Abstract
Combined targeted molecular dynamics (TMD) and potential of mean force (PMF) simulations have been carried out to uncover the detailed pathway and determine the corresponding free energy profile for the structural transformation from the nonprereactive butyrylcholinesterase (BChE)-(−)-cocaine binding to the prereactive BChE-(−)-cocaine binding associated with the (−)-cocaine rotation in the binding pocket of BChE. It has been shown that the structural transformation involves two transition states (TS1rot and TS2rot). TS1rot is mainly associated with the deformation of the nonprereactive complex, whereas TS2rot is mainly associated with the formation of the prereactive complex. It has also been demonstrated that the A328W/Y332G mutation significantly reduces the steric hindrance for (−)-cocaine rotation in the binding pocket of BChE and, thus, decreases the free energy barrier for the structural transformation from the nonprereactive binding to the prereactive binding. The calculated relative free energy barriers are all consistent with available experimental kinetic data. The new mechanistic insights obtained and the novel computational protocol tested in this study should be valuable for future computational design of high-activity mutants of BChE. The general computational strategy and approach based on the combined TMD and PMF simulations may be also valuable in computational studies of detailed pathways and free energy profiles for other similar mechanistic problems involving ligand rotation or another type of structural transformation in the binding pocket of a protein.
Introduction
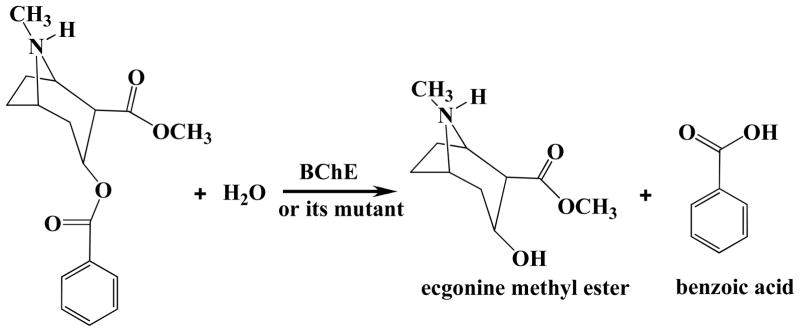
Cocaine is well-known as the most-psychostimulant drug abused by millions of people worldwide.1, 2, 3, 4 The disastrous medical and social consequences of cocaine addiction, such as increasing crimes, medical expense, and loss of lives, have made the development of an effective pharmacological treatment of cocaine abuse a high priority.5,6 It has been found that the rewarding and reinforcing effects of cocaine are predominantly mediated by its inhibition of dopamine transporter (DAT), i.e. blocking the reuptake of neurotransmitter dopamine in the central nervous system (CNS).7,8,9 Traditional pharmacodynamic approach has failed to yield a therapeutically useful antagonist of DAT due to the difficulties inherent in blocking a blocker.2,3,4,6 An alternative to receptor-based approaches is to interfere with the delivery of cocaine to its receptors or accelerate its clearance from the peripheral circulation.10,11,12,13,14,15,16, 17 An ideal molecule for this purpose should be a potent enzyme catalyzing the hydrolysis of cocaine into biologically inactive metabolites. The dominant pathway for cocaine metabolism in primates is butyrylcholinesterase (BChE)-catalyzed hydrolysis at the benzoyl ester group (Chart 1) and the metabolites for this pathway are all biologically inactive.18,19,20 Clearly, BChE-catalyzed hydrolysis of cocaine at the benzoyl ester is the metabolic pathway most suitable for amplification. However, wild-type BChE has a low catalytic efficiency against naturally occurring (−)-cocaine; only (−)-cocaine is biologically active.20 Thus, (−)-cocaine has a plasma half-life of ~47 min or longer even for a (−)-cocaine dose as low as 0.2 mg/kg (i.v.).10 The plasma half-life of (−)-cocaine is expected to be dependent on the dose of (−)-cocaine after the enzyme is saturated. It is highly desirable to develop a mutant of human BChE with a significantly improved catalytic activity against (−)-cocaine.
Chart 1.
BChE-catalyzed Hydrolysis of (−)-cocaine
Generally speaking, rational design of a high-activity enzyme mutant is extremely challenging, particularly when the enzymatic reaction process consists of multiple steps.21,22,23 For rational design of a mutant enzyme with an improved catalytic activity for a given substrate, one needs to design possible amino acid mutations that can accelerate the rate-determining step of the catalytic reaction process19, 24 while the other steps are not slowed down by the mutations. The general reaction pathway for BChE-catalyzed hydrolysis of (−)-cocaine was uncovered by extensive MD simulations21,22,23, 24, 25 and reaction coordinate calculations19,23 using quantum mechanics (QM) and hybrid quantum mechanics/molecular mechanics (QM/MM). These computational studies revealed that the BChE-(−)-cocaine binding involves two different types of complexes, i.e. nonprereactive BChE-(−)-cocaine complex and prereactive BChE-(−)-cocaine complex. The nonprereactive complex is the initial enzyme-substrate (ES) binding complex which is not suitable for chemical reaction. In the nonprereactive complex, (−)-cocaine molecule stays vertically inside the substrate-binding gorge between residues D70 and W82 of the enzyme, while the cationic head of (−)-cocaine is close to W82 and other surrounding residues like E197 and Y440. After the initial, nonprereactive BChE-(−)-cocaine binding, (−)-cocaine needs to rotate in the binding pocket to become the prereactive BChE-(−)cocaine complex ready for the chemical reaction, although the detailed pathway for the (−)-cocaine rotation has not been determined. In the prereactive complex, (−)-cocaine molecule lies horizontally at the bottom of the substrate-binding gorge, with the benzoyl ester group of (−)-cocaine favorably located near the catalytic site consisting of the catalytic triad residues S198-H438-E325 of the enzyme. Starting from the prereactive BChE-(−)-cocaine complex, the chemical reaction process consists of four steps.19,23,24,26,
Our recently reported studies21,22,23,24,25,26, 27, 28, 29, 30 have demonstrated that computational design of a high-activity mutant of BChE is both promising and challenging. It is promising because the catalytic activity of BChE against (−)-cocaine can be improved through structure-and-mechanism-based computational design of site-directed mutagenesis followed by wet experimental tests. It is very challenging because the rate-determining step of the enzymatic reaction process could be changed to another step following amino acid mutations. Particularly, it has been demonstrated19,23,26,27,29,30 that the structural transformation from the nonprereactive BChE-(−)-cocaine complex to the prereactive BChE-(−)-cocaine complex is the rate-determining step of (−)-cocaine hydrolysis catalyzed by wild-type BChE (KM = 4.5 μM, kcat = 4.1 min−1, and kcat/Km = 9.1 × 105 M−1 min−1).30, 31 However, for (−)-cocaine hydrolysis catalyzed by the A328W/Y332G mutant (KM = 17 μM, kcat = 240 min−1, kcat/Km = 1.4 × 107 M−1 min−1),27 the rate-determining step becomes the first step of the chemical reaction process. So, for (−)-cocaine hydrolysis catalyzed by wild-type BChE and its different mutants, the rate-determining step could be completely different. In order to reliably design a BChE mutant with an improved catalytic activity against (−)-cocaine, one needs to determine the transition states and the corresponding free energy barriers for all steps of the enzymatic reaction, including those in the structural transformation from the nonprereactive complex to the prereactive complex and the subsequent four chemical reaction steps.
The background discussed above indicates that the entire catalytic reaction process for BChE-catalyzed hydrolysis of (−)-cocaine should involve at least five transition states: at least one for the structural transformation from the nonprereactive complex to the prereactive complex and four for the subsequent chemical reaction steps. One needs to determine the free energy barriers for all of these five (or more) reaction steps for computational design of high-activity mutants of BChE against (−)-cocaine. QM/MM reaction coordinate calculations have been carried out to uncover the fundamental reaction pathway and determine the corresponding free energy barriers for the chemical reaction steps.19,23,26,29 However, it has been a puzzle how to determine the pathway and free energy profile for the structural transformation from the nonprereactive complex to the prereactive complex, as the reaction coordinate during such a structural transformation does not involve any covalent bond formation or breaking. Thus, the pathway and the free energy profile for the structural transformation from the nonprereactive complex to the prereactive complex still remain to be determined.
In the present study, we have tested an efficient computational approach to uncover the detailed pathway and determine the corresponding free energy profile for the structural transformation from the nonprereactive complex to the prereactive complex. Our computational protocol is based on the combined use of targeted molecular dynamics (TMD) and potential of mean force (PMF) methods; TMD simulation provides more reasonable initial structures for PMF simulations. This computational protocol has been used to uncover the fundamental pathway for the structural transformation from the nonprereactive complex to the prereactive complex and determine the free energy profiles for (−)-cocaine hydrolysis catalyzed by wild-type BChE and the A328W/Y332G mutant. To the best of our knowledge, it is the first time for the combined use TMD and PMF methods to explore the pathway of substrate rotation in the binding site of an enzyme involving two different enzyme-substrate binding structures (i.e. nonprereactive and prereactive complexes). The calculated difference in free energy barrier between wild-type BChE and the A328W/Y332G mutant is consistent with the experimentally kinetic data, suggesting that the computational approach may be valuable in prediction of the relative free energy barriers for the structural transformation from the nonprereactive complex to the prereactive complex associated with various mutants of BChE and in future computational design of high-activity mutants of an enzyme.
Computational Methods
Molecular Dynamics Simulations
The initial structures of both the nonprereactive and prereactive complexes were prepared based on our previous molecular modeling and molecular docking study19,22,25 on wild-type BChE and the A328W/Y332G mutant that were derived from the X-ray crystal structure32 deposited in the Protein Data Bank33 with PDB code 1P0P. In order to further relax all of the constructed binding structures, MD simulations were performed by using the Sander module of Amber 8 program package34 and ff02 force field with the same procedures as used in our previous computational studies.19,21,23,24,25,26,27,29,30 In particular, the partial charges of cocaine atoms were calculated by using the restrained electrostatic potential-fitting (RESP) protocol implemented in the Antechamber module of Amber 8 program,34 following the electrostatic potential (ESP) calculation at ab initio HF/6-31G* level using Gaussian 03 program.35 The geometry used in the ESP calculations was obtained from the previous ab initio reaction coordinate calculations.19 Each of the complex structures was solvated in a rectangular box of TIP3P water molecules36 with a minimum solvent-wall distance of 10 Å. The solvated system for each complex structure was gradually heated to 298.15 K by weak-coupling method37 and equilibrated for 100 ps. Throughout the MD simulations, a 15.0 Å non-bonded interaction cutoff was used and the non-bonded list was updated every 25 steps. The motion for the mass center of the system was removed every 1,000 steps. The particle-mesh Ewald (PME) method38, 39 was applied to treat long-range electrostatic interactions. The lengths of covalent bonds involving hydrogen atoms were fixed with the SHAKE algorithm,40 enabling the use of a 2-fs time step to numerically integrate the equations of motion. Finally, the production MD was kept running for 1 ns or longer with a periodic boundary condition in the NTP ensemble at T = 298.15 K with Berendsen temperature coupling, and at P = 1 atm with isotropic molecule-based scaling.37,41
Targeted MD Simulations
The MD-equilibrat ed structure of the nonprereactive complex of either wild-type BChE or the A328W/Y332G mutant was used as the starting structure to perform TMD simulations by using the Amber 8 program,34 with the MD-simulated structure of the prereactive complex set as the target structure. An additional energy term
| (1) |
was added in the TMD simulations. In Eq. (1), kforce is the additional force constant, Natom is the number of atoms, and RMSD(t) is the mass-weighted root-mean square deviation (RMSD) of the positions of (−)-cocaine atoms in the simulated structure at time t from the corresponding positions in the prescribed target structure. During the TMD simulations, the force constant kforce was set as 0.8 kcal/(mol•Å2) and applied on all atoms of (−)cocaine molecule in order to bias the MD trajectory toward the targeted structure. The RMSD0, i.e. the target RMSD, was set to zero, and the time step for the integration of equations of motion was set to 1 fs. The whole TMD simulation was kept running for 630 ps for the structure of wild-type BChE-(−)-cocaine complex, and 800 ps for the structure of A328W/Y332G BChE-(−)-cocaine complex.
Potential of Mean Force (PMF) Calculations
In order to explore the free energy profile for the structural transformation from the nonprereactive complex to the prereactive complex for wild-type BChE, and to explore the changes of free energy caused by the A328W/Y332G mutations, PMF calculations were carried out by using umbrella-sampling42 MD simulations. The classic PMF definition43 can be represented by a function of reaction coordinate as
| (2) |
In Eq.(2), ρ(χ) is the probability density along the reaction coordinate χ, R is the gas constant, T is the simulation temperature, U(χ) is the biasing potential applied in the umbrella-sampling MD simulations, and F is the normalization constant. According to this approach, the reaction coordinate is usually divided into different regions, i.e., windows, each of which is sampled separately. A biasing (umbrella) potential, i.e. U(χ), is applied for each window in order to obtain nearly uniform sampling of the potential energy surface. In the present study, the reaction coordinate was defined as the distance from the mass center of the benzoyl group of (−)-cocaine to the mass center of the side chain of residue S198 of the enzyme. The total number of windows was 29, each of which was separated by 0.2 Å, covering the reaction coordinate between 9.6 Å ~ 4.0 Å. The biasing force constant applied in different windows of umbrella-sampling ranged from 2.0 to 65.0 kcal/(mol•Å2). For each umbrella-sampling window, the initial complex structure was selected from the trajectory of TMD simulations based on the reaction coordinate to be sampled for the interested window. The selected structure for each window was first equilibrated for 30 ps and then kept running for 500 ps for production sampling. The frequency for data collection was set to 1 fs, which was the same as that of the time step of umbrella-sampling MD. As indicated in our previous study,19,23, 25,26 the binding mode for the cationic head of (−)-cocaine in the nonprereactive complex was very similar as that in the prereactive complex. For convenience, the distance between the nitrogen atom of the cationic head of (−)-cocaine and the aromatic center of the side chain of residue W82 was constrained by a force constant of 20.0 kcal/(mol•Å2) throughout all the umbrella-sampling windows, as this distance was not expected to change during the structural transformation process.
After all the umbrella-sampling MD simulations were finished, the data collected from separate simulation windows were combined along the reaction coordinate. These data were then used to calculate the PMF for the whole structural transformation process with the weighed histogram analysis method (WHAM)44,45 using the code developed by Alan Grossfield (http://membrane.urmc.rochester.edu/Software/WHAM/WHAM.html).
Most of the MD, TMD, and umbrella-sampling MD simulations were performed on a supercomputer (e.g. IBM X-series Cluster with 340 nodes or 1,360 processors) at University of Kentucky Center for Computational Sciences. Some other modeling and computations were carried out on SGI Fuel workstations and a 34-processor IBM x335 Linux cluster in our own lab.
Results and Discussion
Nonprereactive Binding Structures
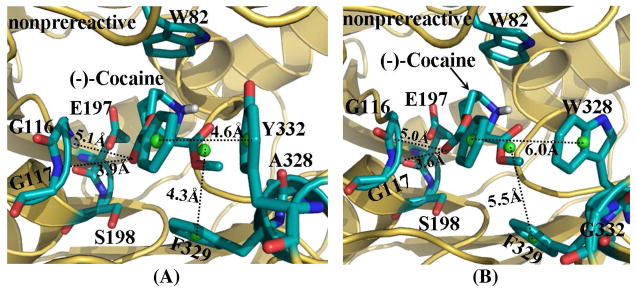
As suggested in our previous studies,19,21,23,25,26 (−)-cocaine can form both the nonprereactive and prereactive complexes with BChE. Once the initial structures were constructed, they were subjected to structural relaxation through MD simulations (see Figures S1 and S2 in Supporting Information). The performed MD simulations were also served to obtain stable complex structures used as the starting structures in subsequent TMD simulations (discussed below). The time-evolved curve (Figure S1 in Supporting Information) of the positional root-mean square deviation (RMSD) for Cα atoms from those in the starting structure indicates that the nonprereactive complex structures for both wild-type BChE and the A328W/Y332G mutant have been well equilibrated and stabilized by the intermolecular interactions between (−)-cocaine and the enzymes. In the nonprereactive complex structures of both wild-type BChE and the A328W/Y332G mutant (Figure 1), the cationic head of (−)-cocaine was located at a sub-binding site around residue W82. It interacts with residue E197 through tight electrostatic attractions; and with aromatic side chain of W82 through a strong cation-π interaction. Other residues, including Y128, G439, Y440, and I442, are also around the bottom of the sub-binding site for the cationic head of (−)-cocaine. The differences between the nonprereactive complex for wild-type BChE-(−)-cocaine binding and the nonprereactive complex for A328W/Y332G BChE-(−)cocaine binding came from the sub-binding site for the benzoyl ester group and the sub-binding site for methyl ester group. For the nonprereactive complex of wild-type BChE-(−)-cocaine binding (Figure 1A), the benzoyl ester group of (−)-cocaine packs tightly with residue Y332 as the distance between the center of benzoyl group and the center of Y332 side chain (denoted by Y332-COCbenzoyl distance here for convenience) fluctuates around ~5.0 Å (Figure S1A in Supporting Information) during the MD simulations, and this Y332-COCbenzoyl distance is ~4.6 Å in the final structure (Figure 1A) derived from MD trajectory. The methyl ester group of (−)-cocaine was situated above residues H438 and Y440 of BChE, and in close contact with residues A328 and F329 (Figure 1A).
Figure 1.
MD-simulated nonprereactive complex structures for (−)-cocaine binding with both wild-type BChE (A) and A328W/Y332A mutant (B). BChE is shown as gold ribbon. Key residues of BChE and (−)-cocaine molecule are shown in stick style and colored by atom type. Dashed lines represent importance distances between different residues of BChE and different groups of (−)-cocaine, including the distance from the center (green ball) of benzoyl group of (−)-cocaine to the center (green ball) of the aromatic side chain of Y332 in wild-type BChE (A) or W328 in the mutant (B); the distance from the center (green ball) of methyl ester group of (−)-cocaine to the center (green ball) of the aromatic side chain of F329; and the distances from the carbonyl oxygen at the benzoyl ester group of (−)-cocaine to the backbone hydrogen atoms of residues G116 and G117, respectively.
However, in the nonprereactive complex of (−)-cocaine with the A328W/Y332G mutant (Figure 1B), both the benzoyl ester group and the methyl ester group of (−)cocaine pack with residues W328 and F329. The distance between the center of benzoyl group of (−)-cocaine to the center of W328 side chain (denoted by W328-COCbenzoyl distance here for convenience) fluctuates around ~6.0 Å in the MD simulations (Figure S1B in Supporting Information), much longer than the Y332-COCbenzoyl distance in the nonprereactive complex with the wild-type (Figure S1A in Supporting Information). Once residue Y332 was replaced by G in the A328W/Y332G mutant, much more space was left around the benzoyl group of (−)-cocaine, making the benzoyl group easier to move around or toward the backbone of G116 and G117. As observed from the MD simulations (Figure S1B in Supporting Information), the fluctuation was very small for the distance from the carbonyl oxygen atom at benzoyl ester group of (−)-cocaine to backbone hydrogen of residue G116.
Prereactive Binding Structures
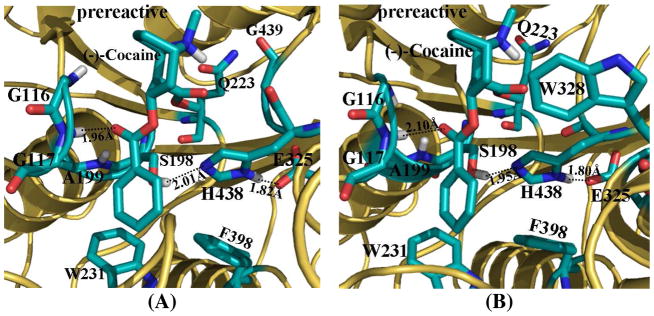
In the MD-simulated prereactive complex structures (Figure 2, and also Figure S2 in Supporting Information) of (−)-cocaine binding with both wild-type BChE (Figure 2A) and the A328W/Y332G mutant (Figure 2B), the orientation of (−)-cocaine at the catalytic site was essentially the same, i.e. lying down at the bottom of the substrate-binding gorge of BChE. The carbonyl oxygen at the benzoyl ester group of (−)-cocaine came very close to the oxyanion hole consisting of the backbone hydrogen atoms of G116, G117, and A199, and formed one hydrogen bond with the backbone of G117 (Figure 2). The methyl ester group of (−)-cocaine rotated down and formed contacts with residues Q223, H438, and G439 of BChE, while the benzoyl group of (−)-cocaine was located to a new position in contact with residues W231 and F398. In the prereactive complex of (−)-cocaine with the A328W/Y332G mutant, residue W328 was also within ~5.0 Å around the cationic head of (−)-cocaine, closely packed with each other. However, residue Y332 in the prereactive complex (−)-cocaine with wild-type BChE became ~8.0 Å away from the cationic head of (−)-cocaine.
Figure 2.
MD-simulated prereactive complex structures for (−)-cocaine binding with both wild-type BChE (A) and the A328W/Y332A mutant (B). BChE is shown as gold ribbon. Key residues of BChE and (−)-cocaine molecule are shown in stick style and colored by atom type. Dashed lines represent the hydrogen bond between the carbonyl oxygen at benzoyl ester group of (−)-cocaine and the backbone of residue G117, and hydrogen bonds within the catalytic triad residues S198-H438-E325.
Transformation of Wild-type BChE-(−)-Cocaine Binding Structures
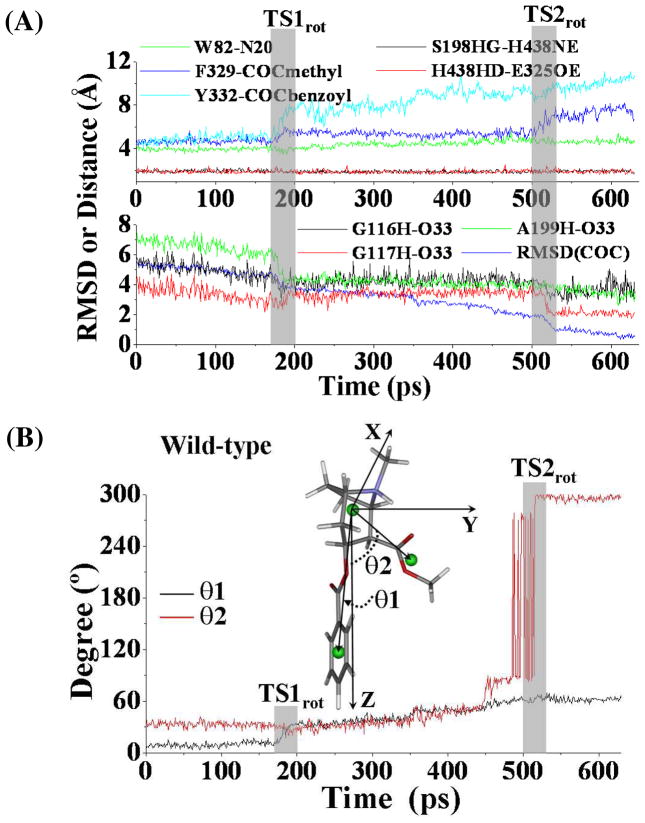
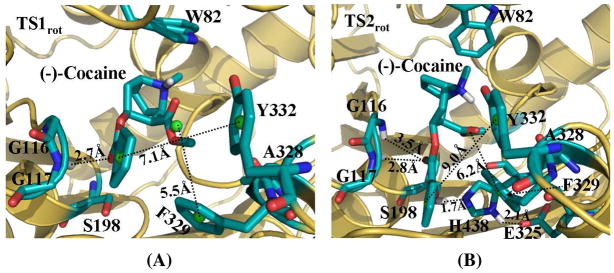
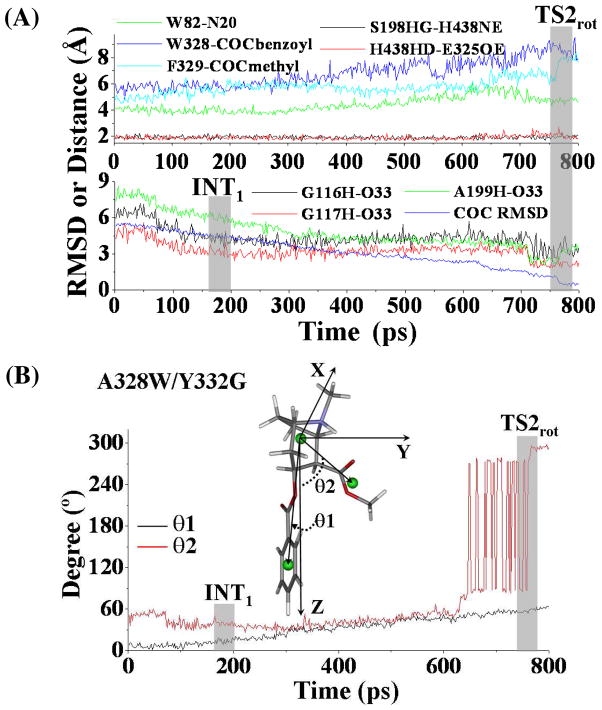
Unlike other studies46,47 using TMD to simulate major molecular motions related to open-close processes of channel proteins, we used TMD to simulate the structural transformation involving (−)-cocaine rotation at the catalytic site of wild-type BChE. Depicted in Figure 3 are the plots for some key distances and the conformational change of (−)-cocaine molecule itself which were tracked during the TMD simulations. As the TMD was started, the MD-equilibrated structure of the nonprereactive complex (Figure 1A) was transformed gradually into the corresponding prereactive complex (Figure 2A). The positional RMSD for (−)-cocaine molecule between the nonprereactive complex and prereactive complex was ~5.7 Å. This RMSD was gradually reduced (lower panel of Figure 3A) via the application of additional energy constraint, resulting in conformational changes of (−)-cocaine molecule itself (Figure 3B). As shown in Figure 3A, the whole transformation process covers two transition states (TS1rot and TS2rot) for the conformational changes. Figure 4 represents the typical structures for TS1rot and TS2rot derived from TMD simulations. The first transition state TS1rot appeared between ~170 ps and ~200 ps of TMD simulation (upper panel of Figure 3A). The TS1rot echoes dramatic enlargement for the Y332G-COCbenzoyl distance (Figure 3A), and the distance from the center of methyl ester group of (−)-cocaine to the center of F329 side chain of BChE (denoted as F329-COCmethyl distance in Figure 3A). Meanwhile, the carbonyl oxygen at the benzoyl ester group of (−)-cocaine came significantly closer to the oxyanion hole, especially approaching to the backbone of residue A199 of BChE (lower panel of Figure 3A). The transition state TS1rot was also associated with the conformational change of benzoyl ester group of (−)-cocaine, as the angle (θ1) between the Z-axis and the vector point from the center of cationic head to the center of benzoyl group of (−)-cocaine increased dramatically (Figure 3B). As the simulation went on, the BChE-(−)-cocaine binding structure came to a time period of plateau with the structural transformation continued slowly and gradually. The second transition state TS2rot showed up between ~500 ps and ~530 ps, featured with second dramatic changes of the above described distances (Figure 3A). The benzoyl ester group of (−)-cocaine left far away from residues F329 and Y332 of BChE, while the carbonyl oxygen at the benzoyl ester group jumped into the oxyanion hole by hydrogen bonding with the backbone of G117 of BChE. Echoing to TS2rot, the angle (θ2) between the Z-axis and the vector pointing from the center of cationic head to the center of the methyl ester group of (−)-cocaine increased from ~90° to ~270° (Figure 3B), indicating that the methyl ester group rotated dramatically.
Figure 3.
(A) Tracked key distances and RMSD for (−)-cocaine molecule of wild-type BChE-(−)-cocaine binding structure along the TMD trajectory. W82-N20 represents the distance from the center of aromatic side chain of W82 to the nitrogen atom at the cationic head of (−)-cocaine; F329-COCmethyl represents the distance from the center of aromatic side chain of F329 to the center of methyl ester group of (−)-cocaine; Y332-COCbenzoyl means the distance from the center of aromatic side chain of Y332 to the center of benzoyl group of (−)-cocaine; G116H-O33 represents the distance between the backbone hydrogen of residue G116 to the carbonyl oxygen at the benzoyl ester of (−)cocaine, and similar meaning for distances as G117H-O33 and A199H-O33, respectively; S198HG-H438NE and H438HD-E325OE represent the distances of hydrogen bonding interactions within the catalytic triad residues S198-H438-E325 of the enzyme. TS1rot and TS2rot represent the transition states (shaded regions) for the transformation process. (B) Conformational change for (−)-cocaine molecule along the TMD trajectory. This is represented by the changes of the two angles (θ1, θ2). θ1 is the angle between the Z-axis and the vector pointing from the center of the cationic head to the center of benzoyl group of (−)-cocaine, and θ2 is the angle between the Z-axis and the vector starting from the cationic head to the center of methyl ester group of (−)-cocaine.
Figure 4.
Typical structures of the transition states derived from TMD simulations. (A) TS1rot complex structure. It was derived from the snapshot at 190 ps of TMD trajectory. (B) TS2rot complex structure. It was derived from the snapshot at 520 ps of the TMD trajectory. BChE is shown as gold ribbon. Key residues of wild-type BChE and (−)cocaine molecule are shown in stick style and colored by atom type. Dashed lines represent importance distances between different residues of BChE and different groups of (−)-cocaine. Those include the distance from the center (green ball) of benzoyl group of (−)-cocaine to the aromatic center (green ball) of the side chain of Y332; the distance from the center (green ball) of methyl ester group of (−)-cocaine to the aromatic center (green ball) of the side chain of F329; and the distance from the carbonyl oxygen at the benzoyl ester group of (−)-cocaine to the backbone hydrogen of G117 (A), and both G116 and G117 (B). Hydrogen bonding within the catalytic triad residues (S198-H328-E325) of BChE is also shown as dashed lines with labeled distances for the TS2rot complex structure in (B).
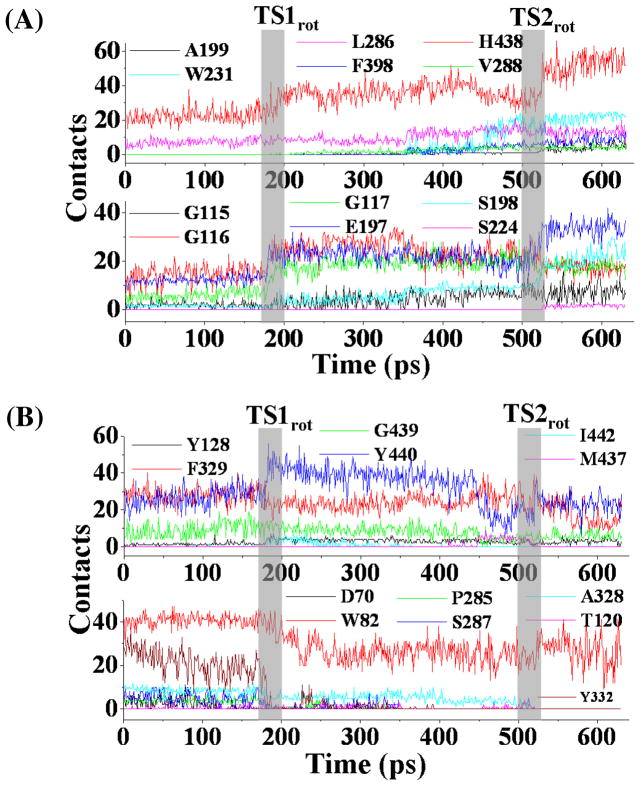
To demonstrate the details of the conformational changes of wild-type BChE during the structural transformation from the nonprereactive complex to the prereactive complex, contacts between BChE and (−)-cocaine molecule were calculated using an atomic distance cutoff of 5 Å and collected at the residue level. For calculating the number of contacts between (−)-cocaine and a residue in a given snapshot, the internuclear distanc es between all non-hydrogen atoms of (−)-cocaine and all non-hydrogen atoms of the residue were evaluated. Any of these distances being shorter than 5 Å was considered as a contact. According to this criterion, a non-hydrogen atom of (−)-cocaine could contact with multiple non-hydrogen atoms of a residue, and a non-hydrogen atom of a residue could also contact with multiple non-hydrogen atoms of (−)-cocaine. Thus, the total number of contacts between (−)-cocaine and a residue could be very large. Residues of BChE having contacts with (−)-cocaine for more than 8% of the TMD trajectory were further analyzed. Based on the calculations, the residue-level contacts (Figure 5, and Table S1 in Supporting Information) can be categorized into three types: type I residues experienced significant increase in the number of contacts with (−)-cocaine during the periods of conformational transitions (Figure 5A); type II residues had stable contacts with (−)-cocaine throughout the TMD trajectory (upper panel of Figure 5B); and type III residues experienced significant decrease in the number of contacts with (−)-cocaine during the conformational transitions (lower panel of Figure 5B).
Figure 5.
Residue-based contacts for wild-type BChE in contacting with (−)-cocaine tracked along the TMD trajectory. (A) Type I residues with significant increase in the number of contacts during the transformation process. (B) Type II and Type III residues. These residues have either continuous contacts (upper panel) or decreased number of contacts (lower panel).
As depicted in Figure 5 and listed in Table S1 (see Supporting Information), residues G116, G117, E197, and H438 in type I group significantly increased their number of contacts with (−)-cocaine during the time periods associated with transition states TS1rot and TS2rot, indicating their vital contributions to the deformation of nonprereactive complex and formation of prereactive complex. Residues G115, S198, A199, S224, V288, W231, and F398 significantly increased the number of contacts, or just came into contact with (−)-cocaine at TS2rot, suggesting these residues only played important role in the formation of prereactive complex (Figure 5A). However, the dramatic decrease in the number of contacts for residues A328, F329, and Y332 (Figure 5B) suggests that their interactions with (−)-cocaine in the nonprereactive complex must be destroyed in order to form prereactive complex. Along the TMD simulations, residue W82 also suffered obvious decrease in the number of contacts with (−)-cocaine at TS1rot period (lower panel of Figure 5B), but still retained a large number of contacts when the prereactive complex was formed near the end of TMD simulations. The change in the contact number indicates that the cationic head of (−)-cocaine adjusted its orientation at its binding site around residue W82. The curve for the number of contacts of residue Y440 (upper panel of Figure 5B) is interesting. This residue (Y440) first experienced an increase in the number of contacts in TS1rot, then a decrease in the number of contacts in TS2rot, and finally retained the number of contacts in the prereactive complex similar to that in the nonprereactive complex, suggesting that this residue acted cooperatively for the change of TS1rot to TS2rot.
Transformation of A328W/Y332G BChE-(−)-Cocaine Binding Structures
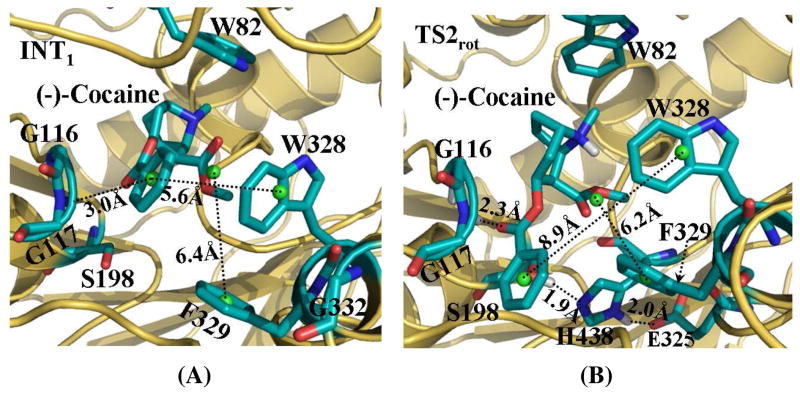
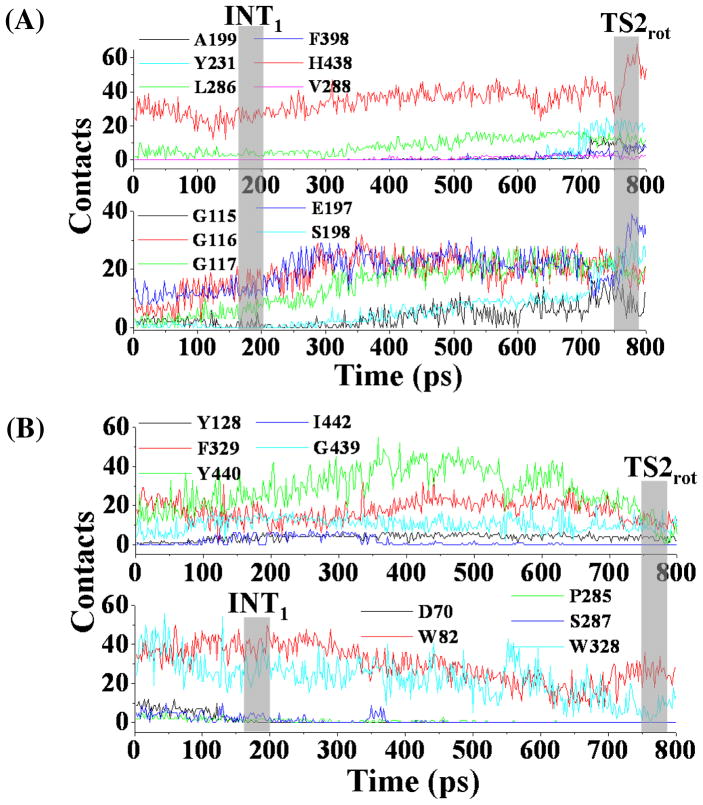
Depicted in Figure 6 are some important changes tracked from TMD simulations, which represents the transformation from the nonprereactive complex to the prereactive complex of the A328W/Y332G BChE-(−)-cocaine binding. Figure 7 represents the typical structures corresponding to the conformational transitions along the transformation pathway. Similar to that in the transformation process of wild-type BChE-(−)-cocaine binding, the positional RMSD for (−)-cocaine molecule at the catalytic site of the A328W/Y332G mutant was gradually reduced due to the application of the additional force. There was also an obvious transition state TS2rot corresponding to the rotation of the methyl ester group of (−)-cocaine (Figures 6B and 7B).
Figure 6.
(A) Tracked key distances and RMSD for (−)-cocaine molecule in the A328W/Y332G BChE-(−)-cocaine binding structure along the TMD trajectory. The definitions for tracked distances (W82-N20, F329-COCmethyl, G116H-O33, G117H-O33, A199H-O33, S198HG-H438NE and H438HD-E325OE) are the same as that in Figure 3A, except that W328-COCbenzoyl represents the distance from the center of aromatic side chain of W328 to the center of benzoyl group of (−)-cocaine. TS2rot represents the transition state (shaded region) for the transformation process. (B) Conformational change for (−)-cocaine molecule along the TMD trajectory. The definitions of θ1 and θ2 are the same as that in Figure 3B.
Figure 7.
Typical structures for A328W/Y332G BChE-(−)-cocaine complex derived from TMD simulations. (A) INT1 structure corresponding to the snapshot at 200 ps of TMD trajectory. (B) TS2rot structure from the snapshot at 760 ps of TMD trajectory. BChE is shown as gold ribbon. Key residues of wild-type BChE and (−)-cocaine molecule are shown in stick style and colored by atom type. Dashed lines represent importance distances between different residues of A328W/Y332G mutant and different groups of (−)-cocaine. Those include the distance from the center (green ball) of benzoyl group of (−)cocaine to the aromatic center (green ball) of the side chain of W328; the distance from the center (green ball) of methyl ester group of (−)-cocaine to the aromatic center (green ball) of the side chain of F329; and the distance from the carbonyl oxygen at the benzoyl ester group of (−)-cocaine to the backbone hydrogen of either G116 or G117, or both. Hydrogen bonding interactions within the catalytic triad residues (S198-H328-E325) of A328W/Y332G mutant are also represented as dashed lines with labeled distances for the TS2rot structure.
However, there were some significant differences in the transformation process between the A328W/Y332G BChE-(−)-cocaine binding and wild-type BChE-(−)-cocaine binding. As shown in Figure 6A, the tracked key distances increased or decreased gradually around the time point of 200 ps during the TMD simulations on A328W/Y332G BChE-(−)-cocaine binding structure. As this time period (around 200 ps of the TMD trajectory) did represent the conformational changes, we still denoted the conformational transition at this time period as INT1 state, and such state can be represented by a typical structure (Figure 7A) selected from the TMD trajectory. Another difference was the conformational change for the benzoyl ester group of (−)-cocaine. As shown in Figure 6B, the θ1 angle increased gradually, indicating that this functional group of (−)-cocaine rotated smoothly. The step-by-step change of θ1 also suggests that the steric hindrance for the rotation of (−)-cocaine was reduced in the A328W/Y332G mutant compared to that in wild-type BChE. This favorable change on the rotation of (−)cocaine is attributed to the A328W/Y332G mutations. When comparing the time period before INT1rot in Figure 7A with the time period before TS1rot in Figure 3A, the W328-COCbenzoyl distance for A328W/Y332G BChE-(−)-cocaine binding structure was much longer than the Y332-COCbenzoyl distance in the wild-type BChE-(−)-cocaine binding structure, and the W328-COCbenzoyl distance had no dramatic change. The shorter distance in the wild-type BChE-(−)-cocaine suggests that W328 residue is not an obstacle for the rotation of (−)-cocaine toward catalytic residue S198 of the enzyme. At the first 200 ps time period of TMD simulations, the F329-COCmethyl distance (Figure 7A) for the A328W/Y332G BChE-(−)-cocaine binding structure was always longer than that in the TMD simulations on the wild-type BChE-(−)-cocaine binding structure (Figure 3A), suggesting that the steric hindrance from residue F329 for the rotation of (−)-cocaine was reduced after the A328W/Y332G mutations. The residue-based contacts along the TMD trajectory for the A328W/Y332G mutant were also analyzed in a similar way as did for wild-type BChE, and the results are shown in Figure 8 and also summarized in Table S2 (see Supporting Information). At this time of TMD simulations on the A328W/Y332G BChE-(−)-cocaine binding structure, type I residues with significantly increased contacts include G115, G116, G117, E197, S198, L286, and H438, suggesting that these residues are critical for the formation of the prereactive complex. Residues A199, Y231, V288, and F398 came into contact with (−)-cocaine when TS2rot showed up, indicating their cooperative role in the formation of prereactive complex for A328W/Y332G BChE-(−)cocaine binding. Residue Y440 experienced significant decrease in the number of contacts around TS2rot, while its number of contacts significantly increased around INT1rot. The change in the contact number indicates that residue Y440 is helpful for the structural transformation toward TS2rot but not important for the formation of the prereactive complex. Type III residues with decreasing number of contacts are D70, P285, S287, and W328, suggesting that these residues may be important only for the existence of the nonprereactive complex.
Figure 8.
Plots for residue-based contacts of A328W/Y332G mutant interacting with (−)cocaine along the TMD trajectory. (A) Type I residues showing significant increase in the number of contacts. (B) Residues with either continuous contacts (upper panel, type II) or decreased number of contacts (lower panel, type III).
In addition, we also analyzed the residue-based contacts of A328W/Y332G BChE with (−)-cocaine along the reaction coordinate (i.e. the distance from the mass center of the benzoyl group of (−)-cocaine to the mass center of the side chain of residue S198 of the enzyme) by using the MD trajectories from all the windows of umbrella sampling. The pattern of residue-based contacts tracked along the reaction coordinate (see Figure S3 in Supporting Information) for each residue is very similar with that from the TMD trajectory (Figure 8), suggesting the observed transformation pathway for A328W/Y332G BChE-(−)-cocaine binding structure is reasonable.
Free Energy Barriers
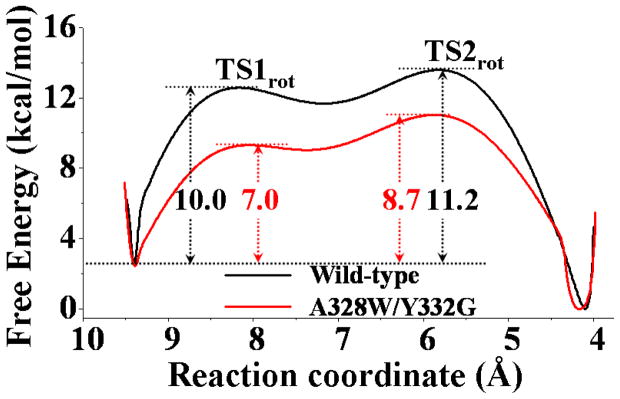
Based on the data collected from the umbrella-sampling MD simulations, the potential of mean force (PMF) for the transformation from the nonprereactive complex to the prereactive complex was determined, and the PMF results for both the wild-type BChE-(−)-cocaine binding and A328W/Y332G BChE-(−)-cocaine binding are depicted in Figure 9. The distance between the mass center of benzoyl group of (−)-cocaine and the mass center of the side chain of residue S198 was used as the reaction coordinate for the PMF calculations. The choice of the reaction coordinate was based on the structural features of our modeled BChE-cocaine binding structures (Figures 1 and 2) and the results of the TMD simulations. We noted that such a distance as the reaction coordinate could more reasonably represent the (−)-cocaine rotation compared to other geometrical parameters such as θ1 or θ2 shown in Figure 3. Along the chosen reaction coordinate, two transition states were identified from the free energy profile for the transformation process of wild-type BChE-(−)-cocaine binding structure (black curve in Figure 9). According to this free energy profile for the transformation process of wild-type BChE-(−)-cocaine binding structure, the first energy barrier (TS1rot in Figure 9) appeared at a position where the value of reaction coordinate was ~8.2 Å. A detailed check on the umbrella-sampling MD simulations revealed that this energy barrier corresponds to the event of unbinding of the benzoyl ester group from its sub-binding site consisting of residues A328, F329, and Y332 of wild-type BChE. This unbinding event also represents the deformation of the nonprereactive complex. Such unbinding event (TS1rot) for benzoyl ester group of (−)-cocaine was also observed at the first conformational transition (Figures 3, 4A, and 5) during the TMD simulations on the wild-type BChE-(−)-cocaine binding, e.g. the significant increase of Y332-COCbenzoyl distance (cyan-colored curve in Figure 3A). The second energy barrier (TS2rot in Figure 8) showed up at a position with a value of ~5.7 Å for the reaction coordinate. This energy barrier occurred when the methyl ester group of (−)-cocaine rotated into its sub-binding site of wild-type BChE. The existence of transition state TS2rot in the free energy profile can also be supported by the observations of structural changes at the second conformational transition through the TMD simulations (Figures 3, 4B, and 5), e.g. the significant decrease of G117H-O33 distance (red curve in the lower panel of Figure 3A) and the significant change of θ2 angle around TS2rot as described in Figure 3B.
Figure 9.
Free energy profiles determined for the structural transformation of binding structures of (−)-cocaine with wild-type BChE (black line) and the A328W/Y332G mutant (red line). The reaction coordinate was defined as the distance from the mass center of benzoyl group of (−)-cocaine to the mass center of the side chain of S198 of wild-type BChE or the A328W/Y332G mutant. The free energy barriers corresponding to TS1rot and TS2rot of the transformation process are also labeled (unit in kcal/mol).
Concerning the transformation process of A328W/Y332G BChE-(−)-cocaine binding, the free energy profile is quite different from that of the transformation process of wild-type BChE-(−)-cocaine binding. As shown in Figure 9, the free energy barriers corresponding to the deformation of the nonprereactive complex and the formation of the prereactive complex for A328W/Y332G BChE-(−)-cocaine binding are all lower than those for wild-type BChE-(−)-cocaine binding. The lower free energy barriers suggest that the rotation of (−)-cocaine in the binding pocket of the A328W/Y332G mutant is much easier than that in the binding pocket of wild-type BChE. In other words, the prereactive complex formation is easier for (−)-cocaine binding with the A328W/Y332G mutant compared to that for the (−)-cocaine binding with wild-type BChE. The features of the free energy profile (Figure 9) for the transformation process of the A328W/Y332G BChE-(−)-cocaine binding are consistent with the features of structural changes observed during the TMD simulations on the A328W/Y332G BChE-(−)-cocaine binding structure (Figures 6 to 8).
Based on the experimental findings in previous reports,19,21,23,24,25,26,,27,28,29,30 the formation of the prereactive complex is the rate-determining step for (−)-cocaine hydrolysis catalyzed by wild-type BChE. So, the experimentally determined catalytic rate constant (kcat) can be used to estimate the free energy barrier ( ) for the structural transformation from the nonprereactive complex to the prereactive complex. According to classic transition state theory,48,49 we have
| (3) |
where ν is the frequency factor and ν = kBT/h, kb is the Boltzmann’s constant, h is the Planck constant, and T is the temperature. In Eq.(3), is the overall free energy barrier for the enzymatic reaction process. only when the structural transformation from the nonprereactive complex to the prereactive complex is the rate-determining step for the enzymatic reaction process. when the structural transformation from the nonprereactive complex to the prereactive complex is not the rate-determining step. According to Eq. (3), can be calculated as
| (4) |
Using the experimental kcat value (4.1 min−1)30,31 for (−)-cocaine hydrolysis catalyzed by wild-type BChE, we obtained for wild-type BChE. Using the experimentally determined kcat (240 min−1) for (−)-cocaine hydrolysis catalyzed by the A328W/Y332G mutant,27 we got and for the A328W/Y332G mutant as the rate-determining step for the A328W/Y332G mutant is not the structural transformation from the nonprereactive complex to the prereactive complex.19,25,26,27,29,30 These experimentally-derived free energy barriers indicate that the difference ( ) in the free energy barrier for the structural transformation between wild-type BChE and the A328W/Y332G mutant should be larger than 19.0 – 16.6 = 2.4 kcal/mol, i.e. .
As shown in Figure 9, the free energy barrier ( ) obtained from the PMF calculations is 11.2 kcal/mol for the wild-type BChE-(−)-cocaine binding, and 8.7 kcal/mol for the A328W/Y332G BChE-(−)-cocaine binding, giving a value of 2.5 kcal/mol. The directly calculated value of 2.5 kcal/mol is consistent with the experimental kinetic data showing . Further, in comparison with the experimental values, the PMF calculations based on the classical MD simulations systematically underestimated the free energy barriers for the structural transformation from the nonprereactive complex to the prereactive complex, although the calculated relative values are qualitatively reasonable. For wild-type BChE, the experimental value of 19.0 kcal/mol is 1.69 times the calculated value of 11.2 kcal/mol. If we can employ the same scaling factor of 1.69 to correct the calculated values for both wild-type BChE and the mutants, the corrected theoretical values should be 19.0 and 14.7 kcal/mol for wild-type BChE and the A328W/Y332G mutant, respectively. Thus, it might be more reasonable to predict that the free energy barrier ( ) for the structural transformation from the nonprereactive complex to the prereactive complex should be ~14.7 kcal/mol for the A328W/Y332G mutant. Based on this prediction, we have , which is also consistent with available experimental kinetic data showing .
The consistency between the predicted free energy barriers and available experimental kinetc data suggests that the computational protocol is feasible and may be valuable for future computational design of high-activity mutants of BChE. Further, the general computational strategy and approach based on the combined use of TMD and PMF simulations may also be used to study the detailed pathway and free energy profile for other similar mechanistic problems involving ligand rotation or any other type of structural transformation in the binding pocket of a protein.
Conclusion
The combined targeted molecular dynamics (TMD) and potential of mean force (PMF) simulations have allowed us, for the first time, to uncover the detailed pathway and determine the corresponding free energy profile for a structural transformation associated with the substrate rotation in the binding pocket of a protein. The combined use of TMD and PMF simulations reveal that the structural transformation from the nonprereactive BChE-(−)-cocaine binding to the prereactive BChE-(−)-cocaine binding associated with the (−)-cocaine rotation in the binding pocket involves two transition states (TS1rot and TS2rot). The transition state TS1rot is mainly associated with the deformation of the nonprereactive complex in which the benzoyl ester group of (−)-cocaine moves toward the catalytic residue S198 of BChE. The transition state TS2rot is mainly associated with the formation of the prereactive complex in which the methyl ester group of (−)-cocaine rotates and the carbonyl oxygen at the benzoyl ester group of (−)-cocaine moves into the oxyanion hole of BChE.
The combined TMD and PMF simulations also demonstrate that the A328W/Y332G mutation do not change the fundamental pathway for the structural transformation from the nonprereactive binding to the prereactive binding. However, the A328W/Y332G mutation significantly reduces the steric hindrance for (−)-cocaine rotation in the binding pocket of BChE and, thus, decreases the free energy barrier for the structural transformation from the nonprereactive binding to the prereactive binding. The calculated relative free energy barriers are all consistent with available experimental kinetic data, suggesting that the computational protocol is reliable for prediction of the relative free energy barriers for wild-type enzyme and the mutants. The new mechanistic insights obtained and the novel computational protocol tested in this study should be valuable for future computational design of high-activity mutants of BChE.
Finally, the general computational strategy and approach based on the combined TMD and PMF simulations may be also valuable in computational studies of detailed pathways and free energy profiles for other similar mechanistic problems involving ligand rotation or another type of structural transformation in the binding pocket of a protein.
Supplementary Material
Acknowledgments
This work was supported by NIH (Grants R01 DA013930, R01 DA025100, and R01 DA021416). The authors also acknowledge the Center for Computational Sciences (CCS) at the University of Kentucky for supercomputing time on IBM X-series Cluster consisting of 340 nodes or 1,360 processors.
Footnotes
Supporting Information Available. Two figures for the results of MD simulations on the binding structures of (−)-cocaine binding with both wild-type BChE and A328W/Y332G mutant; one figure for the tracked residue-based contacts of A328W/Y332G BChE with (−)-cocaine along the reaction coordinate; two tables about the number and fraction of calculated residue-based contacts between (−)-cocaine and BChE. This material is available free of charge via the Internet http://pubs.acs.org.
References
- 1.Mendelson JH, Mello NK. Management of cocaine abuse and dependence. New Eng J Med. 1996;334:965–972. doi: 10.1056/NEJM199604113341507. [DOI] [PubMed] [Google Scholar]
- 2.Sparenborg S, Vocci F, Zukin S. Peripheral cocaine-blocking agents: new medications for cocaine dependence. Drug Alcohol Depend. 1997;48:149–151. doi: 10.1016/s0376-8716(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 3.Singh S. Chemistry, design, and structure-activity relationship of cocaine antagonists. Chem Rev. 2000;100:925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 4.Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ., Jr Three-dimensional quantitative structure-activity relationship modeling of cocaine binding by a novel human monoclonal antibody. J Med Chem. 2004;47:133–142. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 5.Gorelick DA. Enhancing cocaine metabolism with butyrylcholinesterase as a treatment strategy. Drug Alcohol Depend. 1997;48:159–165. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 6.Singh S. Chemistry, design, and structure-activity relationship of cocaine antagonists. Chem Rev. 2000;100:925–1024. doi: 10.1021/cr9700538. [DOI] [PubMed] [Google Scholar]
- 7.Gaintdinov RR, Sotmikova TD, Caron MG. Monoamine transporter pharmacology and mutant mice. Trends Pharmacol Sci. 2002;231:367–373. doi: 10.1016/s0165-6147(02)02044-8. [DOI] [PubMed] [Google Scholar]
- 8.Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- 9.Chen R, Tilley MR, Wei H, Zhou F, Zhou FM, Ching S, Quan N, Stephens RL, Hill ER, Nottoli T, Han DD, Gu HH. Abolished cocaine reward in mice with a cocaine-insensitive dopamine transporter. Proc Natl Acad Sci U S A. 2006;103:9333–9338. doi: 10.1073/pnas.0600905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landry DW, Zhao K, Yang GXQ, Glickman M, Georgiadis TM. Antibody-catalyzed degradation of cocaine. Science. 1993;259:1899–1901. doi: 10.1126/science.8456315. [DOI] [PubMed] [Google Scholar]
- 11.Carrera MRA, Ashley JA, Parsons LH, Wirsching P, Koob GF, Janda KD. Suppression of psychoactive effects of cocaine by active immunization. Nature. 1995;378:727–730. doi: 10.1038/378727a0. [DOI] [PubMed] [Google Scholar]
- 12.Baird TJ, Deng SX, Landry DW, Winger G, Woods JH. Natural and artificial enzymes against cocaine. I. Monoclonal antibody 15A10 and the reinforcing effects of cocaine in rats. J Pharmacol Exp Ther. 2000;295:1127–1134. [PubMed] [Google Scholar]
- 13.Carrera MRA, Ashley JA, Wirsching P, Koob GF, Janda KD. A second-generation vaccine protects against the psychoactive effects of cocaine. Proc Natl Acad Sci U S A. 2001;98:1988–1992. doi: 10.1073/pnas.041610998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng SX, de Prada P, Landry DW. Anticocaine catalytic antibodies. J Immunol Methods. 2002;269:299–310. doi: 10.1016/s0022-1759(02)00237-5. [DOI] [PubMed] [Google Scholar]
- 15.Meijler MM, Kaufmann GF, Qi LW, Mee JM, Coyle AR, Moss JA, Wirsching P, Matsushita M, Janda KD. Fluorescent cocaine probes: a tool for the selection and engineering of therapeutic antibodies. J Am Chem Soc. 2005;127:2477–2484. doi: 10.1021/ja043935e. [DOI] [PubMed] [Google Scholar]
- 16.Zhan CG, Deng SX, Skiba JG, Hayes BA, Tschampel SM, Shields GC, Landry DW. First-principle studies of intermolecular and intramolecular catalysis of protonated cocaine. J Comput Chem. 2005;26:980–986. doi: 10.1002/jcc.20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Gao D, Zhan CG. Modeling the catalysis of anti-cocaine catalytic antibody: competing reaction pathways and free energy barriers. J Am Chem Soc. 2008;130:5140–5149. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxena A, Redman AM, Jiang X, Lockridge O, Doctor BP. Differences in active site gorge dimensions of cholinesterases revealed by binding of inhibitors to human butyrylcholinesterase. Biochem. 1997;36:14642–14651. doi: 10.1021/bi971425+. [DOI] [PubMed] [Google Scholar]
- 19.Zhan CG, Zheng F, Landry DW. Fundamental reaction mechanism for cocaine hydrolysis in human butyrylcholinesterase. J Am Chem Soc. 2003;125:2462–2474. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockridge O, Schopfer LM, Winger G, Woods JH. Large scale purification of butyrylcholinesterase from human plasma suitable for injection into monkeys; a potential new therapeutic for protection against cocaine and nerve agent toxicity. J Med Chem Biol Radiol Def. 2005 Jul;1(3) doi: 10.1901/jaba.2005.3-nihms5095. nihms5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao D, Zhan C-G. Modeling evolution of hydrogen bonding and stabilization of transition states in the process of cocaine hydrolysis catalyzed by human butyrylcholinesterase. Proteins. 2006;62:99–110. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao D, Zhan CG. Modeling effects of oxyanion hole on the ester hydrolyses catalyzed by human cholinesterases. J Phys Chem B. 2005;109:23070–23076. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 23.Gao D, Cho H, Yang W, Pan Y, Yang G, Tai HH, Zhan CG. Computational design of a human butyrylcholinesterase mutant for accelerating cocaine hydrolysis based on the transition-state simulation. Angew Chem Int Ed. 2006;45:653–657. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan CG, Gao D. Catalytic mechanism and energy barriers for butyrylcholinesterase-catalyzed hydrolysis of cocaine. Biophys J. 2005;89:3863–3872. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamza A, Cho H, Tai HH, Zhan CG. Molecular dynamics simulation of cocaine binding with human butyrylcholinesterase and its mutants. J Phys Chem B. 2005;109:4776–4782. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Y, Gao D, Yang W, Cho H, Yang G, Tai HH, Zhan CG. Computational redesign of human butyrylcholinesterase for anticocaine medication. Proc Natl Acad Sci U S A. 2005;102:16656–15661. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y, Gao D, Yang W, Cho H, Zhan CG. Free energy perturbation (FEP) simulation on the transition states of cocaine hydrolysis catalyzed by human butylrycholinesterase and its mutants. J Am Chem Soc. 2007;129:13537–13543. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng F, Zhan CG. Rational design of an enzyme mutant for anti-cocaine therapeutics. J Comput Aided Mol Des. 2008;22:661–671. doi: 10.1007/s10822-007-9144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng F, Yang W, Ko MC, Liu J, Cho H, Gao D, Tong M, Tai HH, Woods JH, Zhan CG. Most efficient cocaine hydrolase designed by virtual screening of transition states. J Am Chem Soc. 2008;130:12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Pan Y, Zheng F, Cho H, Tai HH, Zhan CG. Free-energy perturbation simulation on transition states and redesign of butyrylcholinesterase. Biophys J. 2009;96:1931–1938. doi: 10.1016/j.bpj.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun H, Pang YP, Lockridge O, Brimijoin S. Re-engineering butyrylcholinesterase as cocaine hydrolase. Mol Pharmacol. 2002;62:220–224. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 32.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products. J Biol Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 34.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. University of California; San Francisco: 2004. [Google Scholar]
- 35.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C.02. Gaussian, Inc; Wallingford CT: 2004. [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW. Comparison of simple potential functions for simulating liquidwater. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 37.Morishita T. Fluctuation formulas in molecular dynamics simulations with the weak coupling heat bath. J Chem Phys. 2000;113:2976–2982. [Google Scholar]
- 38.Darden T, York D, Pedersen L. Particle mesh Ewald—an N_log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 39.Toukmaji A, Sagui C, Board J, Darden T. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J Chem Phys. 2000;113:10913–10927. [Google Scholar]
- 40.Ryckaert J, Ciccotti PG, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 41.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 42.Torrie GM, Valleau JP. Nonphysical sampling distribution in monte carlo free-energy estimation: umbrella sampling. J Comput Phys. 1977;23:187–199. [Google Scholar]
- 43.Kirkwood JG. Statistical mechanics of fluid mixtures. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 44.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg J. The weighted histogram analysis method for free-energy calculations on biomolecules. I. Method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 45.Roux B. The calculation of the potential of mean force using computer simulations. Comput Phys Commu. 1995;91:275–282. [Google Scholar]
- 46.Cheng X, Wang H, Grant B, Sine SM, McCammon JA. Targeted molecular dynamics study of C-loop closure and channel gating in nicotinic receptors. PLoS Comput Biol. 2006;2:1173–1184. doi: 10.1371/journal.pcbi.0020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eyring H. The activated complex in chemical reactions. J Chem Phys. 1935;3:107–115. [Google Scholar]
- 49.Truhlar DG, Garrett BC, Klippenstein SJ. Current status of transition-state theory. J Phys Chem. 1996;100:12771–12800. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.