Abstract
A matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) based approach was developed for the rapid analyses of cellular glycerophospholipids. Through multiplexed solvent-enabled optimization of analyte-matrix interactions during the crystallization process, over a 30-fold increase in S/N was achieved using 9- aminoacridine as matrix. The linearity of response (r2=0.99) and dynamic range of this method (over 2 orders of magnitude) were excellent. Moreover, through multiplexing ionization conditions by generating suites of different analyte-matrix interactions in the absence or presence of different alkali metal cations in the matrix, discrete lipid classes were highly and selectively ionized under different conditions resulting in the de facto resolution of lipid classes without chromatography. The resultant decreases in spectral complexity facilitated tandem mass spectrometric analysis through high energy fragmentation of lithiated molecular ions that typically resulted in informative fragment ions. Anionic phospholipids were also detected as singly negatively charged species that could be fragmented using MALDI tandem mass spectrometry leading to structural assignments. Collectively, these results identify a rapid, sensitive and highly informative MALDI-TOF MS approach for analysis of cellular glycerophospholipids directly from extracts of mammalian tissues without the need for prior chromatographic separation.
Introduction
Cellular lipids are comprised of thousands of distinct chemical moieties that are most easily categorized based upon their class (polar head group), subclass (acyl, alkyl or alkenyl) and individual molecular species (aliphatic chain) composition.1 The importance of lipids in facilitating multiple aspects of cellular function has become increasingly appreciated as new technologies have emerged that greatly expand the ability to identify and quantify alterations in cellular lipid constituents in health and disease. Historically, cellular lipids have been analyzed by initial purification steps employing either silica or reverse phase HPLC followed by quantitation using gas chromatography,2–4 gas chromatography/mass spectrometry 5, 6 or fast atom bombardment (FAB)-MS.7–10 However, these techniques are tedious, labor-intensive, relatively insensitive and unfortunately deliver only limited amounts of structural and quantitative information.
During the last two decades robust advances in mass spectrometry have resulted in dramatic improvements in the ionization, detection and fragmentation of lipids leading to unparalleled insight into alterations in the types and amounts of lipids during the evolution of disease processes. As one of the two pronounced soft-ionization technologies of mass spectrometry, electrospray ionization (ESI) MS has been widely used in lipid analyses through the development of two prominent lipidomics approaches including shotgun lipidomics 1, 11–14 and liquid chromatography (LC/MS)-based lipidomics.15, 16
A second low energy “soft ionization” method is MALDI where the energy present in laser pulses results in excited state matrix molecules that facilitate proton transfer and desorption of the analyte. In contrast to ESI MS, the utility of MALDI MS for lipid analysis has been limited due to the presence of matrix clusters obscuring the mass range of interest or difficulties in achieving efficient ionization and desorption of the hydrophobic lipid moieties under study.17, 18 However, MALDI MS has been widely used in proteomics analyses, nucleotide sequencing, polymer analyses19–21 and metabolite analyses.22, 23 In seminal studies, MALDI MS was first used for lipid analysis in 1995.24, 25 However, the utility of MALDI ionization for high throughput lipid analysis has not been fully realized due to continued difficulties imposed by the ionization/desorption of matrix-hydrophobic lipid analyte clusters. In addition, multiple challenges in sensitivity, differential ionization and complications from polydisperse analyte-matrix interactions have slowed further progress.
Continuing efforts to solve these problems have employed various matrix compounds, binary and ternary component matrix systems, and multiple sample preparation strategies. Examples of matrix systems that have been examined to overcome these obstacles have included liquid matrices,26, 27 ionic-liquid matrices,28–30 solid ionic crystal matrices,31 2,4,6-trihydroxyacetophenone (THAP), 32 2,6-dihydroxyacetophenone33 and p-nitroaniline.34 Sample preparation strategies that have been employed include selective extraction of dominant lipid classes that complicate ionization efficiency,35 analyte matrix pairing for promoting ionization and fragmentation and use of different organic solvents for dissolving different lipid classes and matrix preparations.36, 37 Recently, it was reported that a binary mixture of α-cyano-4-hydroxycinnamic acid (CHCA) and 9-aminoacridine as a dual component matrix resulted in greatly improved S/N and decreased matrix cluster ionization while neither CHCA nor 9-aminoacridine alone were effective.38
In the current work, we demonstrate the importance of solvent in optimizing matrix-analyte interactions during the crystallization process to obtain high sensitivity MALDI MS analyses of lipids directly from extracts of biological materials. Multiple comparisons of mass spectra obtained from extracts of biological tissues analyzed using these optimized interactions demonstrated nearly identical ratios of molecular ion intensities in multiparametric comparisons with shotgun lipidomics employing ESI MS.1, 11, 12 Finally, through use of specific analyte-matrix interactions generated by salt doping of the 9-aminoacridine matrix in conjunction with TOF/TOF analysis of molecular ion fragmentation, multiple diagnostic fragment ions were identified allowing molecular species identification and quantitation of lipid molecular species directly from extracts of biological materials. Collectively, through the integrated use of solvent-enabled formation of salutary analyte-matrix interactions, multiplexed ionization conditions, ion pairing with subsequent tandem mass spectrometry and utilization of negative ion MALDI-TOF MS, a new high throughput platform for the quantitative analysis of lipids has been developed.
Experimental Section
Reagents
9-aminoacridine hemihydrate was purchased from Acros Organics (Morris Plains, NJ). 2,5-dihydroxybenzoic acid (DHB), CHCA, and 2,4,6-trihydroxyacetophenone (THAP) were obtained from Sigma-Aldrich (St. Louis, MO). 1,1′,2,2′-tetramyristoylcardiolipin (T14:0 CL), 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (D14:1-14:1 PtdCho), 1,2- dipalmitoleoyl-sn-glycero-3-phosphoethanolamine (D16:1-16:1 PtdEtn), 1,2- dipentadecanoyl-sn-glycero-3-phosphoglycerol (D15:0-15:0 PtdGro), 1,2-dimyristoyl-sn-glycero-3-phosphoserine (D14:0-14:0 PtdSer), 1,2-distearoyl-sn-glycero-3-phosphocholine (D18:0-18:0 PtdCho) and other phospholipids were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Triheptadecenoin was purchased from Nu-Chek Prep, Inc. (Elysian, MN). All solvents used in extraction and MS analysis were purchased from Burdick & Jackson (Muskegon, MI).
Lipid sample preparation
C57BL/6 wild type male mice (4 months of age) were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Studies Committee at Washington University. Mice were sacrificed by asphyxiation with carbon dioxide. Heart and brown adipose tissue were dissected, washed with phosphate-buffered saline to remove blood, blotted with Kim-wipes to remove excess buffer, and immediately freeze-clamped at the temperature of liquid nitrogen. Myocardial wafers were pulverized into a fine powder with a stainless steel mortar and pestle. Heart tissue sample (approximately 10 mg of fine powder) was weighed in a disposable glass test tube. Internal standards containing D14:0-14:0 PtdSer (4.0 nmol/mg of protein), D14:0-14:0 PtdH (0.75 nmol/mg of protein), T14:0 CL (3.0 nmol/mg of protein), D15:0-15:0 PtdGro (2 nmol/mg of protein), D16:1-16:1 PtdEtn (19 nmol/mg of protein), D14:1-14:1 PtdCho (15 nmol/mg of protein), and T17:1 Triacylglyceride (TAG) (7.5 nmol/mg of protein) were added to each myocardial sample on the basis of protein concentration prior to extraction of lipids. Protein assays on the wafers were performed using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) with bovine serum albumin as a standard. For most quantitative measurements, the lipid content was normalized to the protein content as has traditionally been used. These internal standards were selected because they represent <1% of endogenous cellular lipid molecular species present as demonstrated through molecular species profiling by ESI MS without addition of these internal standards. Lipids were extracted using a modified Bligh and Dyer procedure 39 as described previously.40 Next, the lipid extract was dried under a nitrogen stream and reconstituted in a volume of 100 μL chloroform/methanol (1:1, v/v) per mg protein. The lipid extract was flushed with nitrogen, capped, and stored at −20°C for mass spectrometric analyses (typically within one week). Murine brown adipose tissue (approximately 30 mg of fine powder) was weighed in a disposable glass test tube. Next, the internal standard of TAG (triheptadecenoin) was added followed by 2 mL of methanol and 2 mL of hexane. Triglycerides were extracted in hexane, dried under a nitrogen stream, capped, and stored at −20 °C for mass spectrometric analyses. For MALDI-TOF MS analysis, the TAG extract was reconstituted in a volume of 1 mL of isopropanol/acetonitrile (60/40, v/v) per mg protein. For ESI MS analysis, the TAG extract was reconstituted in a volume of 100 μL chloroform/methanol (1/1, v/v) per mg protein and further diluted as necessary.
MALDI-TOF Mass Spectrometric Analyses of Lipids
Murine heart lipid extracts were diluted from 20 μL to 200 μL with isopropanol/acetonitrile (60/40, v/v). After mixing 10 μL of diluted sample with 10 μL of 9-aminoacridine (10 mg/mL; dissolved in isopropanol/acetonitrile (60/40, v/v)), 0.25 μL of the mixture was spotted on an Opti-TOF® 384 well plate. MS analysis was performed on a 4800 MALDI-TOF/TOF Analyzer (Applied Biosystems, Foster City, CA). Mass spectra of inositol glycerophospholipids (PI), phosphatidylglycerol (PG), serine glycerophospholipids (PS), ethanolamine glycerophospholipids (PE), phosphatidic acid (PA) and cardiolipin (CL) molecular species were acquired in the negative ion mode by averaging 1500 consecutive laser shots (50 shots per subspectra with 30 total subspectra) with default calibration and mass spectra of choline glycerophospholipids (PC) acquired in the positive ion mode. MS2 analyses of lipids were accomplished by collision-induced dissociation (CID) using air at medium pressure.
MALDI-TOF Mass Spectrometric Analyses of Mouse Heart Triacylglycerols (TAG)
After drying under a nitrogen stream, 2 mL of hexane, 2 mL of methanol and 200 μL water were added and vortexed for 1 minute. The resultant mixture was centrifuged at 1500 g for 10 minutes, the hexane layer was saved. The methanol layer was subjected to re-extraction twice with 2 mL of hexane following the same procedure as above. The purpose of these extractions is to separate TAG from other polar lipids in the heart lipid extract. The resultant three hexane layers were pooled and reduced to 2 mL under a nitrogen stream prior to addition of 2 mL of methanol and 200 μL of 5 mM lithium chloride (dissolved in water). The mixture was vortexed for 1 minute and subjected to centrifugation at 1500g. The hexane layer was back extracted against 2 mL of methanol twice to remove the residual polar lipids contained in the hexane layer as described above. The resultant hexane layer was dried under a nitrogen stream and re-dissolved in 200 μL of isopropanol/acetonitrile (60/40, v/v). After mixing the reconstituted sample with 9- aminoacridine (10 mg/mL) (1:1,v/v) that was dissolved in isopropanol/acetonitrile (60/40, v/v) containing 15 mM sodium acetate, 0.25 μL of the mixture was spotted on an Opti-TOF® 384 well plate for MALDI-TOF analysis in the positive ion mode as described above.
ESI Mass Spectrometric Analyses of Lipids
Lipid analysis using ESI was performed by shotgun lipidomics as previously described in detail.1, 11, 12, 41 Briefly, lipid extracts of myocardium were diluted in chloroform/methanol/isopropanol (1:2:4, v/v/v) to a final concentration of 10–50 pmol total lipids/μL. A triple-quadrupole mass spectrometer (ThermoFisher TSQ Quantum Ultra, San Jose, CA) equipped with an automated nanospray apparatus (i.e., Nanomate HD, Advion Bioscience Ltd., Ithaca, NY) was employed as described previously. Spray voltage was 1.2 kV for positive ion mode and 1.1 kV for the negative ion mode. Gas pressure was 15.51 Torr for both modes, and the source was controlled by Chipsoft 6.3.2 software (Advion BioSciences Ltd.). After appropriate dilution, lipid extracts were infused at a flow rate of ~150 nL/min. The temperature along the ion transfer capillary was maintained at 150 °C. Typically, a 1 min period of signal averaging in the profile mode was employed for each MS spectrum. For tandem mass spectrometry, a collision gas pressure was set at 1.0 mTorr, but the collision energy was varied with the classes of lipids as described previously.41 Typically, a 2 min period of signal averaging in the profile mode were employed for each tandem MS spectrum.
Results and Discussion
1. Solvent-enabled Analyte-Matrix Interactions Are an Important Determinant of the Ionization/Desorption Properties of Lipids Using MALDI Mass Spectrometry
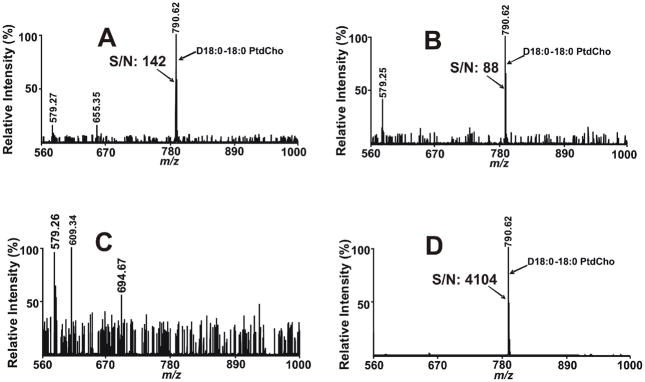
Previously, it has been demonstrated that lipid analyses using binary combinations of CHCA and 9-aminoacridine were markedly superior than the use of either matrix alone.38 This unusual chemical behavior suggested that analyte-matrix interactions were important in the ionization/desorption process during MALDI MS analysis of lipids and that they could potentially be modulated by solvent-mediated alterations in analyte-matrix pairing. To explore this hypothesis, we examined multiple combinations of matrices and solvents to identify a set of optimized conditions for the direct analysis of lipids by MALDI MS. In initial experiments, it became clear that MALDI MS spectra of lipids were critically dependent on the properties of the solvent used for dispersing the matrix and dissolving the lipids. For example, examination of 500 fmol of D18:0-18:0 PtdCho using 9-aminoacridine demonstrated only a very weak signal when the lipids and matrix were dispersed in methanol alone (Fig 1A) or isopropanol alone (Fig 1B). No signal was detectable when acetonitrile was employed as solvent (Fig 1C). In stark contrast, a robust signal (S/N=4104) was routinely observed when lipids and matrix were mixed and spotted using binary mixtures of acetonitrile and isopropanol as solvent for both lipid dilution and matrix emulsion (e.g., Fig 1D). Titration of different ratios of acetonitrile and isopropanol demonstrated that an acetonitrile/isopropanol ratio of 3:2 (v/v) was optimal in enhancing the S/N of the sample (Fig 1D). Previously, Guo and He have reported that 9-aminoacridine dissolved in 1:1 acetonitrile–water solution failed to result in the ionization/desorption of D14:0–18:1 PtdCho in the positive ion mode 38, supporting the importance of solvent in facilitating the productive analyte/matrix interactions during the crystallization process. Collectively, these results demonstrate that solvents profoundly modulate the generation of productive analyte-matrix interactions during MALDI MS resulting in greater than 30-fold increases in S/N.
Figure 1.
MALDI mass spectra of D18:0-18:0 phosphatidylcholine (PtdCho) acquired on a 4800 MALDI-TOF/TOF Analyzer in the positive ion mode using 9-aminoacridine as matrix. Both 9-aminoacridine and D18:0-18:0 PtdCho were dissolved in: A) methanol; B) isopropanol; C) acetonitrile (9-aminoacridine was saturated in acetonitrile); or D) isopropanol/acetonitrile (60/40, v/v) and spotted onto the MALDI plate and spectra were collected as described in the “Experimental Section”. Each spot contained 500 fmol D18:0-18:0 PtdCho. The prefix ‘‘D’’ stands for diacyl (i.e., phosphatidyl-) species. S/N stands for signal/noise.
2. Direct Analysis of choline glycerophospholipid (PC) Molecular Species Using Solvent-enabled MALDI Mass Spectrometry
First, we explored the possibility that ionization and desorption of naturally occurring lipids could be reproducibly obtained in the positive ion mode without the formation of matrix clusters or the generation of multiple ion peaks resulting from the presence of individual lipid molecular species with either protons or different alkali metal ions. Examination of D18:0-18:0 PtdCho by MALDI using a 9-aminoacridine matrix demonstrated a single robust peak at m/z 790.62 that corresponds to the proton adduct with the near absence of ionized matrix clusters. Although 9-aminoacridine has been routinely used in the negative ion mode as a matrix for MALDI MS, this result identifies the potential suitability of 9-aminoacridine as an efficient matrix for positive ion MALDI MS in lipid analysis. Comparisons with multiple other matrices (e.g., CHCA, DHB and THAP) under a constant laser intensity demonstrated the presence of both proton and sodium adducts of the molecular species with a markedly reduced sensitivity and detection limit in comparison to the use of 9-aminoacridine as matrix (Fig S1). The presence of multiple adducts of each lipid molecular species also markedly complicates spectral complexity and data analysis. Thus, through the use of this procedure a single [M+H]+ ion is formed and desorbed with high efficiency in the absence of multiple alkali adducts and matrix clusters in the positive ion mode. Collectively, these results demonstrate that the interaction of analytes with the matrix is solvent dependent providing a novel mechanism to enhance ionization/desorption efficiency. By analogy to other multiphase systems, this likely reflects solvent-mediated alterations in hydrogen bonding, dipole-dipole interactions and altered stereoelectronic interactions of analytes with matrix.
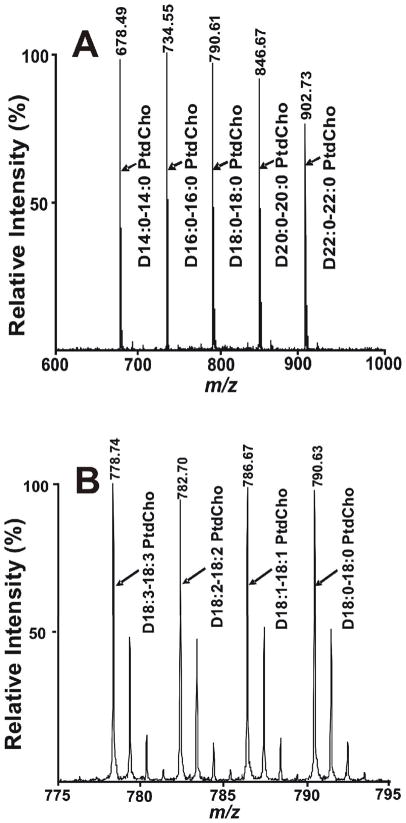
Second, we tested the potential suitability of solvent-enabled analyte matrix interactions for quantitative analyses of phosphatidylcholines containing differing chain lengths. To this end, equimolar mixtures of D14:0-14:0 PtdCho, D16:0-16:0 PtdCho, D18:0-18:0 PtdCho, D20:0-20:0 PtdCho and D22:0-22:0 PtdCho were prepared and analyzed by MALDI MS to determine the effects of chain length and hydrophobicity on the efficiency of ionization and desorption under these conditions. The results clearly demonstrated that the chain length and hydrophobicity of these species of PC do not alter the ionization/desorption efficiency (Fig 2A and Fig S2A). It is important to note that the contributions from 13C isotopologues must be considered during quantitative analysis with MALDI MS. Next, the effect of unsaturation in the aliphatic chains in PC was examined. Through a thousand fold range in concentration (from 15 fmol to 15 pmol), the degree of unsaturation of PC molecular species did not substantially effect the ionization/desorption efficiency of four different PC molecular species (Fig 2B and Fig S2B). Thus, for PC analysis the ionization/desorption efficiency of multiple PC molecular species are nearly identical using the developed methods. Typically, the detection limit of PC using D18:1-18:1 PtdCho is 5 fmol present in the MALDI plate spot, which is 5-fold lower than that previously reported.42 Collectively, these results clearly demonstrate the power of MALDI MS in the analyses of PC mixtures using solvent enabled analyte-matrix interactions with 9-aminoacridine.
Figure 2.
Quantitative analyses of standard PC molecular species using MALDI-TOF MS. A) Mass spectrum of an equimolar mixture of D14:0-14:0 PtdCho, D16:0-16:0 PtdCho, D18:0-18:0 PtdCho, D20:0-20:0 PtdCho and D22:0-22:0 PtdCho; B) Mass spectrum of an equimolar mixture of D18:3-18:3 PtdCho, D18:2-18:2 PtdCho, D18:1-18:1 PtdCho and D18:0-18:0 PtdCho. Mass spectra were acquired on a 4800 MALDI-TOF/TOF Analyzer in the positive ion mode using 9-aminoacridine as matrix as described in the “Experimental Section”. Isopropanol/acetonitrile (60/40, v/v) was used as solvent for dissolving both matrix and PCs. The amount of each individual PC molecular species in each spot is 150 fmol. The prefix ‘‘D’’ stands for diacyl (i.e., phosphatidyl-) species.
Third, the dynamic range for PC quantitation by MALDI-TOF MS was determined by analyses of mixtures of standard PC molecular species at selected concentrations. Plots of the ion peak area ratios of D18:2-18:2 PtdCho to D14:0-14:0 PtdCho versus their molar ratios in the mixture from a range of 17 fmol to 5 pmol per spot resulted in a line with a slope of 1.14 and a correlation coefficient of 0.99 (Fig S2C). Similarly, plots of the ratios of D18:2-18:2 PtdCho to D22:0-22:0 PtdCho versus the actual molar ratios of standards in the mixture resulted in a line with a slope of 0.99 and a correlation coefficient of 0.99 (Fig S2D). Collectively, these results demonstrate that over a 100-fold dynamic range, individual PC molecular species differing widely in chain length and degree of unsaturation could be accurately measured by MALDI MS.
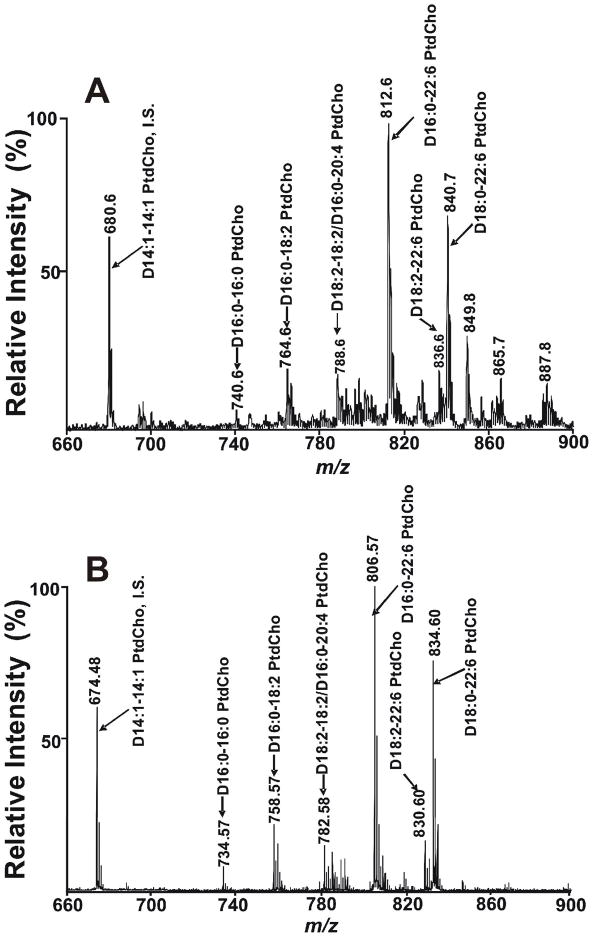
To determine the utility of this method for PC quantitation from biological sources, we compared the types and amounts of PC molecular species present in a mouse heart lipid extract using MALDI-TOF MS with results of the current “gold standard” for quantitation, ESI MS shotgun lipidomics (Fig 3). A total of 38 isobaric peaks of PC molecular species were detected in myocardial lipid extracts using solvent-enabled MALDI MS (Table S1). Comparisons of the mass spectra from Bligh and Dyer extracts of myocardium demonstrated that the peak intensities of PC molecular species in the MALDI mass spectrum under the conditions developed were present in essentially identical ratios as those present using shotgun lipidomics with ESI MS. It should be noted that in the ESI mass spectrum, PC molecular species were present as their lithium adducts (under the conditions employed) while exclusively protonated molecular species of PC were present using MALDI MS as previously discussed. Since quantitation of PC by shotgun lipidomics using ESI MS has been well established,1, 12, 41 these results validate the use of solvent-enabled MALDI MS analysis using 9-aminoacridine as matrix for quantitative analyses of PC. Additionally, Figure 3 shows that mass resolution in MALDI mass spectrum (~12000) is much higher than ESI mass spectrum (~2000). This difference results from the higher resolution inherent in TOF analysis in comparison to the lower resolution present in triple quadrupole instruments. Furthermore, since PC molecular species are readily ionized and TAG molecular species are not ionized (unless oxytropic cations are added to the matrix), MALDI MS leads to complete mass spectrometric resolution of these two lipid classes which have overlapping mass ranges without the need for chromatography.
Figure 3.
Mass spectral comparison of PC molecular species present in mouse heart lipid extracts acquired by either ESI or MALDI. Extracts of murine myocardium were examined by: A) ESI MS or B) MALDI-TOF MS utilizing 9-aminoacridine as matrix dissolved in isopropanol/acetonitrile (60/40, v/v). Myocardial lipid extracts of mice were prepared by a modified Bligh and Dyer procedure as described in the “Experimental Section”. Positive-ion ESI mass spectra of PC was acquired using a TSQ Quantum Ultra Plus triple-quadrupole mass spectrometer as described in the “Experimental Section”. The MALDI-TOF mass spectrum of PC was acquired on a 4800 MALDI-TOF/TOF Analyzer in the positive ion mode as described in the “Experimental Section”. The prefix ‘‘D’’ stands for diacyl (i.e., phosphatidyl-) species. ‘‘IS’’ denotes internal standard.
3. Tandem Mass Spectrometric Analysis of Choline Glycerophospholipids (PC) Using Solvent-enabled Analyte-Matrix Interactions
Tandem mass spectrometric analysis of protonated PC molecular species using ESI tandem mass spectrometry with CID yields an abundant fragment ion at m/z 184 resulting from the facile loss of phosphocholine. Previously, this fragment has been useful as a diagnostic peak in tandem ESI MS analyses to identify choline containing phospholipids with high sensitivity.43 Unfortunately, the extremely rapid loss of phosphocholine from protonated PC results in a near absence of informative fragment ions that can identify the aliphatic chain composition from individual protonated PC molecular species using ESI tandem mass spectrometry. The same facile loss of PC using MALDI-TOF MS also precluded identification of robust informative fragment ions when tandem mass spectrometric analyses of protonated PC molecular species were examined (Fig S3). Tandem mass spectra of protonated PC present in murine heart extracts demonstrated a high intensity phosphocholine ion peak at m/z 184.08 and an abundant precursor ion peak. For example, tandem mass spectra of protonated D18:0–22:6 PtdCho demonstrated a predominant fragment at m/z 184.08 representing phosphocholine derived from the precursor ion. It should be noted that several low abundance ions contained important structural information including ions at m/z 506.27, 524.28, 550.21 and 568.25 (Fig S3A). These extremely low intensity peaks resulted from losses of 22:6 fatty acid, 22:6 ketene, 18:0 fatty acid and 18:0 ketene, respectively. Since tandem mass spectrometry is also required for identification of low abundance peaks, it became necessary to identify conditions that result in easily identifiable fragment ions from low abundance molecular species. Previously, it was demonstrated that the use of alkali metal ions resulted in PC metal ion adducts whose fragmentation by CID led to the formation of structurally informative fragment ions allowing identification of molecular species.44–46 Accordingly, we added 10 mM lithium acetate to the 9-aminoacridine solution used for formation of solvent-enabled metal, analyte and matrix interactions. This resulted in the transformation of 80% of protonated PC to lithiated PC (Fig S4). Fragmentation of lithiated D18:0–22:6 PtdCho molecular species by MALDI tandem mass spectrometry yielded structurally informative fragment ion peaks at m/z 556.27 and m/z 550.28 representing elimination of the stearic acid or the lithium stearate, respectively (Fig S3B). Informative fragment ion peaks were also easily identified at m/z 512.30 and m/z 506.29 representing the loss of the docosahexaenonatic acid or lithium docosahexaenonate from the lithiated D18:0–22:6 PtdCho, respectively (Fig S3B). Moreover, fragment ion peaks at m/z 497.21 and m/z 453.26 corresponding to the neutral loss of N(CH3)3 from fragment ion peaks at m/z 556.27 and m/z 512.30 also provide important structural information for identification of individual PC molecular species.
4. MALDI-TOF Mass Spectrometric Analysis of Triglycerides (TAGs)
Since TAGs do not contain an intrinsically charged moiety and did not ionize/desorb under the previously developed conditions, it was necessary to introduce oxytropic ions into the matrix to facilitate charge separation leading to successful ionization/desorption. This strategy has previously been used successfully with other matrices to enhance TAG ionization/desorption 47. Three additives, 15 mM ammonium acetate, 15 mM lithium acetate and 17 mM sodium acetate, were examined to improve the conditions for TAG analysis. Doping of the matrix with sodium acetate provided the best results. Examination of hexane extracts of brown adipose tissue using 9-aminoacridine doped with sodium acetate resulted in robust signals corresponding to triglyceride molecular species (Fig S5B). Importantly, formation of a single type of ion (i.e., Na+) adduct significantly reduces the complexity of the spectra and facilitates peak assignments and quantitation. Next we compared the ratios of the intensities of ionized triglyceride molecular species by ESI MS (Fig S5A). The results demonstrated nearly identical profiles of triglyceride molecular species using MALDI MS in comparison to ESI MS. Furthermore, dilution or concentration of the lipid extract 10-fold did not alter the profile of TAG molecular species using MALDI MS, thus representing a ≥100-fold linear dynamic range of concentration for each TAG molecular species (data not shown).
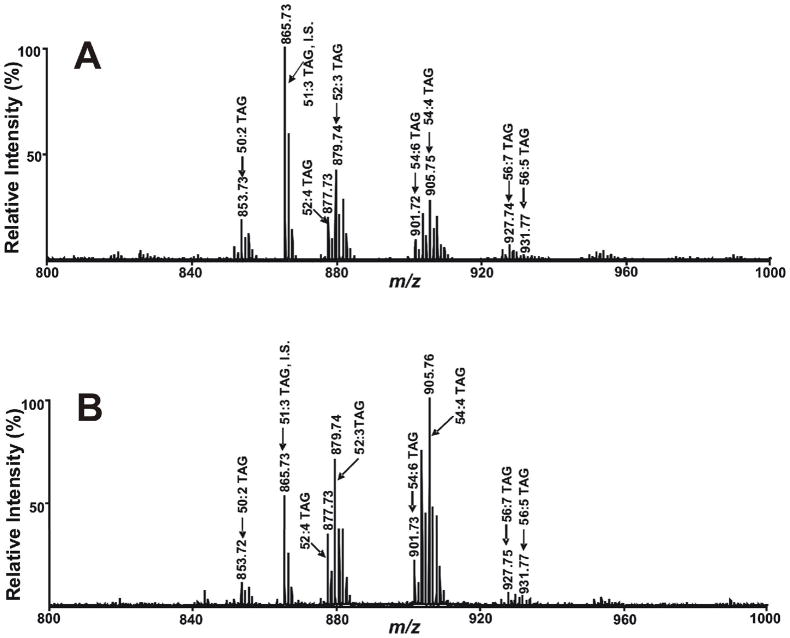
In samples where abundant PC species are present and analyses of both PC and TAG are of interest, it is necessary to first extract the tissue with a routine Bligh and Dyer procedure, evaporate part of the chloroform layer and next selectively extract triglycerides using a two phase hexane/MeOH system as described in Experimental Methods. This procedure resulted in the identification of multiple sodiated TAG molecular species in close agreement with previous results using ESI MS (Fig 4). To further demonstrate the utility of this method to identify alterations in TAG mass and content during pathophysiologic perturbations, we examined the differences in TAG profiles in myocardium using MALDI MS from fed and fasted murine myocardium. The results demonstrated a robust increase in TAG molecular species in the fasted animal similar to results previously demonstrated using ESI based shotgun lipidomics.48 A total of 56 isobaric peaks of TAG molecular species were readily detected. Examination of the sensitivity of this method identified a detection limit (5× background) of 100 fmol per spot using 16:1–18:1–18:2 TAG.
Figure 4.
Comparison of MALDI mass spectra of sodiated TAG molecular species present in mouse heart lipid extract acquired on a 4800 MALDI-TOF/TOF Analyzer in the positive ion mode using 9-aminoacridine as matrix. A) A MALDI mass spectrum of TAG extracted from wild-type mouse heart subjected to normal feeding; and B) A MALDI mass spectrum of TAG extracted from wild-type mouse heart subjected to 24 hours of fasting. Myocardial TAG was extracted by a modified Bligh and Dyer procedure followed by hexane extraction of TAG and methanol back extraction to remove choline glycerophospholipid molecular species as described in the “Experimental Section”. ‘‘IS’’ denotes internal standard.
Since previous work has demonstrated that each isobaric peak is comprised of multiple individual molecular species 49, MALDI tandem mass spectrometric analysis was performed. A tandem mass spectrum of the sodiated TAG molecular species at m/z 879.74 in brown adipose tissue (Fig S6A) demonstrated fragment ions at m/z 597.56, 599.57, 623.56, 625.56 representing neutral losses of oleic acid, linoleic acid, palmitic acid and palmitoleic acid from sodiated TAG molecular species at m/z 879.74. In addition, fragment ions at m/z 575.57, 577.57, 601.57 and 603.59 representing neutral losses of sodium oleate, sodium linoleate, sodium palmitate and sodium palmitoleate from sodiated TAG molecular species at m/z 879.74 were also easily identified. The presence of both groups of fragments unambiguously identifies oleoyl, linoleoyl, palmitoyl and palmitoleoyl chains in this isobaric peak demonstrating the presence of a combination of 18:1-18:1-16:1 TAG and 18:2-18:1-16:0 TAG. Comparisons with CID fragmentation of sodiated TAG molecular species at m/z 879.74 using an ESI mass spectrometer (Fig 6SB) revealed that high energy fragmentation of sodiated TAG molecular species using MALDI tandem mass spectrometry shares essentially the same fragmentation pattern as that obtained using ESI MS with CID.
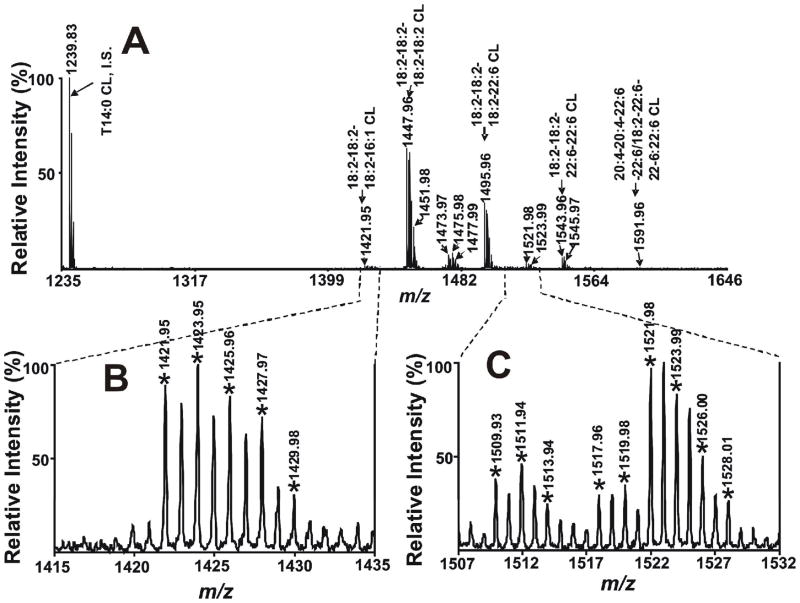
5. Analysis of Cardiolipins in the Negative Ion Mode Using MALDI-TOF Mass Spectrometry
Recent studies demonstrated that CL analyses using MALDI-TOF MS were achievable with DHB and 2,6-dihydroxyacetophenone as matrixes.33, 50–52 Since CLs contain two phosphate groups with pKa1 of ~4.0 which can be easily deprotonated under neutral or basic conditions, we anticipated that the use of a 9-aminoacridine as matrix in a mild basic environment to ionize CLs in the negative ion mode could potentially provide unique information that other matrices cannot. Analyses of CLs extracted from mouse heart tissue were performed by MALDI-TOF MS in the negative ion mode using solvent-enabled analyte-matrix interactions as previously described. The resultant mass spectra demonstrated multiple deprotonated CL molecular species (Fig 5). Remarkably, only deprotonated CL molecular species were detected free from association with other cations (Na+ or Li+) using MALDI-TOF MS in the negative ion mode. As far as we are aware, this is the first time that CL molecular species were observed as single anions derived directly from extracts of biological tissues without prior chromatography. The detection of CL as a singly charged species in a relatively higher mass range can increase the sensitivity of CL analyses and avoid any overlap with other phospholipids. Cardiolipin molecular species were assigned based upon mass accuracy of <12ppm resulting in the detection of 35 peaks of CL molecular species by MALDI-TOF MS. The detection limit of CL assessed with T18:1 CL was 10 fmol per spot under the conditions used. The reasons underlying this surprisingly low detection limit of CL are not known with certainty, but could result from effects relating to either enhanced proton transfer or enhanced desorption of CL due to its unique structure that contains two phosphates and a markedly increased aliphatic chain /polar head group volume in comparison to other phospholipids.
Figure 5.
MALDI mass spectra of Cardiolipins (CLs) present in mouse heart lipid extracts acquired on a 4800 MALDI-TOF/TOF Analyzer in the negative ion mode using 9-aminoacridine as matrix. Mouse myocardial CL was extracted by a modified Bligh and Dyer procedure as described in the “Experimental Section”. Extracts were mixed and spotted onto a 9-aminoacridine matrix and examined in the negative ion mode. * indicates the detected CL ion peaks. ‘‘IS’’ denotes internal standard.
Tandem mass spectrometric analyses of CL molecular species in mouse heart lipid extracts were performed using MALDI tandem mass spectrometry in the negative ion mode with solvent-enabled matrix analyte interactions as described in the Methods Section. The tandem mass spectrum of CL molecular species at m/z 1447.96 demonstrated a single fatty acyl fragment at m/z 279.18 representing linoleate (Fig S7A). These results identify this peak as tetra 18:2 CL. The presence of a 18:2-18:2 PtdH fragment ion at m/z 695.39, a 18:2 lyso-PtdH fragment ion at m/z 433.14, and a dehydrated 18:2 lyso-PtdH fragment ion at m/z 415.15 in conjunction with fragment ions at m/z 751.44 and 831.40 further substantiate the assignment of the peak at m/z 1447.96 to be T18:2 CL. We also observed that the fragmentation pattern of singly charged T14:0 CL obtained by low-energy CAD fragmentation 53 by ESI tandem mass spectrometry is nearly identical to that of singly charged T18:2 CL by high-energy CID fragmentation33 by MALDI tandem mass spectrometry in the negative ion mode. The utility for elucidating the structures of asymmetric CL molecular species by MALDI tandem mass spectrometry was also tested using 9-aminoacridine as matrix in the negative ion mode. Tandem mass spectrum of asymmetric CL molecular species at m/z 1495.97 present in mouse heart lipid extracts (Fig S7B) demonstrated two fragment ions, one at m/z 279.18 representing linoleate and another at m/z 327.19 representing docosahexaenoate, thereby establishing that two types of acyl chains were esterified to this CL molecular species. This conclusion is further confirmed by the identification of a fragment ion at m/z 415.17 (dehydrated 18:2 lyso-PtdH) and another at m/z 463.16 (dehydrated 22:6 lyso-PtdH). Additionally, the detection of the fragment ion at m/z 695.41 representing 18:2-18:2 PtdH and a fragment ion at m/z 743.41 representing 18:2–22:6 PtdH indicate the CL molecular ion is (18:2-18:2)/(18:2–22:6) CL. The identity of the peak at m/z 1495.97 as (18:2-18:2)/(18:2–22:6) CL was also confirmed by the presence of other important fragment ions at m/z 751.35, 799.46, 831.37 and 879.39. Collectively, these results demonstrate the utility of MALDI-TOF MS for the analysis of CL molecular species directly from biological extracts.
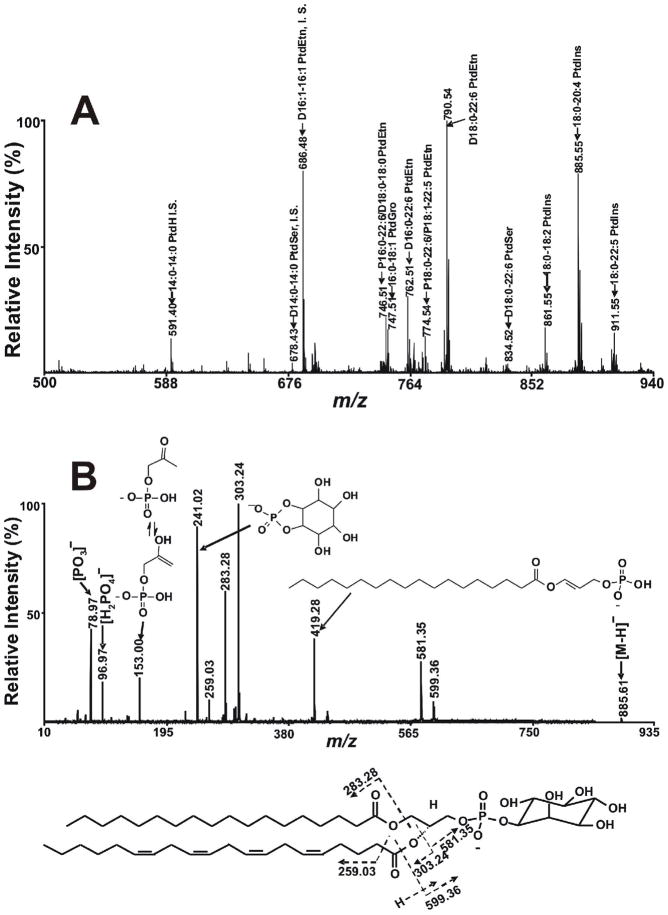
6. Analysis of Other Anionic Phospholipids Extracted from Mouse Heart Tissue
In addition to PC, TAG and CLs, lipid extracts from murine heart tissue contain multiple other abundant phospholipid classes such as the strongly anionic phospholipids PG, PS, PI, and PA and other phospholipid classes that can be induced to contain a negative charge under mildly alkaline conditions (e.g., PE). To examine the utility of the developed method for analysis of these anionic lipids in the negative ion mode, we examined the negative ion MALDI-TOF MS spectra of myocardial lipid extracts. The results demonstrated that PG, PS, PI, PA and PE could also be detected by MALDI-TOF MS in the negative ion mode using the developed method (Fig 6A). As anticipated, multiple molecular species of PG, PS, PI, PA and PE were detected. On the basis of mass accuracy and/or tandem MS analyses, 18 PG molecular species ion peaks, 14 PS molecular species ion peaks, 14 PI molecular species ion peaks, 7 PA molecular species ion peaks and 41 PE molecular species were detected. The detection limit for anionic phospholipids is approximately 1–10 pmol per spot. The advantage of using 9-aminoacridine as matrix in the negative ion mode for efficient ionization/desorption of phospholipids facilitating detection of PE, PG, PS, PI and PA is that interference of matrix peaks in lipid analysis is precluded due to the absence of matrix clusters in the region of interest.
Figure 6.
MALDI-TOF MS analysis of phospholipids in the negative ion mode using 9- aminoacridine as matrix. A) A MALDI mass spectrum of negatively charged phospholipids from murine myocardium was obtained by examining an aliquot of a Bligh and Dyer extract of myocardium by MALDI MS in the negative ion mode using a 4800 MALDI-TOF/TOF Analyzer as described in the “Experimental Section”. The prefix “D” and “P” stand for diacyl (i.e., phosphatidyl-) and alkenyl-acyl (plasmenyl-) species, respectively. ‘‘IS’’ denotes internal standard. B) A MALDI tandem mass spectrum of fragment ions obtained from 18:0–20:4 PtdIns present in mouse heart lipid extracts acquired on a 4800 MALDI-TOF/TOF Analyzer in the negative ion mode. The tandem mass spectrum was recorded on a 4800 MALDI-TOF/TOF Analyzer in the negative ion mode using 9-aminoacridine as matrix using CID with the metastable suppressor on and the timed ion selector enabled. The voltages of source 1, collision cell and collision cell offset were 8.0 kV, 7.0 kV and −0.035 kV, respectively. The tandem MS spectrum was obtained by averaging 2000 consecutive laser shots (50 shots per subspectra with 40 total subspectra).
To determine the utility of tandem mass spectrometric analyses of phospholipids using MALDI tandem mass spectrometry with the developed strategy, extracts of mouse heart tissue were examined in the negative ion mode employing high energy fragmentation of molecular ions. Fragmentation of the molecular ion at m/z 885.55 corresponding to the predicted mass of 18:0–20:4 PtdIns demonstrated two dominate fragment ion peaks at m/z 303.24 and 283.28 that represent arachidonate and stearate, respectively (Fig 6B). As we observed for the fragmentation of PC, and in agreement with data by others 54, 55, the fragment ion peak intensity of arachidonate derived from sn-2 position of PtdIns is significantly higher than that of stearate derived from sn-1 position of PtdIns. This differential fragmentation behavior at the sn-1 and sn-2 positions results from the preferential formation of a 5 vs. 6 member ring after nucleophilic attack by the phosphate O atom. The fragment ion peak at m/z 241.02 represents a cyclized ion resulting from the head group of 18:0–20:4 PI. Similar to PI, fragmentation of molecular species of other phospholipid classes such as PE, PG and PA also yield specific diagnostic fragmentation peaks corresponding to their specific head groups allowing definitive class identification. High energy fragmentation of these anionic lipids also led to the generation of informative fragment ions facilitating structural assignments. Collectively, these results demonstrate the utility of negative ion MALDI-TOF MS in the analysis of negatively charged lipids directly from biological extracts.
Conclusions
In this study a multiplexed solvent-enabled MALDI-TOF mass spectrometry-based approach was developed for the rapid analysis of cellular glycerophospholipids directly from extracts of biological tissues. Through utilizing specific suites of solvent and alkali metal cation interactions with 9-aminoacridine matrix in either the positive or negative ion modes, the highly selective ionization of different lipid classes could be achieved which effectively replaces the need for chromatographic separations of different lipid classes. Judicious use of multiplexed solvent-enabled analyte-matrix interactions during crystallization facilitated subsequent tandem mass spectrometric analysis using high energy fragmentation that led to definitive structural assignments through multiple informative fragment ions. Since MALDI MS analyses can be performed at very high rates in comparison to other MS technologies, this approach provides an extremely sensitive and high throughput platform for lipid analysis directly from extracts of biological samples.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant P01 HL-57278. We thank Mr. Harold Sims, Dr. Christopher Jenkins and Dr. Ari Cedars for their critical reading of the manuscript.
References
- 1.Han X, Gross RW. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 2.Guichardant M, Lagarde M. J Chromatogr. 1983;275:400–406. doi: 10.1016/s0378-4347(00)84386-8. [DOI] [PubMed] [Google Scholar]
- 3.Seewald M, Eichinger HM. J Chromatogr. 1989;469:271–280. doi: 10.1016/s0021-9673(01)96462-3. [DOI] [PubMed] [Google Scholar]
- 4.Watts R, Dils R. Biochem J. 1968;110:51–52. doi: 10.1042/bj1100051p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes H, Smith CV, Horning EC, Mitchell JR. Anal Biochem. 1983;130:431–436. doi: 10.1016/0003-2697(83)90612-7. [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan CV. J Chromatogr. 1974;98:105–128. doi: 10.1016/s0021-9673(00)84782-2. [DOI] [PubMed] [Google Scholar]
- 7.Gross RW. Biochemistry. 1984;23:158–165. doi: 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- 8.Gross RW. Biochemistry. 1985;24:1662–1668. doi: 10.1021/bi00328a014. [DOI] [PubMed] [Google Scholar]
- 9.Pahlsson P, Nilsson B. Anal Biochem. 1988;168:115–120. doi: 10.1016/0003-2697(88)90018-8. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki M, Sekine M, Yamakawa T, Suzuki A. J Biochem. 1989;105:829–833. doi: 10.1093/oxfordjournals.jbchem.a122753. [DOI] [PubMed] [Google Scholar]
- 11.Han X, Gross RW. J Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Gross RW. Expert Rev Proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 13.Schwudke D, Liebisch G, Herzog R, Schmitz G, Shevchenko A. Methods Enzymol. 2007;433:175–191. doi: 10.1016/S0076-6879(07)33010-3. [DOI] [PubMed] [Google Scholar]
- 14.Schwudke D, Oegema J, Burton L, Entchev E, Hannich JT, Ejsing CS, Kurzchalia T, Shevchenko A. Anal Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 15.Hermansson M, Uphoff A, Kakela R, Somerharju P. Anal Chem. 2005;77:2166–2175. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- 16.Sommer U, Herscovitz H, Welty FK, Costello CE. J Lipid Res. 2006;47:804–814. doi: 10.1194/jlr.M500506-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Cohen LH, Gusev AI. Anal Bioanal Chem. 2002;373:571–586. doi: 10.1007/s00216-002-1321-z. [DOI] [PubMed] [Google Scholar]
- 18.McCombie G, Knochenmuss R. Anal Chem. 2004;76:4990–4997. doi: 10.1021/ac049581r. [DOI] [PubMed] [Google Scholar]
- 19.Hillenkamp F, Karas M. Methods Enzymol. 1990;193:280–295. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- 20.Hillenkamp F, Karas M, Beavis RC, Chait BT. Anal Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 21.Karas M, Hillenkamp F. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 22.Edwards JL, Kennedy RT. Anal Chem. 2005;77:2201–2209. doi: 10.1021/ac048323r. [DOI] [PubMed] [Google Scholar]
- 23.Sun G, Yang K, Zhao Z, Guan S, Han X, Gross RW. Anal Chem. 2007;79:6629–6640. doi: 10.1021/ac070843+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey DJ. J Mass Spectrom. 1995;30:1333–1349. [Google Scholar]
- 25.Marto JA, White FM, Seldomridge S, Marshall AG. Anal Chem. 1995;67:3979–3984. doi: 10.1021/ac00117a025. [DOI] [PubMed] [Google Scholar]
- 26.Stubiger G, Pittenauer E, Allmaier G. Phytochem Anal. 2003;14:337–346. doi: 10.1002/pca.724. [DOI] [PubMed] [Google Scholar]
- 27.Zollner P, Stubiger G, Schmid E, Pittenauer E, Allmaier G. Int J Mass Spetrom Ion Processes. 1997;169:99–109. [Google Scholar]
- 28.Darsow KH, Lange HA, Resch M, Walter C, Buchholz R. Rapid Commun Mass Spectrom. 2007;21:2188–2194. doi: 10.1002/rcm.3076. [DOI] [PubMed] [Google Scholar]
- 29.Li YL, Gross ML, Hsu FF. J Am Soc Mass Spectrom. 2005;16:679–682. doi: 10.1016/j.jasms.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Mank M, Stahl B, Boehm G. Anal Chem. 2004;76:2938–2950. doi: 10.1021/ac030354j. [DOI] [PubMed] [Google Scholar]
- 31.Ham BM, Jacob JT, Cole RB. Anal Chem. 2005;77:4439–4447. doi: 10.1021/ac058000a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stubiger G, Belgacem O. Anal Chem. 2007;79:3206–3213. doi: 10.1021/ac062236c. [DOI] [PubMed] [Google Scholar]
- 33.Wang HY, Jackson SN, Woods AS. J Am Soc Mass Spectrom. 2007;18:567–577. doi: 10.1016/j.jasms.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estrada R, Yappert MC. J Mass Spectrom. 2004;39:412–422. doi: 10.1002/jms.603. [DOI] [PubMed] [Google Scholar]
- 35.Johanson RA, Buccafusca R, Quong JN, Shaw MA, Berry GT. Anal Biochem. 2007;362:155–167. doi: 10.1016/j.ab.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 36.Benard S, Arnhold J, Lehnert M, Schiller J, Arnold K. Chem Phys Lipids. 1999;100:115–125. [Google Scholar]
- 37.Schiller J, Arnhold J, Benard S, Muller M, Reichl S, Arnold K. Anal Biochem. 1999;267:46–56. doi: 10.1006/abio.1998.3001. [DOI] [PubMed] [Google Scholar]
- 38.Guo Z, He L. Anal Bioanal Chem. 2007;387:1939–1944. doi: 10.1007/s00216-006-1100-3. [DOI] [PubMed] [Google Scholar]
- 39.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 40.Cheng H, Guan S, Han X. J Neurochem. 2006;97:1288–1300. doi: 10.1111/j.1471-4159.2006.03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X, Yang J, Cheng H, Ye H, Gross RW. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Gellermann GP, Appel TR, Davies P, Diekmann S. Biol Chem. 2006;387:1267–1274. doi: 10.1515/BC.2006.157. [DOI] [PubMed] [Google Scholar]
- 43.Pulfer M, Murphy RC. Mass Spectrom Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 44.Han X, Gross RW. J Am Soc Mass Spectrom. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 45.Ho YP, Huang PC. Rapid Commun Mass Spectrom. 2002;16:1582–1589. doi: 10.1002/rcm.751. [DOI] [PubMed] [Google Scholar]
- 46.Hsu FF, Bohrer A, Turk J. J Am Soc Mass Spectrom. 1998;9:516–526. doi: 10.1016/S1044-0305(98)00012-9. [DOI] [PubMed] [Google Scholar]
- 47.Ayorinde FO, Keith QL, Jr, Wan LW. Rapid Commun Mass Spectrom. 1999;13:1762–1769. doi: 10.1002/(SICI)1097-0231(19990915)13:17<1762::AID-RCM711>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 48.Han X, Cheng H, Mancuso DJ, Gross RW. Biochemistry. 2004;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- 49.Yang K, Zhao Z, Gross RW, Han X. PLoS ONE. 2007;2:e1368. doi: 10.1371/journal.pone.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald-Marsh T, Carroll CA, Robinson NC, Musatov A. Anal Biochem. 2006;359:262–264. doi: 10.1016/j.ab.2006.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shadyro O, Yurkova I, Kisel M, Brede O, Arnhold J. Free Radic Biol Med. 2004;36:1612–1624. doi: 10.1016/j.freeradbiomed.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 52.Shadyro OI, Yurkova IL, Kisel MA, Brede O, Arnhold J. Int J Radiat Biol. 2004;80:239–245. doi: 10.1080/09553002310001655421. [DOI] [PubMed] [Google Scholar]
- 53.Hsu FF, Turk J, Rhoades ER, Russell DG, Shi Y, Groisman EA. J Am Soc Mass Spectrom. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 54.Stubiger G, Pittenauer E, Allmaier G. Anal Chem. 2008;80:1664–1678. doi: 10.1021/ac7018766. [DOI] [PubMed] [Google Scholar]
- 55.Jackson SN, Wang HY, Woods AS. J Am Soc Mass Spectrom. 2007;18:17–26. doi: 10.1016/j.jasms.2006.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.