Abstract
Conjugates of ubiquitin or its homologues to other proteins occur by strictly ordered steps with ordered addition of substrates for each step. High concentrations of E2 were shown to inhibit the formation of E2∼Ubl thioester and Ubl∼target conjugates. We investigated the mechanism of such inhibitory effect of the SUMO E2 and whether the E2 has two binding sites on its E1, one for the inhibitory effect and one for productive SUMOylation. NMR methods in combination with mutagenesis and biochemical assays revealed that Ubc9 binds to two flexible domains of its free E1 simultaneously, suggesting extensive domain movements in the free E1. Further, interaction of free E1 and E2 inhibits SUMO adenylation, and the interfaces responsible for the inhibition were the same as those required for productive transfer of SUMO from E1 to E2. This study indicates a conformational flexibility-dependent mechanism to control the strictly ordered steps in Ubl modifications.
Keywords: Enzyme Mechanisms, NMR, Sumoylation, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination, E1-activating Enzyme, Adenylation
Introduction
Conjugates of ubiquitin or its homologues to other proteins play critical roles in cellular functions (1–3). These modifications require sequential steps; a Ubl (ubiquitin-like modifier) is first activated by an activation enzyme (E1), which catalyzes adenylation of the C-terminal –COOH group of Ubl, and then forms a thioester bond between the –SH group of the active site Cys and the C-terminal –COOH group. Ubl is then transferred to a conjugation enzyme, E2, and forms a thioester bond with the –SH group of the active site Cys residue of the E2. Ubl is then transferred to target proteins. The E1 in the small ubiquitin-like modifier (SUMO)4 pathway is a tight heterodimer of two proteins, known as SAE1 and SAE2 (SUMO activation enzyme 1 and 2), which are homologous to the N- and C-terminal portions of the ubiquitin E1, respectively (4).
Recent advances have shown that the E1 enzyme has extensive conformational flexibility (5–8). Although SUMO adenylation does not appear to require large-scale conformational changes, formation of the SUMO∼E1 thioester requires that the domain containing the catalytic Cys residue (Cys domain) rotates ∼140° accompanied by unfolding of an α-helix and formation of a short β-sheet. Upon formation of the SUMO∼E1 thioester, the Cys domain rotates to its original position to interact with E2 to transfer SUMO to E2 (8, 9). Another critical E2-binding domain, which forms the ubiquitin fold (UFD), rotates 120 ° from a “closed” state found in the crystal structure of NEDD8 E1 in the complex with ATP and NEDD8 (6, 10) to an “open” state shown in the crystal structure of the ternary complex of NEDD8∼E1 thioester with E2 (8). It remains unclear whether these conformational transitions in E1 are triggered by the formation of intermediates or whether they occur continuously.
The E2 inhibitory effect has been described for the formation of E1 catalyzed E2∼ubiquitin and E2∼NEDD8 thioester formation, where high E2 concentrations inhibited formation of the thioester conjugates (11, 12). Such inhibitory effects reflect the ordered nature of the steps in Ubl modifications. The structural mechanism of the inhibitory effect is unclear and was suggested to be due to two E2-binding sites on E1, one inhibitory and one productive as found in well characterized enzymes catalyzing small molecular reactions.
In this study, we investigated the E2 inhibitory effect in SUMO modification. Similar to the ubiquitin and NEDD8 E2s, we found that the SUMO E2 confers an inhibitory effect on its E1. NMR chemical shift perturbation showed that the E2 can bind to both the movable UFD and Cys domains of free E1, in the absence of the E1∼SUMO thioester formation, suggesting that the UFD undergoes large-scale domain movements in free E1. ATP:PPi exchange experiments further revealed that the E1-E2 interaction inhibited E1-catalyzed SUMO adenylation. Furthermore, site-directed mutagenesis in combination with biochemical assays showed that the same E1-E2 interfaces are responsible for both the inhibitory effect and formation of the E2∼SUMO thioester. Because E2 binds to both domains of E1 that undergo large-scale domain movements, and thus would stabilize the conformation of E1, this study indicates that conformational flexibility of E1 is required for SUMO adenylation.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and Protein Preparation
Site-directed mutagenesis of Ubc9 was carried out using QuikChange mutagenesis protocol (Stratagene), using primers designed for each mutation. All plasmid constructs were confirmed by DNA sequencing. Expression and purification of recombinant proteins were carried out as described previously (9, 13).
Protein Labeling and NMR
Full-length SUMO E1 (∼110 kDa) was expressed in lysogeny broth medium. Ubc9 was expressed in M9 medium containing 15NH4Cl as the sole nitrogen source. The M9 medium was also dissolved in 100% D2O to eliminate aliphatic protons that could broaden the resonances of amide protons, which were observed in 1H-15N TROSY, free and in complex with the deuterated SUMO E1. NMR experiments were carried out at 30 °C in 25 mm phosphate buffer, pH 6.0. All NMR experiments were performed on a Brucker Avance 600 MHz spectrometer equipped with cryo-probe, pulse shaping, and pulsed field gradient capabilities.
In Vitro Assays for SUMO Conjugation with E1, Ubc9, and RanGAP1
All conjugation assays were conducted in a mixture containing ATP and its regeneration system (50 mm Tris, pH 7.5, 5 mm MgCl2, 2 mm ATP, 10 mm creatine phosphate, 3.5 units/ml creatine kinase, and 0.6 units/ml inorganic pyrophosphatase). Reaction mixtures were incubated at 37 °C for the indicated times prior to addition of SDS gel loading buffer (with or without reducing reagent dithiothreitol) to stop the reaction. Samples were then resolved on SDS-PAGE gels, and polypeptide bands were visualized by SimpleBlue staining (Invitrogen).
For Ubc9∼SUMO thioester complex formation, SUMO (20 μm) and E1 (0.5 μm) were incubated with serial concentrations of Ubc9 (2.5–20 μm), and aliquots were taken after 5, 10, and 20 min and mixed with nonreducing SDS gel loading solution for electrophoresis and further analysis. For assays of RanGAP1∼SUMO isopeptide complex formation, SUMO (15 μm) and RanGAP1 (15 μm) were incubated in the presence of E1 (0.1 μm) and serial concentrations of Ubc9 (1–12 μm). Aliquots were withdrawn at 5, 10, and 20 min after initiation of the reaction for SDS-PAGE analysis.
Radioactive PPi:ATP Exchange Assays
Radioactive isotope-based PPi:ATP exchange assay was adopted from Burch and Haas (14). Briefly, SUMO (10 μm) was incubated (37 °C for 20 min) with E1 enzyme (0.2 μm) in a 50-μl reaction mixture containing 50 mm Tris, pH 7.5, 10 mm MgCl2, 0.5 mm DTT, 1 mm ATP, and 1 mm [32P]PPi (PerkinElmer Life Sciences). The reaction was then quenched by addition of 5% (w/v) TCA (0.5 ml) containing 4 mm carrier PPi. The isotope exchange reactions were carried out in the presence of various Ubc9 mutants at varying concentrations (0.25–8 μm). The 32P-incorporated ATP was absorbed to a 10% (w/v) slurry of activated charcoal (0.3 ml; Sigma) in 2% TCA, and the charcoal was rinsed three times with 2% TCA (1 ml) prior to Cerenkov counting. For data analysis, the production of radioactive ATP (percentage of CPM over total radioactive input) by the reaction without Ubc9 was used as reference, and relative activity was calculated for all other assays.
E1-E2 Binding and Native Gel Shift Assay
For the E1-E2 binding assay, wild-type or mutant E1 (11.3 μg, 0.1 nmol) was mixed with Ubc9 (20 μg, 1 nmol) in a buffer containing 50 mm Tris, pH 7.6, 150 mm NaCl, and 5 mm dithiothreitol, and the mixture was rocked end-to-end for 1.5 h at room temperature. After being mixed with native gel loading buffer, samples were resolved on nondenaturing 6% Tris-glycine PAGE gel (Invitrogen), and proteins were visualized by SimpleBlue staining.
RESULTS
Substrate Inhibitory Effect of Ubc9
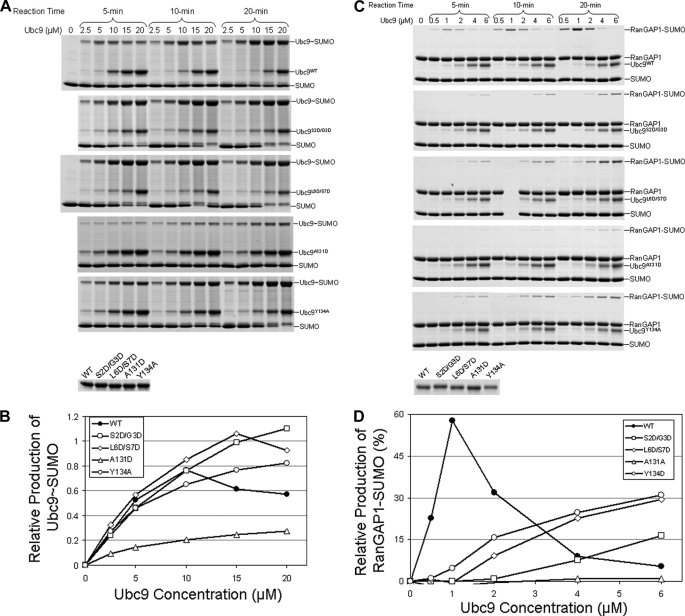
The E2 enzymes of NEDD8 and ubiquitin display classical substrate inhibitory effects on their cognate E1 enzymes: increasing concentrations of the E2s inhibit formation of E2∼Ubl thioesters, in which E2 and Ubl are both the substrates of E1 (11, 12). This effect is thought to be the result of E2 binding to free E1; however, the mechanism for such inhibition has been unclear. We first examined whether the SUMO E2 (also known as Ubc9) confers a similar substrate inhibitory effect for formation of the Ubc9∼SUMO thioester conjugate. Increasing concentrations of E2 lead to the formation of less E2∼SUMO thioester conjugate (Fig. 1A, top panel, and B). This effect was observed under various reaction times and concentrations of the E1 and E2 enzymes. Thus, the E2 inhibitory effect appears to be a conserved property of ubiquitin-like modifications, reflecting the ordered steps of these modifications.
FIGURE 1.
Effect of Ubc9 and Ubc9 mutants that have reduced affinity for E1 on formation of the Ubc9∼SUMO thioester. A, the E1-catalyzed formation of the Ubc9∼SUMO thioester conjugate in the presence of wild-type and mutant Ubc9 at the five indicated concentrations of wild-type and mutant Ubc9 and the three indicated incubation times. B, quantification of the assay results in A. C, formation of RanGAP1-SUMO isopeptide complex in the presence of wild-type and mutant Ubc9. The conjugation assays were carried out at the five indicated concentrations of wild-type and mutant Ubc9 and the three indicated incubation times. All other conditions of the reactions are identical among all reactions in A and C. Details of the assay conditions are as described under “Experimental Procedures.” Loading control gels for wild-type and mutant Ubc9 proteins are shown as the bottom gels of A and C. D, quantification of the assay results in C.
Next, we examined whether the E2 inhibitory effect is limited to formation of the E2∼SUMO thioester, which is “half” of the SUMO modification process, or extends to the overall conjugation reactions. To this end, SUMOylation of RanGAP1 and Sp100 was examined. Although Sp100 SUMOylation is stimulated by the E3 ligase RanBP2, RanGAP1 can be SUMOylated efficiently in the absence of an E3, such that any potential effect of E3 can be eliminated. RanGAP1 modification by SUMO was carried out in the presence of E1 and at increasing concentrations of Ubc9 (Fig. 1, C and D). Again, higher concentrations of the E2 inhibited formation of SUMOylated RanGAP1. Similarly, the E2 inhibitory effect was also observed in Sp100 conjugation in the presence of E3 (data not shown).
The concentrations of Ubc9 when the maximal RanGAP1∼ SUMO conjugates was formed were ∼5-fold less than that when the maximal Ubc9∼SUMO thioesters were achieved. This is likely due to the 5-fold lower E1 concentration (0.1 μm) used in the overall conjugation assays than that used in Ubc9∼SUMO thioester formation assays (0.5 μm), where higher Ubc9 concentrations were used for detection of Ubc9 and Ubc9∼SUMO thioester. Taken together, these data suggest that the E2 inhibitory effect is directly linked to the E1 concentration used in the reactions. In addition, the inhibitory effect of E2 is not limited to the formation of E2∼SUMO thioester but has extended impact on the overall conjugation reactions.
E2 Binds to Both the UFD and Cys Domain of Free E1
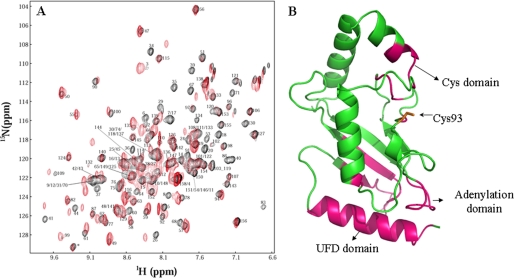
To probe how E2 and E1 interact in the absence of thioester-bonded SUMO, we examined E1-E2 interactions using NMR chemical shift perturbation. 2H/13C/15N-enriched Ubc9 and E1 were used to form a complex (Fig. 2). Fig. 2A shows the superimposed 1H-15N TROSY spectra of Ubc9, free and in complex with the full-length E1. Extensive chemical shift perturbation in Ubc9 occurred upon complex formation with E1. However, the resonances of some residues disappeared upon complex formation (line broadening effect), whereas those of other residues showed significant chemical shift changes. Such chemical shift perturbation is consistent with the affinity between E1 and E2 being ∼100 nm (9).
FIGURE 2.
Identification of the binding interface of free E1 on Ubc9. A, superimposed TROSY spectra of 2H/15N/13C-enriched Ubc9, free (black) and bound to E1 (red). Assignments of amino acid residues of free Ubc9 are shown. B, the three-dimensional structure of Ubc9. Residues that showed the most significant chemical shift perturbation upon binding E1 are indicated in magenta. The active site Cys-93 and the surfaces interacting with the UFD and Cys domains are indicated.
Those residues for which resonances disappeared or shifted significantly (indicated in magenta in the three-dimensional structure of Ubc9; Fig. 2B) occurred within the regions of residues 2–18, 26–44, 64–71, 83–86, 91, 109, and 129–134. Although the surfaces of a protein identified by chemical shift perturbation extend beyond the direct contacting surface, the surface that showed chemical shift perturbation revealed that the E1-E2 interaction in SUMOylation is completely analogous to the E1-E2 interaction in NEDD8 modifications (8, 15); the chemical shift perturbation on Ubc9 indicates that it forms interactions with UFD and the adenylation and the Cys domains of the SUMO E1, as found in the ternary complex of the NEDD8 E2 in complex with the E1∼NEDD8 thioester (8, 9, 15). These data indicate that the E2 can directly bind to all three sites of the free E1 simultaneously, without conjugation of SUMO to E1. Thus, the UFD should be able to rotate between the “open” and “closed” conformation in the absence of SUMO∼E1 thioester formation, because if the UFD stays in the “closed” conformation in the absence of SUMO∼E1 thioester formation, the E2 would not be able to bind to the Cys domain, and chemical shift perturbation of residues 129–134 of Ubc9, which interact with the Cys domain (9), would not have been observed.
E2-E1 Interfaces for Productive and Inhibitory Effects
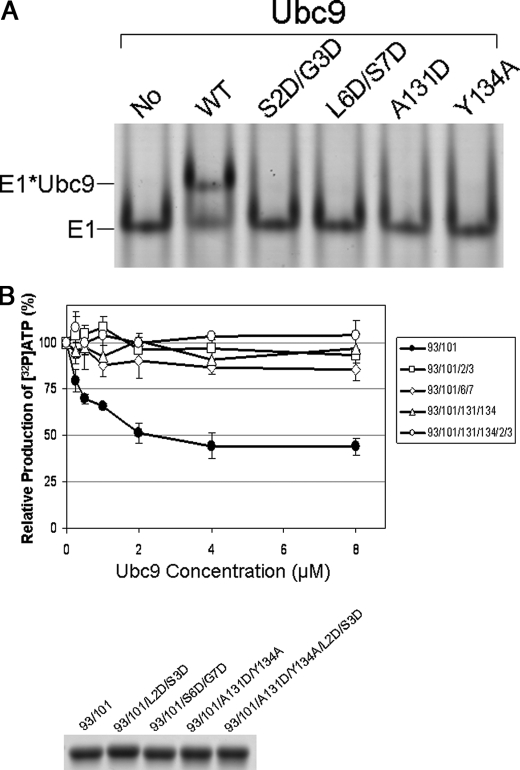
To determine how the different interfaces between E2 and E1 contribute to the productive and inhibitory effects of E2, we analyzed E2 enzymes with mutations that selectively disrupt the interfaces with UFD or the Cys domain. Because E2 has overlapping surfaces for noncovalent binding of SUMO and the UFD of E1, we designed specific mutations that disrupt only the E2-E1 interaction but not the E2 and SUMO interaction, based on the structure of the noncovalent complex between Ubc9 and SUMO (16, 17). Two Ubc9 mutants Ser-2 and Gly-3 to Asp (S2D/G3D) and Leu-6 and Ser-7 to Asp (L6D/S7D) disrupted the interaction with E1 (Fig. 3A). However, their structural integrities and affinities for SUMO-1 or RanGAP1 were the same as for wild-type Ubc9, as revealed by NMR spectra and NMR chemical shift perturbation (data not shown). These mutations are located at the surface next to the SUMO-binding site but are >30Å from the RanGAP1 binding surface (18). Furthermore, these mutant proteins were less efficient at forming E2∼SUMO thioester conjugates at low Ubc9 concentrations (Fig. 1, A and B) and were less active than wild-type Ubc9 in the overall conjugation reactions (Fig. 1, C and D). However, unlike wild-type E2, the E1-binding deficient Ubc9 mutants S2D/G3D and L6D/S7D were not inhibitory toward formation of thioester with SUMO, or toward overall conjugation of SUMO to RanGAP1, as the amount of conjugated product increased as the E2 concentration increased (Fig. 1). Thus, at high Ubc9 concentrations, these mutants appear to be more active than wild-type E2. The lack of inhibitory effects of these mutants ruled out the possibility that the inhibitory effect of Ubc9 in the overall conjugation reactions is due to competition of free Ubc9 with SUMO-loaded Ubc9 for binding RanGAP1 because the Ubc9 S2D/G3D and L6D/S7D mutants have similar affinity for RanGAP1 as wild-type Ubc9.
FIGURE 3.
Binding of Ubc9 to E1 inhibits E1-catalyzed adenylation. A, native gel analysis of the binding between E1 and the WT and mutant Ubc9 proteins. The far left lane shows E1 alone, and other lanes show mixtures of E1 with the wild-type and mutated Ubc9 proteins. Positions of E1 and the E1-Ubc9 complex are indicated. B, ATP:PPi exchange assays in the presence of the E1 binding-capable Ubc9 mutant C93A/K101R that cannot accept SUMO from E1 and Ubc9 mutants that are E1 binding-deficient as well as SUMO conjugation-deficient mutants. E1 binding-capable Ubc9 inhibited the E1 catalyzed PPi:ATP exchange reaction, but E1 binding-deficient Ubc9 mutants significantly reduced the inhibitory effect of E1 activity. The SDS-PAGE gel below the graph shows that equal quantities of the wild-type and mutant Ubc9 proteins were used in the assays.
In addition to the mutants that disrupted Ubc9 binding to the UFD domain of E1, we also tested Ubc9 mutants that had been previously shown to disrupt the interaction between Ubc9 and the Cys domain during productive transfer of SUMO from E1 to E2 (9). Because Ubc9 residues Ala-131 and Tyr-134 are important for the interaction with the Cys domain of E1, we mutated these residues. The A131D and Y134A mutants had greatly reduced ability to bind E1 (Fig. 3A), as well as reduced activity in forming E2∼SUMO thioester conjugates and conjugates of RanGAP1 at low concentrations of the E2 (Fig. 1). Similar results were observed using Sp100 as a substrate (data not shown). However, neither mutant displayed the inhibitory effect, and as concentration of the mutants increased, they formed monotonically increasing amounts of SUMO∼E2 thioester and SUMO∼RanGAP1 conjugate (Fig. 1), suggesting that the productive interaction between Ubc9 and the Cys domain of E1 is also responsible for the inhibitory effects of the E2.
Binding of E2 to E1 Inhibits SUMO Adenylation
To investigate how E2 confers the substrate inhibitory effect, we used radioactivity-based pyrophosphate group (PPi) exchange (19) to investigate whether binding of Ubc9 to E1 inhibited the first step of E1-catalyzed reactions – SUMO adenylation. When 32P-labeled (“hot”) PPi is mixed with unlabeled (“cold”) ATP in the presence of E1 and SUMO, ATP gradually becomes radioactive, whereas PPi gradually loses radioactivity because of the reversibility of the SUMO adenylation reaction. To prevent SUMO from transferring to the E2, we mutated the active site Cys-93 to Ala (C93A). Small amounts of C93A mutants formed isopeptides with SUMO at Lys-101, which is the closest Lys to Cys-93; therefore, we also generated and incorporated a K101R mutation. In addition, the four previously described mutations with reduced ability to bind E1 (i.e. S2D/G3D, L6D/S7D, A131D, and Y134A) also were incorporated into the Cys-93/Lys-101 mutant. All mutant proteins maintained their structural integrity, as shown by NMR spectra (data not shown). The design of these thioester-deficient E2 variants was necessary to examine the specific effects on adenylation, because transthiolation between E1 and E2 is reversible (20); thus, high concentrations of wild-type Ubc9, which binds E1 with higher affinity than the mutants, could shift the equilibrium to E1∼SUMO thioester, thereby inhibiting SUMO adenylation. The C93A/K101R mutant inhibited E1 catalyzed SUMO adenylation, and the inhibitory effect increased as Ubc9 concentration increased (Fig. 3, B and C). However, none of the four mutants with reduced affinity for E1 were inhibitory to ATP:PPi exchange in comparison, indicating that the noncovalent interaction between E1 and E2 inhibits E1-catalyzed SUMO adenylation. The inhibitory effect of wild-type Ubc9 was less pronounced on SUMO adenylation (Fig. 3) than on the overall conjugation reactions, likely due to the added effect from reversal of transthiolation between E1 and E2 (20).
DISCUSSION
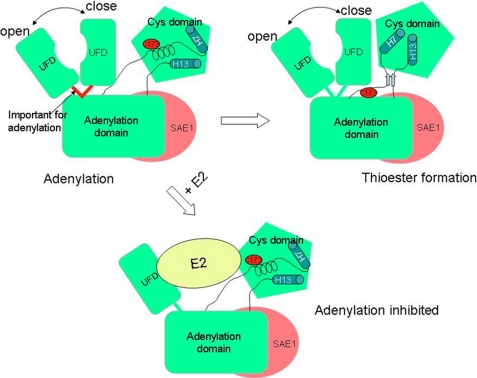
Using NMR chemical shift perturbation data, we have shown that in the absence of SUMO thioester conjugation to E1, E2 not only binds to the UFD but also simultaneously binds to the Cys domain of SUMO E1 (Fig. 2). In the previously determined x-ray structures of the NEDD8 and SUMO E1s (6, 7, 10), the UFD is oriented in a closed conformation that prevents binding of E2 to the Cys domain. However, the crystal structure of the NEDD8 E1 in a ternary complex with thioester-conjugated SUMO and E2 showed that the UFD domain rotated 120 °, leading to an open conformation that allows simultaneous interactions of E2 with both the UFD and the Cys domain (8). The importance of the UFD and Cys domains of the ubiquitin and SUMO E1s in recruiting their cognate E2 for transferring their cognate Ubl from E1 to E2 was confirmed by site-directed mutagenesis and enzyme kinetic analysis (9, 21). It was not clear whether rotation of UFD to the open conformation is triggered by E1∼Ubl thioester conjugation. Results described here suggest that the UFD undergoes domain movement between the open and closed states (Fig. 4) in free E1. Consistent with this, the x-ray structure of yeast ubiquitin E1, without ubiquitin conjugation, showed that UFD is crystallized in the open conformation (22).
FIGURE 4.
Model of the E2 inhibitory effect on E1-catalyzed SUMO adenylation. The SAE1 and SAE2 subunits are shown in red and green, respectively. The domain movements of the UFD and Cys domain are illustrated, and the crossover loop connecting UFD to the rest of E1, which is important for adenylation, is highlighted in red. Catalytic Cys-173 is indicated with an oval, and the two helices in the Cys are depicted as cylinders to illustrate the Cys domain movement. Binding of E2 to free E1 is expected to restrict the conformational flexibility and thus adenylation as well.
We have also shown that binding of E2 to the free E1 leads to inhibition of SUMO adenylation. E2 does not bind in the vicinity of the adenylation active site of E1 and therefore should not alter adenylation in a direct way (8). However, E2 binds to both of the E1 domains that undergo large-scale conformational changes. Thus, the E1-E2 interaction should stabilize both domains in the open conformations and restrict their conformational changes (Fig. 4). It is unlikely, however, that restriction of the conformational flexibility of the Cys domain is responsible for inhibition of SUMO adenylation because the crystal structure of E1 in complex with SUMO, ATP, and Mg2+ is very similar to that of E1 in complex with a SUMO andenylate mimic (5). In addition, mutation of the catalytic Cys to Ala in the ubiquitin and SUMO E1 does not affect ATP:PPi exchange (19). Therefore, it is likely that restricting the movements of UFD is responsible for inhibition of SUMO adenylation. We have recently found that the crossover loop linking the UFD to the rest of E1 is important for the adenylation activity of E1 (Fig. 4).5 Thus, restricted conformational flexibility of UFD likely restricted the flexibility of the crossover loop necessary for optimal SUMO adenylation activity.
We have shown here that the same interfaces between E1 and E2 are responsible for both the productive transfer of SUMO from E1 to E2 and the inhibitory effect of E2 on E1 activity. These studies suggest that the E2 inhibitory mechanism is due to alteration of the conformational flexibility of E1 required for adenylation. A similar mechanism is likely responsible for the inhibitory effect displayed by the E2s of ubiquitin, NEDD8, and possibly other Ubls.
This study underscores the importance of using a wide range of enzyme concentrations when characterizing the activities of ubiquitin-like modifications. As shown in Fig. 1, at low E2 concentrations, before the onset of the inhibitory effect, the E2 mutants that had significantly reduced affinities for E1 also were less efficient at forming the E2∼SUMO thioester and at overall SUMO conjugation. However, at higher concentrations of E2, greater than those that caused E1 inhibition with wild-type E2, these mutants had higher apparent activities, which can be misleading.
Ubiquitin-like modifications require multiple protein factors and steps. This study highlights the highly ordered steps of such processes, and a conformational flexibility-dependent mechanism for regulating the ordered nature of such processes.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM074748 and R01GM086171.
J. Wang, S. Cai, and Y. Chen, submitted for publication.
- SUMO
- small ubiquitin-like modifier
- UFD
- ubiquitin fold
- TROSY
- transverse relaxation optimized spectroscopy.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. (1997) Trends Biochem. Sci. 22, 383–387 [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O., Felberbaum R., Hochstrasser M. (2006) Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 4.Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olsen S. K., Capili A. D., Lu X., Tan D. S., Lima C. D. (2010) Nature 463, 906–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lois L. M., Lima C. D. (2005) EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walden H., Podgorski M. S., Schulman B. A. (2003) Nature 422, 330–334 [DOI] [PubMed] [Google Scholar]
- 8.Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., Schulman B. A. (2007) Nature 445, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Hu W., Cai S., Lee B., Song J., Chen Y. (2007) Mol. Cell 27, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L., Jr., Holton J. M., Schulman B. A. (2003) Mol. Cell 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 11.Siepmann T. J., Bohnsack R. N., Tokgöz Z., Baboshina O. V., Haas A. L. (2003) J. Biol. Chem. 278, 9448–9457 [DOI] [PubMed] [Google Scholar]
- 12.Wee K. E., Lai Z., Auger K. R., Ma J., Horiuchi K. Y., Dowling R. L., Dougherty C. S., Corman J. I., Wynn R., Copeland R. A. (2000) J. Protein Chem. 19, 489–498 [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Lee B., Cai S., Fukui L., Hu W., Chen Y. (2009) J. Biol. Chem. 284, 20340–20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burch T. J., Haas A. L. (1994) Biochemistry 33, 7300–7308 [DOI] [PubMed] [Google Scholar]
- 15.Huang D. T., Miller D. W., Mathew R., Cassell R., Holton J. M., Roussel M. F., Schulman B. A. (2004) Nat. Struct. Mol. Biol. 11, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q., Jin C., Liao X., Shen Z., Chen D. J., Chen Y. (1999) J. Biol. Chem. 274, 16979–16987 [DOI] [PubMed] [Google Scholar]
- 17.Capili A. D., Lima C. D. (2007) J. Mol. Biol. 369, 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 19.Haas A. L., Rose I. A. (1982) J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 20.Haas A. L., Bright P. M. (1988) J. Biol. Chem. 263, 13258–13267 [PubMed] [Google Scholar]
- 21.Huang D. T., Zhuang M., Ayrault O., Schulman B. A. (2008) Nat. Struct. Mol. Biol. 15, 280–287 [DOI] [PubMed] [Google Scholar]
- 22.Lee I., Schindelin H. (2008) Cell 134, 268–278 [DOI] [PubMed] [Google Scholar]