Abstract
D. radiodurans single stranded (ss) DNA binding protein (DrSSB) originates from a radiation-resistant bacterium and participates in DNA recombination, replication and repair. Although it functions as a homodimer, it contains four DNA binding domains (OB folds) and thus is structurally similar to the E. coli SSB (EcoSSB) homotetramer. We examined the equilibrium binding of DrSSB to ssDNA to compare with EcoSSB. We find that the occluded site size of DrSSB on poly(dT) is ~45 nucleotides in low salt (<0.02M NaCl) but increases to 50–55 nucleotides at [NaCl] ≥ 0.2M. This suggests that DrSSB undergoes a transition between ssDNA binding modes as is observed for EcoSSB, although the site size difference between modes is not as large as for EcoSSB, suggesting that the pathways of ssDNA wrapping differ for these two proteins. The occluded site size corresponds well to the contact site size (52 nucleotides) determined by Isothermal Titration Calorimetry (ITC). Electrophoretic studies of complexes of DrSSB with phage M13ssDNA indicate the formation of stable, highly cooperative complexes at low salt conditions. Using ITC we find that DrSSB binding to oligo(dT)s with lengths close to the determined site size (50–55 nts) is stoichiometric with ΔHobs ≈-94±4 kcal/mole, somewhat smaller than for EcoSSB (≈-130 kcal/mole) under the same conditions. The observed binding enthalpy shows a large sensitivity to NaCl concentration, similar to that observed for EcoSSB. With the exception of the less dramatic change in occluded site size, the behavior of DrSSB is similar to that of EcoSSB protein (although, clear quantitative differences exist). These common features for SSB proteins having multiple DNA binding domains enable versatility of SSB function in vivo.
Single-stranded DNA binding proteins are essential in nearly all organisms and play central roles in all aspects of genome maintenance, including DNA replication, recombination and repair (1–3). They bind selectively single stranded DNA with high affinity and independent of sequence specificity, protecting ssDNA intermediates from degradation and preventing them from the formation of secondary structures. The other functions of SSB proteins have become recognized only recently and include the regulation of the activity of many other DNA metabolic proteins through direct protein-protein interactions (4) as well as by modulating the accessibility of ssDNA by either different modes of binding (3, 5) or by the ability to translocate along ssDNA as shown recently for EcoSSB (6). Although SSB proteins from different organisms can differ in architecture, for example, the bacteriophage T4 gene 32 protein is a monomer (7, 8), EcoSSB protein is a homotetramer (9–11) and eucaryotic RPA proteins are heterotrimers (12, 13), they all contain oligonucleotide/oligosaccharide binding folds (OB-fold), which form the primary site for ssDNA binding (14, 15). The number of OB folds varies significantly in these proteins, from one in T4 gene 32 protein to six in the hetero-trimeric RPAs. Most of the bacterial SSBs are homotetramers and contain 4 OB-folds (one per monomer) (see for example (16–19), with the EcoSSB tetramer being the most studied. In contrast to the homotetrameric SSB proteins, SSB proteins from Deinococcus-Thermus group function as homodimers; however, each monomer encodes two OB-folds linked by a conserved spacer sequence (20–23). Therefore, the functional form of these proteins still is composed of four OB-folds, although the sequence differences between the two OB-folds within each dimer impose an asymmetry that is likely to affect its DNA binding properties. Furthermore, these dimers possess only two unstructured C-termini, rather than four as is the case for the homo-tetrameric SSB proteins.
DrSSB protein originates from one of the most radiation resistant organisms known (24). D. radiodurans, which is a soil bacterium featuring a D37 γ irradiation dose of approximately 6,000 Gy, making this organism roughly 200 times more radiation resistant than Escherichia coli (24). DrSSB forms a stable homodimer in solution (22) and its quantitative estimates in the D. radiodurans cell indicate approximately 2500–3000 dimers/cell, independent of the level of irradiation (25).
A crystal structure of the full length DrSSB was determined with 1.8A resolution (23) and shows that each DrSSB subunit comprises two OB-folds linked by a β-hairpin motif and the protein assembles a four-OB-fold arrangement by means of symmetric dimerization. The C-terminal tail residues 234–301 are missing from the model due to the lack of electron density suggesting that the extended C-terminal region of this protein is highly unstructured similar to the C-terminal tails of EcoSSB (11).
In order to better understand the functional consequences arising from these differences, we have examined the binding of Deinococcus radiodurans SSB (DrSSB) to a variety of ssDNA to compare its DNA binding properties with those of the well-studied homo-tetrameric EcoSSB.
MATERIALS AND METHODS
Reagents and Buffers
Buffers were prepared with reagent grade chemicals and distilled water that was subsequently treated with a Milli Q (Millipore, Bedford, MA) water purification system. Buffer T is 10 mM Tris, pH8.1, and buffer P is 10 mM Phosphate pH 7.0. All buffers contained 0.1 mM Na3EDTA and 1mM BME. The concentrations of NaCl used in the experiments are specified in the text.
DrSSB and ssDNA
DrSSB protein was expressed and purified as described (22, 26) and its concentration determined spectrophotometrically in Tris buffer (pH 8.1, 0.2 M NaCl) using an extinction coefficient of 8.2×104 M−1 cm−1 (dimer) (22). Single-stranded circular phage M13 mp18 DNA was purchased from New England Biolabs, Inc. (Ipswich, MA). The oligodeoxynucleotides, (dT)L, (L=30,35,40,45,50,55,60,70) were synthesized and purified as described (27) and were ≥ 98% pure as judged by denaturing gel electrophoresis and autoradiography samples that was 5′ end-labeled with 32P using polynucleotide kinase. The poly(dT) (Midland Certified Reagent Co., Midland, TX) had an average length of ~1100 nucleotides. Concentrations of nucleic acids were determined spectrophotometrically in buffer T (pH 8.1), 100 mM NaCl using an extinction coefficient ε260=8.1×103 M−1 (nucleotide) cm−1 for oligo(dT) and poly(dT) (8) and ε259=7370 M−1 (nucleotide) cm−1 for single stranded M13 ssDNA (28).
Fluorescence titrations
Equilibrium binding of DrSSB protein to oligodeoxynucleotides, (dT)L and poly(dT) was performed by monitoring quenching of the intrinsic Trp fluorescence of DrSSB upon addition of ssDNA using a PTI QM-2000 spectrofluorometer (Photon Technologies, Inc., Lawrenceville, NJ) [λex=296 nm (2 nm excitation band-pass) and λem=345 nm (2–4 nm emission band-pass)] with corrections applied as described (29, 30). The binding isotherms shown in Fig. 2 for the interaction of (dT)L (L=45,50 and 55) (X, ligand) with DrSSB dimer (M, macromolecule) were analyzed with a 1:1 binding model using eq. 1:
| (1) |
where Qobs is the observed fluorescence quenching and Qmax is a fluorescence quenching at saturation; n and Kobs are the stoichiometry of binding and equilibrium association constant. The concentration of free (dT)L, X, was determined from the mass conservation equation 1a:
| (1a) |
Figure 2.
Determination of occluded site size of DrSSB using oligonucleotides of different length (Buffer T, pH 8.1, 25°C). (A) - Results of fluorescence titrations of DrSSB (~1µM) with (dT)45, (dT)50 and (dT)55 in the presence of 0.2M NaCl. (B) - Fluorescence titration of DrSSB (~1µM) with (dT)45, in 1.0 M NaCl. The smooth curve represents the best fit of the data to a 1:1 model (see eq. 1 in Materials and Methods) with n=0.98±0.01, Qmax=0.72±0.01 and Kobs=(1.48±0.06)×107 M−1.
The binding isotherms shown in Fig. 3 for the interaction of two (dT)25 molecules (ligand, X) with DrSSB dimer (macromolecule, M) were analyzed using a two-site sequential binding model and eq. 2:
| (2) |
where Qobs is the observed fluorescence quenching and Q1 and Q2 are the fluorescence quenchings corresponding to one and two (dT)25 molecules bound, respectively; K1,obs and K2,obs are the observed step-wise macroscopic association constants for the binding of the first and the second DNA molecule. The concentration of free DNA, X, was determined from the mass conservation equation 2a:
| (2a) |
In eqs 1 and 2 Xtot and Mtot are total concentrations of the (dT)25 and the protein, respectively. The fitting of the data were performed using the nonlinear regression package in Scientist (MicroMath Scientist Software, St. Louis, MO).
Figure 3.
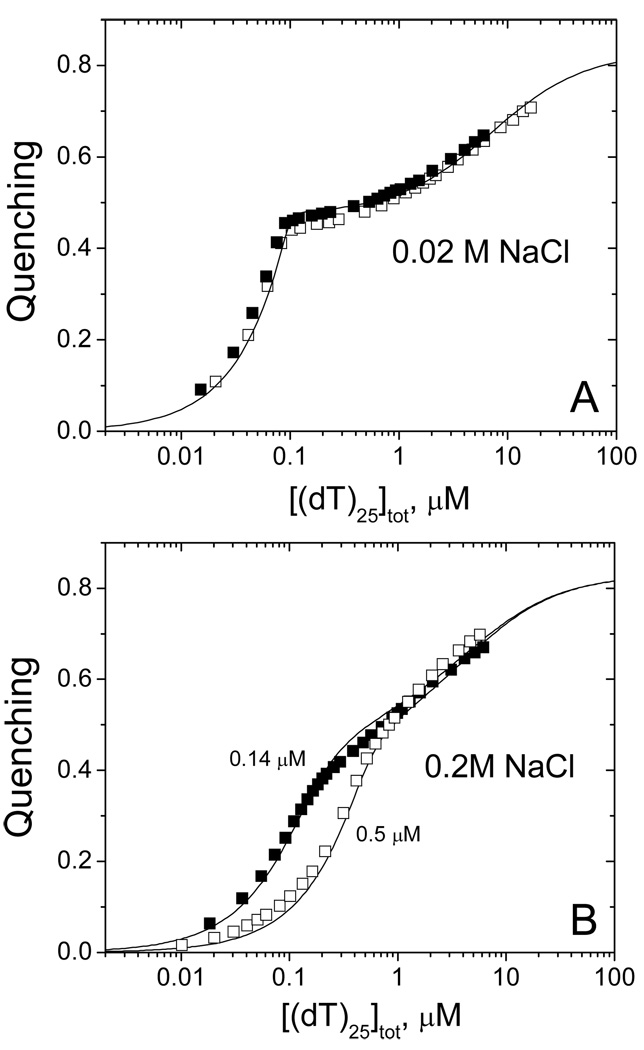
DrSSB binds two molecules of (dT)25 with negative cooperativity, which decreases with increasing salt concentration (Buffer T, pH 8.1, 25°C). (A) – Two titration of DrSSB (0.1µM (open and closed squares) with (dT)25 0.02M NaCl. Solid curve represent the result of global nonlinear least squares fitting of both sets of data to a two site model (eq, 2 in Materials and Methods) with best fit parameters: Q1 = 0.48±0.01, K1,obs =(4.5±6.2)×109 M−1, K2,obs =(1.4±0.1)×105 M−1 (value of Q2=0.83 was fixed according to maximum quenching observed in poly(dT) and (dT)L experiments). (B) - Titration of DrSSB (0.14µM (closed squares) and 0.5µM (open squares)) with (dT)25 in 0.2M NaCl. Solid curves represent the result of global nonlinear least squares fitting of both sets of data to the two site model (eq, 2 in materials and Methods) with best fit parameters: Q1 = 0.51±0.01, K1,obs =(3.1±0.6)×107 M−1, K,obs =(2.1±0.3)×105 M−1 (value of Q2=0.83 was fixed according to maximum quenching observed in poly(dT) and (dT)L experiments).
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a VP-ITC titration microcalorimeter (MicroCal Inc., Northhampton, MA) (31). Generally, experiments were carried out by titrating DrSSB (1–2µM (dimer) in the cell) with oligo(dT) solutions (10–25 µM in the syringe). The heats of dilution were usually obtained from a reference titration in which the species in the syringe is titrated into the cell containing only buffer solution. All corrections for heats of dilution were applied as described (32). Oligo(dT) and protein samples were dialyzed extensively vs. the buffer containing the indicated salt concentration used in the ITC experiments.
The DrSSB binds one molecule of (dT)L, when L=35,40,45,50,55 and 60. The stoichiometry of binding, n, and the values of Kobs (where measurable) and ΔHobs were obtained by fitting the ITC titration curves to a model of ligand (X= (dT)L) binding to n-identical and independent sites on the macromolecule (M=DrSSB) using eq. 3,
| (3) |
where is the total heat after the i-th injection and V0 is the volume of the calorimetric cell. The concentration of free ligand (X) was obtained by solving eq. 1a. When the length of (dT)L is shorter than 35 nucleotides, or longer than 60 nucleotides the observed binding isotherms display two-step behavior indicative of weak binding of a second (dT)L molecule (X) to DrSSB (M) (when L<35) or a second DrSSB dimer (X) to (dT)L (macromolecule M) (when L>60), respectively. For these cases we used a 2 site sequential binding model, characterized by two macroscopic binding constants (K1,obs and K2,obs) and two binding enthalpies (ΔH1,obs and ΔH 2,obs) as described by eq. 4,
| (4) |
where the concentration of free ligand (X) was obtained by solving eq. 2a. In eqs 3 and 4 Xtot and Mtot are the total concentrations of the ligand and macromolecule, respectively, in the calorimetric cell after the i-th injection. Non-linear least squares fitting of the data was performed using the “ITC Data Analysis in Origin” software provided by the manufacturer. The conversion of integral heats () to differential heats (heats per injection observed in the experiment) and the fitting routine including corrections for heat displacement effects and ligand and macromolecule dilutions in the calorimetric cell were performed as described (32) (see also the ITC Data Analysis in Origin Tutorial Guide (Microcal Inc.)). Additional fittings and simulations were also performed using the nonlinear regression package in Scientist (MicroMath Scientist Software, St. Louis, MO).
Agarose gel electrophoresis of DrSSB-ssM13 DNA complexes
The distribution of DrSSB molecules bound to circular phage M13 ssDNA was monitored by separating the DNA molecules by gel electrophoresis based on the amount of DrSSB bound using 0.5% agarose gels as described (33). In these experiments the amount of DNA loaded into each lane was held constant ≈1.5 nmole (nucleotides) while the amount of DrSSB in the solution was varied from 0 to 0.03 nmole (dimer) in order to saturate the M13mp8 with DrSSB, assuming that DrSSB binding site size is 45–50 nucleotides. The complexes were formed in buffer T (10 mM Tris, pH 8.1, 0.1mM EDTA) at two NaCl concentrations (0.02M and 0.3M); the total reaction volume of each sample was 30 µl. The time of incubation was varied as noted in the text. The electrophoresis running buffer was 20 mM Tris-Acetate (pH 8.1), 0.5 mM Na3EDTA. Loading dye (2 µl of 50% (v/v) glycerol, 0.04% (w/v) bromophenol blue) was added to each 30 µl sample. Electrophoresis was carried out at room temperature (22° C) at constant voltage (8 V/cm) for ~3 hours. The gels were then stained for 15 min with ethidium bromide (2 µg/ml solution) and destained for 2 hours at 4° C in buffer T + 1 M NaCl.
RESULTS
Occluded ssDNA binding site size of DrSSB on poly(dT)
Characterization of the ssDNA binding properties of a nonspecific ssDNA binding protein such as DrSSB requires a determination of the occluded site size (n), defined as the number of ssDNA nucleotides that are occluded by one protein and thus prevented from interacting with a second protein (34). For oligomeric SSB proteins containing multiple OB folds and thus potential sites for ssDNA binding the occluded site size will be affected by the number of OB-folds that are involved in the interaction. For example, the EcoSSB homotetramer, possessing four OB-folds (one per subunit), can bind ssDNA in multiple binding modes. Two of the major binding modes differ in that ssDNA occupies either two or four subunits with corresponding occluded site sizes of ~35 and 65 nucleotides (3, 35). Both modes involve at least partial wrapping of ssDNA around the tetramer (10). The relative populations of these binding modes are influenced by protein binding density, salt type and valence and salt concentration (5, 35–39) with the SSB35 binding mode preferred at low salt (<0.02M NaCl) and high protein binding densities, and the SSB65 binding mode favored at high salt (≥0.2M NaCl). Similar effects have been observed for the Saccaromyces cerivisiae (sc)RPA where the occluded site size increases from ~18 nucleotides at low salt to ~27 nucleotides at high salt as a result of the interaction of ssDNA with an additional OB-fold within scRPA heterotrimer (40).
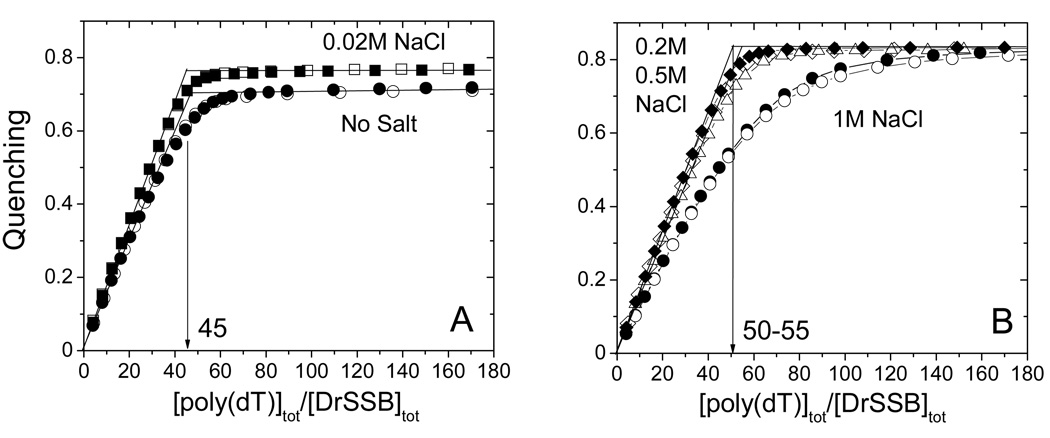
We measured the occluded site size for the DrSSB dimer binding to the homopolynucleotide poly(dT) as a function of salt concentration to determine if multiple ssDNA binding modes could be detected for this protein. The results of DrSSB titrations with poly(dT) at different NaCl concentrations (Tris-buffer, pH 8.1, 25°C), monitoring the quenching of drSSB tryptophan fluorescence, are presented in Fig. 1 and Table 1. In the absence of added salt and at low [NaCl] concentration (20mM) (see Fig. 1A), the Trp fluorescence quenching increases linearly with increasing poly(dT) concentration, reaching a quenching plateau of 72% (no salt added) and 76% (20mM NaCl) at saturating poly(dT). The intersection of the initial linear region with the plateau occurs at n≈45 nucleotides per DrSSB dimer providing an estimate of the occluded site size of the DrSSB dimer on poly(dT). The observed stoichiometries were independent of DrSSB concentration (titrations performed at 70 and 200 nM DrSSB), indicating that binding is stoichiometric under these conditions. Similar stoichiometric binding is observed at 0.2M and 0.5M NaCl (see Fig. 1B); however, both the occluded site size and the maximum Trp fluorescence quenching increase to n≈50–55 and 83–84%, respectively. Titrations performed at a still higher [NaCl] of 1 M results in a binding isotherm indicating that binding is no longer stoichiometric (Fig. 1B), making it impossible to determine an occluded site size by the extrapolation method. Thus, binding at 1 M NaCl is no longer stoichiometric. This is different from what is observed for the binding of EcoSSB with poly(dT), where binding remains stoichiometric up to 3M NaCl and is characterized by an occluded site size n≈65 with 90–92% quenching (36). Hence, DrSSB appears to have a smaller high salt occluded site size and lower affinity for poly(dT) compared to EcoSSB.
Figure 1.
Determination of occluded site size of DrSSB on poly(dT) at different NaCl concentrations (Buffer T, pH 8.1, 25°C). (A) - Results of fluorescence titrations of DrSSB (0.07µM – open squares; 0.2µM - closed squares) with poly(dT) in 0.02M NaCl and in the absence of salt (0.07µM – open circles; 0.2µM - closed circles). (B) - Results of fluorescence titrations of DrSSB with poly(dT) in 0.2 M NaCl (0.07µM – open diamonds; 0.2µM - closed diamonds), 0.5M NaCl (0.1 µM open triangles) and in 1.0 M NaCl (0.1µM – open circles; 0.2µM - closed circles). The linear increase in relative quenching indicates stoichiometric binding of the protein to poly(dT) until the point of saturation (plateau value) is reached. The occluded site sizes were determined by extrapolation of the linear parts of the titration curves to the point of intersection with the corresponding plateau value as described (35) and yields n≈45 nucleotides per DrSSB dimer (panel A) and n≈50–55 per DrSSB dimer (panel B). For the concentration of [NaCl]= 1M the binding is not stoichiometric, so the determination of the occluded site size is not posssible.
Table 1.
Results of Equilibrium Titrations of DrSSB with ssDNA Monitoring Trp Fluorescence Quenchinga.
| [NaCl] | Poly(dT) | (dT)50 (1:1) | (dT)45 (1:1) | (dT)25 (2:1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Nb (nts) |
Qmax | Kobs,c (M−1) |
Qmax | Kobs,c (M−1) |
Qmax | Kobs, (M−1)c | Q1, Q2 | K1,obs (M−1),c K2,obs (M−1) |
|
| no salt | 46 | 0.72 | st | - | - | - | - | - | - |
| 0.02 M | 45 | 0.76 | st | 0.84 | st | 0.76 | st | 0.48±0.01 0.83(fixed) |
(4.5±6.2)×109 (1.4±0.1)×105 |
| 0.2 M | 50 | 0.84 | st | 0.83 | st | 0.76 | st | 0.51±0.01 0.83(fixed) |
(3.1±0.6)×107 (2.1±0.3)×105 |
| 0.5 M | 55 | 0.83 | st | - | - | - | - | - | - |
| 1.0 M | N/A | 0.84 | N/A | - | - | 0.72 | (1.5±0.1)×107 | - | - |
Solution conditions: 10mM Tris (pH 8.1), 0.1 mM EDTA.
Occluded site size in nucleotides determined as described in the text.
st – stoichiometric binding; the errors are shown as S.D.
As a further test of whether the high [NaCl] site size of the DrSSB dimer on ssDNA does not exceed 50–55 nucleotides we performed titrations of DrSSB with oligodeoxythymidylates of different length ((dT)L, L=45, 50 and 55 nucleotides) at different NaCl concentrations (buffer-T, pH 8.1, 25°C). Fig.2 and Table 1 show the results obtained at 0.2M NaCl indicating that DrSSB dimer binds one molecule of (dT)45, (dT)50 or (dT)55, stoichiometrically as expected based on the DrSSB-poly(dT) results. Furthermore, the extent of Trp fluorescence quenching is ~74% for (dT)45, but increases to 83–85% for both (dT)50 and (dT)55 consistent with a high [NaCl] site size of ~ 50 nucleotides. Titrations performed at a lower [NaCl] (0.02M NaCl (data not shown)) also show stoichiometric binding behavior with quenching values of ~76% for (dT)45 and ~84–85% for (dT)50, (dT)55). The results of the titrations of DrSSB with the different (dT)L suggests that the different behavior observed for DrSSB binding to poly(dT) reflects a real difference in the mode of ssDNA binding, possible reflecting a difference in the mode of ssDNA wrapping and/or inter-dimer protein-protein cooperativity (see Discussion). It is also clear from the isotherm in Fig. 2B that in 1M NaCl (dT)45 interacts with DrSSB much more weakly than at lower [NaCl]. The titration curve in Fig. 2B can be fitted to an n-identical-independent sites model (eq. 1, Materials and Methods) with the following parameters, n=0.98±0.02, Qmax=0.73±0.01, K=(1.48±0.06)×107 M−1. This suggests that the more shallow isotherm observed in the titration of DrSSB with poly(dT) at 1M NaCl (Fig. 1B) reflects a lower affinity due to the increase in NaCl concentration.
DrSSB binds two molecules of (dT)25 with a [NaCl]-dependent negative cooperativity
The EcoSSB tetramer displays intra-tetramer negative cooperativity for binding two molecules of (dT)35 and this negative cooperativity is a strong function of salt concentration (41, 42). Each subunit of DrSSB dimer contains two OB folds and thus we wanted to determine whether a similar negative cooperativity might exist for ssDNA binding to the DrSSB dimer. Assuming that the full occluded site size for ssDNA binding to the DrSSB dimer is ~50 nucleotides we examined the equilibrium binding of two molecules of (dT)25 to the DrSSB dimer. Figure 3A and 3B shows the results of equilibrium titrations of DrSSB with (dT)25, monitoring Trp fluorescence at low (0.02M NaCl) and moderate (0.2M NaCl) salt concentrations (Tris buffer, pH 8.1, 25°C), respectively. From these results it is clear that at both salt concentrations the second molecule of (dT)25 binds with weaker affinity than the first molecule, indicating negative cooperativity. Furthermore, the binding of the first (dT)25 to DrSSB occurs with much stronger affinity at the lower [NaCl]. In order to estimate the binding parameters we used a 2-site sequential binding model (see eq. 2 in Materials and Methods) with two macroscopic binding constants K1,obs and K2,obs for the first and the second site with Q1 and Q2 characterizing the extent of DrSSB Trp quenching for one and two molecules of (dT)25 bound, respectively. For each salt concentration we globally fit two titration curves by constraining Q2 = 0.83 corresponding to the maximum fluorescence quenching observed upon binding poly(dT) and (dT)50. The following parameters were obtained: Q1 = 0.48±0.01, K1 =(4.5±6.2)×109 M−1, K2 =(1.4±0.1)×105 M−1, for 20mM NaCl and Q1 = 0.51±0.01, K1 =(3.1±0.6)×107 M−1, K2 =(2.1±0.3)×105 M−1, for 0.2M NaCl. Interestingly the affinity for binding the first (dT)25 to DrSSB decreases by more than a factor of 100 as the salt concentration increases 10 fold, whereas the binding of the second (dT)25 is essentially unchanged. In general this observed decrease in negative cooperativity with increasing [NaCl] follows the same trend as observed for the EcoSSB tetramer (41, 42).
Calorimetric determination of DrSSB contact site size
The contact site size, m, is defined as a number of contiguous nucleotides required to form all interactions with the protein. This may differ from the occluded site size, n, which is a measure of the number nucleotides that are occluded by one protein molecule upon binding to ssDNA, such that the second protein molecule is unable to access these nucleotides, therefore, m ≤ n. In order to estimate the contact site size for DrSSB binding to ssDNA we performed ITC titrations of DrSSB with the a series of (dT)L (L=30, 35, 40, 45, 50, 55, 60, 70 nucleotides) at 0.2M NaCl (buffer T, pH 8.1, 25°C) to determine how the equilibrium binding constant (Kobs) and the binding enthalpy (ΔHobs) are affected by L. The magnitude of ΔHobs is expected to increase as the number of nucleotides involved in the protein interaction increases until m is reached.
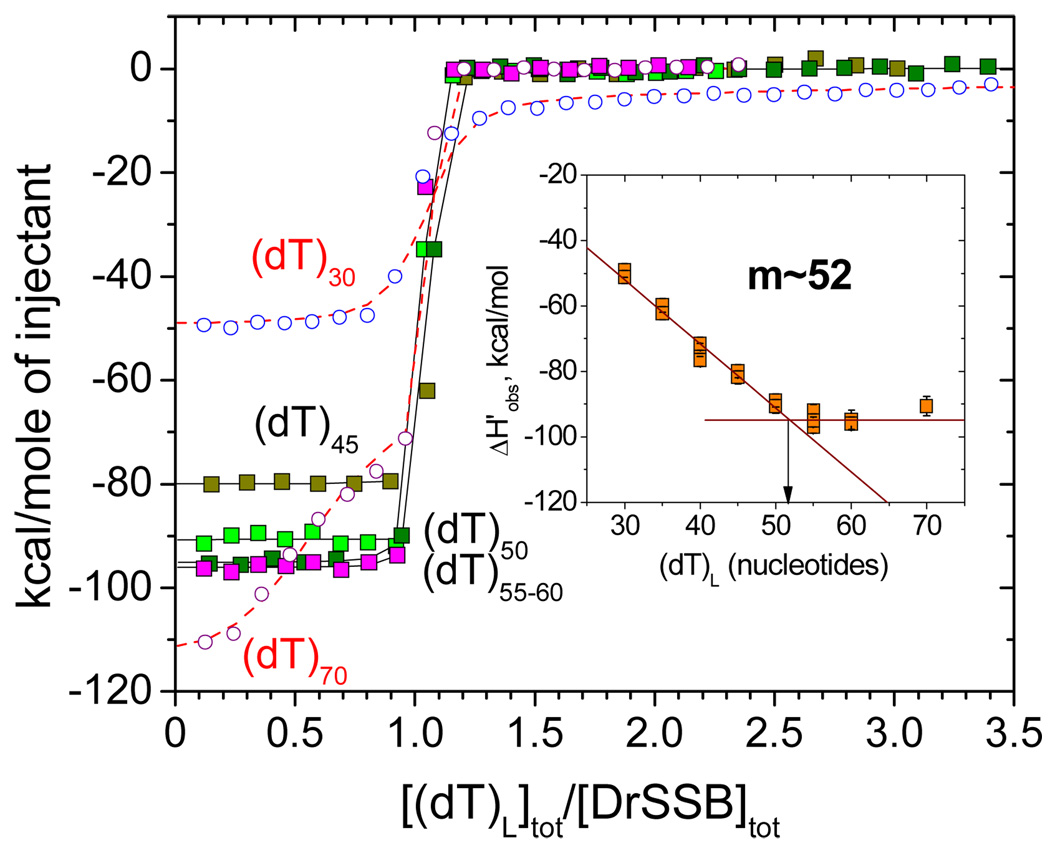
Figure 4 shows a series of representative ITC isotherms for experiments in which (dT)L was titrated into DrSSB (0.5–0.8 µM) in conditions mentioned above. These isotherms indicate that when 40 ≤ L ≤ 60 nucleotides, the DrSSB dimer binds one molecule of (dT)L stoichoiometrically, so that the equilibrium binding constant cannot be determined due to the very high affinity (Kobs>109 M−1). Under these conditions only the binding enthalpy (ΔHobs) can be estimated reliably. When L is less than 35 nucleotides an interaction of a second molecule of (dT)L with DrSSB can be detected, although this binds with much weaker affinity (see Fig. 4). At the same time, weak binding of a second DrSSB dimer to (dT)L becomes detectable when the length of oligo(dT) exceeds the binding site size (e.g., see L ≥ 70 nucleotides in Fig. 4). Although in both cases determination of the binding parameters for the second molecule of (dT)L or DrSSB is not reliable, the estimates of the binding enthalpy for the first molecule (ΔH1,obs) can be obtained accurately. The inset to Figure 4 shows that the values of ΔHobs for the interaction of DrSSB with one molecule of (dT)L are all negative and increase in magnitude linearly with increasing L up to L≈52 nucleotides leveling off at ΔHobs = 94±4 kcal/mol for L >50. These data allow an estimate of m≈52 nucleotides for the contact site size, which is the same as the estimate of the occluded site size obtained from fluorescent experiments with poly(dT) under the same conditions.
Figure 4.
Determination of the contact site size of DrSSB and corresponding binding enthalpy using ITC titrations of DrSSB with (dT)L (L=30,35,40,45, 50 55 60 and 70) in 0.1M NaCl (Buffer T, pH 8.1, 25°C). The insert shows the decrease in binding ehthalpy with increasing of oligonucleotide length until the number of nucleotides corresponding to the contact site size (m≈52) is reached. The binding enthalpy ΔHobs=94±4 kcal/mol was estimated from the plateau value as an average for DrSSB titrations with (dT)55, (dT)60 and (dT)70. In the case of (dT)30 and (dT)70 (open circles and triangles) the shape of the isotherms indicate weak binding the second ssDNA molecule to DrSSB dimer and second DrSSB dimer to ssDNA, respectively. The reliable estimates for the binding of one ssDNA to one DrSSB dimer (K1,obs and ΔH1,obs) can be obtained for either case fitting the data to a two-sequential sites binding model (eq 4 in Materials and Methods). The solid curve through experimental data points represents simulation based on the best fit parameters.
Salt and temperature dependencies of the interaction of DrSSB with (dT)45
Solution conditions and salt concentration and type have significant effects on the magnitude of binding enthalpy (ΔHobs) for the binding of the EcoSSB tetramer to ssDNA (32, 43) as well as on its temperature dependence (ΔCpobs) (44). These effects are linked primarily to the release of anions from EcoSSB upon ssDNA binding (44). In order to investigate the generality of these effects we examined the interaction of DrSSB with ssDNA at different NaCl concentrations and temperatures using ITC.
Fig.5A shows the results of 1:1 binding of DrSSB to (dT)45 at different NaCl concentrations (buffer T, pH 8.1, 25°C). The binding is stoichiometric at 0.02M and 0.2M NaCl, but is weakened at 1 M NaCl. Hence, a determination of both ΔHobs and Kobs is possible at 1 M NaCl, and the estimated value of Kobs=(1.7±0.1)×107 M−1 is in a good agreement with the value determined by monitoring Trp fluorescence (Kobs=(1.5±0.1)×107 M−1, see Fig. 2B). The ΔHobs values are all negative, but exhibit a nonlinear decrease in magnitude as a function of [NaCl] (see inset in Fig. 5A). This overall dependence of ΔHobs on [NaCl] is very similar to that observed for EcoSSB binding to (dT)70 (32) indicating large effect of salt on ΔHobs.
Figure 5.
The effects of salt concentration and temperature on the observed enthalpy change for DrSSB binding to (dT)45 . (A) – ITC titrations of DrSSB with (dT)45 in Tris buffer, pH 8.1 at 0.02 (green), 0.2 (olive) and 1M NaCl (cyan). Solid curves represent the fits to a 1:1 binding model (see eq. 3 in Materials and Methods). The binding is stoichiometric at 0.02 and 0.2M NaCl so only the values of ΔHobs can be determined, although in 1M NaCl all binding parameters (n=0.99±0.01, Kobs=(1.66±0.14)×107 M−1 and of ΔHobs = −64.8±1.4 kcal/mol) can be estimated. The dependence of ΔHobs on [NaCl] is shown in the insert. (B) – the dependence of ΔHobs on temperature obtained within the range from 15 to 30°C in Phosphate buffer, (pH 7.5, 0.2M NaCl). The value of ΔCpobs =0.29±0.07 kcal/mol deg was determined from the linear least square fit of the data.
We next examined the temperature dependence of ΔHobs for DrSSB binding to (dT)45 in 0.2M NaCl (buffer P, pH 7.5) (see Figure 5B). Phosphate buffer was used since it has a very low ionization enthalpy and thus minimizes the potential contributions to ΔHobs from any protonation events that may accompany ssDNA binding as has been observed for EcoSSB (45). Figure 5B shows that ΔHobs becomes more negative with increasing temperature. Linear least square analysis of the data in Figure 5B yields a value of ΔCpobs = −290 ± 70 cal/mol deg which is approximately four times smaller in magnitude than that measured for the EcoSSB-(dT)70 interaction under similar conditions (44)
Cooperativity of DrSSB binding to M13ssDNA probed by agarose gel electrophoresis
Single-stranded binding proteins have been observed to bind long ssDNA cooperatively under some conditions to form extended protein clusters. The T4 gene 32 protein binds ssDNA with high cooperativity independent of solution conditions (7, 8). On the other hand the EcoSSB tetramer only binds ssDNA with high cooperativity in its (SSB)35 binding mode (27, 33, 37–39, 46) which is favored at low monovalent salt concentrations and high protein/DNA ratios. In contrast, the fully wrapped (SSB)65 binding mode which dominates at high salt concentrations displays little cooperativity (33, 37, 46–48). We were therefore interested to determine if DrSSB binds to ssDNA with detectable cooperativity and whether salt concentration influences this cooperativity. In general such information can be described in terms of intrinsic binding constant (K) and a nearest neighbor cooperativity parameter (ω) using the models of protein binding to infinite (34, 49, 50) or finite lattices (51). Unfortunately, under most conditions (up to 0.5 M NaCl) the binding of DrSSB to poly(dT) is stoichiometric so that accurate determination of the affinity (K) and cooperativity (ω) is not possible. Therefore in order to investigate this we used an agarose gel electrophoresis assay as used in studies of the cooperativity of EcoSSB binding to M13ssDNA (33).
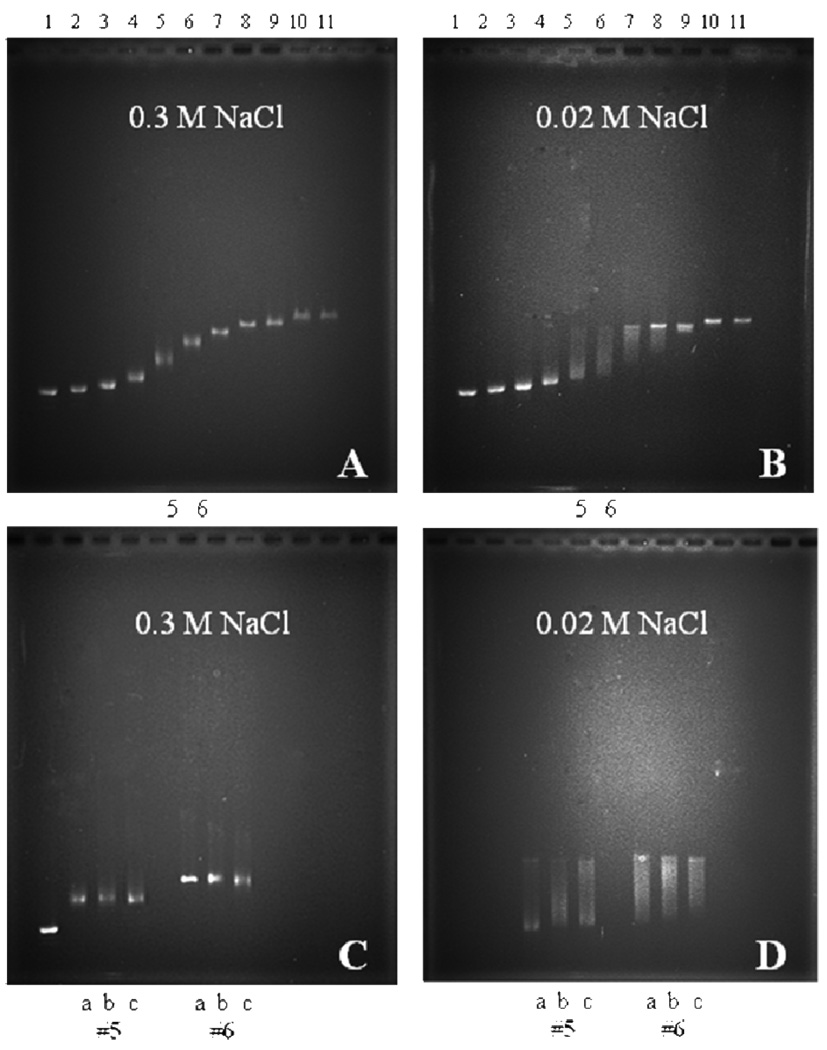
We examined the binding cooperativity of DrSSB to circular M13ssDNA using an agarose gel electrophoresis assay at two different NaCl concentrations (0.02 and 0.3M). The complexes were formed at varying DrSSB to DNA ratios by directly mixing the DrSSB and M13ssDNA in buffer T, pH 8.1, and equilibrating for one hour before electrophoresis (see Materials and Methods). At 0.3 M NaCl (Figure 6A) the ssDNA patterns show a single, somewhat diffuse band at each DrSSB concentration indicating that all ssDNA molecules have close to the same DrSSB binding densitiy consistent with low cooperativity of DrSSB binding to M13ssDNA. However, at 0.02 M NaCl (Figure 6B) the ssDNA pattern shows a bimodal distributions at intermediate saturation levels (lines 5 and 6, in particular, with 20 and 30% saturation of DNA) indicating of high degree of cooperativity. These non-random binding density distributions at low DrSSB to DNA ratios indicate the presence of ssDNA that has high amounts of DrSSB bound in the same population with ssDNA that has much lower amounts of DrSSB bound. At higher ratios of DrSSB to DNA (≥50% saturation, lanes 8–11) only single bands are observed indicating a fairly uniform population of highly saturated DNA molecules.
Figure 6.
Electrophoresis of DrSSB-M13ssDNA complexes (buffer T, pH 8.1, 22°C) shows that at low salt concentration DrSSB forms stable cooperative complexes on ssDNA. The complexes were formed adding DrSSB to single stranded M13mp8 at different protein to ssDNA ratios: R=0 (free DNA) (#1), R=0.025 (#2), R=0.05 (#3), R=0.1 (#4), R=0.2 (#5), R=0.3 (#6), R=0.4 (#7), R=0.5 (#8), R=0.6 (#9), R=0.8 (#10) and R=1.0 (#11). The ratios R=n×(DrSSB(dimer)/nucleotide), were calculated using site size n=45 and n=50 nucleotides per dimer for 0.02M and 0.3M NaCl, respectively. (A) – 0.3M NaCl, gel patterns show only single band at all protein DNA ratios indicative of low cooperative binding; (B) – 0.02 M NaCl, non-random distribution of protein among the DNA molecules (lanes #4–8) is indicative of cooperative binding; (C) – noncooperative complexes in 0.3M NaCl (R=0.2 (#5) and R=0.3 (#6)) after different preincubation times: 18 hours (a), 2 hours (b) and 10 minutes (c); (D) – cooperative complexes formed in 0.02M NaCl (all conditions as in (C)).
Interestingly, at low protein to DNA ratios (20 and 30% of saturation) the DNA molecules with intermediate binding densities slowly redistribute to form fully saturated DNA and free DNA as indicated by the appearance of corresponding more distinct and sharp bands on the gel (see Figure 6D). Similar experiments performed for the same protein to DNA ratios in 0.3M NaCl (Figure 6C) show the same random distributions independent of incubation time. Therefore, it appears that DrSSB is able to bind to M13ssDNA with either very low cooperativity at high salt (0.3M NaCl), but with much higher cooperativity at lower salt concentrations of 0.02M. This behavior is similar to that observed for EcoSSB However it is important to point out that in the case of EcoSSB the highly cooperative complexes formed at low salt are metastable and slowly redistribute to random low cooperative complexes (33), while DrSSB does show the opposite trend, indicative of high stability of its cooperative complexes.
DISCUSSION
The dimeric DrSSB protein has a four OB-fold structural organization (two OB folds per subunit) (23) that is similar to its EcoSSB bacterial analog. Although the latter forms homotetramers with one OB-fold per subunit (9, 10) they both possess four OB-folds that can provide potential sites for interaction with ssDNA. Structural similarities aside, certain differences observed in the crystal structures of these proteins may affect their interaction with ssDNA. First, DrSSB possesses a structural asymmetry arising from sequence differences between its N- and C-terminal OB domains. In fact, the DrSSB polypeptide sequence of the C-terminal OB-fold shares more homology to the OB-fold in EcoSSB as compared with the N-terminal OB-fold (23). This raises the possibility that the affinity of the N- and C- terminal OB-folds of DrSSB for ssDNA can differ. Another observation that originates from the analysis of the crystal structure is that the orientation of the two monomers in DrSSB places its OB-folds in positions different from that seen in the EcoSSB tetrameric structure. In order to fit the EcoSSB arrangement, one of the DrSSB monomers would need to be rotated ≈40° relative to the other (23). Moreover, inspection of the protein-protein interfaces in the crystal structures of DrSSB reveals a large interaction surface linking neighboring dimers through β-hairpin connectors and the N-terminal OB-folds of adjacent molecules (23). The latter interactions have been suggested as potentially responsible for cooperative interactions between DrSSB molecules. However, those interactions are not observed in the EcoSSB crystal structures. Therefore, based on the observed differences in crystal structures, it is possible that the ssDNA binding affinities, cooperativities and modes of ssDNA binding and wrapping may differ for these two proteins and display a different dependence on solution conditions.
DrSSB-ssDNA binding mode transition and intra-dimer negative cooperativity
At moderate and high salt conditions (≥0.2M NaCl) EcoSSB forms an (SSB)65 binding mode (occluded site size ~65 nucleotides) in which ssDNA wraps around all four subunits (OB folds) of the tetramer, whereas at low salt concentrations (≤20mM NaCl) and high protein-DNA binding ratios EcoSSB binds to ssDNA in highly cooperative (SSB)35 binding mode in which on average only two subunits of the tetramer (2 OB folds) interact with ssDNA (5, 27, 33, 35, 36, 38, 46). In this latter mode ~65–70 nucleotides of ssDNA can accommodate two EcoSSB tetramers, which bind with positive cooperativity (27).
We show here that DrSSB does undergo a small change in its occluded ssDNA binding site size as a function of [NaCl], although the change is not as extreme as the ssDNA binding mode transition observed with EcoSSB (27, 35, 36, 46). At moderate and high NaCl concentrations (from 0.1M to 1M) the occluded site size of DrSSB on poy(dT) is 50–53 nucleotides with a Trp fluorescence quenching of ~ 82%, which agrees well with the contact site size (m≈52) determined by ITC (Fig. 4). However, this is significantly smaller than the high [NaCl] induced occluded site size of 65 nucleotides determined for the EcoSSB tetramer (> 0.2 M NaCl). However, it is interesting to note that the EcoSSB tetramer displays a more similar occluded site size of ~56 nucleotides on poly(dT) at intermediate [NaCl] (40 – 100 mM) as well as intermediate [MgCl2] (3 – 60 mM) (36).
At lower NaCl concentrations ((≤20mM), DrSSB displays both a lower extent of Trp fluorescence quenching and lower occluded site size (Qmax≈0.72–0.76 and n≈45) upon binding to poly(dT) as also observed at 1mM NaCl by Witte et al. (52). However, this is still much larger than the low salt site size of ~35 nucleotides observed for EcoSSB (35, 36, 46). Further evidence that DrSSB dimers do not adopt a very small site size mode at lower [NaCl] is that only one DrSSB dimer is observed to bind to oligonucleotides ((dT)L) with lengths of 45–55 nucleotides. However, the observed difference in site size (45 vs. 52 nucleotides) and fluorescence quenching (0.72–0.76 vs. 0.83) for the DrSSB dimers binding to ssDNA may result from either a difference in ssDNA wrapping or a difference in cooperative interactions or both.
The DrSSB binding affinity for ssDNA is strongly dependent on oligodeoxynucleotide length. Under moderate salt concentrations DrSSB binds stoichiometrically to only one molecule of (dT)L if L is in the range from 40 to 60 nucleotides. For these oligonucleotides both the Trp fluorescence quenching and enthalpy change increases (in magnitude) with increasing oligonucleotide length up to n≈50–55 and does not increase further (Qmax≈83% and ΔHobs ≈-94±4 kcal/mole). This can be compared to values of Qmax≈90% (41) and ΔHobs ≈ −130 kcal/mol (44) for EcoSSB binding to (dT)65. We note that the same extent of Trp fluorescence quenching is observed for DrSSB binding to (dT)L at both low and high NaCl concentrations (with a maximum of 83–85% quenching for L >50 nucleotides), whereas a [NaCl]-dependence of Trp quenching is observed for DrSSB binding to poly(dT) (increasing from 72–76% to 84%). This suggests that there are [NaCl] dependent effects on DrSSB binding to poly(dT) that are not observed for oligodeoxynucleotide binding. This could reflect effects on the binding mode and/or cooperativity of DrSSB binding to longer ssDNA.
It is also clear from our data that the binding affinity of ssDNA for DrSSB is weaker than for EcoSSB. This is emphasized by the fact that the binding constant for oligonucleotides with a length close to its site size is measurable for DrSSB at a [NaCl] concentration of 1M, whereas the binding to EcoSSB is still stoichiometric up to 3.0 M NaCl (41). The binding of a second DrSSB dimer (although with much lower affinity) is detectable only on (dT)70 (Fig. 4), an oligo that provides 20 additional nucleotides for interaction with the protein, which is in good agreement with results from previous electrophoretic mobility shift assays (25). On the other hand the DrSSB dimer is able to bind a second molecule of ssDNA if its length is below 35 nucleotides. Furthermore, just as for EcoSSB, the binding of a second molecule of such a shorter oligonucleotide occurs with a significant negative cooperativity that increases with decreasing salt concentration, just as for EcoSSB (41, 42, 53).
DrSSB forms stable highly cooperative complexes on ssDNA at low [NaCl]
Agarose gel electrophoresis have shown that at low salt concentrations (20mM NaCl) EcoSSB can form highly cooperative complexes on ssDNA characterized by a nonrandom distributions of EcoSSB bound to M13ssDNA (33). However, at higher [NaCl] (0.3M) EcoSSB was bound to the ssDNA population in a more random distribution indicative of low or non-cooperative binding. Our experiments with DrSSB performed at the same salt conditions show very similar patterns (Fig.5) indicating that at low salt (20 mM NaCl) DrSSB binds to ssDNA cooperatively, whereas at higher [NaCl], a low cooperativity binding distribution is observed. However, in contrast to the EcoSSB behavior, the DrSSB-ssDNA complexes remain stable and do not redistribute with time to the lower cooperativity complexes. These results contrast with the sedimentation velocity results of Witte et al. (52) who reported that cooperative complexes of DrSSB on poly(dT) are metastable. Analysis of DrSSB crystal structures obtained in different solution conditions reveals a large interaction surface that links neighboring dimers and comprises β-hairpin connectors and N-terminal OB domains of adjacent molecules (23). This interface includes a number of van der Waal’s, hydrogen and ionic interactions and is different from the L45 loop mediated interface that links EcoSSB tetramers (10). Although it is still not clear whether the inter-tetramer interfaces observed in the crystal structures represent the cooperative interactions that exist when bound to ssDNA, these differences raise the possibility that the cooperative complexes formed on ssDNA with EcoSSB vs DrSSB may be distinct with different numbers of OB folds involved in ssDNA wrapping.
Thermodynamics of DrSSB interaction with oligo(dT)
The thermodynamics of EcoSSB binding to ssDNA is quite complex and was studied extensively using ITC (32, 43–45, 54). The binding of EcoSSB tetramer to ssDNA is stoichiometric under most of the conditions and is characterized by an extremely large and negative enthalpy change (up to −160 kcal/mol (32), the largest enthalpy reported so far for protein-DNA interactions) and a large and negative heat capacity change, ΔCp,obs (up to −1.2 kcal/mol °K (44)). This thermodynamics is explained in large measure by multiple contributions from linked equilibrium reactions that accompany SSB-DNA binding. Among these the effects of salt concentration and type (32, 43, 44), protonation (45), conformational transitions within DNA (54) and protein (44) have been quantified.
Using ITC we find that at moderate salt concentrations (0.2M NaCl) DrSSB binds to oligodeoxythymidylates with a contact site size of ~52 nucleotides and is characterized by a large and negative ΔHobs= −94+4 kcal/mole, although both of these are significantly smaller than has been measured for EcoSSB (≈ −130 kcal/mole, n=65) under the same conditions (44). Interestingly, the values of ΔHobs for these two proteins are similar if normalized by their respective contact sizes (−1.8 and −2.0 kcal/mol nucleotide). The dependence of ΔHobs on [NaCl] for DrSSB-(dT)45 binding shows nonlinear behavior (Fig. 5A) with the largest effect at 1M NaCl. This behavior is analogous to that observed for EcoSSB and related to contribution to observed enthalpy change due to anion release from the protein upon interaction with ssDNA (32, 44), which also affects ΔCp,obs (44). Temperature dependence of ΔHobs for DrSSB obtained at 0.2M NaCl (Fig.5B) provides an estimate of heat capacity change ΔCp,obs= −0.29+0.07 kcal mol−1 K−1, which is significantly lower than the value ΔCp,obs≈ −1.3 kcal mol−1 K−1 for EcoSSB-(dT)70 binding under the same conditions (44). A large part of the heat capacity change measured for EcoSSB is due to contributions from protonation (~ −0.5 kcal mol−1 K−1) and the effects of salt (~ -(0.2–0.3) kcal mol−1 K−1). The remaining contribution ~ -(0.5–0.6) kcal mol−1 K−1, that likely reflects changes in accessible surface area upon complexation, but may have contributions from protein conformational changes (44), is still almost twice as large as the total heat capacity change measured here for DrSSB. In the absence of a more detailed investigation, it is not possible to understand the origins of the lower value of ΔCp observed for DrSSB, since this could result from a combination of a number of factors such as differences in accessible surface area, vibrational modes hydration or other coupled equilibria.
ACKNOWLEDGMENT
We thank T. Ho for the synthesis and purification of all oligodeoxynucleotides.
Abbreviations
- drSSB
Deinococcus radiodurans Single Stranded DNA Binding protein
- ssDNA
single stranded DNA
- ITC
isothermal titration calorimetry
- Tris
tris(hydroxymethyl)aminomethane
- EDTA
ethylenediaminetetraacetic acid
Footnotes
This research was supported in part by the NIH (GM30498 to TML and GM32335 to MMC)
REFERENCES
- 1.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 2.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 4.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alberts B, Frey L, Delius H. Isolation and characterization of gene 5 protein of filamentous bacterial viruses. J Mol Biol. 1972;68:139–152. doi: 10.1016/0022-2836(72)90269-0. [DOI] [PubMed] [Google Scholar]
- 8.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 9.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E-coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 11.Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G. The C-terminal domain of full-length E-coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–1947. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 13.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 14.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suck D. Common fold, common function, common origin? Nat Struct Biol. 1997;4:161–165. doi: 10.1038/nsb0397-161. [DOI] [PubMed] [Google Scholar]
- 16.Purnapatre K, Handa P, Venkatesh J, Varshney U. Differential effects of single-stranded DNA binding proteins (SSBs) on uracil DNA glycosylases (UDGs) from Escherichia coli and mycobacteria. Nucleic Acids Res. 1999;27:3487–3492. doi: 10.1093/nar/27.17.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, Varshney U, Vijayan M. Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J Mol Biol. 2003;331:385–393. doi: 10.1016/s0022-2836(03)00729-0. [DOI] [PubMed] [Google Scholar]
- 18.Saikrishnan K, Manjunath GP, Singh P, Jeyakanthan J, Dauter Z, Sekar K, Muniyappa K, Vijayan M. Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr D Biol Crystallogr. 2005;61:1140–1148. doi: 10.1107/S0907444905016896. [DOI] [PubMed] [Google Scholar]
- 19.Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J Mol Biol. 2009;388:508–519. doi: 10.1016/j.jmb.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 20.Dabrowski S, Olszewski M, Piatek R, Brillowska-Dabrowska A, Konopa G, Kur J. Identification and characterization of single-stranded-DNA-binding proteins from Thermus thermophilus and Thermus aquaticus - new arrangement of binding domains. Microbiology. 2002;148:3307–3315. doi: 10.1099/00221287-148-10-3307. [DOI] [PubMed] [Google Scholar]
- 21.Fedorov R, Witte G, Urbanke C, Manstein DJ, Curth U. 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 2006;34:6708–6717. doi: 10.1093/nar/gkl1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eggington JM, Haruta N, Wood EA, Cox MM. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 2004;4:2. doi: 10.1186/1471-2180-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein DA, Eggington JM, Killoran MP, Misic AM, Cox MM, Keck JL. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc Natl Acad Sci U S A. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battista JR. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 25.Eggington JM, Kozlov AG, Cox MM, Lohman TM. Polar destabilization of DNA duplexes with single-stranded overhangs by the Deinococcus radiodurans SSB protein. Biochemistry. 2006;45:14490–14502. doi: 10.1021/bi061178m. [DOI] [PubMed] [Google Scholar]
- 26.Shan Q, Cox MM, Inman RB. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J Biol Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- 27.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 28.Berkowitz SA, Day LA. Molecular weight of single-stranded fd bacteriophage DNA. High speed equilibrium sedimentation and light scattering measurements. Biochemistry. 1974;13:4825–4831. doi: 10.1021/bi00720a022. [DOI] [PubMed] [Google Scholar]
- 29.Ferrari ME, Lohman TM. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–12910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- 30.Lohman TM, Mascotti DP. Nonspecific ligand-DNA equilibrium binding parameters determined by fluorescence methods. Methods Enzymol. 1992;212:424–458. doi: 10.1016/0076-6879(92)12027-n. [DOI] [PubMed] [Google Scholar]
- 31.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 32.Kozlov AG, Lohman TM. Calorimetric studies of E-coli SSB protein single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. J Mol Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 33.Lohman TM, Overman LB, Datta S. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 34.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 35.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 36.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 37.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffith JD, Harris LD, Register J., 3rd Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 39.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem. 1988;263:4629–4640. [PubMed] [Google Scholar]
- 40.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J Mol Biol. 1989;207:249–268. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- 42.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J Mol Biol. 1989;207:269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 43.Lohman TM, Overman LB, Ferrari ME, Kozlov AG. A highly salt-dependent enthalpy change for Escherichia coli SSB protein-nucleic acid binding due to ion-protein interactions. Biochemistry. 1996;35:5272–5279. doi: 10.1021/bi9527606. [DOI] [PubMed] [Google Scholar]
- 44.Kozlov AG, Lohman TM. Effects of monovalent anions on a temperature-dependent heat capacity change for Escherichia coli SSB tetramer binding to single-stranded DNA. Biochemistry. 2006;45:5190–5205. doi: 10.1021/bi052543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozlov AG, Lohman TM. Large contributions of coupled protonation equilibria to the observed enthalpy and heat capacity changes for ssDNA binding to Escherichia coli SSB protein. Proteins Suppl. 2000;4:8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 46.Lohman TM, Bujalowski W. E. coli SSB protein: multiple binding modes and cooperativities. In: Revzin A, editor. The Biology of Nonspecific DNA-Protein Interactions. CRC Press; 1990. pp. 131–170. [Google Scholar]
- 47.Bujalowski W, Lohman TM. Limited co-operativity in protein-nucleic acid interactions. A thermodynamic model for the interactions of Escherichia coli single strand binding protein with single-stranded nucleic acids in the "beaded", (SSB)65 mode. J Mol Biol. 1987;195:897–907. doi: 10.1016/0022-2836(87)90493-1. [DOI] [PubMed] [Google Scholar]
- 48.Overman LB, Bujalowski W, Lohman TM. Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry. 1988;27:456–471. doi: 10.1021/bi00401a067. [DOI] [PubMed] [Google Scholar]
- 49.Kowalczykowski SC, Paul LS, Lonberg N, Newport JW, McSwiggen JA, von Hippel PH. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 50.Bujalowski W, Lohman TM, Anderson CF. On the cooperative binding of large ligands to a one-dimensional homogeneous lattice: the generalized three-state lattice model. Biopolymers. 1989;28:1637–1643. doi: 10.1002/bip.360280912. [DOI] [PubMed] [Google Scholar]
- 51.Epstein IR. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice. A model for ligand-oligonucleotide interactions. Biophys Chem. 1978;8:327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- 52.Witte G, Urbanke C, Curth U. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 2005;33:1662–1670. doi: 10.1093/nar/gki310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohman TM, Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988;27:2260–2265. doi: 10.1021/bi00407a002. [DOI] [PubMed] [Google Scholar]
- 54.Kozlov AG, Lohman TM. Adenine base unstacking dominates the observed enthalpy and heat capacity changes for the Escherichia coli SSB tetramer binding to single-stranded oligoadenylates. Biochemistry. 1999;38:7388–7397. doi: 10.1021/bi990309z. [DOI] [PubMed] [Google Scholar]