washed with 1X PBS three times, and then incubated with primary antibody (monoclonal anti-FLAG antibody from Sigma-Aldrich, clone M2) for 1 h at room temperature. The cells were washed with 1X PBS three times and incubated with FITC-conjugated goat anti-mouse IgG for 30 min. The cells were washed with 1X PBS three times and the cover slips were mounted with mounting medium (Vector shield's mounting medium with DAPI). Cells were visualized using an Olympus fluorescence microscope, and photographs were generated using a Kodak digital camera.
Western blot
Cells were transfected with Gly-PUFwt or Gly-PUF531 constructs, respectively and harvested 24 h after transfection. The total cell pellets were boiled in 2X SDS-PAGE loading buffer for 10 min and then resolved by 12% SDS-PAGE and transferred to nitrocellulose membrane. The following antibodies were used in this study: Caspase-3 (#9668) and PARP (#9542) antibodies were purchased from Cell Signaling Technology, Bcl-x antibody (610211) was purchased from BD Bioscience, and antibodies against actin (A5441), alpha-tubulin (T5168) and FLAG M5 (F4042) were purchased from Sigma-Aldrich. Bound antibodies were visualized with the ECL kit (GE Healthcare).
Propidium iodide staining and flow cytometry
Cells were harvested at 24 h after transfection and stained for 5 min in a PBS solution containing a final concentration of 2 μg/ml propidium iodide (PI). The PI-stained cells were analyzed with a FACSCalibur fluorescence-activated cell sorter (FACS) using CELLQuest software (Becton Dickinson,NJ).
Lentivirus infection and cell viability assay
The full-length ESFs were PCR amplified from original expression vectors and integrated in the lentiviral expression vector pWPXLd between MluI/SpeI sites. Lentiviruses were generated by co-transfecting 293T cells with packaging vectors pPAX2 and pMD2.G with either pWPXLd-Gly-Puf531, pWPXLd-Gly-Pufwt (Control), or pWPXLd-GFP (mock) using the standard calcium phosphate precipitation method. The titer of lentivirus was determined by infecting 293 cells with serial dilutions of virus preparation.
Cell viability was determined with the WST-1 assay (Roche) following the manufacturer's instructions. MDA-MB-231, A549, and HeLa cells were infected with lentivirus expressing GFP (as non ESF control), control ESF or designer ESF, and then seeded 72 h after infection in a 96-well plate for overnight incubation. Cisplatin (Sigma, P4394), Paclitaxel (Sigma, T7191), TNF-alpha (GeneScript, Z01001) and TRAIL (GenWay, 10663-45267) were added to each cell line at their respective concentrations. After 24 h incubation at 37 in the presence of 5% CO2, 10 μl of cell proliferation reagent WST-1 was added to each well. The cells were incubated with WST-1 reagent for 0.5 h and the absorbance at 450 nm was measured using a Benchmark microplate reader (Bio-Rad, CA). At least three independent experiments were performed and the means and standard deviations were calculated and plotted in Fig. 5.
- 1.Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiao X, Wang Z, Jang M, Burge CB. Coevolutionary networks of splicing cis-regulatory elements. Proc Natl Acad Sci U S A. 2007;104:18583–18588. doi: 10.1073/pnas.0707349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 4.Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 7.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai Y, Lee D, Yu T, Chasin LA. Control of 3' splice site choice in vivo by ASF/SF2 and hnRNP A1. Nucleic Acids Res. 1999;27:1126–1134. doi: 10.1093/nar/27.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eperon IC, et al. Selection of alternative 5' splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Contreras R, et al. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 15.Keryer-Bibens C, Barreau C, Osborne HB. Tethering of proteins to RNAs by bacteriophage proteins. Biol Cell. 2008;100:125–138. doi: 10.1042/BC20070067. [DOI] [PubMed] [Google Scholar]
- 16.Philipps D, Celotto AM, Wang QQ, Tarng RS, Graveley BR. Arginine/serine repeats are sufficient to constitute a splicing activation domain. Nucleic Acids Res. 2003;31:6502–6508. doi: 10.1093/nar/gkg845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boise LH, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 18.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat Biotechnol. 1999;17:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- 20.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 21.Wilusz JE, Devanney SC, Caputi M. Chimeric peptide nucleic acid compounds modulate splicing of the bcl-x gene in vitro and in vivo. Nucleic Acids Res. 2005;33:6547–6554. doi: 10.1093/nar/gki960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gendron D, et al. Modulation of 5' splice site selection using tailed oligonucleotides carrying splicing signals. BMC Biotechnol. 2006;6:5. doi: 10.1186/1472-6750-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou A, Ou AC, Cho A, Benz EJ, Jr., Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5' splice site selection. Mol Cell Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 25.Shoemaker AR, et al. A small-molecule inhibitor of Bcl-XL potentiates the activity of cytotoxic drugs in vitro and in vivo. Cancer Res. 2006;66:8731–8739. doi: 10.1158/0008-5472.CAN-06-0367. [DOI] [PubMed] [Google Scholar]
- 26.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 Exon 7 Inclusion by Antisense Oligonucleotides Targeting the Exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- 28.Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci U S A. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. Embo J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchison S, LeBel C, Blanchette M, Chabot B. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5' splice site selection in the hnRNP A1 mRNA precursor. J Biol Chem. 2002;277:29745–29752. doi: 10.1074/jbc.M203633200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of supplementary items
Supplementary Figure 1 Correlation of ESF activities with the binding affinity of their PUF domains to the target sequences
Supplementary Figure 2 Activities of ESFs in HeLa cells
Supplementary Figure 3 Increased amount of ESF expression can modulate splicing in sequence non-specific fashion
Supplementary Figure 4 PUF domain alone does not affect splicing
Supplementary Figure 5 Binding affinity between PUF531 and Bcl-x RNA sequence
Supplementary Figure 6 Detection of Bcl-xL and Bcl-xS protein in same Western blot after the ESF transfection
Supplementary Table 1 Primers used in constructing ESF expression vectors