What is cryptic genetic variation?
Cryptic genetic variation refers to unexpressed, bottled-up genetic potential. It is not normally seen, but is expressed under abnormal conditions such as in a new environment or a different genetic background. In a sense, the measurable component of normal variation is just the tip of an iceberg of genetic possibilities that are hidden below the visible surface.
Examples?
Antennapedia is a mutation in Drosophila melanogaster flies that transforms the antennae into legs. When this mutation is placed in a dozen different wild-type genetic backgrounds, each strain will show a different phenotype, ranging from almost perfect antennae to almost perfect legs where the antennae should be. This is cryptic genetic variation modifying the mutant phenotype, even though it is unobservable in normal flies. For a human example, consider type 2 diabetes, the prevalence of which is increasing dramatically worldwide along with the obesity epidemic. This is a highly heritable disease, but largely due to exposure of cryptic genetic variation by modern culture (Figure 1).
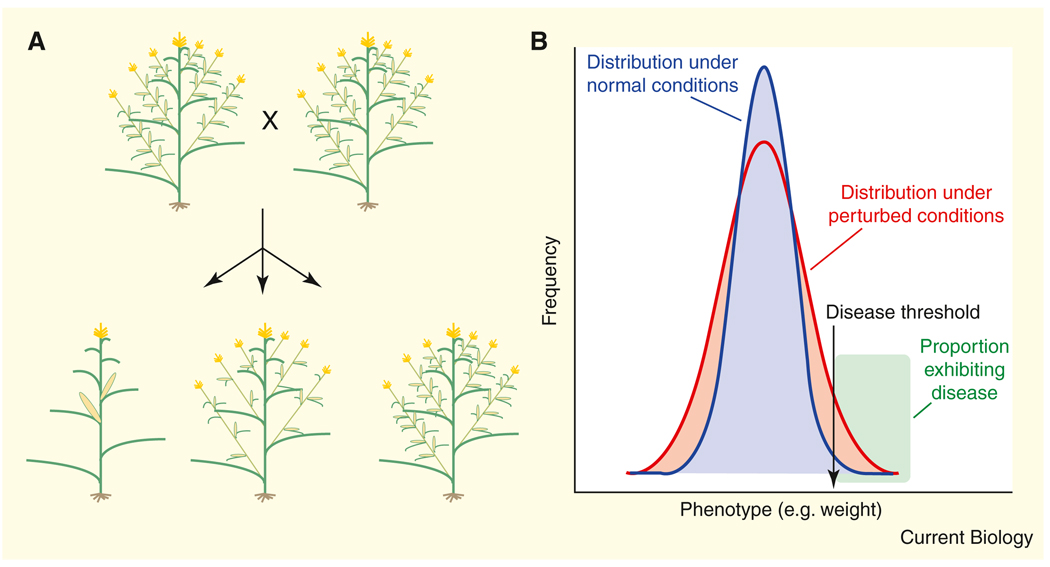
Figure 1. Two examples of cryptic genetic variation.
(A) Two lines of teosinte that are phenotypically very similar were crossed, and their progeny were then testcrossed to an inbred line of maize. Each cross gave rise to a range of phenotypes ranging from maize-like (bottom left) to teosinte-like (bottom right), indicating the expression of cryptic genetic variation hidden in the parents. Redrawn with permission from Lauter and Doebley (2002). (B) If a disease, such as diabetes, is only seen in a small fraction of individuals, perturbation can change the distribution of susceptibility such that a much larger fraction of people are affected (green proportion above a liability threshold).
Why should I care about cryptic genetic variation?
You shouldn’t — unless you have an interest in the genetic basis of complex diseases, improving crop plants or understanding how organisms will adapt to climate change. Plant breeders can improve crops by selecting on cryptic genetic variation that modifies transgenes or introduced germ plasm. Cryptic variation may also have a major influence on the capacity of organisms to adapt to ecological perturbation. Cancer, depression, asthma and many other diseases are all affected by interactions between our genetic legacy and the contemporary environment, and to some extent their increasing incidence is due to genetic variants that may not have influenced disease for most of human evolution.
If it’s cryptic, how can it be detected?
Introgression is the most dramatic way to detect cryptic genetic variation. A mutation with a visible phenotype, such as Antennapedia, is introduced into different wild-type genetic backgrounds by many generations of backcrossing. This approach routinely results in the production of a series of lines that differ markedly in the expression of the phenotype. Alternatively, cryptic genetic variation can be inferred from the profile of changes in mean phenotypes of a set of lines grown under normal and extreme conditions, such as a high-sugar diet or unusual temperature.
Is the architecture of cryptic genetic variation the same as that of visible genetic variation?
We don’t know enough yet about the extent to which cryptic genetic variation consists predominantly of a few loci of large effect or of many loci of small effect, to make such a comparison. Examples of both architectures have been described: More than half of the dramatic difference between the enhanced and suppressed phenotypes of two strains of the homeotic mutant Ultrabithorax of Drosophila is attributable to a single cryptic polymorphism, and major effect modifiers have also been shown to influence various abnormalities caused by Hsp90 mutations in flies. Similarly, some of the genetic loci that affect the extent of transformation of teosinte toward maize-like structures are also of large effect (Figure 1). On the other hand, in Drosophila small effect mutations in Egfr can subtly modify the cryptic eye-roughening phenotype of a gain-of-function allele of the same gene.
How is cryptic genetic variation related to canalization?
Canalization refers to the evolution of phenotypic robustness that occurs under conditions of long-term stabilizing selection. It leads to suppression of the effects of genetic variation under normal, unperturbed circumstances. Because of canalization, cryptic genetic variation can build up in a population, only to be released as visible phenotypic variation when the environment changes or a new mutation appears, or in hybrid zones where novel genotypes are introduced. Canalization itself can either be modelled as discrete loci that suppress variation (so-called ‘capacitors of evolutionary change’), or as the network of interactions among genes that constrain the phenotype at an optimum, hiding the underlying genetic variation from natural selection. Cryptic genetic variation can also be uncovered without de-canalization: Any change that shifts the phenotype mean can potentially change the architecture of the trait.
Why haven’t I heard more about cryptic genetic variation if it so prevalent and important?
This may be a matter of the politics and psychology of science. It is hard enough to study regular quantitative traits, and the study of cryptic genetic variation requires novel experimental prowess. Also, most of evolutionary and quantitative genetic theory assumes additive gene effects, and cryptic genetic variation directly challenges this paradigm.
Then why study cryptic genetic variation now?
Because it is one of the keys to unlocking the secrets of human disease, animal and plant breeding and biological evolution. New genomic strategies are revolutionizing genetic mapping, making it possible to identify major effect loci influencing cryptic genetic variation. In the face of unprecedented environmental change, how can we ignore such a relatively unexplored scientific niche, with so much to learn?
Further reading
- Flatt T. The evolutionary genetics of canalization. Q Rev. Biol. 2005;80:287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- Gibson G, Wemple M, van Helden S. Potential variance affecting homeotic Ultrabithorax and Antennapedia phenotypes in Drosophila melanogaster. Genetics. 1999;151:1081–1091. doi: 10.1093/genetics/151.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat. Rev. Genet. 2004;5:681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Wagner GP. The population genetic theory of hidden variation and genetic robustness. Genetics. 2004;168:2271–2784. doi: 10.1534/genetics.104.029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Doebley J. Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics. 2002;160:333–342. doi: 10.1093/genetics/160.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC, Kaiser M, Kawecki TJ. The differential genetic and environmental canalization of fitness components in Drosophila melanogaster. J. Evol. Biol. 1995;8:539–557. [Google Scholar]