Abstract
Chromosomal instability during cell division frequently causes cell death or malignant transformation. Orderly chromosome congression at the metaphase plate, a paramount process to vertebrate mitosis and meiosis, is controlled by a number of molecular regulators, including kinesins. Kinesin-8 (Kif18A) functions to control mitotic chromosome alignment at the mid-zone by negative regulation of kinetochore oscillation. Here the authors report that disrupting Kif18a function results in complete sterility in male but not in female mice. Histological examination reveals that Kif18a−/− testes exhibit severe developmental impairment of seminiferous tubules. Testis atrophy in Kif18a−/− mice is caused by perturbation of microtubule dynamics and spindle pole integrity, leading to chromosome congression defects during mitosis and meiosis. Depletion of KIF18A via RNAi causes mitotic arrest accompanied by unaligned chromosomes and increased microtubule nucleating centers in both GC-1 and HeLa cells. Prolonged depletion of KIF18A causes apoptosis due to perturbed microtubule dynamics. Further studies reveal that KIF18A silencing results in degradation of CENP-E and BubR1, which is accompanied by premature sister chromatid separation. KIF18A physically interacts with BubR1 and CENP-E, and this interaction is modulated during mitosis. Combined, the studies indicate that KIF18A is essential for normal chromosome congression during cell division and that the absence of KIF18A function causes severe defects in microtubule dynamics, spindle integrity, and checkpoint activation, leading to germinal cell aplasia in mice.
Keywords: KIF18A, testis development, chromosome congression, knockout mice, BubR1
Introduction
It has been estimated that male infertility accounts for up to 7% of infertility in human couples.1 Given a limited number of treatment options for most male infertility cases, efforts have been directed to searching for its genetic basis. Extensive research in the past has identified genes whose functions are essential for spermatogenesis,2 the process by which male spermatogonia develop into mature spermatozoa. Spermatogenesis is a highly organized cyclic process that includes the mitotic proliferation of spermatogonia, the meiotic division that gives rise to the maturation of spermatids, which in turn develop into motile spermatozoa. Chromosomal abnormalities, including Klinefelter syndrome (XXY), specific translocations, and Y chromosome microdeletions, are well-described cases of male infertility.3,4 Point mutations in genes coding for androgen receptor (AR), follicle-stimulating hormone (FSH), and cystic fibrosis transmembrane conductance regulator (CFTR) are responsible for spermatogenesis failure.5-8 These represent a few well-established gene defects associated with male infertility. Recently, mouse genetic studies have revealed that gene products affecting chromatin structures play an essential role in regulating male fertility. For example, ablation of mouse JHDM2A, a JmjC-domain-containing histone demethylase 2A, causes defects in spermatogenesis by affecting packaging and condensation of sperm chromatin.9 In addition, deficiency in Sirt1, a member of the sirtuin family of deacetylases, markedly compromises spermatogenesis but not oogenesis.10
KIF18A encodes a molecular motor protein of the kinesin-8 family.11-13 It is essential for chromosome congression during mitosis and meiosis because it regulates proper assembly and positioning of the spindles.11-13 During mitosis and meiosis, the spindle is a microtubule-based structure that controls the proper partitioning of chromosomes or sister chromatids. Microtubules facilitate chromosomal congression to the metaphase plate before their accurate segregation at the onset of anaphase. Before all chromosomes are aligned at the equator, chromosomes oscillate along the mid-zone, which appears to be a necessary step for proper alignment of paired chromosomes or sister chromatids and for attachment by the spindles. Microtubules that coordinate chromosome congression and segregation are dynamic by elongating or shortening at the plus end. During mitosis, KIF18A is concentrated at the plus ends of microtubules, facilitating microtubule depolymerization as a loss of its function results in the formation of elongated microtubules.13 KIF18A reduces the amplitude of preanaphase oscillations and negatively controls the movement of chromosomes toward the spindle poles during anaphase.12 Mitotic regulators, including CENP-E and SGO1, also affect chromosome alignment and segregation because disruption of their functions causes the appearance of unaligned chromosomes and induces chromosome missegregation.14-17
Mammalian spermatogenesis is a classic adult stem cell–dependent process. It is supported by constant self-renewal (division) and differentiation of the spermatogonial stem cell compartment. Given that Drosophila Klp67A, a kinesin-8 family member, is essential for mitotic division and male meiosis,18 we studied Kif18a function through the mouse genetic approach. We observed that disruption of Kif18a function causes infertility only in male mice, which was accompanied by severe testis atrophy. Extensive in vivo and in vitro studies revealed that Kif18a−/− depletion caused impaired microtubule dynamics and spindle pole integrity, leading to compromised chromosome congression during mitosis and meiosis.
Results
Disruption of Kif18a Causes Testis Atrophy in Mice
Through transgenic knockout mouse approaches, we obtained two independent lines of mice deficient in Kif18a (one approach shown in Supplementary Fig. S1). Genotype analyses of live births revealed that heterozygotes and homozygotes with a mutation at the Kif18a locus were capable of surviving to term. Newborn Kif18a−/− mice had no gross abnormalities in major organs examined except for undersized testes in male mice. Eight weeks after birth, Kif18a−/− testes were markedly underdeveloped compared with that of the wild-type and Kif18a+/− mice (Fig. 1A). Testes in wild-type and Kif18a+/− mice grew at the faster rate than the whole body did during the first 8 weeks, resulting in a net increase in the testis/body weight ratio; however, Kif18a−/− testes grew at a significantly slower rate, leading to a net decrease in the testis/body weight ratio (Fig. 1B). Kif18a−/− mutant male mice were unable to give rise to offspring, although these mice were capable of producing plugs (Supplementary Fig. S2A). The reproductive defect caused by the Kif18a mutation was rather specific to male mice because Kif18a−/− female mice were fertile and gave live pups at a rate slightly lower than that of wild-type mice (Supplementary Fig. S2A).
Figure 1.
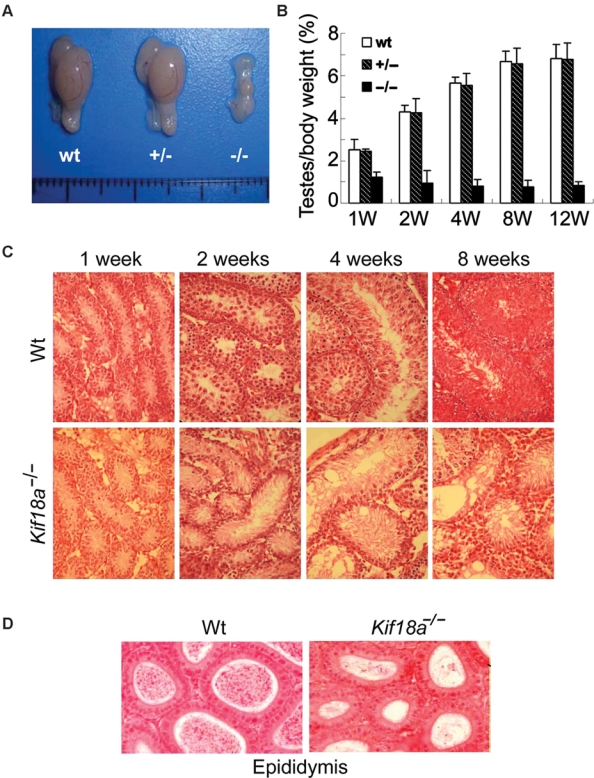
Disruption of Kif18a causes testis atrophy and germ cell aplasia. (A) Morphologies of representative testes from 8-week-old mice of various genotypes. (B) Testes of individual mice, as well as their bodies, of various genotypes were weighed during various stages of development (weeks 1, 2, 4, 8, and 12). The testes/body weight ratios were calculated for wild-type mice (wt), heterozygotes (+/−), and homozygotes (−/−). (C) Sections of wt and Kif18a−/− testes at different developmental stages were stained with H&E. Representative images are shown. (D) Sections of wt and Kif18a−/− epididymis at week 4 were stained with H&E, and representative images are shown.
Histological analyses confirmed that Kif18a−/− testes exhibited severe developmental defects. At week 1, Kif18a−/− testes had relatively normal seminiferous tubules that were single cell layered (Fig. 1C). Mitotic spermatogonial cells were observed in both wild-type and Kif18a−/− testes. Starting from week 2, Kif18a−/− testes started to show developmental retardation. Whereas seminiferous tubules of wild-type testes contained multilayered spermatocytes, Kif18a−/− testes remained largely single cell layered. At week 4, the growth arrest of Kif18a−/− seminiferous tubules became more evident. A majority of inner seminiferous tubules were single cell layered with vacuolated, degenerative changes in the luminal side. Focal Leydig cell hyperplasia was visible. At week 8, degenerative seminiferous tubules became atrophic coupled with Leydig cell hyperplasia in the stroma. In contrast, Kif18a−/− epididymis was rather normal compared with that of the wild-type one (Fig. 1D). Likewise, Kif18a−/− ovaries exhibited no apparent histological defects (Fig. S2B), consistent with the fact that female Kif18a−/− mice were fertile (Supplementary Fig. S2A).
Mitotic and Meiotic Defects in Kif18a−/− Mouse Testes
During the first week of testis development, spermatogonial cells in seminiferous tubules were marked by a high rate of proliferation. Severe atrophy of Kif18a−/− testes implicated their growth impairment. As Klp67A, a Drosophila ortholog of mammalian Kif18A, plays an essential role in mitosis,18 we reasoned that KIF18a deficiency would compromise expansion of the stem cell compartment of seminiferous tubules in mice. Supporting this notion, the number of seminiferous tubules in Kif18a−/− testis was indeed significantly smaller than that of the wild-type testis (Fig. 2 A and B). Moreover, the average number of cells per tubule section was also greatly reduced in Kif18a mutant testis (Fig. 2 A and C). To further confirm the role of Kif18a in supporting stem/progenitor cell proliferation in the testes, c-Kit-positive cells isolated from individual testes of Kif18a−/− and wild-type mice at week 1 were subjected to cell cycle analysis by flow cytometry. The total number of c-Kit-positive cells was significantly decreased in Kif18a−/− testes compared with that of wild-type ones (Fig. 2D). Whereas a significant number of wild-type c-Kit-positive cells were active in the cell cycle, only a small fraction of Kif18a−/− cells were in S and G2/M stages (Fig. 2D). Further histological analysis revealed that active mitotic cells were easily visible in wild-type seminiferous tubules at week 1 and that most of them were in the metaphase stage (Fig. 2 E and F). However, significantly fewer mitotic cells were present in age-matched Kif18a−/− seminiferous tubules; those cells in mitosis exhibited abnormal chromosome congression manifested as the presence of unaligned chromosomes (Fig. 2 E [arrows] and F). Taken together, these results indicate mitotic defects caused by the absence of Kif18a function in the testis compartment.
Figure 2.
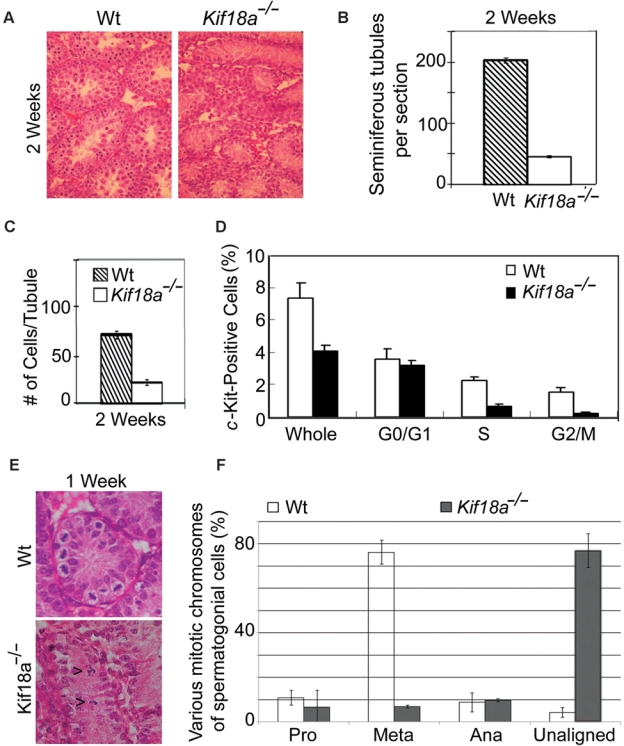
Mitotic defects in Kif18a−/− mouse testes. (A) Representative sections of wt and Kif18a−/− at week 2 were stained with H&E. (B) Seminiferous tubules were counted from 6 random fields of each wt or Kif18a−/− testis. The data were summarized from 3 independent mice. (C) Cells in each seminiferous wt or Kif18a−/− tubule were counted from 6 random fields of each testis. The data were summarized from 3 independent mice. (D) The c-Kit-positive cells isolated from wt or Kif18a−/− testes were subjected to the DNA content analysis by flow cytometry. Representative results are shown. (E) Representative wt and Kif18a−/− seminiferous tubules at week 1 are presented. Arrowheads denote mitotic cells with unaligned chromosomes. (F) Mitotic cells in week 1 wt or Kif18a−/− seminiferous tubules were examined under microscope. Cells of various mitotic stages or with unaligned chromosomes were quantified.
At week 4, wild-type testes contained spermatocytes of various developmental stages, forming multilayered seminiferous tubules (Fig. 3 A and B). However, a majority of age-matched Kif18a−/− seminiferous tubules remained single cell layered and contained few, if any, spermatocytes. A majority of Kif18a−/− seminiferous tubules contained no actively dividing cells (Fig. 3A). At week 6, a few seminiferous tubules in the periphery of Kif18a−/− testes contained cells morphologically similar to spermatids but with multiple nuclei (Fig. 3C, arrows), suggesting a dysregulated meiotic process in Kif18a−/− testes that failed to yield haploid spermatozoa. This was also consistent with the observation that Kif18a−/− epididymis only contained nonmotile spermatids, many of which were bi-nucleated (Fig. 3D, arrows). Flow cytometry analysis revealed that more than 50% of testicular cells isolated from week 8 testes of Kif18a−/− mice remained diploid, whereas a majority of these cells isolated from wild-type testes were haploid (Fig. 3E). Together, these results strongly suggest a crucial role of Kif18a in mouse spermatogenesis by regulating meiosis.
Figure 3.
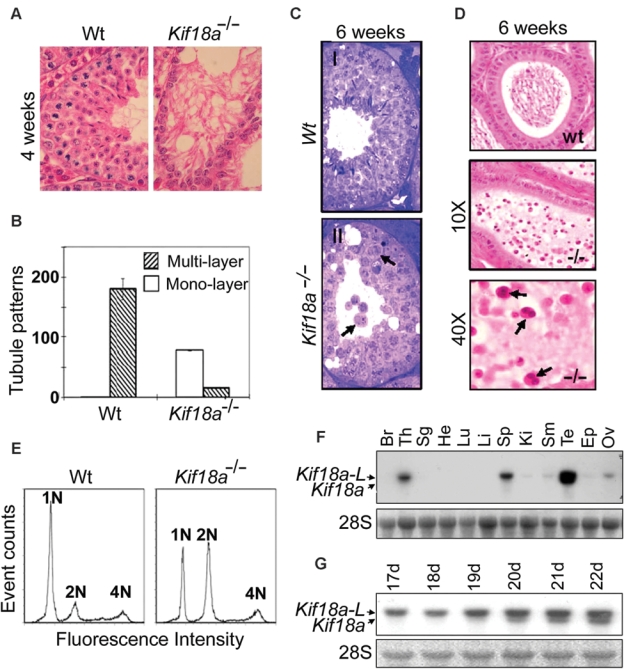
Meiotic defects in Kif18a−/− mouse testes and tissue-specific expression of Kif18a. (A) Wt and Kif18a−/− seminiferous tubules at week 4 were stained with H&E. Representative tubules are shown. (B) Seminiferous tubules with monolayer or multilayers of cells were counted from wt or Kif18a−/− testes. The data were summarized from 6 individual testes. (C) H&E-stained wt (I) and Kif18a−/− (II) seminiferous tubules at week 6. Arrows denote the spermatids with multiple or bi-nuclei. (D) H&E-stained wt and Kif18a−/− epididymis. Arrows denote the nonmotile bi-nucleated spermatozoa. (E) Testicular cells isolated from wt and Kif18a−/− testes at week 6 were stained with propidium iodide and subjected to DNA content analysis by flow cytometry. (F) Equal amounts of total RNA isolated from various mouse organs were subjected to Northern blotting analysis using Kif18a cDNA as a probe. Br, Th, Sg, He, Lu, Li, Sp, Ki, Sm, Te, Ep, and Ov stand for brain, thymus, salivary glands, heart, lung, liver, spleen, kidney, small intestine, testes, epididymis, and ovaries, respectively. (G) Equal amounts of total RNA isolated from wild-type testes of various developmental stages (days 17-22) were subjected to Northern blotting using Kif18a cDNA as a probe. Kif18a-L stands for the large form of Kif18a transcript.
To understand the differential effect of Kif18a on the development of testes and ovaries in mice, we examined its expression in various mouse organs. As expected, adult testes contained the highest level of Kif18a, which was detected as 2 discrete mRNA species. The longer form, designated as Kif18a-L, was about 3.8 kb. The shorter one, designated as Kif18a, was about 3.5 kb (Fig. 3 F and G). Kif18a mRNA was undetectable until about 3 weeks after birth (Fig. 3G), and this was correlated with meiotic division that gave rise to spermatocytes. Kif18a mRNA expression was testis specific and temporally controlled (Fig. 3 F and G; Supplementary Fig. S3A), whereas Kif18a-L mRNA was present at various levels in testes and several other organs, including thymus, spleen, and ovaries (Fig. 3F and Supplementary Fig. S3A). Further studies revealed that a high level of Kif18a was detected in wild-type but not Kif18a−/− testes (Supplementary Fig. S3 B and C).
KIF18A Depletion Causes Spindle Pole and Chromosome Congression Defects
To elucidate the cellular and molecular basis by which KIF18A controls cell division, we first examined the consequence of Kif18a depletion in the mouse spermatogonial cell line GC-1. Whereas Kif18a localized to kinetochore and mid-zone areas during mitosis, its depletion via RNAi caused a chromosome congression defect characterized by the presence of unaligned (lagging) chromosomes (Fig. 4 A and B). Kif18a depletion also caused the formation of extra γ-tubulin foci that were capable of nucleating microtubules (Fig. 4 C and D). Moreover, compared with those of the wild-type, bipolar meiotic spindles and the metaphase plate were largely absent from Kif18a−/− testes (Supplementary Fig. S4A). These results suggest that deregulated microtubule dynamics may be partly responsible for chromosome congression defects.
Figure 4.
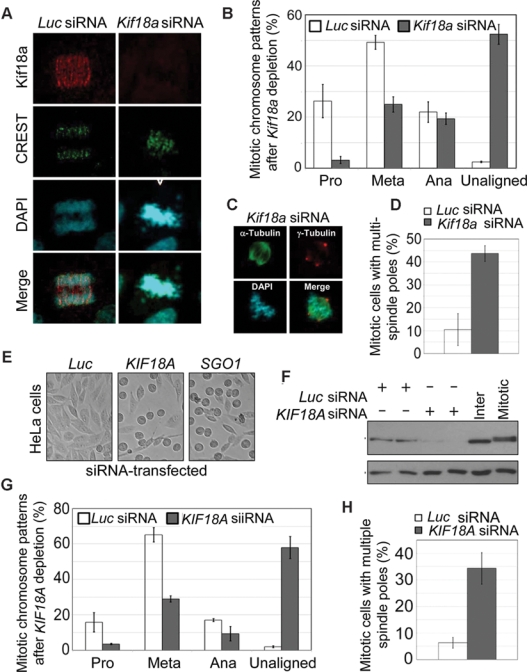
KIF18A depletion causes spindle pole and chromosome congression defects in vitro. (A) Mouse spermatogonial GC-1 cells transfected with Kif18a or luciferase (Luc) siRNA for 24 h were fixed and stained with antibodies to KIF18A (green) and CREST (red). DNA was stained with 4′6′-diamidino-2-phenylindole (DAPI; blue). Representative results are shown. (B) Cells at various mitotic stages or with unaligned/misaligned chromosomes were summarized from GC-1 cells transfected with Kif18a or luciferase siRNA. The data were summarized from 3 independent experiments. (C) GC-1 cells transfected with Kif18a or luciferase siRNA for 24 h were fixed and stained with antibodies to α-tubulin (green) and γ-tubulin (red). DNA was stained with DAPI (blue). Representative images are shown. (D) GC-1 cells transfected with Kif18a or luciferase siRNA for 24 h were fixed and stained with antibodies to α-tubulin (green) and γ-tubulin (red). Mitotic cells with multiple spindle poles were summarized from 3 independent experiments. (E) HeLa cells transfected with KIF18A or luciferase siRNA for 24 h were examined under a light microscope. Representative images are shown. (F) Equal amounts of proteins from HeLa cells transfected with KIF18A or luciferase siRNA for 24 h were blotted for KIF18A and β-actin. (G) Mitotic cells of various stages or with unaligned/misaligned chromosomes were counted from HeLa cells transfected with KIF18A or luciferase siRNA for 24 h. The data were summarized from 3 independent experiments. (H) HeLa cells transfected with KIF18A or luciferase siRNA for 24 h were fixed and stained with antibodies to α-tubulin (green) and γ-tubulin (red). Mitotic cells with multiple spindle poles were summarized from 3 independent experiments.
To determine whether human KIF18A functioned in the same way as the mouse counterpart, we studied HeLa cells depleted of KIF18A via RNAi. KIF18A depletion caused significant mitotic arrest that was evidenced by the formation of rounded-up cells (Fig. 4E). As expected, SGO1 depletion also induced mitotic arrest. RNAi-mediated depletion of KIF18A in HeLa cells was confirmed by immunoblotting (Fig. 4F). Again, the absence of KIF18A function resulted in the formation of multiple spindle poles in mitotic cells as well as defective chromosome congression characterized by the presence of unaligned chromosomes (Fig. 4 G and H). Consistent with these observations, multiple spindle poles were also detected in Kif18a−/− testes after staining with the antibody to NuMA (Supplementary Fig. S4B, arrow), a spindle pole–specific marker.
Kif18A Depletion-Induced Mitotic Arrest Leads to Apoptosis
Time lapse confocal microscopy confirmed that KIF18A depletion induced a prolonged mitotic arrest, during which distorted spindle microtubules, multiple microtubule nucleation centers, and spindle pole rotation were observed (Fig. 5A). The prolonged mitotic arrest frequently leads to mitotic catastrophe, a specialized case of apoptosis. HeLa cells transfected with KIF18A siRNA for more than 24 h exhibited the morphology of typical apoptosis (Fig. 5B). Consistent with a role of KIF18A in microtubule dynamics by limiting elongation of spindle microtubules,12 KIF18A-depleted cells struggled during mitosis, resulting in elongated, bundled spindle microtubules during mitotic catastrophe (Fig. 5C, arrow). Apoptosis induced by KIF18A depletion was confirmed by the appearance of p89 (Fig. 5D), a PARP-1 cleavage product due to caspase-3 activation.19 The enhanced apoptosis was also recapitulated in mouse Kif18a−/− testes. Terminal deoxynucleotidyl transferase–mediated dUTP-biotin nick end labeling (TUNEL) staining revealed that a significant number of cells in Kif18a−/− seminiferous tubules underwent apoptosis, but this did not occur in the wild-type (Fig. 5E). Together, these results indicate that KIF18A is essential for both mitosis and meiosis and that the absence of its function leads to apoptosis.
Figure 5.
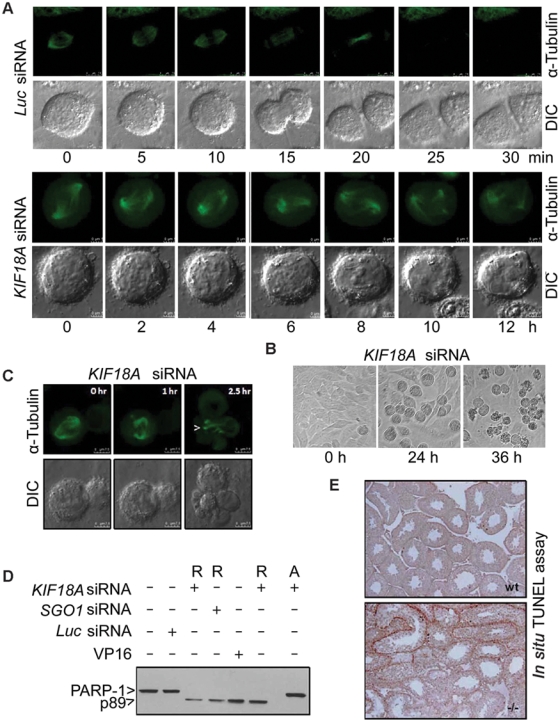
KIF18A depletion induces prolonged mitotic arrest, leading to mitotic catastrophe. (A) HeLa cells constitutively expressing green fluorescent protein (GFP)–tubulin were transfected with KIF18A or luciferase siRNA for 24 h. These cells were then subjected to time lapse confocal microscopy analysis. Representative differential interference contrast (DIC) and α-tubulin images of metaphase cells undergoing mitosis are shown. (B) HeLa cells transfected with KIF18A or luciferase siRNA for 0, 24, or 36 h were observed under a light microscope. Representative images are shown. (C) HeLa cells constitutively expressing GFP-tubulin were transfected with KIF18A siRNA for 36 h. Mitotic cells were then subjected to time lapse confocal microcopy. DIC and α-tubulin images of a representative cell undergoing mitosis are shown. Arrows indicate the distorted, bundled microtubules. (D) HeLa cells transfected with KIF18A, SGO1, or luciferase siRNA for 24 h were collected and lysed. HeLa cells treated with VP16 overnight were also used for lysate preparation. Equal amounts of proteins were blotted for PARP-1 or its degradation product p89. R and A denote rounded-up cells and adherent cells, respectively. (E) Sections of wt and Kif18a−/− seminiferous tubules were subjected to terminal deoxynucleotidyl transferase–mediated dUTP-biotin nick end labeling (TUNEL) analysis. Representative images are shown.
KIF18A Interacts with BubR1 and CENP-E and Regulates Their Functions
Kif18A accumulates as a gradient on kinetochore microtubules during mitosis.12,13 To study whether spindle checkpoint components or kinetochore-associated proteins were regulated by KIF18A, we examined the subcellular localization of BubR1 and CENP-E in KIF18A-depleted cells. BubR1, a major spindle checkpoint component, was activated and concentrated at the kinetochores before the anaphase onset (Fig. 6A). KIF18A knockdown abolished BubR1 signals on chromosomes aligned at the metaphase plate; however, these signals were high on unaligned kinetochores clustered at spindle pole regions (Fig. 6A). CENP-E, a motor protein also having a role in spindle checkpoint control,20 was highly enriched at the kinetochores during mitosis. KIF18A depletion caused the disappearance of CENP-E from the kinetochores (Fig. 6B). Subcellular localization of other mitotic regulators and motor proteins, including Aurora B, survivin, and Eg5, was apparently affected as well, although their levels were not significantly changed as the result of KIF18A silencing (Supplementary Fig. S5).
Figure 6.
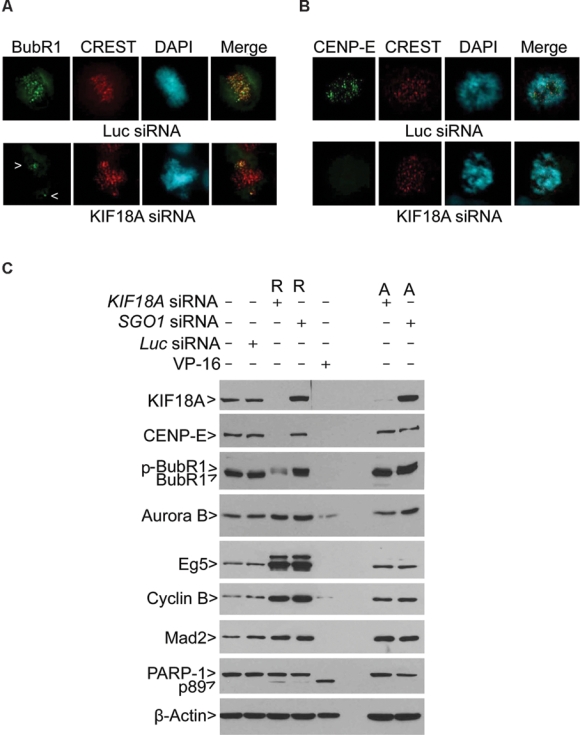
KIF18A regulates the stability of BubR1 and CENP-E during mitosis. (A) HeLa cells transfected with KIF18A or luciferase siRNA for 24 h were fixed and stained with antibodies to α-BubR1 (green) and CREST (red). DNA was stained with 4′6′-diamidino-2-phenylindole (DAPI; blue). Representative images are shown. Arrows indicate BubR1 signals on unaligned chromosomes. (B) HeLa cells transfected with KIF18A or luciferase siRNA for 24 h were fixed and stained with antibodies to CENP-E (green) and CREST (red). DNA was stained with DAPI (blue). Representative images are shown. (C) HeLa cells transfected with KIF18A, SGO1, or luciferase siRNA for 24 h were collected and lysed. HeLa cells treated with VP16 overnight were also used for lysate preparation. Equal amounts of proteins from various treatments were blotted for KIF18A, CENP-E, BubR1, Aurora B, Eg5, cyclin B, Mad2, PARP-1, and β-actin. R and A denote rounded-up cells and adherent cells, respectively. Arrow p-BubR1 denotes the phosphorylated BubR1. R and A denote rounded-up and adherent cells, respectively.
To further confirm a role for KIF18A in regulating BubR1 and CENP-E, both rounded-up and adherent cells transfected with KIF18A siRNA or SGO1 siRNA were collected for lysate preparation. Western blot analyses showed that depletion of KIF18A, but not of SGO1, caused degradation of CENP-E in both rounded-up and adherent cells (Fig. 6C). Silencing KIF18A, but not SGO1, also led to a greatly reduced level of BubR1 in rounded-up but not adherent cells. In contrast, Mad2, another important player in the spindle checkpoint,21 was not affected by KIF18A depletion. Likewise, neither Aurora B nor Eg5 levels were negatively affected by the loss of KIF18A function. In fact, Aurora B and Eg5, as well as cyclin B, were upregulated in rounded-up cells, consistent with the fact that they were mitotic. Moreover, KIF18A depletion for 24 h had already caused the appearance of the PARP-1 cleavage product (p89).
As BubR1 and CENP-E are important components of the spindle checkpoint,22,23 we then examined the sister chromatid cohesion in cells depleted of KIF18A. Different from normal mitotic cells and those induced by Taxol treatment, more than 50% of mitotic cells induced by KIF18A depletion contained sister chromatids that were prematurely separated (Fig. 7 A and B), strongly suggesting a significantly weakened spindle checkpoint due to the absence of KIF18A function. To further understand the role of KIF18A-mediated degradation of BubR1 and CENP-E, experiments were carried out in which both interphase and mitotic cell lysates were immunoprecipitated with the KIF18A antibody or with a control IgG, followed by immnoblotting for BubR1, as well as for KIF18A. BubR1, but not Eg5, was co-immunoprecipitated with KIF18A; more BubR1 was brought down from the mitotic lysates than the interphase ones by the KIF18A antibody (Fig. 7C). KIF18A also interacted with CENP-E; in contrast to BubR1, less CENP-E was brought down by the KIF18A antibody during mitosis (Fig. 7D). Thus, our studies demonstrate that KIF18A physically interacts with both BubR1 and CENP-E and that their interactions are regulated during cell cycle progression.
Figure 7.
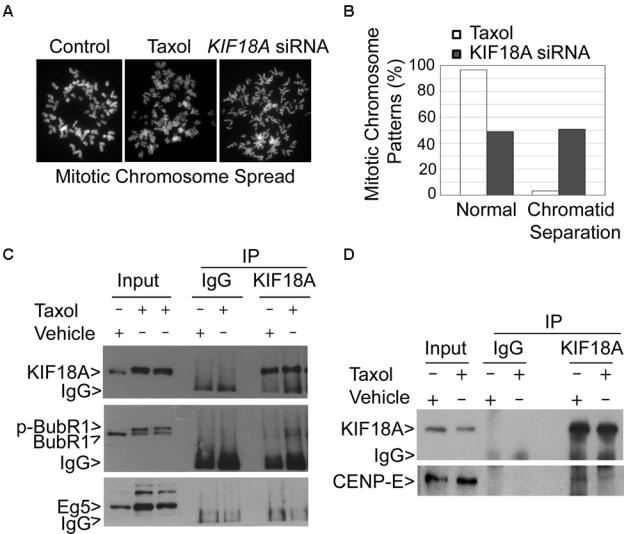
KIF18A physically interacts with BubR1 and CENP-E. (A) Control HeLa cells or the cells treated with paclitaxel or transfected with KIF18A siRNA for 24 h were subjected to chromosome spread analysis. Representative mitotic chromosomes from each treatment are shown. (B) HeLa cells treated with paclitaxel or transfected with KIF18A siRNA for 24 h were subjected to chromosome spread analysis. The percentages of normal chromosomes and chromosomes with premature sister chromatid separation were summarized from 3 independent experiments. (C) Equal amounts of interphase cell or mitotic cell (Taxol-treated) lysates were subjected to immunoprecipitation using the KIF18A antibody or a control IgG. Immunoprecipitates, along with the lysate inputs, were blotted for KIF18A, BubR1, and Eg5. Arrow p-BubR1 denotes the phosphorylated BubR1. (D) Equal amounts of interphase cell or mitotic cell (Taxol-treated) lysates were subjected to immunoprecipitation using the KIF18A antibody or a control IgG. Immunoprecipitates and the lysate inputs were blotted for KIF18A and CENP-E. Each experiment was repeated for at least 3 times. Representative data are shown.
Discussion
In this report, we demonstrate that a targeted disruption of Kif18a causes complete infertility in male mice, which is correlated with a severe developmental defect in the seminiferous tubules. Female mice are fertile, although they may exhibit a feature of hypofertility (Supplementary Fig. S2A). Given the histology of Kif18a−/− seminiferous tubules as well as overall atrophy of the testes, Kif18a mutant male mice can be described as a case of germinal cell aplasia. Infertility in men is a major problem, affecting a significant percentage of couples of reproductive age. To date, treatment of male factor infertility has been largely unsuccessful because of unknown genetic factors. The human KIF18A gene localizes to chromosome 11, where structural abnormalities are linked to defective spermatogenesis and male infertility in clinics.24,25 Given that relatively little is known about defects in molecular motor function in male reproductive failure in mammals, this study is of high significance in terms of helping us understand the molecular defects that are associated with male infertility and provides appropriate strategies for future clinical intervention.
Kif18a directly participates in mitotic expansion of spermatogonial cells and their differentiation (meiosis) during testis development and maturation. Kif18a is not only highly expressed in testes but also localizes to male germ cells, including spermatogonia, spermatocytes, and spermatids. The function of Kif18a in mice is highly specific. Kif18a mutation primarily affects the fertility of male but not female mice. Furthermore, Kif18a-null male mice grow relatively normally and exhibit defects in testes but not in other organs. The temporal and tissue-specific manifestations of Kif18a function as revealed by the mouse genetic study can be explained by its tightly regulated expression. KIF17β, a testis-specific kinesin,26 localizes to chromatoid bodies that are thought to act as male germ cell–specific apparatuses for storing and processing transcripts.27 Therefore, it would be also interesting to examine potential functional interactions between KIF18A and KIF17β.
KIF18A plays a major role in regulating chromatid congression during mitosis in vitro. This is likely due to perturbed microtubule dynamics. KIF18A is enriched at the plus end of kinetochore microtubules. It is conceivable that KIF18A depletion would significantly affect the integrity of the kinetochores. We have demonstrated that KIF18A depletion also disrupts the integrity of spindle poles during mitosis. This is not surprising because recent studies show that kinetochore or spindle checkpoint components also have centrosomal functions.28,29 The defective kinetochores and spindle poles due to a lack of KIF18A function would have a severe consequence on chromosome congression and segregation. Supporting this, more than 50% of cells that are depleted of KIF18A contain unaligned/misaligned chromosomes. Failed chromosome congression causes a lengthy mitotic delay, eventually leading to mitotic catastrophe.
KIF18A physically and functionally interacts with spindle checkpoint components BubR1 and CENP-E, thus regulating the spindle checkpoint integrity. At the cellular level, the BubR1 signal is absent from or greatly reduced at the kinetochores of chromosomes congregated at the mid-zone. More strikingly, CENP-E is undetectable in KIF18A-depleted mitotic kinetochores. At the molecular level, BubR1 and CENP-E, but not Mad2, Eg5, and Aurora B, are significantly downregulated after KIF18A depletion, indicating that KIF18A is essential for stabilization of CENP-E and BubR1 at the kinetochores. Supporting this notion, several early studies have shown that deregulated CENP-E function leads to chromosome congression errors,17,30 phenotypically resembling KIF18A depletion. One scenario to explain the regulation of CENP-E and BubR1 by KIF18A is that these checkpoint components are cargos of the molecular motor or that a major cargo molecule of KIF18A plays an important role in controlling BubR1 and CENP-E stability. During mitosis, the kinetochore components, including BubR1, are highly dynamic.31 It is conceivable that KIF18A maintains a constant supply of BubR1 and CENP-E at the kinetochores, which are essential for spindle checkpoint activation until the anaphase entry. Our observation that BubR1 and CENP-E physically interact with KIF18A is consistent with this possibility. Homozygous deletion of BubR1 results in embryonic lethality.23 Given that Kif18a expression appears to be confined to testes during development (Fig. 3 F and G; Supplementary Fig. S3A), it is likely that KIF18A protein may interact with BubR1 and CENP-E and regulate their stability via the unique domain encoded by Kif18a mRNA.
Although the molecular mechanism by which KIF18A depletion causes instability of BubR1 and CENP-E remains unclear, the motor protein may keep BubR1 and CENP-E confined to the kinetochores during early mitosis, thus preventing the access of negative regulator(s). Alternatively, KIF18A may ship an unknown factor that positively regulates or stabilizes these checkpoint proteins at the kinetochores. For example, among KIF18A, cargos can be a factor that regulates CENP-E and BubR1 sumoylation because both proteins are shown to be sumoylated.32 In fact, CENP-E sumoylation is critical for the maintenance of its activity.32 It is conceivable that sumoylation stabilizes CENP-E and BubR1 by competing for lysine residues that are otherwise the targets of polyubiquitination followed by proteosomal degradation. It is a rather attractive possibility that KIF18A may interact with sumoylation modification enzyme(s) that stabilizes BubR1 and CENP-E given existing lines of supporting experimental evidence. (1) Mouse genetic studies indicate that sumoylation is essential for chromosome congression and mitotic progression.33 (2) SUMO-1, SUMO-2, and SUMO-3 are all localized at the kinetochores during mitosis.32 (3) MG132 treatment partially rescues CENP-E in cells depleted of KIF18A (data not shown). Therefore, one line of future studies should focus on the identification of factors that mediate stability BubR1 and CENP-E in a KIF18A-dependent manner during cell division.
Materials and Methods
Cell culture
The GC-1 and HeLa cell lines were obtained from the American Type Culture Collection. Cells were cultured under 5% CO2 in dishes or on Lab-Tek II chamber slides (Fisher Scientific) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 µg of penicillin and 50 µg of streptomycin sulfate per mL).
Histology and immunohistochemistry analysis
Wild-type and Kif18a mutant testes were fixed in Bouin’s or formalin fixative solutions and embedded in paraffin. Testis sections (8-µm thickness) were de-waxed and stained with hematoxylin and eosin. For immunohistochemistry analysis, testes were fixed in paraformaldehyde. Antigens were unmasked by boiling slides in a 0.01-M citrate solution. Immunohistochemistry was performed using a commercially available kit according to the instructions provided by the supplier (Santa Cruz).
Flow cytometric analysis
Testis single cells were obtained according to previous literature. Briefly, after removing tunica, the testes were placed in DMEM buffered with HEPES (20 mM, Sigma) and containing collagenase type I (1 mg/mL, Sigma). After incubation at 37°C for 15 min, the testes were washed with calcium and magnesium-free phosphate-buffered saline (PBS) and subjected to additional incubation for 10 min in the same buffer containing 0.25% trypsin with 1 mM EDTA. The trypsin digestion was terminated by the addition of FBS. Testis single cells were counted and stained with FITC-c-kit. After washing, testis cells were suspended in 300 µL of PBS containing 0.1% NaN3 and 50% FBS and then fixed overnight by addition of 70% ethanol drop-wise. After washing with calcium and magnesium-free PBS, cells were stained with propidium iodide for flow cytometry.
Reverse transcriptase-mediated polymerase chain reaction (RT-PCR)
Total RNA was extracted from wild-type and Kif18a−/− testes, spleen, and thymus of 8-week-old mice using Trizol reagents (Invitrogen). RNA was also extracted from 1-week-old wild-type and Kif18a−/− testes. Reverse transcription was carried out using AMV reverse transcriptase (Takara). Isoform-specific primers were designed for detecting expression of Kif18a (5′-GAACGGCAGCCAATGAGATG-3′ and 5′-AGATGGCCTTCTTTCCCAGACT-3′) and Kif18a-L (5′-TTCCCAGGTATTCATGTAACAG-3′ and 5′-TTTGCAATGTAAAGACTGGTAG-3′) isoforms, respectively.
Western blot analysis
HeLa cells were treated with VP-16 (inducing apoptosis) or transfected with KIF18A, Sgo1, or control (luciferase) siRNAs for 24 or 36 h. Rounded-up (mitotic) cells were collected by shake-off. The adherent fraction of cells was collected by trypsinization. Equal amounts (50 µg) of protein lysates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting with antibodies to KIF18A (Bethyl), BubR1, CENP-E (Sigma), Aurora B, Mad2 (Abcam), Eg5 (Abcam), cyclin B1 (Santa Cruz), PARP-1 (Cell Signaling), Plk1 (Invitrogen), or β-actin (Sigma, 1:1500). Specific signals were detected with horseradish peroxidase–conjugated goat secondary antibodies (Cell Signaling) and enhanced chemiluminescence reagents (Pierce Biotechnology).
Fluorescence microscopy
Testes dissected from wild-type and homozygous Kif18a−/− mice at various ages were fixed in 10% neutral-buffered formalin. After dehydration in increasing concentrations of ethanol, the tissues were embedded in paraffin and sectioned (5-µm thickness). Tissue sections were stained with H&E using a standard protocol (Fisher Scientific). For fluorescence detection, tissue sections were deparaffinized in xylene and rehydrated in descending concentrations of ethanol followed by antigen retrieval in a sodium citrate buffer (10 mM sodium citrate; 0.05% Tween-20, pH 6.0). Sections were then blocked in 5% bovine serum albumin(BSA) for 1 h and incubated with antibodies to KIF18A and α-tubulin at 4°C overnight. After washing with PBS containing 0.1% Tween-20, slides were incubated for 1 h at room temperature with Alexa488 or Alexa594 conjugated species-specific secondary antibodies (Invitrogen). After additional washes, tissue sections were mounted with fluorescence mounting medium (Dako). Fluorescence microscopy was performed on a Nikon microscope.
GC-1 and HeLa cells cultured on chamber slides with various treatments were quickly washed with PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. Fixed cells were treated with 0.1% Triton X-100 in PBS for 10 min and then washed 3 times with ice-cold PBS. After blocking with 2% bovine serum albumin (BSA) in PBS for 15 min on ice, cells were incubated for 1 h at room temperature with antibodies against CENP-E, BubR1, CREST, KIF18A, Aurora B, Eg5, survivin, NuMA, α-tubulin, or γ-tubulin in a 2% BSA solution. Cells were then washed with PBS and incubated with Rhodamine red X–conjugated antirabbit (or antihuman) IgGs and/or fluorescein isothiocyanate-conjugated antimouse IgGs (Jackson ImmunoResearch) at room temperature for 1 h in the dark. Cells were finally stained with 4′6′-diamidino-2-phenylindole (DAPI; Fluka, 1 µg/mL) for 5 min. Fluorescence microscopy was performed and images were captured using a digital camera (Optronics) using Optronics MagFire and Image-Pro Plus software.
RNA interference
Small interfering RNAs (siRNAs) of human KIF18A were synthesized from Dharmacon, which corresponded to the following sequences: 5′ACCAACAACAGTGCCATAAA3′ (designated as hKIF18A siRNA-1) and 5′ACAGATTCGTGATCTCTTA3′ (hKIF18A siRNA-2). These sequences are capable of silencing human KIF18A.13 Mouse Kif18a siRNAs were obtained from Dharmacon, targeting the following sequences: 5′GCAAGAGTATCTGAAGTTA3′ (mKif18a siRNA-1), 5′CAAATGAGTTCTACATCAA3′ (mKif18a siRNA-2), 5′CGGGATAATTCAAGCGTTA3′ (mKif18a siRNA-3), and 5′TAAAGGGTCACGATTTGTA3′ (mKif18a siRNA-4). hKIF18A or mKif18a siRNA duplexes were transfected into HeLa or GC-1 cells with Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, cells seeded at 60% confluency in an antibiotic-free culture medium were transfected with siRNA duplexes at a final concentration of 100 nM for 24 h (unless otherwise specified). The negative controls were cells transfected with 100 nM siRNA duplex targeting firefly (Photinus pyralis) luciferase (5′UUCCTACGCTGAGTACTTCGA3′, GL-3 from Dharmacon).
Live-cell time lapse imaging
The HeLa cell line expressing GFP-tubulin was kindly provided by Dr. Xiaoqi Liu at Purdue University. HeLa GFP-tubulin cells grown in a 35-mm glass-bottom dish (MatTek) for 24 h at 60% confluence were transfected with 100 nM luciferase siRNA or human KIF18A siRNA. Cell culture medium was changed to CO2-independent medium (Invitrogen) supplemented with 10% FBS and 10 mM glutamine during live-cell time lapse imaging. From 18 to 36 h after transfection, confocal GFP fluorescence and DIC time lapse sequences were collected on a Leica TCS SP5 microscope equipped with a heated incubation chamber, a motorized Z-positioning device, and 60xNA1.4 or 40xNA1.25 DIC optics. One DIC image and a stack of 5 fluorescence images (0.5-µm steps) were simultaneously acquired at 30-sec intervals.
Statistical analysis
Data were expressed as mean ± SD. The statistical differences were analyzed using Student t test.
Supplementary Material
Acknowledgments
We thank Dr. Rulong Shen for his assistance in evaluating the histological data, Dr. Xiaoqi Liu for providing GFP-tubulin HeLa cells, and Dr. Qiang-su Guo for his excellent technical assistance in the use of the confocal microscope.
Footnotes
The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
This study was supported in part by US Public Service Awards to WD (CA090658 and CA113349). This work was also partially supported by the grants from National Natural Science Foundation of China (30871420, 30530390), Ministry of Science and Technology of China (2006BAI23B02), and the Science and Technology Commission of Shanghai Municipality (06DZ05907, 07DZ22929) to ZGW, E-Institutes of Shanghai Municipal Education Commission (E03003).
Supplementary material for this article is available on the Genes & Cancer Web site at http://ganc.sagepub.com/supplemental.
References
- 1. Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol 2002;4(suppl):s41-9 [DOI] [PubMed] [Google Scholar]
- 2. Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med 2008;14:1197-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Kretser DM. Male infertility. Lancet 1997;349:787-90 [DOI] [PubMed] [Google Scholar]
- 4. Meschede D, Horst J. The molecular genetics of male infertility. Mol Hum Reprod 1997;3:419-30 [DOI] [PubMed] [Google Scholar]
- 5. Kulczycki LL, Kostuch M, Bellanti JA. A clinical perspective of cystic fibrosis and new genetic findings: relationship of CFTR mutations to genotype-phenotype manifestations. Am J Med Genet 2003;116A:262-7 [DOI] [PubMed] [Google Scholar]
- 6. Martens JW, Verhoef-Post M, Abelin N, Ezabella M, Toledo SP, Brunner HG, et al. A homozygous mutation in the luteinizing hormone receptor causes partial Leydig cell hypoplasia: correlation between receptor activity and phenotype. Mol Endocrinol 1998;12:775-84 [DOI] [PubMed] [Google Scholar]
- 7. Saunders PT, Padayachi T, Tincello DG, Shalet SM, Wu FC. Point mutations detected in the androgen receptor gene of three men with partial androgen insensitivity syndrome. Clin Endocrinol (Oxf) 1992;37:214-20 [DOI] [PubMed] [Google Scholar]
- 8. Tapanainen JS, Aittomaki K, Min J, Vaskivuo T, Huhtaniemi IT. Men homozygous for an inactivating mutation of the follicle-stimulating hormone (FSH) receptor gene present variable suppression of spermatogenesis and fertility. Nat Genet 1997;15:205-6 [DOI] [PubMed] [Google Scholar]
- 9. Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007;450:119-23 [DOI] [PubMed] [Google Scholar]
- 10. Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS ONE 2008;3:e1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luboshits G, Benayahu D. MS-KIF18A, new kinesin; structure and cellular expression. Gene 2005;351:19-28 [DOI] [PubMed] [Google Scholar]
- 12. Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell 2008;14:252-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stumpff J, Wordeman L. Chromosome congression: the kinesin-8-step path to alignment. Curr Biol 2007;17:R326-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol 2008;181(3):411-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 2004;118:567-78 [DOI] [PubMed] [Google Scholar]
- 16. Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA 2004;101:18012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanudji M, Shoemaker J, L’Italien L, Russell L, Chin G, Schebye XM. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol Biol Cell 2004;15:3771-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell 2004;15:121-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dukers DF, Meijer CJ, ten Berge RL, Vos W, Ossenkoppele GJ, Oudejans JJ. High numbers of active caspase 3-positive Reed-Sternberg cells in pretreatment biopsy specimens of patients with Hodgkin disease predict favorable clinical outcome. Blood 2002;100:36-42 [DOI] [PubMed] [Google Scholar]
- 20. Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol 2000;2:484-91 [DOI] [PubMed] [Google Scholar]
- 21. Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science 1996;274:246-8 [DOI] [PubMed] [Google Scholar]
- 22. Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol 2008;10:825-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res 2004;64:440-5 [DOI] [PubMed] [Google Scholar]
- 24. Gianotten J, van der Veen F, Alders M, Leschot NJ, Tanck MW, Land JA, et al. Chromosomal region 11p15 is associated with male factor subfertility. Mol Hum Reprod 2003;9:587-92 [DOI] [PubMed] [Google Scholar]
- 25. Zhang S, Qiu W, Wu H, Zhang G, Huang M, Xiao C, et al. The shorter zinc finger protein ZNF230 gene message is transcribed in fertile male testes and may be related to human spermatogenesis. Biochem J 2001;359:721-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kimmins S, Kotaja N, Fienga G, Kolthur US, Brancorsini S, Hogeveen K, et al. A specific programme of gene transcription in male germ cells. Reprod Biomed Online 2004;8:496-500 [DOI] [PubMed] [Google Scholar]
- 27. Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci 2006;119:2819-25 [DOI] [PubMed] [Google Scholar]
- 28. Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 2006;442:947-51 [DOI] [PubMed] [Google Scholar]
- 29. Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell 2008;14:331-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McEwen BF, Chan GK, Zubrowski B, Savoian MS, Sauer MT, Yen TJ. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell 2001;12:2776-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 2004;14:953-64 [DOI] [PubMed] [Google Scholar]
- 32. Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell 2008;29:729-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 2005;9:769-79 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


