Abstract
Invasive pneumococcal disease (IPD) has been reduced in the US following conjugate vaccination (PCV7) targeting seven pneumococcal serotypes in 2000. However, increases in IPD due to other serotypes have been observed, in particular 19A. How much this “serotype replacement” will erode the benefits of vaccination and over what timescale is unknown. We used a population genetic approach to test first whether the selective impact of vaccination could be detected in a longitudinal carriage sample, and secondly how long it persisted for following introduction of vaccine in 2000. To detect the selective impact of the vaccine we compared the serotype diversity of samples from pneumococcal carriage in Massachusetts children collected in 2001, 2004 and 2007 with others collected in the pre-vaccine era in Massachusetts, the UK and Finland. The 2004 sample was significantly (p >0.0001) more diverse than pre-vaccine samples, indicating the selective pressure of vaccination. The 2007 sample showed no significant difference in diversity from the pre-vaccine period, and exhibited similar population structure, but with different serotypes. In 2007 the carriage frequency of 19A was similar to that of the most common serotype in pre-vaccine samples. We suggest that serotype replacement involving 19A may be complete in Massachusetts due to similarities in population structure to pre-vaccine samples. These results suggest that the replacement phenomenon occurs rapidly with high vaccine coverage, and may allay concerns about future increases in disease due to 19A. For other serotypes, the future course of replacement disease remains to be determined.
Keywords: Streptococcus pneumoniae, Infectious disease epidemiology, Nasopharyngeal carriage, Population genetics
Introduction
Conjugate vaccination against seven serotypes of S. pneumoniae (the pneumococcus) has had great benefits for public health. In the US between 1998-9 and 2003, following vaccine licensure and despite vaccine shortages, the incidence of invasive pneumococcal disease (IPD) due to vaccine serotypes among children under 5 fell by 94% (2005). Moreover, the interruption of pneumococcal transmission within this core group has led to a substantial reduction in IPD due to vaccine types among unvaccinated older adults and the population at large (Lexau et al., 2005).
Despite this success, concern has been raised that these public health benefits are being eroded by the phenomenon of serotype replacement (Hanage, 2008; Spratt and Greenwood, 2000). This refers to the fact that the vaccine targets only seven of the more than 90 known pneumococcal serotypes. As well as protecting against IPD, vaccination also prevents carriage of vaccine serotypes (Ghaffar et al., 2004) and so removes these from the population. If vaccine and non-vaccine serotypes compete in the human nasopharynx (their normal anatomical niche) then this will produce an opportunity for those serotypes not in the vaccine to benefit from the removal of any such competitors, and increase in prevalence.
Such serotype replacement has been documented in carriage (Ghaffar et al., 2004; Huang et al., 2009; Huang et al., 2005), and a number of disease contexts (Byington et al., 2005; Eskola et al., 2001). However replacement has, thus far, been limited in invasive disease (Hicks et al., 2007). The incidence of a small number of non-vaccine serotypes, notably 19A and serogroup 15 strains has been increasing in IPD (Hicks et al., 2007). Prior to vaccination, the incidence of IPD due to 19A was estimated at 2.5 /100,000 per year in the vulnerable under 5 age group. By 2004 in the same age group it was estimated to have increased to 7.8/100,000 per year, equivalent to a relative risk for IPD due to 19A of 3.2 (95% CI 2.3-4.4) in comparison with the pre-vaccine era (Hicks et al., 2007). In some particularly vulnerable populations, the overall 19A disease rate can be much higher (Singleton et al., 2007).
A question of pressing public health importance is whether we are only seeing the start of this phenomenon. Will the increase in IPD due to replacement serotypes continue as the pneumococcal population responds and adapts to the presence of vaccination? Or has the population already reached a new equilibrium, and so future changes will be relatively small in comparison with those that have already occurred?
Previous analyses have focussed on longitudinal trends in carriage or invasive disease rates to detect and analyse serotype replacement (Hicks et al., 2007; Huang et al., 2005). Here we present an alternative approach based on changes in the observed diversity of serotypes, hypothesising that this is a sensitive measure of the selective effects of the vaccine on the pneumococcal population as a whole, rather than individual serotypes. We use longitudinal data from Massachusetts (MA) to show the effect of vaccination on the diversity and rank-frequency distribution of serotypes in carriage.
Methods
Samples
Pre-vaccine communities
We use three datasets. As an indicator of the baseline serotype distribution in Massachusetts we use a sample of 71 carried isolates from children between 3 months and 6 years of age with acute otitis media (AOM). These were collected in 1998 and 1999 from practices in two Massachusetts communities (Pelton et al., 2004). They are taken as representative of the serotypes in NP carriage (since there are small differences in the ability of colonizing serotypes to cause AOM (Hanage et al., 2004)). Serotyping was carried out in the Maxwell Finland laboratory at Boston University School of Medicine. For further comparison we use two additional samples, comprising isolates from nasopharyngeal (NP) swabs from healthy children <2 years of age in Oxford, UK and Tampere, Finland (Brueggemann et al., 2003; Hanage et al., 2004). To ensure that a single isolate was included per episode of carriage, we used multilocus sequence typing (MLST) (Enright and Spratt, 1998) data which was available for all three pre-vaccine datasets. Where more than one isolate with the same serotype and sequence type (ST) was retrieved from the same child, all but one was excluded (leading to the exclusion of 18 isolates from the 1998-9 MA sample; 69 isolates from the Tampere, Finland sample, and 69 isolates from the Oxford, UK dataset. For the latter two datasets the resulting samples are as described previously (Fraser et al., 2005)). The sizes of the datasets used in this work are Tampere N=216; Oxford N=228; MA 1998-9 N=71.
Longitudinal sampling of post vaccine cohorts
Serial samples of children <7 years of age were collected in 2001, 2004 and 2007 (described in detail in (Hanage et al., 2007; Hicks et al., 2007; Huang et al., 2005)) from children seen in pediatric practices in 8 Massachusetts communities. 8 additional communities were sampled in 2001 and 2004 but have been excluded from this analysis to ensure consistency. The overall carriage prevalence of pneumococcus did not change greatly over the three study periods (Huang et al., 2009). Final sample sizes were as follows: 2001 N=124; 2004 N=220; 2007 N=295.
For samples to be comparable, they must be serotyped to the same level of discrimination. For example, we cannot compare datasets which pool all serogroup 19 isolates together with those that do not. Because 15B and 15C strains have been found to interconvert with high frequency (van Selm et al., 2003), these have been pooled together as 15B/C in all samples including those from post-vaccine communities (see below). For those cases in the MA 1998-9 dataset where isolates were unavailable for testing to a finer level of discrimination, the MLST data were used to determine the most likely serotype based on the relationship of the isolate to the rest of the sample and the MLST database. For example, isolates recorded as serogroup 15 with no further discrimination, and ST 199 by MLST, were assumed to be 15B/C because in all previously recorded cases of this ST expressing serogroup 15 capsule, the subtype has been found to be 15B/C. In one case where this was not possible (ST 1910 is a singleton which does not have any close relatives in the MLST database), the isolate was removed from the dataset.
Statistical analysis
The diversity of each dataset was estimated using Simpson's index of diversity D (Ref (Simpson, 1949)); defined here as where is the fraction of the sample with serotype i, m is the total number of serotypes and N is the sample size. The choice of measure was motivated by analogy with population genetics, where D is equivalent to the heterozygosity of a sample, and here is the probability that two randomly selected isolates have different serotypes. is a correction for sample size. Variances and 95% confidence intervals for each sample were calculated as previously described (Simpson, 1949). Since diversity did not differ between the baseline pre-vaccine era samples (see Table 1), we pooled the pre-vaccine samples by summing the number of isolates with each serotype across the three samples, and dividing by the combined sample size of 515 isolates (from the 1998-9 MA sample combined with the Tampere and Oxford datasets) to compare diversity D between the pre and post vaccine samples. To test whether estimates of D were significantly different p-values were calculated by two tailed Welch's t-test. We also tested the effect of pooling by rank (ie combining the numbers of isolates in the category of most common serotype, second most common serotype, etc) and obtained similar results to those reported below (not shown).
Table 1.
Diversity of samples used in this study.
| Sample | N | No of serotypes | D (95% CI) |
|---|---|---|---|
| Finland | 216 | 28 | 0.911 (0.896 – 0.926) |
| Oxford | 228 | 34 | 0.922 (0.905 – 0.938) |
| MA 1998-9 | 71 | 20 | 0.918 (0.889 – 0.946) |
| Combined pre vaccine | 515 | 40 | 0.919 (0.909 – 0.929) |
| MA 2001 | 124 | 24 | 0.941 (0.918 – 0.964) |
| MA 2004 | 220 | 29 | 0.946 (0.934 – 0.958) |
| MA 2007 | 295 | 32 | 0.923 (0.912 – 0.935) |
N is the total number of isolates after censoring as described in the methods section. D is the observed diversity. 95% CIs were calculated by the method of Simpson (Simpson, 1949).
To further explore the selective impact of vaccination, we calculated the rank frequency distribution of each sample. Specifically, if {x1, x2,…, xm} denotes the distribution of serotype frequencies, then we denote the permutation of this distribution re-ordered from largest to smallest as {x[1], x[2],…, x[m]}, which is called the rank-frequency distribution. We do this ranking in turn for each sample, and note the strong similarity between all the pre-vaccine era samples despite differences in serotype composition (Figs 1A and B). To obtain a range of rank-frequency distributions to compare with the vaccine era samples, we simulated 1000 Monte-Carlo replicates drawn from the empirically observed rank-frequency distribution of the pooled pre-vaccine samples. All procedures were carried out in Microsoft Excel 2007.
Figure 1.
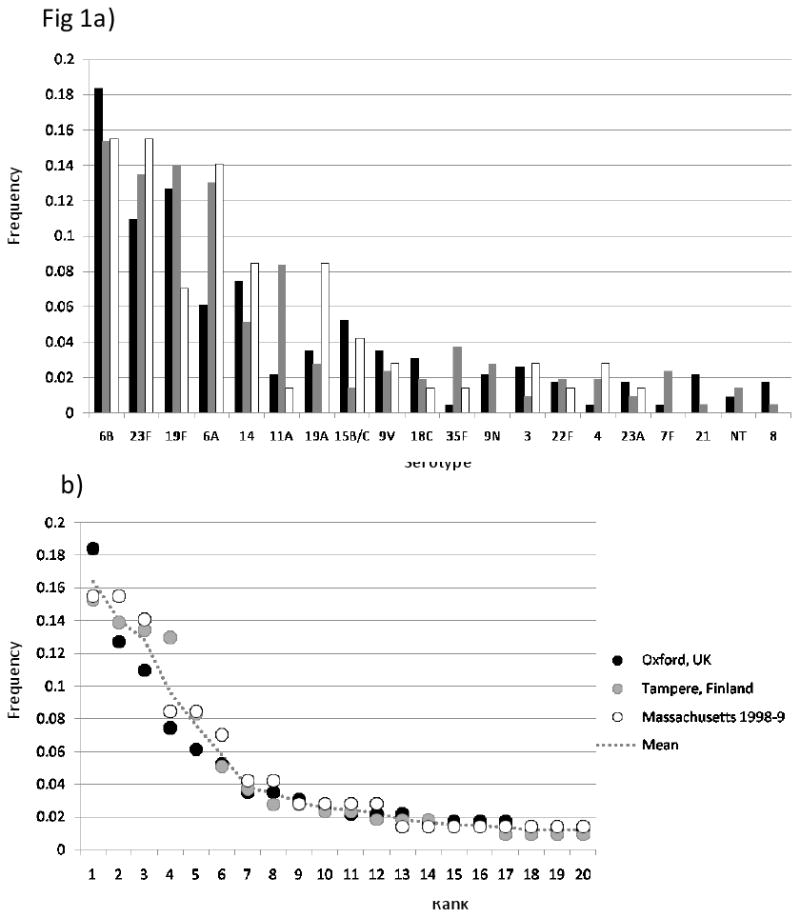
A) Frequencies of individual serotypes in the three pre-vaccine datasets. Sample details are described in the main text and Table 1. For reasons of space only the 20 most common serotypes in the combined dataset are shown. Vaccine serotypes are underlined. B) The same data arranged by rank (most-least common).
Results
Comparison of serotype diversity in pre-vaccine populations
To characterise the pneumococcal population prior to vaccination, we used three independent samples from similar age groups typed to the same level of discrimination (namely Oxford, UK (Meats et al., 2003); Tampere, Finland (Hanage et al., 2004), and Massachusetts 1998-9). Details of all samples used in this work are provided in Table 1. Fig 1A compares the frequency of individual serotypes in the different locations (for reasons of clarity, only the 20 most common are shown). While 6B was the most common in all three, marked differences were observed (for example in the frequencies of 19F, 6A, 11A and 19A). However the rank frequency distribution of serotypes, shown in Fig 1B, is strikingly similar. In none of these samples does the most common serotype make up more than 20% of the population. Also shown in Figure 1 is the mean rank frequency distribution, calculated as the sum of each serotype in the combined dataset, divided by the combined sample size of 515, and arranged by rank. The diversity of this combined dataset was 0.919 (95% CI: 0.909 – 0.929), with the diversity of individual datasets shown in Table 1.
Diversity and rank frequency serotype distributions following vaccination
All three samples collected following vaccination are more diverse than the pooled pre vaccination dataset (see Table 1), however only the 2004 sample is significantly so (p>0.0001). While a similar increase in diversity is apparent in 2001, this is not significant, possibly due to the small size of this sample. The rank frequency distributions for each sample, calculated as previously, are shown in figure 2 B-D, along with that from the 1998-9 MA sample which is reproduced from Fig 1B for purposes of comparison (Fig 2A). For each sample, the median and 95% intervals of 1000 samples of the same size drawn from the empirical pre-vaccine distribution are also shown. The greater diversity of the 2004 sample is reflected by the pronounced flattening in the rank frequency distribution (figure 2C). Although serotype replacement has led to changes in the specific serotypes which compose the 2007 sample (the largest single sample N= 295), its diversity is not significantly different from that of the pre-vaccine samples (see Table 1), while the frequency of the most common serotype (19A) and the rank frequency distribution appear similar to that expected from the pre-vaccine datasets (Fig 2D).
Figure 2.
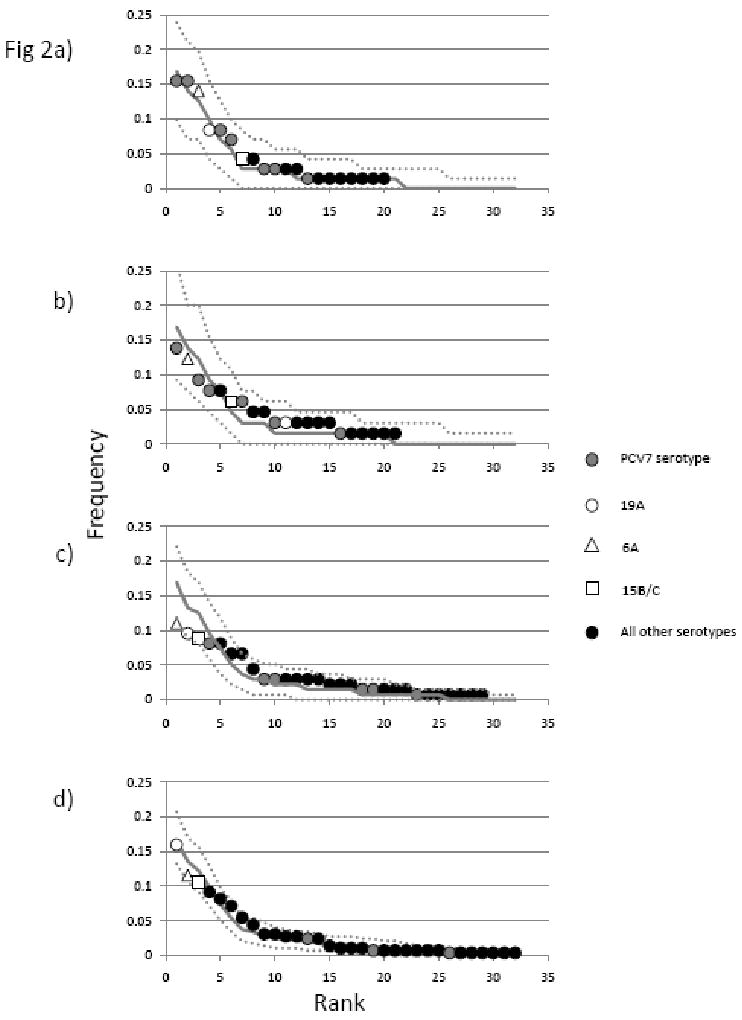
Rank frequency distributions of serotypes in the MA samples from 1998-9 (a), 2001 (b), 2004 (c) and 2007 (d). Details of the samples are as described in the text and table. Serotypes are shown as indicated in the legend. The median values obtained from 1000 samples of the same size drawn from the combined pre-vaccine distribution (as described in the text) are shown as a solid line with dashed lines either side representing the 5th and 95th percentiles for each point. Larger numbers of serotypes in the later samples reflect larger sample size.
Replacement
Serotype replacement refers to the expansion of non vaccine serotypes as a result of the removal from the population of vaccine types which compete with them to colonise new hosts. The major replacement serotypes in the carriage studies described here, thus far, have been serotypes 6A, 19A and 15B/C. The serial increase of these has been previously documented in these communities (Huang et al., 2009; Huang et al., 2005), and their expansion may be clearly observed in Figure 2. In contrast, by 2007 the pooled vaccine serotypes (4, 6B, 9V, 14, 18C, 19F and 23F and shown in gray) were responsible for a small fraction of carriage. Note that over the period of the study the overall carriage prevalence did not change (1998, 21.6%; 2001, 25%; 2004, 24%; 2007, 31%) (Huang et al., 2009).
Discussion
We have found that we can detect the impact of selection, due to conjugate vaccination, on the population structure of carried pneumococci by comparing three longitudinal samples collected post vaccination with samples collected in the absence vaccination. The samples collected from MA in 2001, 2004 and 2007 document the change produced by vaccination. The 2004 sample is significantly more diverse than the pre-vaccine samples (with a non-significant change in the same direction in 2001). The pre-vaccine samples are similar to each other terms of serotype diversity (Table 1). Data from the National Immunisation Survey (http://www.cdc.gov/vaccines/stats-surv/imzcoverage.htm) show that vaccine coverage in MA was relatively low in 2001 and 2002, as most toddlers were born before the vaccine became available and “catch up” vaccination was incomplete. The first time point for which data are available is July 2001 – June 2002, when coverage with three doses of vaccine was 35.8 ± 5.1% among children aged 19-35 months. By 2004 coverage in the same age group had reached 89.7 ± 3.8% and has not dropped below 90% since. It is reasonable therefore to suggest that the data from 2004 reflect the period at which the pneumococcal population was in the process of responding to the novel selective pressure of the vaccine, and it is this which produces the comparatively flat distribution observed as vaccine serotypes, initially common, are selected against (clearly evident in the decline of the gray points in Fig 2) and non-vaccine serotypes experience a relative fitness advantage. The results show that the 2004 sample is more diverse than expected from samples in the absence of vaccination (D = 0.946; 95% CI: 0.934 – 0.958).
In contrast by 2007 the distribution has regained many of its pre-vaccine characteristics, in both serotype diversity and the rank frequency distribution of serotypes (Table 1 and Figure 2d). It should be noted however that although we did not find the 2007 sample to be significantly different from the pre-vaccine observations, this could be because of limitations in the power of the test used here. Nevertheless the similarity between the rank frequency serotype distributions of the 2007 sample and those observed in pre-vaccine samples is pronounced (Fig 2d). In this most recent sample, the most common serotype in 2007 is 19A, now a relatively common cause of invasive disease (Hicks et al., 2007). The vaccine serotypes, which together were responsible for the majority of isolates in all pre-vaccine samples (56% in Oxford; 53% in both Finland and MA) have dwindled to a small fraction of their previous prominence. There has been considerable concern about the future trajectory of replacement disease due to 19A strains: do we expect that in the absence of vaccination against 19A they would continue to expand? Based on these observations we suggest that, at least in MA, this is unlikely. Given that carriage rates remain similar to the pre-vaccine era (data not shown), by 2007 19A is already as common as the most common serotypes in the pre-vaccine population, making up ∼20% of the carriage population. If this empirically observed limit in the frequency of the most common serotype is maintained in the post vaccine era, further large increases in the carriage prevalence of this serotype are unlikely. And in consequence, large increases, compared to current rates, in invasive disease due to 19A should not be expected in coming years.
This study has several shortcomings. It would be preferable to have a larger sample for 2001. The relatively small sample size at this time point and 2004 is due to the exclusion of data from the 8 additional communities not sampled in 2007, in order to ensure consistency. Inclusion of these communities does not alter the results (not shown). Another concern is that the only pre-vaccine sample available for MA is from NP carriage in children with AOM. Those serotypes causing AOM may not be representative of those in carriage, but the differences are expected to be minor in the light of a study showing that the serotypes in AOM are a relatively good reflection of NP carriage, in comparison with IPD (Hanage et al., 2004). Furthermore the children in question are not necessarily suffering from pneumococcal AOM. We hence use this sample as an approximation to the pre-vaccine carriage population in MA. It is gratifying to note that the serotype diversity D is similar to other pre-vaccine populations and the fit to the empirical rank frequency distribution is a good one (Table 1 and figure 2).
In addition to this it should be noted that the clonal composition of 19A could continue to change to include larger numbers of antibiotic resistant or non-susceptible strains (Hanage et al., 2007). This could have a substantial clinical impact. It must also be emphasized that small fluctuations in the carriage prevalence of a rarely carried but highly invasive serotype (such as serotype 7F) could have a substantial impact on overall IPD rates, and that such changes would not be apparent in analyses of carriage serotype diversity such as this one. We have made no assumptions in this study regarding the reasons for the success of 19A, but it may be related to the relatively high frequency of penicillin non-susceptibility in clones of this serotype (Pelton et al., 2007). We suggest that 6A strains, which in Figure 2 appear well positioned to expand, have not done so because of confusion with the newly discovered serotype 6C (Nahm et al., 2009). Until recently, it was not possible to distinguish these serotypes by usual quellung reaction, but they nevertheless differ markedly in terms of vaccine efficacy: 6A appears to cross react with the 6B component of PCV7 and has been declining since vaccine introduction whereas no such decline has been noted for 6C (Park et al., 2008). We suggest therefore that our data reflect a decrease in 6A, and replacement with 6C, but because the methods used here were unable to distinguish between these two serotypes 6A prevalence apparently remains reasonably constant. Finally, the data shown in figure 1 clearly show that even prior to vaccination, considerable variation in the prevalence of individual serotypes could exist among different communities.
New vaccines with higher valency which target 19A strains (among other serotypes) will be available in the near future. We would predict that the impact of these will be similar to that which has been seen with the current conjugate vaccine, removing vaccine serotypes over a period of 4-6 years, with replacement occurring on the same timescale. The effect on IPD will depend on the invasive nature of the replacing serotypes though we propose that no serotype is likely to increase to cause >20% of carriage episodes. The precise rank order of the serotypes cannot be predicted with confidence at this stage, but it is reasonable to suppose that presently common non-vaccine serotypes will be well represented. Finally, we do not wish to suggest that once replacement has occurred, substantial secular changes are not possible in terms of the precise serotype composition of different communities, such as have given rise to the differences between unvaccinated communities seen in Fig1a.
In conclusion, we have used a simple test to demonstrate the selective impact of vaccination on the carried population of pneumococci. In 2004 differences in population structure are evident, but by 2007 the distribution of serotype frequencies is not significantly different from that observed in three pre-vaccination samples. We suggest that this means we are unlikely to see any great further increase in the common carriage serotypes, either in carriage or disease. Of course, this hypothesis will be tested through ongoing monitoring of IPD serotypes in Massachusetts. The impact of the next generation of vaccines, which will provide an opportunity for rarer serotypes, remains unknown.
Acknowledgments
WPH and CF acknowledge funding from the Royal Society. The work of the remaining authors and data collection for the samples from 2001 and beyond were supported by a grant (R01 AI066304, J Finkelstein) from the US National Institute of Allergy and Infectious Diseases. The funding agencies had no involvement in the design and conduct of this work or the preparation of the manuscript. We would like to thank Marc Lipsitch, Jukka Corander and Helen Jenkins for helpful discussions.
Footnotes
Potential conflicts of interest: WPH has acted as an advisor to Glaxo Smithkline. SIP receives research support from Wyeth Lederle. The other authors have no conflicts of interest to report
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease--United States, 1998-2003. MMWR Morb Mortal Wkly Rep. 2005;54:893–897. [PubMed] [Google Scholar]
- 2.Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis. 2003;187:1424–1432. doi: 10.1086/374624. [DOI] [PubMed] [Google Scholar]
- 3.Byington CL, Samore MH, Stoddard GJ, Barlow S, Daly J, Korgenski K, Firth S, Glover D, Jensen J, Mason EO, Shutt CK, Pavia AT. Temporal trends of invasive disease due to Streptococcus pneumoniae among children in the intermountain west: emergence of nonvaccine serogroups. Clin Infect Dis. 2005;41:21–29. doi: 10.1086/430604. [DOI] [PubMed] [Google Scholar]
- 4.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144(Pt 11):3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 5.Eskola J, Kilpi T, Palmu A, Jokinen J, Haapakoski J, Herva E, Takala A, Kayhty H, Karma P, Kohberger R, Siber G, Makela PH. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N Engl J Med. 2001;344:403–409. doi: 10.1056/NEJM200102083440602. [DOI] [PubMed] [Google Scholar]
- 6.Fraser C, Hanage WP, Spratt BG. Neutral microepidemic evolution of bacterial pathogens. Proc Natl Acad Sci U S A. 2005;102:1968–1973. doi: 10.1073/pnas.0406993102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghaffar F, Barton T, Lozano J, Muniz LS, Hicks P, Gan V, Ahmad N, McCracken GH., Jr Effect of the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae in the first 2 years of life. Clin Infect Dis. 2004;39:930–938. doi: 10.1086/423379. [DOI] [PubMed] [Google Scholar]
- 8.Hanage WP. Serotype-specific problems associated with pneumococcal conjugate vaccination. Future Microbiol. 2008;3:23–30. doi: 10.2217/17460913.3.1.23. [DOI] [PubMed] [Google Scholar]
- 9.Hanage WP, Auranen K, Syrjanen R, Herva E, Makela PH, Kilpi T, Spratt BG. Ability of pneumococcal serotypes and clones to cause acute otitis media: implications for the prevention of otitis media by conjugate vaccines. Infect Immun. 2004;72:76–81. doi: 10.1128/IAI.72.1.76-81.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanage WP, Huang SS, Lipsitch M, Bishop CJ, Godoy D, Pelton SI, Goldstein R, Huot H, Finkelstein JA. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J Infect Dis. 2007;195:347–352. doi: 10.1086/510249. [DOI] [PubMed] [Google Scholar]
- 11.Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, Jackson D, Thomas A, Beall B, Lynfield R, Reingold A, Farley MM, Whitney CG. Increase in Non-vaccine-Type Pneumococcal Disease in the Era of Widespread Pneumococcal Conjugate Vaccination, United States, 1998-2004. Journal of Infectious Diseases. 2007;196:1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 12.Huang SS, Hinrichsen VL, Stevenson AE, Rifas-Shiman SL, Kleinman K, Pelton SI, Lipsitch M, Hanage WP, Lee GM, Finkelstein JA. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics. 2009;124:e1–11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang SS, Platt R, Rifas-Shiman SL, Pelton SI, Goldmann D, Finkelstein JA. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005;116:e408–413. doi: 10.1542/peds.2004-2338. [DOI] [PubMed] [Google Scholar]
- 14.Lexau CA, Lynfield R, Danila R, Pilishvili T, Facklam R, Farley MM, Harrison LH, Schaffner W, Reingold A, Bennett NM, Hadler J, Cieslak PR, Whitney CG. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. Jama. 2005;294:2043–2051. doi: 10.1001/jama.294.16.2043. [DOI] [PubMed] [Google Scholar]
- 15.Meats E, Brueggemann AB, Enright MC, Sleeman K, Griffiths DT, Crook DW, Spratt BG. Stability of serotypes during nasopharyngeal carriage of Streptococcus pneumoniae. Journal of Clinical Microbiology. 2003;41:386–392. doi: 10.1128/JCM.41.1.386-392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nahm MH, Lin J, Finkelstein JA, Pelton SI. Increase in the prevalence of the newly discovered pneumococcal serotype 6C in the nasopharynx after introduction of pneumococcal conjugate vaccine. J Infect Dis. 2009;199:320–325. doi: 10.1086/596064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park IH, Moore MR, Treanor JJ, Pelton SI, Pilishvili T, Beall B, Shelly MA, Mahon BE, Nahm MH. Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J Infect Dis. 2008;198:1818–1822. doi: 10.1086/593339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelton SI, Huot H, Finkelstein JA, Bishop CJ, Hsu KK, Kellenberg J, Huang SS, Goldstein R, Hanage WP. Emergence of 19A as virulent and multidrug resistant Pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2007;26:468–472. doi: 10.1097/INF.0b013e31803df9ca. [DOI] [PubMed] [Google Scholar]
- 19.Pelton SI, Loughlin AM, Marchant CD. Seven valent pneumococcal conjugate vaccine immunization in two Boston communities: changes in serotypes and antimicrobial susceptibility among Streptococcus pneumoniae isolates. Pediatr Infect Dis J. 2004;23:1015–1022. doi: 10.1097/01.inf.0000143645.58215.f0. [DOI] [PubMed] [Google Scholar]
- 20.Simpson EH. Measurement of diversity. Nature. 1949;163 [Google Scholar]
- 21.Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. Invasive pneumococcal disease caused by nonvaccine serotypes among alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. Jama. 2007;297:1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 22.Spratt BG, Greenwood BM. Prevention of pneumococcal disease by vaccination: does serotype replacement matter? Lancet. 2000;356:1210–1211. doi: 10.1016/S0140-6736(00)02779-3. [DOI] [PubMed] [Google Scholar]
- 23.van Selm S, van Cann LM, Kolkman MA, van der Zeijst BA, van Putten JP. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect Immun. 2003;71:6192–6198. doi: 10.1128/IAI.71.11.6192-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


