Abstract
Alcohol abuse and alcoholism are serious and costly problem in USA. Thus, the development of anti-alcoholism agents could be very significant. The understanding of the neurochemical basis underlying the addictive properties of drugs of abuse is imperative for the development of new pharmacological means to reverse the addictive state, prevent relapse or to reduce the intake of addictive compounds. The nicotinic acetylcholine receptors (nAChRs) are important therapeutic targets for various diseases. Recent studies have revealed that the α3β2, α3β3, and α6 subunits of nAChR protein family might be pharmacological targets for developing new drugs in the treatment of alcoholism. We have performed computational homology modeling of the α3β2, α3β3, and α6 subunits of human nACHRs based upon the recently determined crystal structure of the extracellular domain (ECD) of the mouse nAChR α1 subunit complexed with α-bungarotoxin at 1.94 Å resolution. For comparison, we also built the ECD models of α4β2, and α7 subunits of human nACHRs which are neurochemical targets for cessation of smoking. The three-dimensional (3D) models of the ECD of the monomer, and pentamer of these human nAChR were constructed. The docking of the agonist in the ligand-binding pocket of the human nAChR dimers was also performed. Since the nAChR ligand-binding site is a useful target for mutagenesis studies and the rational design of drugs against various diseases, these models provide useful information for future investigation.
Keywords: homology modeling, nicotinic acetylcholine receptor, molecular modeling, docking, ligand-binding interface, alcoholism
1 Introduction
Alcohol abuse and alcoholism are among the most serious and costly problems of Western society. In the United States, about 10% of the population abuse alcohol. The economic cost is more than $185 billion every year (Gao et al., 2003). Thus, the development of safe and effective anti-alcoholism agents is highly desirable. The understanding of the neurochemical basis underlying the addictive properties of alcohol abuse is imperative for the development of new pharmacological means to reverse the addictive state, prevent relapse or to reduce the intake of addictive compounds. During the last few years new pharmacological strategies, most notably naltrexone and acamprosate (Spanagel and Kiefer, 2008), have been introduced for reducing alcohol consumption and preventing relapse in alcoholic patients.
Accumulating evidence from electrophysiological, pharmacological and neurochemical studies suggest that ethanol may interact with the nAChRs. It has been shown that the ethanol-induced stimulation of the mesolimbic dopamine system and of locomolor activity as well as ethanol intake and preference in rodents may involve central nAChRs. Additionally, data has been presented that nAChRs located in the ventral tegmental area may be of particular importance for these effects of ethanol (Larsson and Engel, 2004).
Recently studies aimed at defining the nAChRs sub-units involved in mediating ethanol-induced locomotor stimulation and accumbal dopamine overflow as well as ethanol intake have revealed that the α3β2, α3β3, and/or α6 subtypes of nAChR protein family could constitute neurochemical targets for developing new drugs in the treatment of alcoholism (Jerlhag et al., 2008; Jerlhag et al., 2006; Narahashi et al., 1999), which arouses our interest to design small molecule agonists of human nAChRs subunits using structure-based design methods. It is highly desirable to find drugs that can selectively interact with different nAChR subtypes. In order to perform structure-based drug discovery for treatment of alcoholism, it is vital important to understand the 3D structures of nAChR α3β2, α3β3 and/or α6 subtypes, particularly their ligand-binding domain. However, so far no crystal structures for human nAChRs are available yet. Lack of information on the nAChR 3D structures has prevented attempts to design nAChR agonists with diverse specificity profiles.
nAChR is a well studied, pharmacologically important member of the Cys-lop superfamily of oentamiric ligand-gated ion channels (Albuquerque et al., 2009). They are composed of five membrane-spanning sub-units arranged around a central pore (Wells, 2008). There are two groups of nAChRs: the muscle type and neuronal type, consisted of a variety of subunits in different combinations. A variety of nAChR subtypes are known to exist, depending on different subunit assemblies (α1-α10, β1-β4, δ, γ and ε) (Alkondon and Albuquerque, 2004). They are composed of a large N-terminal ECD (also called ligand-binding domain, LED), four hydrophobie transmembrane regions (M1-M4), one intracellular domain joining M3 and M4 and a small extracellular C-terminal domain.
The current interest in nAChRs stems from the fact they are important pharmaceutical targets for many human diseases, such as cognitive and attention deficits, Alzheimer’s disease, Parkinsons’s disease, epilepsy, schizophrenia, anxiety, pain management, as well as for cessation of smoking and alcohol drinking (Steinlein and Bertrand, 2008). In order to treat these diseases, it would be helpful to design drugs that can selectively interact with different nAChR subtypes. For this purpose, it is important to have a detailed knowledge of nAChRs ligand binding site.
The first breakthrough in the investigation of the structure of ECD of nAChRs was the elucidation of the X-ray structure of a soluble acerylcholine-binding protein (AChBP) which is a functional homologue of the ECD of nAChRs (Brejc et al., 2001). Since then, several crystal structures of AChBP have been reported, providing structural details of the interaction between the ECD and variety of agonists and antagonists (Celie et al., 2004; Bourne et al., 2005; Hansen et al., 2005; Celie et al., 2005; Unwin, 2005; Ihara et al., 2008). AChBP has the same pentameric assembly as nAChRs and shares ~24% sequence identity with nAChRs. The discovery of AChBP has paved the way to the construction of structural models of the nAChR’s LED using homology modeling (Krieger et al., 2003) and has been extensively used as a model to investigate structural and dynamic features of nAChRs. Models of nAChR subtypes α7 (Schapira et al., 2002; Le Novère et al., 2002; Huang et al., 2008; Mordvitsev et al., 2007; Amiri et al., 2005; Chou, 2004; Bisson et al., 2008), α4β2 (Schapira et al., 2002; Le Novère et al., 2002; Bisson et al., 2008; Huang et al., 2005; Haddadian et al., 2008), α4β4 (Schapira et al., 2002), α3β4 (Costa et al., 2003), α3β2 (Hu and Southerland, 2007; Schapira et al., 2002), α9α10 (Pérez et al., 2009), and (α1)2(β1)γδ (Le Novère et al., 2002; Mordvitsev et al., 2007; Mordvintsev et al., 2005) have been reported.
A further breakthrough in the structure studies of the nAChRs was the recent crystal structure determination of the entire N-terminal extracellular domain of the mouse nAChR a1 subunit bound to α-bungarotoxin at 1.94 Å resolution (Dellisanti et al., 2007). Since this structure provides the first high-solution view of the ECD of nAChRs and is better than AChBP to be used to model the ECD of the human nAChR subunits due to the high degree of homology between them, it is a good template for the modeling of the ECD of the human nAChRs. Model of the ECD of the human α7 nAChR based on the 3D structure of the mouse α1 nAChR ECD has been reported (Konstantakaki et al., 2008).
Using the crystal structure of the mouse α1 nAChR ECD as a template, we have built both monomer and pentamer models of the LED of human nAChR α3β2, α4β2, α3β3, α6, and α7 subunits by computational homology modeling. The resulting models provide molecular targets for structure-based design of subtype-specific nAChR agonists. Computational docking study also was carried out to gain understanding on the interactions between nicotine and nAChR.
2 Methods
2.1 Preparation of the Sequence and Template
The amino acid sequences of the human nAChR α3 (hA3), α4 (hA4), α6 (hA6), α7 (hA7), β2 (hB2), and β3 (hB3) were obtained from the National Center for Biotechnology Institute (NCBI). The Uniprot accession number for hA3, hA4, hA6, hA7, hB2, and hB3, are P32297, P43681, Q15825, P36544, P17787, and Q05901, respectively. The sequences were first edited to remove the signal peptide segments. All subsequent amino acid numbering is based on the mature sequences without the signal peptide. The sequences were then edited to remove all of the residues beyond LED. The X-ray structure of the ECD of the α1 subunit of the mouse nAChR (Dellisanti et al., 2007), the first X-ray structure obtained of a region of the nAChR (PDB: 2QC1), was used as the template and was edited so that it contained only the chain B, and hereafter it was referred to as 2QC1B.
2.2 Sequence Alignments
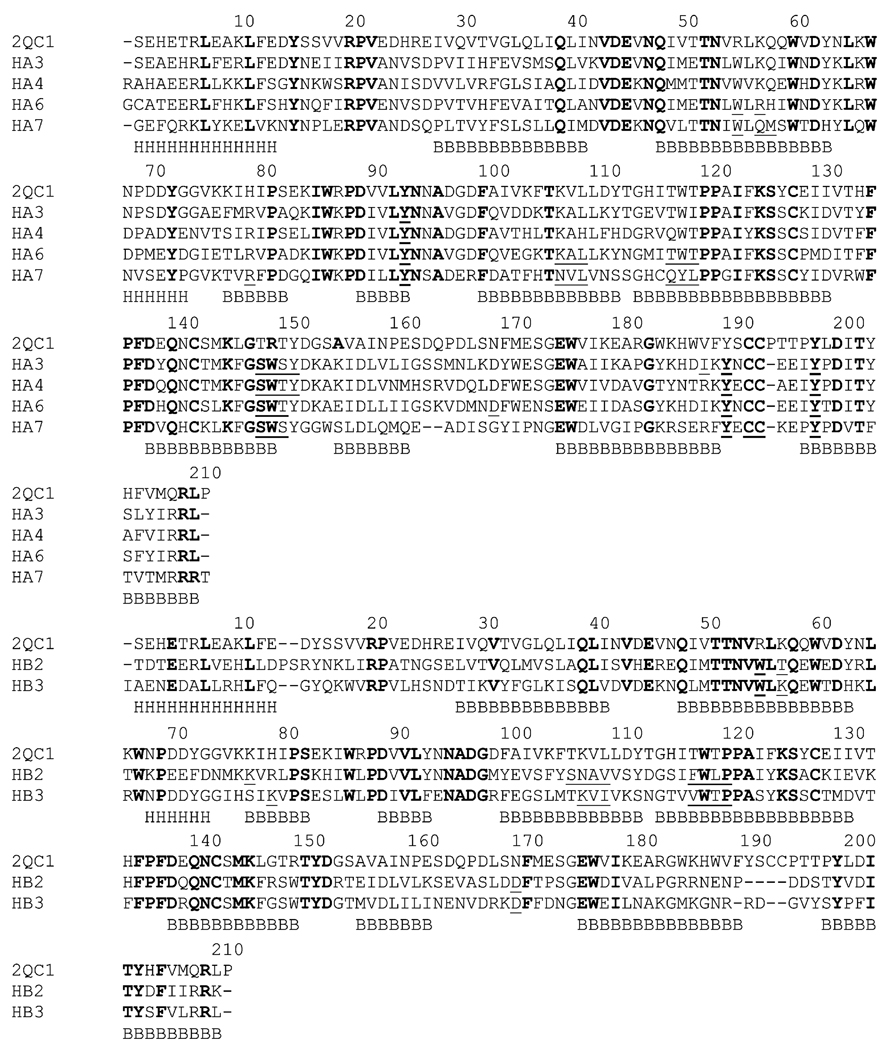
The edited sequences of six human nAChR momomers (hA3, hA4, hA6, hA7, hB2, and hB3) were first aligned to the ECD of the mouse nAChR α1 subunit template respectively using the BLAST server at NCBI (http://blast.ncbi.nlm.nih.gov/). They were further refined by aligning these six sequences with the structure of 2QC1B based on a dynamic programming algorithm present within the MODELLER software package (Sali and Blundell, 1993), which is different from standard sequence-sequence alignment methods because it takes into account structural information from the template when constructing an alignment. This task is achieved through a variable gap penalty function that tends to place gaps outside secondary structure segments (Fig. 1). This improvement becomes more important as the similarity between the sequences decreases and the number of gaps increases.
Fig. 1.
Multiple sequence alignment of the ECDs of the mouse α1 with human nAChR monomers. The alignment was divided into subunit 1 monomers (top, α3, α4, α6, and α7) and subunit 2 monomers (bottom, β2, and β3). Identical residues are shown in bold, while residues involved in ligand binding are shown in underline. Secondary structure elements are shown under the sequences: H= a-helix, B=b-strand.
2.3 Homology Modeling
Based on the sequence alignment with the ECD of the α1 mouse subunit, the three-dimensional model of the LBD of six human nAChR monomers were built using the program MODELLER (Sali and Blundell, 1993). The MODELLER’s automodel command was invoked by a script (model-single.py) to automatically assign atomic coordinates to regions structurally aligned with the template, build intervening loops, optimize the rotamers of amino acid side chains, and perform an initial energy optimization of the structure, using the 2QC1B template structure and the alignment obtained in the previous sequence alignment stage (file: model-tem.ali). For each monomer, 5 models were generated, and the model with the lowest value of the MODELLER objective function was selected for further refinement.
The pentameric structures of the ECD of five human nAChR subunits, α3β2, α4β2, α3β3, α6, and α7, were modeled using the X-ray structure of the AChBP pentamer (PDB: 1I9B) (Brejc et al., 2001) as the template. The 3D structures were derived using Chimera program (Meng et al., 2006) to superimpose corresponding human nAChR monomers to the A-, B-, C-, D, and E-chains of 1I9B consecutively.
The models generated were subjected to an overall energy minimization with respect to all atoms to finalize the entire pentamer structure using CHARMM program (Miller et al., 2008; Kim et al., 2008; Brooks et al., 1983). The energy calculation and minimization was performed with GBMV (Generalized Born using Molecular Volume) implicit solvation model. This model mimics the Poisson-Boltzmann (PB) electrostatic solvation energy calculation using a generalized Born method, which allows the computation of solvation energies very similar to the PB equations. The PB method has been considered a benchmark for implicit solvation calculations (Lee et al., 2003). The resulting models were used for next molecular docking and virtual screening step without any further refinement.
2.4 Docking of nicotine with the homodimer
The docking of nicotine was performed with the program WinDock developed in our laboratory (Hu and Southerland, 2007), which uses the widely distributed DOCK searching engine (Ewing et al., 2001; Moustakas et al., 2006) to dock flexible small molecules to macomolecular sites, and to evaluate the binding affinity of ligands. WinDock’s SPHBOC module was used to determine the binding site and produce a set of spheres for binding site characterization. Contact scores and energy scores were calculated using an energy cutoff distance of 6.0 Å and a van der Waals repulsive exponent of 8.0 Å. Ligands were oriented to the spheres with a distance tolerance of 0.5 Å and distance minimum of 2.0 Å. A minimum anchor size of 50 was used with an internal energy repulsive exponent of 8.0 Å and clash overlap of 0.25 Å. All other parameters were left as their defaults.
3 Results
3.1 Sequence Alignments
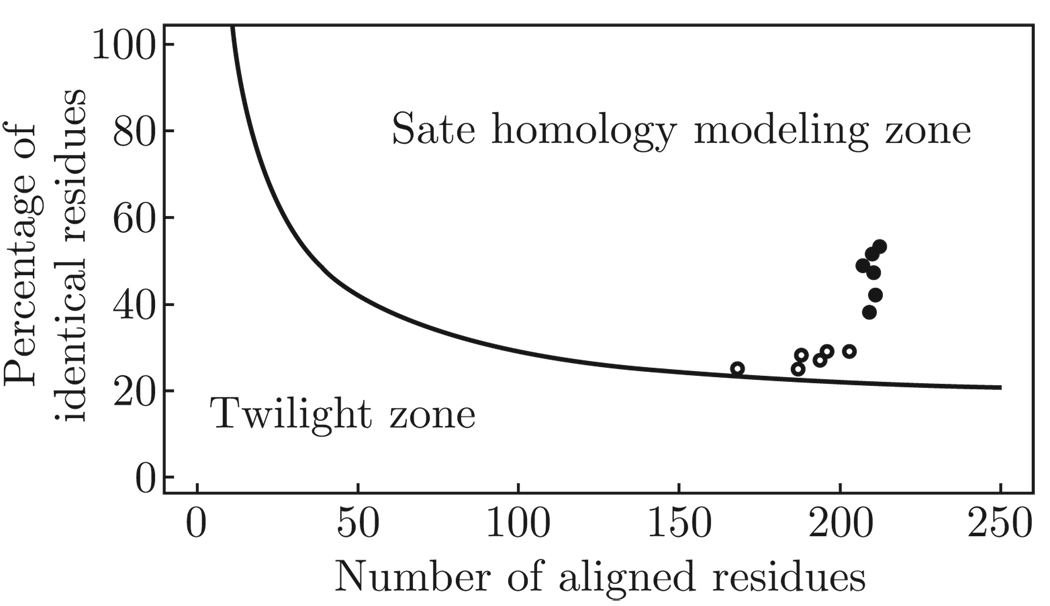
Since the structure of a protein is uniquely determined by its sequence and similar sequence fold into similar structures, it is possible to obtain the structure of a protein by sequence alignment of its sequence with knowing protein structure (Krieger et al., 2003; Rost, 1999), as long as the length of two sequences and the percentage of identical residues fall in the region marked as “safe” in Fig. 2. Protein sequence alignments thus unambiguously distinguish between protein pairs of similar and non-similar structure (Rost, 1999). Based on the sequence alignment, the structure of the ECD of the mouse nAChR α1 subunit is better than AChBP to be used to model the ECD of the human nAChR subunits due to the high degree of homology between them (Table 1 and Fig. 2), which is about 50% higher than the degree of identity between the ECDs of the human nAChR subunits and AChBP.
Fig. 2.
The two zones of protein sequence alignments. Two sequences are practically guaranteed to adopt a similar structure if their length and percentage sequence identity fall into the region marked as “safe”. A heavy black dot represents the degree of homology between the ECDs of the mouse α 1 and human nAChR monomers; a cycle represents the degree of homology between the ECDs of AChBP and human nAChR monomers.
Table 1.
The degree of residue identity between the ECDs of the mouse α 1 nAChR subunit and the human nAChR monomers (identity of residues with AchBP is included for comparison)
| 2QC1B | 2BR7 | |||
|---|---|---|---|---|
| TYPE | ||||
| Identities | Gaps | Identities | Gaps | |
| α3 | 108/210 (51%) | 1/210 (0%) | 54/188 (28%) | 4/188 (2%) |
| α4 | 110/211 (52%) | 1/211 (0%) | 59/203 (29%) | 6/203 (2%) |
| α6 | 102/207 (49%) | 1/207 (0%) | 57/196 (29%) | 6/196 (3%) |
| α7 | 80/209 (38%) | 3/209 (1%) | 54/194 (27%) | 14/194 (7%) |
| β2 | 89/211 (42%) | 6/211 (2%) | 42/168 (25%) | 8/168 (4%) |
| β3 | 99/210 (47%) | 3/210 (1%) | 47/187 (25%) | 6/187 (3%) |
3.2 Human nAChR monomers
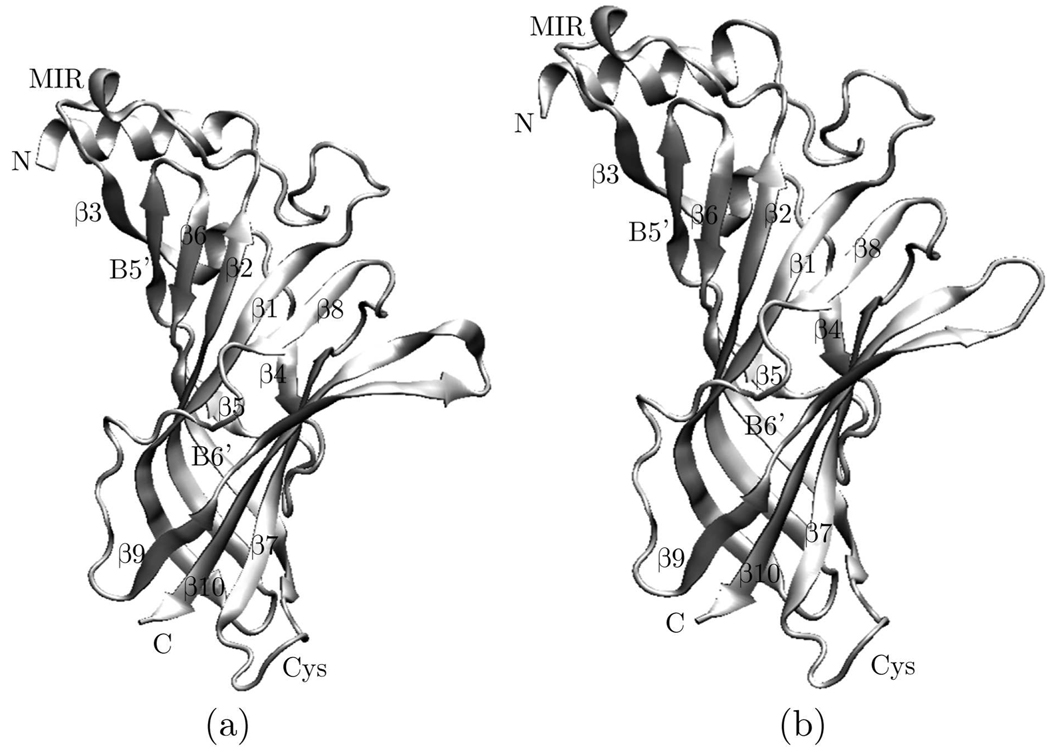
The overall 3D structural model of the ECD for human nAChR monomers were given in Fig. 3. As expected from the low number of insertions/deletions, the model of the ECD of the human nAChR monomers does not differ largely from that of the template. It consists of an N-terminal α-helix followed by ten strands that form a β-sandwich. The inner sheet is made of strands β1, β2, β3, β6, and β8, whereas the outer sheet is made of strands β4, β7, β9, and β10. A disulphide bond is formed between Cys128 on the inner sheet and Cys142 on the outer sheet, liking the two sheets together. Investigation has shown that loops β4-β5 (A), β7-β8 (B), and β9-β10 (C) serves as the principle ligand-binding elements (Brejc et al., 2001; Sin and Engel, 2006; Unwin, 2005), while loops β1-β2, β6-β7 (Cys loop), and β8-β9, three membrane-facing loops, has the important role in the interaction between the ECD and the transmembebrane domain during ligand-induced gating (Dellisanti et al., 2007).
Fig. 3.
Model of the ECD of the human nAChR monomer based on the sequence alignment shown in Fig. 1 and subsequent homology modeling. (a) α3 monomer, (b) β2 monomer.
The 13-residue Cys loop, which is highly conserved in the nAChRs, adopts a similar structure to that of the mouse α1 ECD as a type VIb turns. Phe135, located the tip of the Cys-loop, has the structural role to maintain this unique conformation and has been shown to be a key residue for the function of nAChR (Dellisanti et al., 2007).
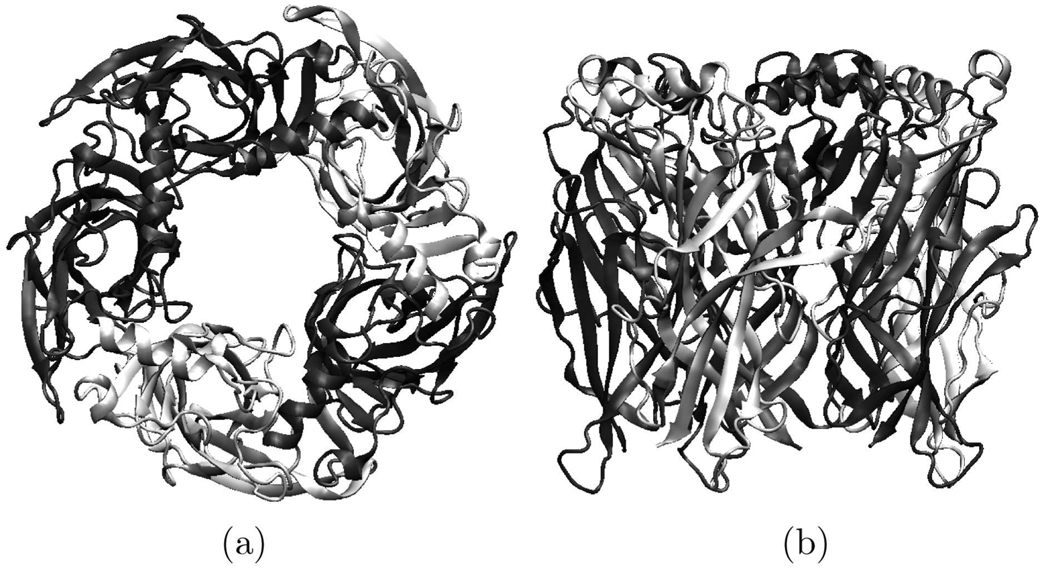
3.3 Human nAChR pentamers
Like the AChBP pentamer, the resulting 3D structural model of the ECD of human nAChR shows fivefold symmetry when viewed along the axis (Fig. 4). It was a barrel of 80 ´Å diameter and 63 ´Å height with a central irregular pore (10–15 ´Å).
Fig. 4.
Model of the ECD of the human nAChR pentamer. (a) side view and (b) top view.
The comparison of the structure of the mouse α1 ECD with that of AChBP and the electron microscopic model of the Torpedo nAChR pentamer indicated that monomeric state of the mouse α1 ECD crystal structure does not significantly affects its structure, especially in regions that are expected to interact with the adjacent subunits (Dellisanti et al., 2007). Therefore, the modeled structures of the ECD of human nAChR pentamer could be used to investigate the ligand binding.
3.4 The ligand binding site
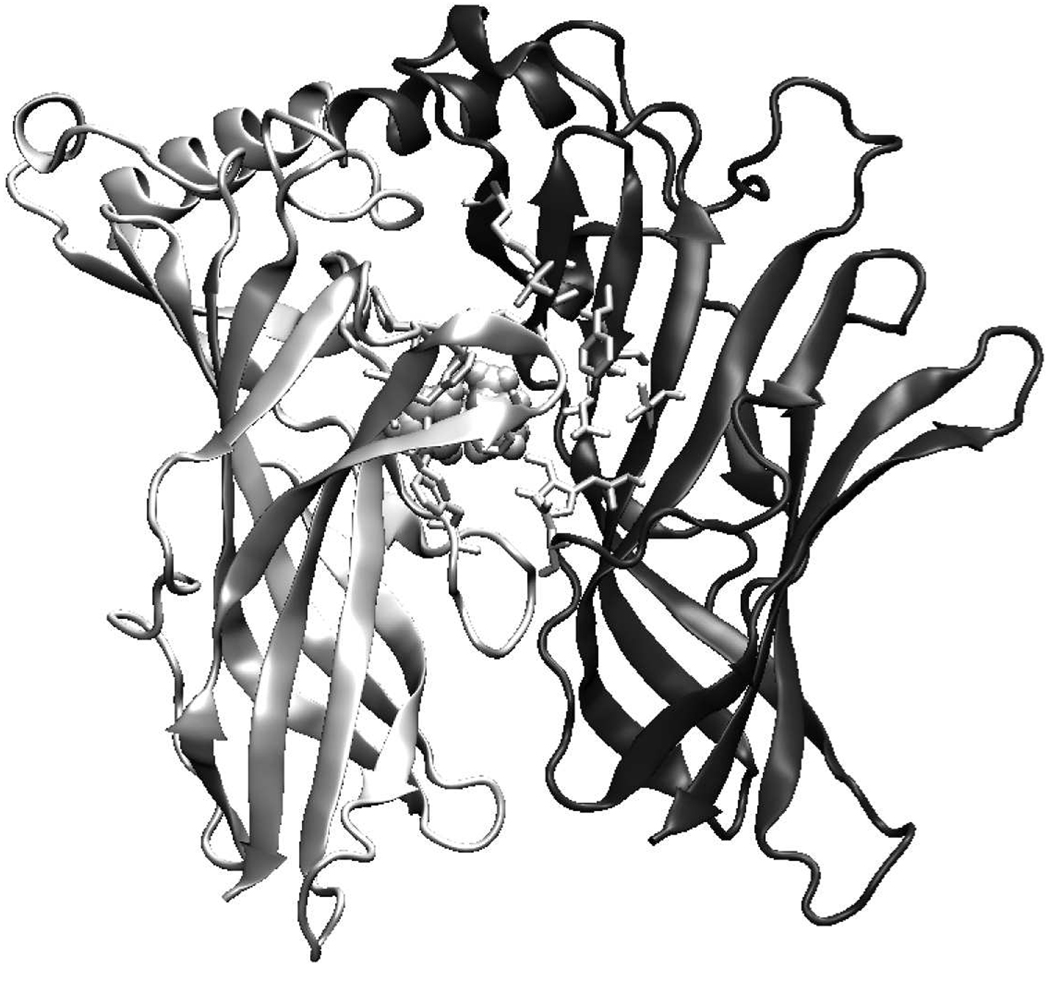
The model of the ligand-binding pocket of the human α3 β2 nAChR dimer and its complex with nicotine is shown in Fig. 5. The dimmer interface is formed by an interlocking array of neighboring chain secondary structure elements. The residues involved in the dimmer interface are listed in Table 2. The interface consists of 18 residues (16 for α6 dimer), of which 8 or 7 are from chain A and rest from chain B.
Fig. 5.
Ligand-binding site of the human α3β2 nAChR dimer model with nicotine bound (shown in sphere). The principal component (α3) is shown in light grey and the complementary component (β2) in dark grey. All key residues discussed in the text are shown in stick.
Table 2.
Residues involved in the ligand binding at human nAChR dimer interface
| Chain | α3β2 | α3β3 | α4β2 | α6α6 | α7α7 |
|---|---|---|---|---|---|
| A | TYR93 | TYR93 | TYR98 | TYR98 | TYR93 |
| SER148 | SER148 | SER153 | SER153 | SER148 | |
| TRP149 | TRP149 | TRP154 | TRP154 | TRP149 | |
| SER150 | SER150 | THR155 | THR155 | SER150 | |
| TYR151 | TYR156 | ||||
| ILE188 | ILE188 | ILE193 | |||
| TYR190 | TYR190 | TYR195 | TYR195 | TYR188 | |
| CYS190 | |||||
| CYS191 | |||||
| TYR197 | TYR197 | TYR202 | TYR202 | TYR195 | |
| B | TRP57 | TRP56 | TRP57 | TRP60 | TRP55 |
| THR59 | LYS58 | THR59 | ARG62 | GLN57 | |
| MET58 | |||||
| LYS79 | |||||
| LYS80 | |||||
| ARG79 | |||||
| SER108 | |||||
| ASN109 | LYS108 | ASN109 | LYS112 | ASN107 | |
| ALA110 | VAL109 | ALA110 | ALA113 | VAL108 | |
| VAL111 | ILE110 | VAL111 | LEU114 | LEU109 | |
| PHE119 | VAL118 | PEH119 | THR122 | GLN117 | |
| TRP120 | TRP119 | TRP120 | TRP123 | TYR118 | |
| LEU121 | THR120 | LEU121 | THR124 | LEU119 | |
| PRO121 | PRO122 | ||||
| ASP171 | ASP170 | ASP171 | ASP174 |
The ligand-binding pockets of the human nAChRs are formed by loops A, B, and C of the principal component (chain A) and loops D and E of the complementary component (chain B) of the adjacent subunit. The key residues of the loops involved in the formation of the ligand-binding site are Tyr93 from loop A, Ser 148 and Trp149 from loop B, and Tyr190 and Tyr195 from loop C (all these residues belong to chain A and use mouse α1 numbering), and Trp57 from loop D (residues from chain B).
Identification of all residues involved in the ligand binding (e.g. agonists, competitive antagonists, and noncompetitive agonists) is a primary objective to understand which strctureal components are related to the physiological function of nAChR. The position of the ligand in the current models is in good agreement with the results of biochemical experiments performed on Torpedo nAChR. According to the mutagenesis experiments, residue Trp149 is able to establish a π-cation interaction with the ammonium group of acetylcholine (Zhong et al., 1998). Moreover, photolabeling experiments have shown that residues Tyr190 and Tyr198 in α subunit were the principal amino acids labeled by [3H]nicotine (Middleton and Cohen, 1991), residue Trp55 was the primary site of [3H]nicotine photoincorporation within a non-α-subunit (Chiara et al., 1998), and residues Tyr190 and Tye198 involved primarily in the interaction with the ester moiety of acetylcholine (Grutter et al., 2000). In addition, residue Tyr93 within α subunit was identified as contributing to the cation-binding domain of the AChR agonist-binding site (Cohen et al., 1991). All these residues mentioned are key residues in the ligand-binding pockets of our ECD models of the human nAChR α3β2, α3β3, α4β2, α6, and α7 subunits (Table 2). Furthermore, the ligand-binding site of our human nAChR α7 model is formed essentially by the same residues as those in the chicken α7 nAChR modeled previously (Le Novère et al., 2002). The key residues involved in the formation of the ACh-binding site are also involved in the formation of nicotine-binding site: Tyr93, Trp149, Tyr151, Tyr188, Cys190 and Tyr195 from chain A, and Trp55, Leu109, Gln117, and Leu119 from chain B.
Investigations have indicated that the major role of α subunits of nAChRs in the channel gating process is proving the principle binding surface (the plus side) (Brejc et al., 2001; Sin and Engel, 2006; Unwin, 2005). It could be shown (Fig. 1 and Table 2) that residues from the principle subunit involved in ligand binding are generally conserved (the residues in bold in Table 2), whereas the residues in the complementary part (minus side) of the binding site shown more variation. Previous studies have indicated that the β subunits confer agonist selectivity to the nAChRs (Cohen et al., 1995; Luetje and Patrick, 1991; Parker et al., 1998; 2001), it is therefore possible to design nAChR subtype-specific drugs according to the difference between ligand binding sites of human nAChRs.
4 Conclusion
Alcoholism and alcohol abuse are one of the most prevalent neuropsychiatric diseases and have an enormous health and socioeconomic impact. The human nAChR α3β2, α3β3, and α6 subtypes have been shown to be neurochemical targets for developing new drugs in the treatment of alcoholism. To discover new drugs using structure-based method, it is important to find the 3D structures of these human nAChR subtypes. Based on the crystal structure of the ECD of the mouse α1 nAChR, the ECD models of the human nAChR α3β2, α3β3, α4β2, α6, and α7 subunits was constructed using comparative modeling. The 3D models of the ECD of the monomer, and pentamer of these human nAChRs were constructed. The docking of the agonist nicotine in the ligand-binding pocket of the human nAChR dimers was also performed. Since the nAChR ligand-binding site is a useful target for mutagenesis studies and the rational design of drugs that can selectively activate different human nAChR subtypes against various diseases, these models provide structural frames for future investigation.
Acknowledgments
This work is supported by grant RCMI-NIH 2G12RR03048.
References
- 1.Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkondon M, Albuquerque EX. The nicotinic acetylcholine receptor subtypes and their function in the hippocampus and cerebral cortex. Prog Brain Res. 2004;145:109–120. doi: 10.1016/S0079-6123(03)45007-3. [DOI] [PubMed] [Google Scholar]
- 3.Amiri S, Tai K, Beckstein O, Biggin PC, Sansom MS. The α7 nicotinic acetylcholine receptor: molecular modelling, electrostatics, and energetics. Mol Membr Biol. 2005;22:151–162. doi: 10.1080/09687860500063340. [DOI] [PubMed] [Google Scholar]
- 4.Bisson WH, Westera G, Schubiger PA, Scapozza L. Homology modeling and dynamics of the extracellular domain of rat and human neuronal nicotinic acetylcholine receptor subtypes alpha4beta2 and alpha7. J Mol Model. 2008;14:891–899. doi: 10.1007/s00894-008-0340-x. [DOI] [PubMed] [Google Scholar]
- 5.Bourne Y, Talley TT, Hansen SB, Taylor P, Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 7.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 8.Celie PH, Kasheverov IE, Mordvintsev DY, Hogg RC, van Nierop P, van Elk R, van Rossum-Fikkert SE, Zhmak MN, Bertrand D, Tsetlin V, Sixma TK, Smit AB. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat Struct Mol Biol. 2005;12:582–588. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- 9.Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 10.Chiara DC, Middleton RE, Cohen JB. Identification of tryptophan 55 as the primary site of [3H]nicotine photoincorporation in the gamma-subunit of the Torpedo nicotinic acetylcholine receptor. FEBS Lett. 1998;423:223–226. doi: 10.1016/s0014-5793(98)00093-3. [DOI] [PubMed] [Google Scholar]
- 11.Chou KC. Insights from modelling the 3D structure of the extracellular domain of α7 nicotinic acetylcholine receptor. Biochem Biophys Res Commun. 2004;319:433–438. doi: 10.1016/j.bbrc.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Cohen BN, Figl A, Quick MW, Labarca C, Davidson N, Lester HA. Regions of beta 2 and beta 4 responsible for differences between the steady state dose-response relationships of the alpha 3 beta 2 and alpha 3 beta 4 neuronal nicotinic receptors. J Gen Physiol. 1995;105:745–764. doi: 10.1085/jgp.105.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen JB, Sharp SD, Liu WS. Structure of the agonist-binding site of the nicotinic acetylcholine receptor. [3H] acetylcholine mustard identifies residues in the cation-binding subsite. J Biol Chem. 1991;266:23354–23364. [PubMed] [Google Scholar]
- 14.Costa V, Nistri A, Cavalli A, Carloni P. A structural model of agonist binding to the alpha3beta4 neuronal nicotinic receptor. Br J Pharmacol. 2003;140:921–931. doi: 10.1038/sj.bjp.0705498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 ? resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 16.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 17.Gao GY, Li DJ, Keung WM. Synthesis of daidzin analogues as potential agents for alcohol abuse. Bioorg Med Chem. 2003;11:4069–4081. doi: 10.1016/s0968-0896(03)00397-3. [DOI] [PubMed] [Google Scholar]
- 18.Grutter T, Ehret-Sabatier L, Kotzyba-Hibert F, Goeldner M. Photoaffinity labeling of Torpedo nicotinic receptor with the agonist [3H]DCTA: identification of amino acid residues which contribute to the binding of the ester moiety of acetylcholine. Biochemistry. 2000;39:3034–3043. doi: 10.1021/bi992393o. [DOI] [PubMed] [Google Scholar]
- 19.Haddadian EJ, Cheng MH, Coalson RD, Xu Y, Tang P. In silico models for the human alpha4beta2 nicotinic acetylcholine receptor. J Phys Chem B. 2008;112:13981–13990. doi: 10.1021/jp804868s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–3646. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Z, Southerland W. WinDock: structure-based drug discovery on Windows-based PCs. J Comput Chem. 2007;28:2347–2351. doi: 10.1002/jcc.20756. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Zheng F, Crooks PA, Dwoskin LP, Zhan CG. Modeling multiple species of nicotine and deschloroepibatidine interacting with alpha4beta2 nicotinic acetylcholine receptor: from microscopic binding to phenomenological binding affinity. J Am Chem Soc. 2005;127:14401–14414. doi: 10.1021/ja052681+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Zheng F, Stokes C, Papke RL, Zhan CG. Modeling binding modes of alpha7 nicotinic acetylcholine receptor with ligands: the roles of Glnll7 and other residues of the receptor in agonist binding. J Med Chem. 2008;51:6293–6302. doi: 10.1021/jm800607u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ihara M, Okajima T, Yamashita A, Oda T, Hirata K, Nishiwaki H, Morimoto T, Akamatsu M, Ashikawa Y, Kuroda S, Mega R, Kuramitsu S, Sattelle DB, Matsuda K. Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin. Invert Neurosci. 2008;8:71–81. doi: 10.1007/s10158-008-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerlhag E, Egecioglu E, Dickson SL, Svensson L, Engel JA. Alpha-conotoxin MII-sensitive nicotinic acetylcholine receptors are involved in mediating the ghrelin-induced locomotor stimulation and dopamine overflow in nucleus accumbens. Eur Neuropsychopharmacol. 2008;18:508–518. doi: 10.1016/j.euroneuro.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Jerlhag E, Grøtli M, Luthman K, Svensson L, Engel JA. Role of the subunit composition of central nicotinic acetylcholine receptors for the stimulatory and dopamine-enhancing effects of ethanol. Alcohol. 2006;41:486–493. doi: 10.1093/alcalc/agl049. [DOI] [PubMed] [Google Scholar]
- 27.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 28.Konstantakaki M, Tzartos SJ, Poulas K, Eliopoulos E. Model of the extracellular domain of the human alpha7 nAChR based on the crystal structure of the mouse alpha1 nAChR extracellular domain. J Mol Graph Model. 2008;26:1333–1337. doi: 10.1016/j.jmgm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Krieger E, Nabuurs SB, Vriend G. Homology modeling. In: Bourne PE, Weissig H, editors. Structural bioinformatics. New Jersey: Wiley-Liss, Hoboken; 2003. pp. 509–524. [Google Scholar]
- 30.Larsson A, Engel JA. Neurochemical and behavioral studies on ethanol and nicotine interactions. Neurosci Biobehav Rev. 2004;27:713–720. doi: 10.1016/j.neubiorev.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Le Novère N, Grutter T, Changeux JP. Models of the extracellular domain of the nicotinic receptors and of agonist- and Ca2+ binding sites. Proc Natl Acad Sci USA. 2002;99:3210–3215. doi: 10.1073/pnas.042699699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MS, Feig M, Salsbury FR, Jr, Brooks CL., 3rd New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. J Comput Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 33.Luetje CW, Patrick J. Both alpha- and beta-subunits contribute to the agonist sensitivity of neuronal nicotinic acetylcholine receptors. J Neurosci. 1991;11:837–845. doi: 10.1523/JNEUROSCI.11-03-00837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339–349. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 36.Miller BT, Singh RP, Klauda JB, Hodoscek M, Brooks BR, Woodcock HL., 3rd CHARMMing: a new, flexible web portal for CHARMM. J Chem Inf Model. 2008;48:1920–1929. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mordvintsev DY, Polyak YL, Levtsova OV, Tourleigh YV, Kasheverov IE, Shaitan KV, Utkin YN, Tsetlin VI. A model for short alpha-neurotoxin bound to nicotinic acetylcholine receptor from Torpedo californica: Comparison with long-chain alpha-neurotoxins and alpha-conotoxins. Comput Biol Chem. 2005;29:398–411. doi: 10.1016/j.compbiolchem.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Mordvitsev DY, Polyak YL, Kuzmin DA, Levtsova OV, Tourleigh YV, Utkin YN, Shaitan KV, Tsetlin VI. Computer modeling of binding of diverse weak toxins to nicotinic acetylcholine receptors. Comput Biol Chem. 2007;31:72–81. doi: 10.1016/j.compbiolchem.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. Development and validation of a modular, extensible docking program: DOCK 5. J Comput Aided Mol Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 40.Narahashi T, Aistrup GL, Marszalec W, Nagata K. Neuronal nicotinic acetylcholine receptors: a new target site of ethanol. Neurochem Int. 1999;35:131–141. doi: 10.1016/s0197-0186(99)00055-8. [DOI] [PubMed] [Google Scholar]
- 41.Parker MJ, Beck A, Luetje CW. Neuronal nicotinic receptor beta2 and beta4 subunits confer large differences in agonist binding affinity. Mol Pharmacol. 1998;54:1132–1139. [PubMed] [Google Scholar]
- 42.Parker MJ, Harvey SC, Luetje CW. Determinants of agonist binding affinity on neuronal nicotinic receptor beta subunits. J Pharmacol Exp Ther. 2001;299:385–391. [PubMed] [Google Scholar]
- 43.Pérez EG, Cassels BK, Zapata-Torres G. Molecular modeling of the alpha9alpha10 nicotinic acetylcholine receptor subtype. Bioorg Med Chem Lett. 2009;19:251–254. doi: 10.1016/j.bmcl.2008.10.094. [DOI] [PubMed] [Google Scholar]
- 44.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 45.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 46.Schapira M, Abagyan R, Totrov M. Structural model of nicotinic acetylcholine receptor isotypes bound to acetylcholine and nicotine. BMC Struct Biol. 2002;2:1–8. doi: 10.1186/1472-6807-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 48.Spanagel R, Kiefer F. Drugs for relapse prevention of alcoholism: ten years of progress. Trends Pharmacol Sci. 2008;29:109–115. doi: 10.1016/j.tips.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Steinlein OK, Bertrand D. Neuronal nicotinic acetylcholine receptors: from the genetic analysis to neurological diseases. Biochem Pharmacol. 2008;76:1175–1183. doi: 10.1016/j.bcp.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 51.Wells GB. Structural answers and persistent questions about how nicotinic receptors work. Front Biosci. 2008;13:5479–5510. doi: 10.2741/3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong W, Gallivan JP, Zhang Y, Li L, Lester HA, Dougherty DA. From ab initio quantum mechanics to molecular neurobiology: a cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]