Abstract
Several missense mutations in the protein kinase Cγ (γPKC) gene have been found to cause spinocerebellar ataxia type 14 (SCA14), an autosomal dominant neurodegenerative disease. We previously demonstrated that the mutant γPKC found in SCA14 is susceptible to aggregation, which induces apoptotic cell death. The disaccharide trehalose has been reported to inhibit aggregate formation and to alleviate symptoms in cellular and animal models of Huntington disease, Alzheimer disease, and prion disease. Here, we show that trehalose can be incorporated into SH-SY5Y cells and reduces the aggregation of mutant γPKC-GFP, thereby inhibiting apoptotic cell death in SH-SY5Y cells and primary cultured Purkinje cells (PCs). Trehalose acts by directly stabilizing the conformation of mutant γPKC without affecting protein turnover. Trehalose was also found to alleviate the improper development of dendrites in PCs expressing mutant γPKC-GFP without aggregates but not in PCs with aggregates. In PCs without aggregates, trehalose improves the mobility and translocation of mutant γPKC-GFP, probably by inhibiting oligomerization and thereby alleviating the improper development of dendrites. These results suggest that trehalose counteracts various cellular dysfunctions that are triggered by mutant γPKC in both neuronal cell lines and primary cultured PCs by inhibiting oligomerization and aggregation of mutant γPKC.
Keywords: Fluorescence, Neurodegeneration, Protein Kinase C (PKC), Signal Transduction, Synapses, Purkinje Cells, Aggregation, Oligomerization, Spinocerebellar Ataxia Type 14, Trehalose
Introduction
Autosomal dominant spinocerebellar ataxias (SCAs)2 are a heterogeneous group of neurological disorders that are clinically characterized by various symptoms of cerebellar dysfunction, including progressive ataxia of gait and limbs, cerebellar dysarthria, and abnormal eye movement. SCAs are classified into at least 28 types according to the chromosomal location of the causal genes (1–3). CAG trinucleotide repeat expansions in coding regions were the first types of mutations identified in SCAs (SCA1, -2, -3, -6, -7, and -17 and dentatorubral pallidoluysian atrophy), which are considered polyglutamine diseases (4, 5). Thereafter, missense mutations and deletions were found in other SCAs (SCA5, -11, -13, -14, -15, and -27) (3, 6, 7). SCA14 is caused by missense mutations in the PRKCG gene encoding protein kinase Cγ (γPKC), which was first identified by Chen et al. in 2003 (8). To date, 23 mutations have been identified in different SCA14 families, including a 2-amino acid-deletion mutant (ΔK100-H101) (9–15).
PKC is a family of serine/threonine kinases that plays important roles in various cellular functions by participating in diverse signal transduction pathways. The subtype γPKC is specifically present in the central nervous system and is especially abundant in cerebellar Purkinje cells (PCs) (16). γPKC knockout mice show mildly impaired motor coordination and an incomplete elimination of synapses between Purkinje cells and climbing fibers during development (17, 18). These ataxic symptoms in γPKC knockout mice are milder than those found in SCA14 patients, however. Moreover, SCA14 is inherited in an autosomal dominant fashion, raising the possibility that a toxic gain of function of mutant γPKC, rather than a loss of function, underlies the pathogenesis of SCA14.
We have previously demonstrated that mutant versions of γPKC tend to form aggregates in cultured cells (19) and mouse primary cultured PCs (20). This aggregation causes apoptotic cell death by inhibiting the ubiquitin proteasome system and inducing endoplasmic reticulum stress (21). Furthermore, mutant γPKC forms soluble oligomers and induces the improper development of PC dendrites (20). Aggregation and oligomerization of a mutant or misfolded protein is also frequently observed in various other neurodegenerative diseases, including Parkinson disease, Alzheimer disease, amyotrophic lateral sclerosis, and polyglutamine diseases (22, 23), suggesting that aggregation is a common part of the pathogenesis of neurodegenerative diseases like SCA14. Therefore, we believe that drugs that inhibit the aggregation of mutant γPKC might be useful to treat SCA14 and related neurodegenerative diseases.
Trehalose is a natural disaccharide of two glucose molecules in an α,α-1,1-glycosidic linkage that is resistant to cleavage by acid or glycosidases. It is present in various non-mammalian species, including bacteria, yeast, fungi, insects, invertebrates, and plants, but no trehalose is found in mammals (24). It has been shown to protect proteins from denaturation and aggregation, helping the cell maintain homeostasis and handle various environmental stresses (25). Trehalose has also been shown to inhibit the aggregation of disease-related proteins, including polyglutamine-expanded huntingtin in Huntington disease (26), β-amyloid protein in Alzheimer disease (27), and protease-resistant prion protein in prion disease (28).
In the present study, we examined whether trehalose could inhibit the aggregation and cytotoxic effects of mutant γPKC in SH-SY5Y cells and primary cultured cerebellar PCs. We demonstrate that intracellular trehalose directly inhibits the aggregation of mutant γPKC without affecting protein turnover. By inhibiting aggregation in both cell types, trehalose also prevents the apoptotic cell death that is triggered by the presence of mutant γPKC. We also show that trehalose reverses the impairment of dendritic development as well as the attenuated mobility and insufficient translocation of mutant γPKC in PCs lacking mutant γPKC aggregates.
EXPERIMENTAL PROCEDURES
Materials
Trehalose, Hoechst 33342, Ham's F-12 medium, Dulbecco's modified Eagle's medium (DMEM), and anti-β-tubulin 1 mouse monoclonal antibody were obtained from Sigma-Aldrich. SUMITOMO Nerve-Cell Culture System (Neuron culture medium and Dissociation solutions) was from Sumitomo Bakelite (Tokyo, Japan). The anti-GFP mouse monoclonal antibody was from Nacalai Tesque (Kyoto, Japan). The anti-γPKC rabbit polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-calbindin D28k antibody was from Swant (Bellinzona, Switzerland). The anti-activated (cleaved) capsase-3 and anti-glutamate receptor δ1 and δ2 (GluRδ1/2) rabbit polyclonal antibodies were from Millipore (Billerica, MA). The anti-heat shock protein 40 (Hsp40) rabbit polyclonal antibody and anti-heat shock proteins 70 (Hsp70) and 90 (Hsc90) mouse monoclonal antibodies were from Enzo Life Sciences (Plymouth Meeting, PA). The anti-heat shock cognate protein 70 (Hsc70) mouse monoclonal IgM antibody was from Abcam (Cambridge, United Kingdom). The anti-horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, anti-rabbit IgG, and anti-mouse IgM antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Alexa Fluor 546 (Alexa546)-conjugated goat anti-mouse IgG and anti-rabbit IgG antibodies, normal goat serum, and Hanks' balanced salt solution were from Invitrogen (Carlsbad, CA). Glass-bottomed culture dishes (35-mm diameter) were from MatTek (Ashland, MA). Glass-bottomed culture dishes with grids (35-mm diameter) were from Matsunami Grass (Kishiwada, Japan).
Cell Culture
SH-SY5Y cells were cultured in DMEM/F-12 mixture (1:1) medium, supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C.
Mouse cerebellar primary culture was prepared as described previously (20). Briefly, E18 embryos from pregnant ICR mice were dissociated with the dissociation solutions of the SUMITOMO Nerve-Cell Culture System according to the manufacturer's protocol. Dissociated cerebellar cells were suspended in the neuron culture medium of the SUMITOMO Nerve-Cell Culture System and plated at 2 × 105 cells/100 μl on the center of a 35-mm diameter glass-bottomed culture dish. Cells were cultured for 28–29 days in vitro (DIV) in a humidified atmosphere containing 5% CO2 at 37 °C. One-half of the medium was changed every 3–4 days.
Construction of Adenoviral Vectors to Express γPKC-GFP Using a Tet-regulated System
We constructed two types of adenoviral vectors to regulate the expression of γPKC-GFP with a tetracycline (Tet)-regulated system in promoter- and Tet-dependent manners (supplemental Fig. 1). The first type vector encodes Tet transactivator (tTA) cDNA under the control of the cytomegaloviral (CMV) promoter for SH-SY5Y cells (Ad-CMV-tTA) or L7 promoter for primary-cultured PCs (Ad-L7-tTA). The second type of vector, Ad-TetOp-γPKC-GFP, encodes WT or mutant γPKC-GFP cDNA under the control of the TetOp minimal promoter, which is transactivated by tTA.
Ad-L7-tTA and Ad-TetOp-γPKC-GFP were constructed as previously described (20). Ad-CMV-tTA was constructed using an AdEasy adenoviral vector system (Stratagene, La Jolla, CA, USA) according to the manufacturer's protocol. Briefly, tTA cDNA was subcloned from a pTet-tTAk plasmid (Invitrogen) into pShuttle-CMV (Stratagene) downstream of the CMV promoter. The shuttle vector was recombined with pAdEasy-1, an adenoviral backbone cosmid vector, in the E. coli strain BJ5183. The recombinant adenoviral genome was removed from the cosmid vector using PacI and transfected into HEK293 cells. Adenoviral vectors that proliferated in HEK293 cells were extracted and concentrated with a cesium chloride ultracentrifugation.
Expression and Live Imaging of γPKC-GFP
SH-SY5Y cells were spread on 3.5-cm-diameter glass-bottom (2 × 105 cells/dish) or 6-cm-diameter (5 × 105 cells/dish) dishes. After a 24-h cultivation, cells were infected with two adenoviral vectors, Ad-CMV-tTA and Ad-TetOp-γPKC-GFP, at a multiplicity of infection (m.o.i.) of 10 and cultured for another 2 days. Various concentrations of trehalose (10–500 μm) were added at the time of the adenoviral infection.
On DIV14 or DIV22, primary-cultured PCs were infected with Ad-L7-tTA (m.o.i. of 20) and Ad-TetOp-γPKC-GFP (m.o.i. of 3) and further cultured for 7–14 days. Trehalose (100 μm) was added at the time of adenoviral infection.
The GFP fluorescence in the living cells on the glass-bottom dishes was detected using a 488-nm argon laser excitation with a 505- to 530-nm band-pass barrier filter on a confocal scanning fluorescent microscope (LSM 510 META, Carl Zeiss). In the primary cultured cells, PCs were morphologically distinguished by their relatively large somata and highly branched dendrites. The number of cells with γPKC-GFP aggregates from a total of 100–150 cells expressing γPKC-GFP was counted in each experiment. The areas of γPKC-GFP-expressing PCs were calculated from Z-stack projected images using Image-Pro Plus 5.1 (Media Cybernetics, Bethesda, MD).
Observation of γPKC-GFP Translocation and Fluorescent Recovery after Photobleaching
On DIV28, culture medium was replaced with 950 μl of HEPES buffer (NaCl, 165 mm; KCl, 5 mm; CaCl2, 1 mm; MgCl2, 1 mm; HEPES, 5 mm; glucose, 10 mm; pH 7.4). In trehalose-treated cells, the medium was replaced with HEPES buffer containing 100 μm trehalose. Translocation of WT and mutant γPKC-GFP was induced with the direct application of 50 μl of HEPES buffer containing high KCl (KCl, 100 mm; NaCl, 70 mm; CaCl2, 1 mm; MgCl2, 1 mm; HEPES, 5 mm; glucose, 10 mm; pH 7.4). With a confocal laser microscope, fluorescent images of γPKC-GFP in PC dendrites were recorded every 0.5 s for 5 min before and after stimulation. To quantitatively analyze the translocation amplitude, changes in fluorescence of cytosolic γPKC-GFP were measured using LSM510 software and normalized to the total fluorescence in the image.
For fluorescent recovery after photobleaching, fluorescent images of γPKC-GFP in the somata and dendrites of PCs were obtained at lower resolution to rapidly monitor changes in GFP fluorescence. To photobleach γPKC-GFP, repetitive irradiation with the maximal excitation laser was applied to a small circular region of the somata and dendrites of PCs. Sequential fluorescent images were recorded every 0.1 s for 2 min before and after photobleaching. Changes in fluorescence in the bleached area were normalized to the total fluorescence of the cell and analyzed using Prism (GraphPad, San Diego, CA).
In these experiments, we observed PCs without any aggregates of γPKC-GFP. All experiments were performed at room temperature.
Immunoblotting and Chase Assay
Cells on 6-cm-diameter dishes were analyzed by immunoblotting using an anti-GFP antibody as described previously (19). Briefly, cells were lysed in radioimmune precipitation assay buffer (1% Nonidet P40, 0.1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 1 mm EDTA, 20 μg/ml leupeptin, 1 mm phenylmethanesulfonyl fluoride (PMSF), 1 mm sodium orthovanadate, 1 mm NaF, 100 nm Calyculin A and 10 mm Tris/HCl, pH 7.4) by sonication (UR-20P, TOMY SEIKO, Tokyo; output, 4; duty, 50%). Protein concentration in the cell lysate was quantified using a BCA protein assay kit (Pierce Biotechnology). Protein samples were subjected to SDS-PAGE, and separated proteins were then electrophoretically transferred onto polyvinylidine difluoride (PVDF) filters (Millipore, Bedford, MA). Nonspecific binding sites on the PVDF filters were blocked by incubating in 5% skim milk in PBS-T (0.01 m phosphate-buffered saline, pH 7.4, containing 0.03% Triton X-100) for >1 h at room temperature. After washing with PBS-T, the PVDF filters were incubated with PBS-T containing the anti-GFP (diluted 1:2,000), anti-activated caspase-3 (diluted 1:1,000), anti-Hsp40 (diluted 1:5,000), anti-Hsp70 (diluted 1:1,000), anti-Hsp90 (diluted 1:1,000), or anti-Hsc70 (diluted 1:5,000) antibodies and 1% normal goat serum for >1 h at room temperature. After further washing, the filters were incubated with PBS-T containing the HRP-conjugated anti-mouse IgG, anti-rabbit IgG, or anti-mouse IgM antibodies (diluted 1:10,000) for >30 min at room temperature. After three more washes, the immunoreactive bands were visualized with a chemiluminescence detection kit (Chemi-Lumi One, Nacalai Tesque) and detected with a luminescent image analyzer, LAS-1000plus (Fuji Photo Film, Tokyo, Japan).
To assess the degradation rates of the WT and mutant γPKC-GFPs, chase assays were conducted using the Tet-regulated system. One day after the adenoviral infection, SH-SY5Y cells were treated with Tet (1 μg/ml) to arrest the expression of γPKC-GFP. The degradation rates of γPKC-GFPs were analyzed by measuring the amount of residual γPKC-GFPs, which were detected with immunoblotting and 1 and 2 days after the Tet treatment. In this experiment, trehalose (100 μm) was added during the Tet treatment.
Analysis of Solubility of Recombinant Mutant GST-γPKC
Recombinant baculoviruses encoding glutathione S-transferase (GST)-tagged S119P (GST-S119P) and G128D (GST-G128D) mutant γPKC were prepared according to the manufacturer's instruction (Merck, Darmstadt, Germany). About 3–4 days after infection, Sf9 cells were collected and washed once with ice-cold PBS, and resuspended in ice-cold TBS-T (150 mm NaCl, 0.5% Triton X-100, and 50 mm Tris-HCl, pH 7.4) containing 20 μg/ml leupeptin and 1 mm PMSF. After homogenization using a sonicator, the cell lysates were cleared by centrifugation for 15 min at 15,000 × g at 4 °C. The supernatants were incubated for 4 h at 4 °C with glutathione-Sepharose 4B resin (Amersham Biosciences). After washing six times with ice-cold TBS-T, the bound proteins were eluted with 50 mm Tris-HCl and 20 mm reduced glutathione at pH 8.0. The recombinant proteins were dialyzed three times against 50 mm Tris-HCl at pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1 mm DTT and stored at −80 °C until use.
Recombinant GST-S119P and GST-G128D were mixed with an equal volume of PBS with or without trehalose (100 or 500 μm), followed by incubation at 37 °C for 1 h. The incubated samples were centrifuged at 15,000 × g for 10 min at 4 °C, and the supernatants were harvested as the soluble (S) fractions. The pellets were resuspended in the same volumes of radioimmune precipitation assay buffer, sonicated, and used as the insoluble (I) fractions. Half of each fraction was subjected to 7.5% SDS-PAGE; the amount of mutant GST-S119P in the S and I fractions was quantified by immunoblotting with anti-γPKC antibody (diluted in 1:2000) as described above.
Detection of Apoptotic Cells by Nuclear Staining
The extent of apoptotic cell death triggered by mutant γPKC-GFP was evaluated using nuclear fluorescent Hoechst 33342 staining as described previously (29). Briefly, in SH-SY5Y cells, adenoviral-infected cells were cultured for 3 days with various concentrations of trehalose, stained with 50 μg/ml Hoechst 33342 for 30 min, and harvested using a cell scraper. In primary-cultured PCs, cells infected on DIV14 were fixed with 4% paraformaldehyde in PBS and stained with 0.5 μg/ml Hoechst 33342; they were subsequently stained with anti-calbindin D28k mouse monoclonal antibody (diluted in 1:1000) and Alexa546 anti-mouse IgG antibody (diluted to 1:500). The fluorescence of Hoechst 33342 and Alexa546 were monitored with a confocal laser scanning fluorescent microscope using a 364 nm UV laser excitation and a 546 nm HeNe laser, respectively, and a 385- to 470 nm band pass barrier filter or 560 nm long pass barrier filter, respectively. Cells with condensed or fragmented nuclei were considered to be apoptotic.
Measurements of Intracellular Trehalose
The levels of intracellular trehalose were measured as described previously (30, 31). Briefly, SH-SY5Y cells, spread on 6-cm-diameter (1 × 106 cells/dish) dishes, were cultured for 48 h in the absence or presence of trehalose (1 and 10 mm). After three washes with ice-cold PBS, cells were harvested and vortexed with 0.1 mg of sorbitol as an internal standard in 1 ml of 90% ethanol. After centrifugation (18,000 × g, 15 min), the supernatant was desiccated completely using a vacuum concentrator. The dried residue was dissolved in ∼500 μl of MilliQ water, and the amount of trehalose was analyzed using high performance liquid chromatography.
RESULTS
Trehalose Inhibits Aggregation of Mutant γPKC-GFP Without Affecting Its Rate of Turnover in SH-SY5Y Cells
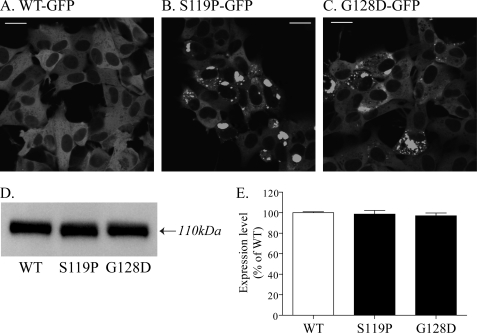
Of the 23 mutations that have been identified in SCA14 families, two mutant versions of γPKCs (S119P and G128D) were selected for this study; these mutants have been demonstrated to form aggregates at a high frequency (19). Wild-type and mutant γPKC-GFPs (WT-GFP, S119P-GFP, and G128D-GFP) were expressed in SH-SY5Y cells, a neuronal cell line, using two adenoviral vectors: Ad-CMV-tTA, which carries the tetracycline transactivator (tTA) gene downstream of the CMV promoter, and Ad-TetOp-γPKC-GFP, which uses the tTA-operated promoter (TetOp promoter) to transactivate γPKC-GFP expression (supplemental Fig. 1). 1 or 2 days after infection with Ad-CMV-tTA and Ad-TetOp-WT-γPKC-GFP at an m.o.i. of 10, 70–80% of SH-SY5Y cells were found to express WT γPKC-GFP in the cytoplasm (Fig. 1A). In contrast, in many of the cells infected with Ad-CMV-tTA and Ad-TetOp-S119P/G128D-γPKC-GFP, there were aggregates of S119P-GFP and G128D-GFP (Fig. 1, B and C). Several cells with aggregates of mutant γPKC-GFP were round and presumably dead. Few dead cells were observed in cells expressing WT-GFP (Fig. 1A), suggesting that the infection with these adenoviral vectors is not toxic. Furthermore, the expression levels were similar for WT-GFP-, S119P-GFP-, and G128D-GFP-γPKC in transfected SH-SY5Y cells (Fig. 1, D and E), so it cannot be true that aggregation is induced by higher expression levels of mutant γPKC-GFP.
FIGURE 1.
Expression and aggregation of WT and mutant γPKC-GFP in SH-SY5Y cells infected with adenoviral vectors. A–C, representative images of WT-GFP (A), S119P-GFP (B), and G128D-GFP (C) fluorescence in SH-SY5Y cells at 2 days post-adenoviral infection. No aggregates were found in cells expressing WT-GFP, whereas aggregates of S119P-GFP and G128D-GFP were frequently observed. Bar = 20 μm. D, representative anti-GFP immunoblot of SH-SY5Y cells expressing WT-GFP, S119P-GFP, and G128D-GFP. Cells were harvested 2 days after adenoviral infection. γPKC-GFP was detected with a molecular mass of ∼110 kDa. E, quantitative analysis of immunoblotting in D. Each column represents the mean ± S.E. of three independent experiments. The expression levels of WT-GFP, S119P-GFP, and G128D-GFP were similar in SH-SY5Y cells.
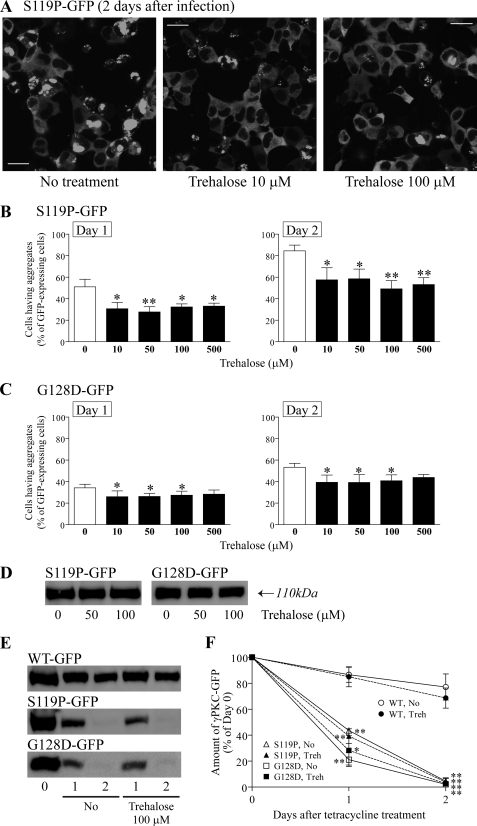
To assess the effect of trehalose on the aggregation of mutant γPKC-GFP, various concentrations of trehalose (10–500 μm) were added during adenoviral infection. After a 2-day cultivation, it was evident that trehalose prominently inhibits the aggregation of mutant γPKC-GFP (Fig. 2A). To quantitate this effect, the percentage of cells containing aggregates in the entire population of cells expressing mutant γPKC-GFP was calculated at 1 and 2 days after adenoviral infection (Fig. 2A). When trehalose was not added, ∼30–50% and 50–80% of cells expressing mutant γPKC-GFP contained aggregates at 1 and 2 days of cultivation, respectively (Fig. 2, B and C). Trehalose at all concentrations (10–500 μm) significantly decreased or tended to decrease the percentage of cells containing aggregates both in cells expressing S119P-GFP and G128D-GFP (Fig. 2, B and C). Moreover, in each fluorescent image obtained from experiments in Fig. 2 (B and C), the percentage of total aggregation area in GFP-expressing cells was quantified. Trehalose decreased the total aggregation area of mutant γPKC-GFP, more prominently than the number of cells having aggregation of mutant γPKC-GFP (supplemental Fig. 2), suggesting that trehalose reduces the size of mutant γPKC-GFP aggregates. However, trehalose at lower concentration (<10 μm) did not inhibit aggregation of mutant γPKC-GFP (supplemental Fig. 3), suggesting that trehalose at >10 μm is necessary to inhibits aggregation of mutant γPKC.
FIGURE 2.
Trehalose inhibits the aggregation of mutant γPKC-GFP in SH-SY5Y cells without affecting its expression level or degradation rate. A, representative images of S119P-GFP fluorescence in SH-SY5Y cells at 2 days post-adenoviral infection without (left) or with trehalose (center: 10 μm, right: 100 μm). Trehalose was added during the adenoviral infection. Trehalose was found to strongly reduce the number of cells with aggregates of S119P-GFP. Bar = 20 μm. B and C, quantitative analyses of the effect of trehalose on aggregation of S119P-GFP (B) and G128D-GFP (C). Left and right graphs indicate the results at 1 and 2 days after adenoviral infection, respectively. We evaluated 100–200 GFP-positive cells in each experiment. Each column represents the mean ± S.E. of the percentages of cells with aggregates. Various concentrations of trehalose significantly inhibit the aggregation of mutant γPKC-GFP in SH-SY5Y cells (*, p < 0.05; **, p < 0.01 versus without trehalose, Dunnett's multiple comparison test, n = 5 in S119P-GFP, n = 4 in G128D-GFP). D, representative anti-GFP immunoblot of SH-SY5Y cells expressing S119P-GFP (left) and G128D-GFP (right). Cells were cultured for 2 days after adenoviral infection without or with trehalose (50 or 100 μm). Trehalose does not affect the expression levels of S119P-GFP and G128D-GFP. E, representative anti-GFP immunoblot results of a chase assay for WT-GFP (upper), S119P-GFP (middle), and G128D-GFP (lower). One day after adenoviral infection, the expression of γPKC-GFP was arrested with the addition of tetracycline (Tet, 1 μg/ml). Trehalose (100 μm) was added at the time of Tet treatment. The number of elapsed days after Tet treatment is indicated below the panels. F, quantitative analyses of the chase assay in E. Each symbol represents the mean ± S.E. of the residual amount of WT-GFP (circle), S119P-GFP (triangle), or G128D-GFP (square), indicated as the percentage of day 0. Open and closed symbols indicate the results of cells without (No) and with 100 μm trehalose (Treh), respectively. S119P-GFP and G128D-GFP were found to be more rapidly degraded than WT-GFP after expression arrest, but trehalose does not significantly affect the degradation rates of WT-GFP, S119P-GFP, or G128D-GFP (*, p < 0.05; **, p < 0.01, unpaired t test, n = 3).
Recently, trehalose has been reported to promote degradation of mutant proteins by stimulating autophagy, a major protein degradation pathway (32); thus, it seemed possible that trehalose could inhibit aggregation by reducing the expression level or accelerating the degradation of mutant γPKC-GFP. Immunoblot experiments revealed that trehalose (50 and 100 μm) does not significantly affect the expression levels of the WT-GFP, S119P-GFP, and G128D-GFP after 2 days of cultivation, however (Fig. 2D). The degradation rate of mutant γPKC-GFP was also evaluated using a chase assay. In this assay, the expression of γPKC-GFP was arrested after 1 day of cultivation using tetracycline (Tet, 1 μg/ml); the residual amount of γPKC-GFP was quantified by immunoblotting at 1 and 2 days after Tet treatment. Although S119P-GFP and G128D-GFP were more rapidly degraded than WT-GFP, consistent with our recent study (33), trehalose was not found to affect the degradation rates of WT-GFP, S119P-GFP, or G128D-GFP (Fig. 2, E and F). These results indicate that trehalose inhibits the aggregation of mutant γPKC-GFP without affecting its turnover.
Trehalose Is Taken Up by SH-SY5Y Cells and Inhibits Aggregation by Directly Stabilizing Mutant γPKC
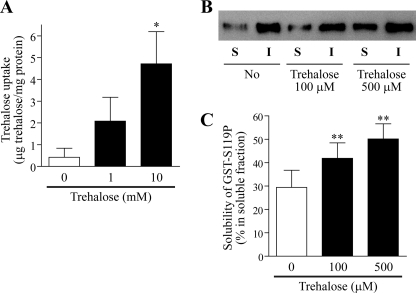
Because trehalose is considered to be a chemical chaperone that directly stabilizes the conformation of misfolded proteins (34), it is possible that the chemical is taken up by cells and inhibits aggregation by directly stabilizing protein structures. To address this possibility, we investigated whether trehalose could be incorporated into SH-SY5Y cells. Significant trehalose uptake was confirmed when SH-SY5Y cells were cultured in the presence of 10 mm trehalose for 48 h (Fig. 3A). Intracellular trehalose was slightly, but not significantly, detected in cells cultured with 1 mm trehalose.
FIGURE 3.
Trehalose is incorporated into SH-SY5Y cells and inhibits transition of recombinant mutant γPKC into the insoluble fraction. A, quantitative analysis of trehalose uptake in SH-SY5Y cells. Cells were cultured for 48 h in the presence of 0–10 mm trehalose, as indicated below the graph. The amount of intracellular trehalose was quantified as described under “Experimental Procedures.” The levels of intracellular trehalose were significantly higher in SH-SY5Y cells cultured with 10 mm trehalose for 48 h (*, p < 0.05 versus without trehalose, Dunnett's multiple comparison test, n = 5). B, representative anti-γPKC-immunoblot results of soluble (S) and insoluble (I) fractions of recombinant GST-S119P after a 1-h incubation at 37 °C without or with trehalose (100 or 500 μm). C, quantitative analysis of immunoblot results after a 1-h incubation in B. Each column represents the mean ± S.E. of the percentage of GST-S119P found in S fraction relative to the total amount of GST-S119P (S + I fraction). Trehalose significantly inhibited insolubilization of recombinant GST-S119P by a 1-h incubation at 37 °C (**, p < 0.01, Dunnett's multiple comparison test, n = 4).
To further investigate whether trehalose directly stabilizes mutant γPKC, we examined the effect of trehalose on the solubility of recombinant mutant γPKC in vitro. Recombinant GST-fused mutant S119P γPKC (GST-S119P) was constructed using a baculovirus system as described under “Experimental Procedures.” Without incubation, more than half of recombinant GST-S119P was detected in a soluble fraction (59.9 ± 14.3% in a soluble fraction). After a 1-h incubation at 37 °C, ∼70% of recombinant GST-S119P was found to accumulate in an insoluble fraction, probably due to aggregation (Fig. 3, B and C). However, incubation with trehalose (100 or 500 μm) significantly improved the solubility of recombinant GST-S119P (Fig. 3, B and C). Similar results were obtained by using recombinant GST-fused mutant G128D γPKC (GST-G128D) (supplemental Fig. 4). These results indicate that trehalose directly stabilizes recombinant mutant GST-γPKC and inhibits its aggregation.
Trehalose Inhibits Apoptosis Induced by Mutant γPKC-GFP in SH-SY5Y Cells
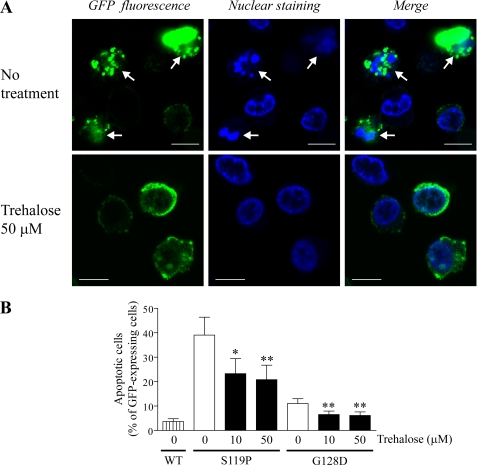
Apoptotic cell death induced by mutant γPKC-GFP was assessed by chromatin condensation detected with nuclear staining by Hoechst 33342 (50 μg/ml) (29). Three days after infection, we found a significantly higher percentage of apoptotic cells containing condensed nuclei in cells expressing S119P-GFP (39.1 ± 7.3%, n = 4, p < 0.01 versus WT-GFP, unpaired t test, Fig. 4, A and B) and G128D-GFP (11.0 ± 2.0%, n = 4, p < 0.05 versus WT-GFP, unpaired t test, Fig. 4B) compared with those expressing WT γPKC-GFP (3.6 ± 1.2%, n = 4). Although apoptotic cells induced by G128D-GFP were less than those by S119P-GFP, it would be reflected by the different tendency of these mutants to from aggregates; S119P-GFP more frequently formed aggregates than G128D-GFP (Fig. 2B). Trehalose (10 and 50 μm) was found to significantly lower the percentage of apoptotic cells in a population expressing S119P-GFP and G128D-GFP (Fig. 4, A and B). Moreover, trehalose (10 and 100 μm) slightly reduced the amount of activated caspase-3 in cells expressing S119P-GFP (supplemental Fig. 5). These results suggest that trehalose inhibits apoptotic cell death triggered by mutant γPKC-GFP.
FIGURE 4.
Trehalose alleviates cytotoxicity induced by mutant γPKC-GFP in SH-SY5Y cells. Effect of trehalose on apoptosis induced by mutant γPKC. Three days after adenoviral infection, SH-SY5Y cells were stained with Hoechst 33342 (50 μg/ml). Apoptosis was evaluated by examining chromatin condensation and fragmentation of the nuclei stained by Hoechst 33342. A, representative images of GFP (left), Hoechst 33342 (center), and merged fluorescence of cells expressing S119P-GFP in the absence (upper) or presence (lower) of 50 μm trehalose. Arrows in the upper images indicate apoptotic cells with condensed or fragmented nuclei. Bar = 10 μm. B, quantitative analyses of the apoptotic cells by nuclear staining in B. Each column represents the mean ± S.E. of the percentage of apoptotic cells in the GFP-positive population. Trehalose (10 and 50 μm) significantly reduces the number of apoptotic cells expressing S119P-GFP and G128D-GFP (*, p < 0.05; **, p < 0.01 versus without trehalose, Dunnett's multiple comparison test, n = 4).
Trehalose Inhibits Aggregation of Mutant γPKC-GFP and Improper Dendrite Morphology Induced by Mutant γPKC-GFP in Primary Cultured PCs
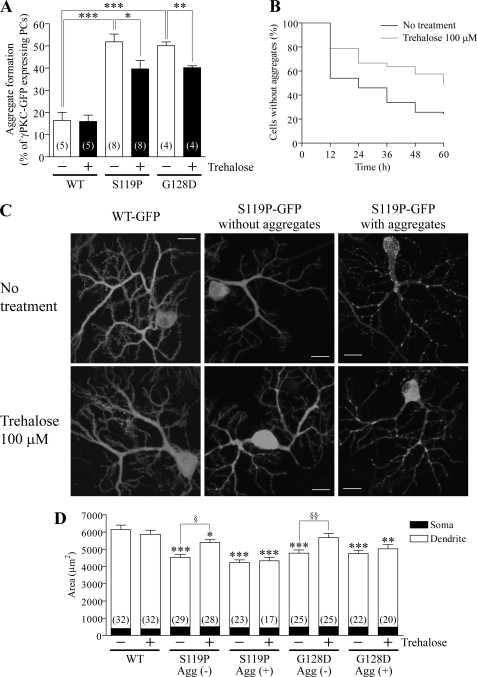
We recently demonstrated that mutant γPKC-GFP frequently forms aggregates and induces the incorrect development of dendrites in primary cultured PCs (20). We first examined whether trehalose could inhibit the aggregation of S119P-GFP in primary cultured PCs. PC-specific expression of WT-GFP, S119P-GFP, and G128D-GFP was achieved with adenoviral infection of Ad-L7-tTA, which express tTA in a PC-specific manner, and Ad-TetOP-γPKC-GFP (supplemental Fig. 1). Trehalose (100 μm) was added at the time of adenoviral infection on DIV14. After another 14 days of cultivation, aggregation of S119P-GFP and G128D-GFP was significantly inhibited in the trehalose-treated group; aggregation of WT-GFP was not affected by trehalose treatment (Fig. 5A). To precisely measure this inhibitory effect of trehalose on aggregate formation, we performed long term time-lapse imaging using an incubation imaging system (LCV100, Olympus). GFP fluorescence in PCs expressing S119P-GFP was observed every 30 min for 60 h from 3 to 6 days after adenoviral infection and trehalose (100 μm) treatment on DIV23. S119P-GFP that was diffuse in PCs at beginning of the observation was aggregated after 12 h in many PCs that were not treated with trehalose (34 of 74 observed cells, Fig. 5B and supplemental movie 1); most PCs (56 of 74 observed cells) had aggregates by end of the observation period (Fig. 5B). Trehalose was found to significantly delay aggregation of S119P-GFP in PCs (Fig. 5B, p < 0.01 versus no treatment, log rank test); more than half of the PCs (17 of 33 observed cells) did not appear to have any aggregates throughout the observation period (supplemental movie 2). These results indicate that trehalose inhibits aggregation of mutant γPKC-GFP in primary cultured PCs.
FIGURE 5.
Trehalose inhibits aggregation of mutant γPKC-GFP and mutant γPKC-GFP-mediated incorrect development of dendrites in primary cultured PCs. A, effect of trehalose on aggregation of WT-GFP, S119P-GFP, and G128D-GFP in primary cultured PCs. Adenoviral vectors were infected on DIV14, at the same time of trehalose (100 μm) treatment, and GFP fluorescence was observed on DIV28. Each column represents the mean ± S.E. of the percentage of cells with aggregates of γPKC-GFP in GFP-positive PCs. The numbers of experiments are indicated in the columns. S119P-GFP and G128D-GFP forms aggregates in PCs more frequently than WT-GFP, and trehalose significantly inhibits of the aggregation of S119P-GFP and G128D-GFP in PCs (*, p < 0.05; ***, p < 0.001, unpaired t test). B, effect of trehalose on aggregation of S119P-GFP in PCs evaluated by long term time-lapse imaging. Adenoviral vectors were infected on DIV22, at the same time of trehalose (100 μm) treatment, and the GFP fluorescence was acquired every 30 min for 60 h from DIV26 to DIV29. Aggregation of S119P-GFP was evaluated every 6 h. The black line (no treatment, 73 cells) and gray line (trehalose 100 μm, 33 cells) indicate the percentages of PCs that never formed aggregates during the observation period. Trehalose significantly delays aggregate formation of S119P-GFP in PCs (p < 0.01, log rank test). C and D, effect of trehalose on dendrite morphology in PCs expressing WT-GFP, S119P-GFP, and G128D-GFP. Adenoviral vectors were infected on DIV14, at the same time of trehalose (100 μm) treatment, and GFP fluorescence was observed on DIV28. C, representative images of GFP fluorescence in PCs expressing WT-GFP (left), S119P-GFP without aggregates (center), and S119P-GFP with aggregates (right). Upper and lower images show PCs without and with 100 μm trehalose treatment. D, quantitative analysis of areas of the PCs expressing WT-GFP, S119P-GFP, and G128D-GFP. Each column represents the mean ± S.E. of the PC areas, which were quantified by measuring the distribution of GFP fluorescence. The numbers of observed PCs are indicated in the columns. Open and closed portions of the columns indicate the areas of the PC dendrites and somata, respectively. Trehalose significantly prevents the decrease in area, especially in the dendrites, of PCs expressing S119P-GFP and G128D-GFP without aggregates; the areas of PCs with S119P-GFP and G128D-GFP aggregates were not affected by trehalose (*, p < 0.05; **, p < 0.01; ***, p < 0.001 versus WT-GFP with no treatment, §, p < 0.05; §§, p < 0.01, unpaired t test).
Next, we investigated the effect of trehalose on the incorrect development of PC dendrites triggered by the expression of mutant γPKC-GFP. In live PCs infected on DIV14, dendrites of PCs expressing S119P-GFP were found to be less expanded than those in PCs expressing WT-GFP, regardless of the presence or absence of S119P-GFP aggregates (Fig. 5C, upper panels). Trehalose treatment was found to improve dendrite development in PCs lacking S119P-GFP aggregates; trehalose does not affect dendrites of PCs expressing WT-GFP or PCs containing S119P-GFP aggregates (Fig. 5C, lower panels). To quantitatively measure this effect of trehalose, the areas of the PCs were measured using the distribution of the GFP fluorescence. As shown in Fig. 5D, the areas of PCs expressing S119P-GFP and G128D-GFP were significantly smaller than those of PCs expressing WT-GFP. Trehalose was found to significantly prevent this reduction in area, especially in the dendritic areas, of PCs lacking aggregates of S119P-GFP and G128D-GFP, whereas it did not affect the areas of PCs with aggregates. No effect of trehalose was observed in PCs containing aggregates or PCs expressing WT-GFP (Fig. 5D). We focused on two parameters, length and number of branch points on the longest dendrites, in PCs lacking S119P-GFP aggregates. After we confirmed the identity of the PCs by immunostaining with an anti-calbindin antibody, the length and number of branch points on the longest dendrite was measured according to the calbindin staining. Trehalose was found to significantly prevent the reduction in branch point number without altering the length of the longest dendrite in PCs expressing S119P-GFP; these two parameters were unaffected by trehalose in PCs expressing WT-GFP (supplemental Fig. 6). These results indicate that trehalose prevents improper dendritic development in PCs lacking aggregates of mutant γPKC-GFP but not in PCs containing aggregates.
Furthermore, we investigate the effect of trehalose on dendritic spines of PCs. As we have previously demonstrated (20), S119P-GFP reduced GluRδ2-positive spines of PC dendrites, compared with those in PCs expressing WT-GFP (supplemental Fig. 7). Trehalose restored the reduction of GluRδ2-positive dendritic spines in PCs expressing S119P-GFP without its aggregates (supplemental Fig. 7). This result suggests that trehalose prevents the loss of dendritic spines triggered by mutant γPKC-GFP.
Trehalose Improves the Mobility and Translocation of Mutant γPKC-GFP in Primary Cultured PCs
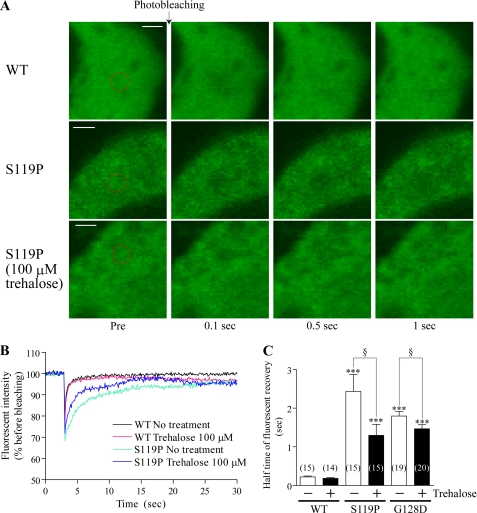
We have previously demonstrated that oligomerization of mutant γPKC reduces its mobility and translocation ability in primary cultured PCs (20). Therefore, we investigated whether trehalose affects these properties of mutant γPKC-GFP. Fluorescent recovery after photobleaching revealed that trehalose enhances the rate of fluorescence recovery of S119P-GFP in the PC somata (Fig. 6, A and B) but not that of WT-GFP. The mobility of the fluorescent protein can be quantitatively evaluated as the half time of fluorescence recovery. Trehalose significantly decreases the half times of fluorescence recovery of S119P-GFP and G128D-GFP but not that of WT-GFP (Fig. 6C), suggesting that trehalose improves mobility of mutant γPKC-GFP in PCs.
FIGURE 6.
Trehalose increases the mobility of mutant γPKC-GFP in primary cultured PCs. A, representative images of GFP fluorescence in PC somata expressing WT-GFP (upper panels), S119P-GFP (middle panels), or S119P-GFP cultured with 100 μm trehalose (lower panels) before (Pre) and 0.1, 0.5, and 2 s after photobleaching. Bleached areas are shown as red circles. Cells were infected on DIV14 and observed on DIV28. Bar = 2 μm. B, temporal changes in fluorescence intensities in bleached areas of PC somata expressing WT-GFP and S119P-GFP with or without trehalose. C, half time of fluorescence recovery in bleached area of PC somata expressing WT-GFP, S119P-GFP, and G128D-GFP with or without trehalose. Half time of recovery was calculated by fitting the changes in fluorescence intensities after photobleaching to single exponential functions. Data represent means ± S.E. of half times obtained from 14–20 PCs. Trehalose significantly decreases the half times of S119P-GFP and G128D-GFP in PC somata (***, p < 0.001 versus WT; §, p < 0.05, unpaired t test).
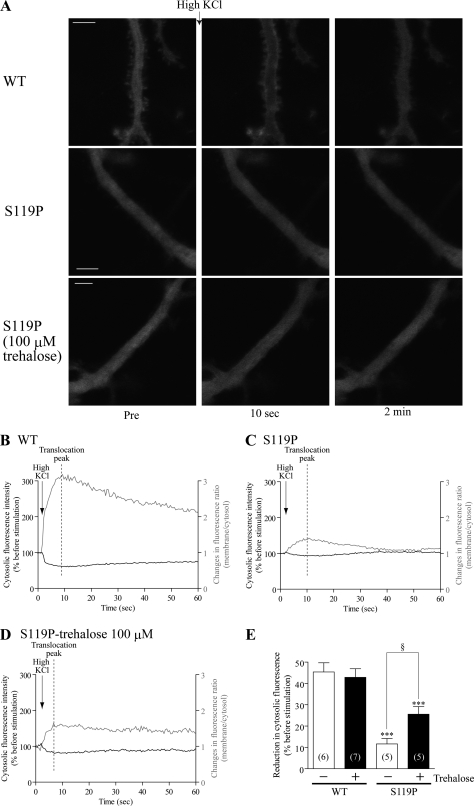
We recently demonstrated that the reduced mobility of mutant γPKC causes attenuated translocation of mutant γPKC in response to K+-induced depolarization (20). Therefore, we also examined whether trehalose ameliorates the attenuated translocation of mutant γPKC-GFP in primary cultured PCs. Similar to our previous findings, high KCl (100 mm) stimulation was found to trigger rapid and pronounced translocation of WT-GFP from the cytosol to the plasma membrane of PC dendrites. WT-GFP almost completely returns to the cytosol within 2 min after stimulation (Fig. 7, A (upper panels) and B, and supplemental movie 3). Only a faint translocation of the S119P-GFP was seen, however (Fig. 7, A (middle panels) and C, and supplemental movie 4), and trehalose slightly enhances the translocation of S119P-GFP (Fig. 7, A (lower panels) and D, and supplemental movie 5). To quantitate this effect of trehalose, we measured the translocation amplitude by quantifying the reduction in cytosolic γPKC-GFP fluorescence at the translocation peak, the time point when the fluorescence ratio of plasma membrane/cytosol reached maximum (Fig. 7, B–D). The amplitude of S119P-GFP was found to be significantly lower than that of WT-GFP, and trehalose significantly increases this value (Fig. 7E). These results suggest that trehalose alleviates the attenuated translocation of mutant γPKC by improving its mobility in primary cultured PCs.
FIGURE 7.
Trehalose prevents attenuated translocation of mutant γPKC-GFP in primary cultured PCs. A, representative images of GFP fluorescence in PC dendrites expressing WT-GFP (upper panels) and S119P-GFP (middle panels) and S119P-GFP cultured with 100 μm trehalose (lower panels) before (Pre) or 10 s and 2 min after high KCl (100 mm KCl, 50 μl) stimulation. PCs were infected on DIV14 and observed on DIV28. Bar = 5 μm. B–D, temporal changes in fluorescent intensity (black line) and fluorescent ratio (membrane/cytosol, gray line) of WT-GFP (B), S119-GFP (C), and S119P-GFP in the presence of 100 μm trehalose (D) in PC dendrites shown in A. We evaluated the reduction in fluorescence intensity of cytosolic γPKC-GFP at the translocation peak, the time point when the fluorescence ratio reached maximum, as the index for translocation amplitude (B–D). E, translocation amplitude was quantitatively evaluated by measuring the reduction in fluorescence of cytosolic γPKC-GFP. Data represent means ± S.E. of amplitudes obtained from five to seven PCs. Trehalose significantly improves the translocation amplitude of S119P-GFP in PCs (***, p < 0.001 versus WT; §, p < 0.05, unpaired t test).
Trehalose Inhibits Mutant γPKC-GFP-mediated Apoptosis of Primary Cultured PCs
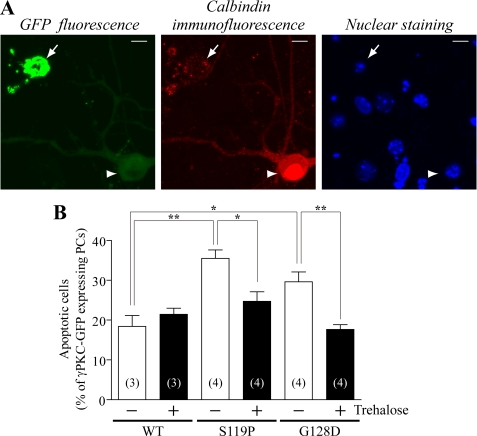
Apoptosis of PCs was assessed by chromatin condensation in calbindin-positive PCs (20). Trehalose (100 μm) significantly reduces the amount of apoptotic PCs expressing S119P-GFP and G128D-GFP; no effect was observed for PCs expressing WT-GFP (Fig. 8B). Apoptotic PCs with fragmented or condensed nuclei were observed only in PCs with aggregates of mutant γPKC-GFP (Fig. 8A). These cells were immunostained with anti-activated caspase-3 antibody, another apoptotic marker (supplemental Fig. 8). Trehalose obviously reduced cells stained with this antibody in PCs expressing S119P-GFP (supplemental Fig. 8). Trehalose was found to decrease the percentage of PCs containing aggregates of S119P-GFP (50.1 ± 5.1% in non-treated group, n = 4 and 40.2 ± 5.5% in the trehalose-treated group, n = 4) and G128D-GFP (54.4 ± 1.2% in non-treated group, n = 4 and 42.3 ± 2.9% in the trehalose-treated group, n = 4). Therefore, trehalose does not significantly affect apoptosis in PCs containing aggregates of S119P-GFP (72.6 ± 6.4% in non-treated group, n = 4 and 64.9 ± 10.6% in trehalose-treated group, n = 4) and G128D-GFP (50.8 ± 5.2% in non-treated group, n = 4 and 42.0 ± 2.8% in trehalose-treated group, n = 4). To more precisely evaluate the effect of trehalose on apoptosis in PCs containing aggregates, we observed the fate of the same PCs cultured on glass-bottomed dishes with a grid in the presence or absence of 100 μm trehalose. PCs were infected on DIV22, and PCs with aggregates of S119P-GFP were identified on DIV26. After another 3-day cultivation (DIV29), apoptosis in the same PCs, marked with the grids, was assessed. The percentage of apoptotic PCs was similar in trehalose-treated and untreated groups (Table 1), indicating that trehalose cannot inhibit apoptosis in PCs after aggregates of mutant γPKC have formed. These results suggest that trehalose inhibits mutant γPKC-mediated apoptosis in primary cultured PCs, probably by preventing aggregation.
FIGURE 8.
Trehalose inhibits apoptotic cell death in PCs expressing a mutant γPKC-GFP. A, representative GFP fluorescence (left), calbindin immunostaining (center), and nuclear staining (right) in PCs expressing S119P-GFP. Cells were infected on DIV14, fixed on DIV28, and immunostained with an anti-calbindin antibody, concomitantly with nuclear staining with Hoechst 33342 (0.5 μg/ml). Cells with fragmented or condensed nuclei were regarded as apoptotic cells (arrow). All apoptotic PCs had aggregates of mutant γPKC-GFP and stained with the anti-calbindin antibody in a dot-like manner. The arrowhead indicates a surviving PC lacking aggregates of S119P-GFP and containing a normal nucleus. Bar = 10 μm. B, quantitative analysis of apoptotic cells by nuclear staining in A. Each column represents the mean ± S.E. of the percentage of apoptotic cells in GFP-positive PCs. The number of experiments is indicated in each column. S119P-GFP and G128D-GFP triggers apoptosis in PCs, but trehalose (100 μm) significantly reduces the number of apoptotic cells in the population expressing mutant γPKC-GFP (*, p < 0.05; **, p < 0.01, unpaired t test).
TABLE 1.
Trehalose does not inhibit apoptosis in PCs with aggregates of mutant γPKC-GFP
On DIV26 and DIV29, we observed the same PCs with aggregates of S119P-GFP cultured on a glass-bottomed dish with a grid. PCs were infected with adenoviral vectors on DIV22 with or without trehalose (100 μm) treatment. On DIV26, live PCs with aggregates of S119P-GFP were identified; thereafter, the same PCs were subsequently fixed and immunostained with the anti-calbindin antibody (diluted 1:1000) and stained with Hoechst 33342 (0.5 μg/ml) on DIV29. The table shows the number of PCs with aggregates of mutant γPKC-GFP that were identified on DIV26, the number of apoptotic PCs on DIV29, and the percentage of apoptotic PCs.
| PC aggregates observed on DIV26 | Apoptotic PCs on DIV29 | Apoptotic PCs | |
|---|---|---|---|
| cells | cells | % | |
| No treatment | 33 | 13 | 39.4 |
| Trehalose (100 μm) | 32 | 13 | 40.6 |
DISCUSSION
To identify novel therapeutic agents for SCA14 that inhibit the aggregation and cytotoxic effects of mutant γPKC, we attempted to establish a drug-screening system using a cell line model of SCA14. Adenovirus-infected SH-SY5Y cells can express enough mutant γPKC-GFP to induce aggregation and apoptotic cell death within a 3-day observation after infection (Figs. 1 and 4). Using this system, we determined that the disaccharide trehalose inhibits aggregate formation and apoptosis induced by mutant γPKC (Figs. 2 and 4). This result is consistent with previous findings showing that trehalose inhibits aggregation and cytotoxicity of disease-causing proteins in cellular and animal models of diseases (26, 27, 35, 36). This result highlights the benefits of using adenovirus-infected SH-SY5Y cells as a drug-screening system for SCA14.
In addition to its inhibitory effects on aggregation and apoptosis, trehalose was found to alleviate the improper development of PC dendrites and apoptosis triggered by mutant γPKC in primary cultured PCs (Figs. 5 and 8). Our work is the first demonstration of trehalose preventing the aberrant neuronal morphology that is triggered by a mutant protein causing neurodegenerative disease. Incorrect development of PC dendrites was prevented by trehalose treatment only in PCs lacking mutant γPKC aggregates. In these cells, mutant γPKC has reduced intracellular mobility and attenuated translocation upon stimulation (Figs. 6 and 7), probably due to soluble oligomerization of mutant γPKC. We have already described the oligomerization of mutant γPKC in SH-SY5Y cells and PCs (20). Oligomers of mutant proteins have recently been recognized as being more toxic than aggregates in various neurodegenerative disorders (37, 38). Because trehalose reduces these aberrant molecular properties of mutant γPKC in PCs without aggregates (Figs. 6 and 7), trehalose could prevent oligomerization of mutant γPKC as well as aggregation. Incorrect development of dendrites and apoptosis still occurs in PCs with aggregates of mutant γPKC even with trehalose treatment (Fig. 5, C and D). These results suggest that trehalose cannot alter PCs after mutant γPKC has aggregated. These findings provide new insights into how a chemical chaperone such as trehalose can affect the intracellular behavior of a mutant protein that causes neurodegenerative disease.
The mechanism by which trehalose inhibits the aggregation of mutant proteins is not yet known. Because we observed that trehalose inhibits the rapid insolubilization of mutant γPKC in vitro (Fig. 3, B and C), we conclude that trehalose directly stabilizes mutant γPKC. This conclusion is consistent with a previous report demonstrating that trehalose inhibits the in vitro aggregation of a purified protein with an expanded polyglutamine region (26). We believe the most reasonable explanation of trehalose activity involves the direct stabilization of mutant proteins, as in the case of chemical chaperones (25, 34). Three major hypotheses have been proposed for the molecular mechanism by which trehalose stabilizes mutant proteins (39): 1) trehalose molecules directly interact with protein molecules through hydrogen bonds and replace water molecules (water-replacement hypothesis), 2) trehalose traps water molecules close to the protein surface (water-layer hypothesis), and 3) trehalose traps the protein surface in a highly viscous trehalose matrix (mechanical-entrapment hypothesis). In all three hypotheses, trehalose reduces the interaction between water molecules and the hydrophobic regions of mutant proteins that are exposed on the surface and prevents hydrophobic interactions between mutant protein molecules, thereby preventing the formation of aggregates and oligomers. We have preliminary data showing that aggregates of mutant γPKC form via the C1A region, a hydrophobic phospholipid-interacting domain. Thus, trehalose may prevent aggregation of mutant γPKC by inhibiting the intermolecular association of γPKC mediated by the hydrophobic domain. However, the effective concentrations of trehalose in the present study (10–500 μm) is markedly less than those as a chemical chaperone (∼100 mm), providing the possibility that the inhibitory effect of trehalose on aggregation of mutant γPKC results from enhancement of protein degradation or potentiation of molecular chaperones by trehalose. Indeed, trehalose has been reported to promote degradation of mutant proteins by stimulating macroautophagy (32). We have recently revealed that the degradation of mutant γPKC is selectively accelerated by the induction of autophagy (33). However, the expression and degradation of mutant γPKC were not affected by trehalose in the present study (Fig. 2), and macroautophagy was not activated by trehalose in SH-SY5Y cells (supplemental Fig. 9A). As to the second possibility, trehalose has been reported to positively regulate the transcription of heat shock proteins in yeast (41). However, in SH-SY5Y cells, trehalose did not up-regulate molecular chaperones, including heat shock cognate 70, which is involved in chaperone-mediated autophagy, another lysosomal protein degradation system (supplemental Fig. 9B). These results suggest that trehalose did not affect molecular chaperones and lysosomal protein degradation, including macroautophagy and chaperone-mediated autophagy. These findings support the idea that trehalose inhibits aggregation of mutant γPKC as a chemical chaperone. Indeed, in the present study, trehalose inhibits insolubilization of recombinant mutant γPKC in vitro (Fig. 3, B and C). Because our previous findings demonstrated that mutant γPKC did not form tight aggregates and easily disappeared by the arrest of its expression (20, 33), weak chemical chaperone activity of trehalose at micromolar order might be enough to inhibit aggregation of mutant γPKC.
To directly interact with intracellular proteins, trehalose must be incorporated into cells. In this work, we first confirmed that trehalose is incorporated into neuronal cell lines (Fig. 3A), in which the aggregation of mutant proteins is inhibited by trehalose. The concentrations of trehalose used in uptake assay were higher (1 and 10 mm, Fig. 3A) than other experiments (<500 μm). Although we could not detect the trehalose uptake because of the detection limit (1 μg/sample) when the concentration was <500 μm, we assume that a nanogram order of trehalose would be incorporated into SH-SY5Y cells, because ∼2 μg of trehalose per mg of protein was likely taken up by 1 mm trehalose. This finding strongly supports a direct interaction between trehalose and mutant proteins. Although a trehalose transporter has been identified in insects (31), a mammalian homolog has not been identified yet. Alternatively, trehalose may be internalized by endocytosis (42) or the P2X7 purinergic receptor (43). To apply trehalose as a therapeutic agent for SCA14, it should pass through the blood-brain barrier and be taken up into Purkinje cells. It has been reported that the ingestion of trehalose inhibits aggregation and improves motor dysfunction in a mouse model of Huntington disease (26), suggesting that trehalose is taken up via an endogenous pathway in mammalian neurons through the blood-brain barrier and may be available for the treatment of SCA14.
Since missense mutations in γPKC were first identified as the genetic cause of SCA14 (8), several researchers have focused on the molecular mechanism by which this mutant γPKC triggers the neurodegeneration of cerebellar Purkinje cells and cerebellar dysfunction. Verbeek et al. reported that certain mutations (G118D and C150F) increase the kinase activity of γPKC (44). In contrast, Lin et al. reported that other mutant γPKCs (H101Y, S119P, and G128D) have reduced kinase activities and inhibited hydrogen peroxide-induced activation of endogenous γPKC, making the expressed cells vulnerable to oxidative stress (45). It has also been demonstrated that the kinase activities of mutant γPKCs (G118D, V138E, and C142S) at the plasma membrane are lower when activated by a phorbol ester, leading to aberrant MAPK signaling (46). More recently, Asai et al. has revealed that mutant γPKC with higher activity induces aberrant phosphorylation and localization of aprataxin, a DNA repair protein (47). Although we have shown that most of the SCA14 mutant γPKCs have increased basal kinase activities, several mutations actually have lower or unchanged activities (48). In contrast to controversial results concerning the kinase activity of mutant γPKC, we confirmed that most of the mutant γPKCs found in SCA14 tend to form aggregates (19). Furthermore, we and another group have recently demonstrated that the aggregation of mutant γPKC triggers apoptosis by impairing the ubiquitin proteasome system and inducing endoplasmic reticulum stress (21, 49). Therefore, the ability of mutant γPKCs to aggregate must be a part of the molecular pathogenesis of SCA14, as is the case for other neurodegenerative disorders (22, 23). Alternatively, based on the idea that the conformational instability of mutant γPKC might cause its aberrant kinase activity (46) as well as its aggregation, it is possible that trehalose could normalize the kinase activity of mutant γPKC by stabilizing its conformation. In summary, our data suggest that trehalose may be applicable to the treatment of various other neurodegenerative diseases in addition to SCA14. Trehalose is widely used in foods as a sweetener and in cosmetics as a humectant; thus its safety has already been established (40). We expect that trehalose has the potential to become a useful drug for treatment of neurodegenerative diseases, including SCA14.
Supplementary Material
Acknowledgments
We thank Takeshi Hamada, Biosignal Research Center, Kobe University, for the technical support. This work was carried out using equipment at the Analysis Center of Life Science, Hiroshima University and the Research Center for Molecular Medicine, Faculty of Medicine, Hiroshima University.
This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Sports and Culture and by grants from the Takeda Science Foundation, the Uehara Memorial Foundation, the Naito Foundation, the Suzuken Memorial Foundation, the Tokyo Biochemical Research Foundation, and the Japanese Smoking Research Association.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–9 and Movies 1–5.
- SCA
- spinocerebellar ataxia
- SCA14
- SCA type 14
- PKC
- protein kinase C
- PC
- Purkinje cell
- DIV
- days in vitro
- tTA
- tetracycline transactivator
- m.o.i.
- multiplicity of infection
- Tet
- tetracycline.
REFERENCES
- 1.Schöls L., Bauer P., Schmidt T., Schulte T., Riess O. (2004) Lancet Neurol. 3, 291–304 [DOI] [PubMed] [Google Scholar]
- 2.Cagnoli C., Mariotti C., Taroni F., Seri M., Brussino A., Michielotto C., Grisoli M., Di Bella D., Migone N., Gellera C., Di Donato S., Brusco A. (2006) Brain 129, 235–242 [DOI] [PubMed] [Google Scholar]
- 3.Dueñas A. M., Goold R., Giunti P. (2006) Brain 129, 1357–1370 [DOI] [PubMed] [Google Scholar]
- 4.Martin J. B. (1999) N. Engl. J. Med. 340, 1970–1980 [DOI] [PubMed] [Google Scholar]
- 5.Everett C. M., Wood N. W. (2004) Brain 127, 2385–2405 [DOI] [PubMed] [Google Scholar]
- 6.van de Leemput J., Chandran J., Knight M. A., Holtzclaw L. A., Scholz S., Cookson M. R., Houlden H., Gwinn-Hardy K., Fung H. C., Lin X., Hernandez D., Simon-Sanchez J., Wood N. W., Giunti P., Rafferty I., Hardy J., Storey E., Gardner R. J., Forrest S. M., Fisher E. M., Russell J. T., Cai H., Singleton A. B. (2007) PLoS Genet. 3, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houlden H., Johnson J., Gardner-Thorpe C., Lashley T., Hernandez D., Worth P., Singleton A. B., Hilton D. A., Holton J., Revesz T., Davis M. B., Giunti P., Wood N. W. (2007) Nat. Genet. 39, 1434–1436 [DOI] [PubMed] [Google Scholar]
- 8.Chen D. H., Brkanac Z., Verlinde C. L., Tan X. J., Bylenok L., Nochlin D., Matsushita M., Lipe H., Wolff J., Fernandez M., Cimino P. J., Bird T. D., Raskind W. H. (2003) Am. J. Hum. Genet. 72, 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D. H., Cimino P. J., Ranum L. P., Zoghbi H. Y., Yabe I., Schut L., Margolis R. L., Lipe H. P., Feleke A., Matsushita M., Wolff J., Morgan C., Lau D., Fernandez M., Sasaki H., Raskind W. H., Bird T. D. (2005) Neurology 64, 1258–1260 [DOI] [PubMed] [Google Scholar]
- 10.Dalski A., Mitulla B., Bürk K., Schattenfroh C., Schwinger E., Zühlke C. (2006) J. Neurol 253, 1111–1112 [DOI] [PubMed] [Google Scholar]
- 11.Hiramoto K., Kawakami H., Inoue K., Seki T., Maruyama H., Morino H., Matsumoto M., Kurisu K., Sakai N. (2006) Mov Disord. 21, 1355–1360 [DOI] [PubMed] [Google Scholar]
- 12.Vlak M. H., Sinke R. J., Rabelink G. M., Kremer B. P., van de Warrenburg B. P. (2006) Mov Disord. 21, 1025–1028 [DOI] [PubMed] [Google Scholar]
- 13.Klebe S., Faivre L., Forlani S., Dussert C., Tourbah A., Brice A., Stevanin G., Durr A. (2007) Arch. Neurol. 64, 913–914 [DOI] [PubMed] [Google Scholar]
- 14.Nolte D., Landendinger M., Schmitt E., Müller U. (2007) Mov. Disord. 22, 265–267 [DOI] [PubMed] [Google Scholar]
- 15.Wieczorek S., Arning L., Gizewski E. R., Alheite I., Timmann D. (2007) Mov. Disord. 22, 2135–2136 [DOI] [PubMed] [Google Scholar]
- 16.Saito N., Kikkawa U., Nishizuka Y., Tanaka C. (1988) J. Neurosci. 8, 369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C., Kano M., Abeliovich A., Chen L., Bao S., Kim J. J., Hashimoto K., Thompson R. F., Tonegawa S. (1995) Cell 83, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 18.Kano M., Hashimoto K., Chen C., Abeliovich A., Aiba A., Kurihara H., Watanabe M., Inoue Y., Tonegawa S. (1995) Cell 83, 1223–1231 [DOI] [PubMed] [Google Scholar]
- 19.Seki T., Adachi N., Ono Y., Mochizuki H., Hiramoto K., Amano T., Matsubayashi H., Matsumoto M., Kawakami H., Saito N., Sakai N. (2005) J. Biol. Chem. 280, 29096–29106 [DOI] [PubMed] [Google Scholar]
- 20.Seki T., Shimahara T., Yamamoto K., Abe N., Amano T., Adachi N., Takahashi H., Kashiwagi K., Saito N., Sakai N. (2009) Neurobiol Dis. 33, 260–273 [DOI] [PubMed] [Google Scholar]
- 21.Seki T., Takahashi H., Adachi N., Abe N., Shimahara T., Saito N., Sakai N. (2007) Eur. J. Neurosci. 26, 3126–3140 [DOI] [PubMed] [Google Scholar]
- 22.Taylor J. P., Hardy J., Fischbeck K. H. (2002) Science 296, 1991–1995 [DOI] [PubMed] [Google Scholar]
- 23.Ross C. A., Poirier M. A. (2004) Nat. Med. 10, (suppl.) S10–S17 [DOI] [PubMed] [Google Scholar]
- 24.Elbein A. D. (1974) Adv. Carbohyd.r Chem. Biochem. 30, 227–256 [DOI] [PubMed] [Google Scholar]
- 25.Singer M. A., Lindquist S. (1998) Mol. Cell. 1, 639–648 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M., Machida Y., Niu S., Ikeda T., Jana N. R., Doi H., Kurosawa M., Nekooki M., Nukina N. (2004) Nat. Med. 10, 148–154 [DOI] [PubMed] [Google Scholar]
- 27.Liu R., Barkhordarian H., Emadi S., Park C. B., Sierks M. R. (2005) Neurobiol Dis. 20, 74–81 [DOI] [PubMed] [Google Scholar]
- 28.Béranger F., Crozet C., Goldsborough A., Lehmann S. (2008) Biochem. Biophys. Res. Commun. 374, 44–48 [DOI] [PubMed] [Google Scholar]
- 29.Seki T., Irie N., Nakamura K., Sakaue H., Ogawa W., Kasuga M., Yamamoto H., Ohmori S., Saito N., Sakai N. (2006) Genes Cells. 11, 1051–1070 [DOI] [PubMed] [Google Scholar]
- 30.Watanabe M., Kikawada T., Minagawa N., Yukuhiro F., Okuda T. (2002) J. Exp. Biol. 205, 2799–2802 [DOI] [PubMed] [Google Scholar]
- 31.Kikawada T., Saito A., Kanamori Y., Nakahara Y., Iwata K., Tanaka D., Watanabe M., Okuda T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11585–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar S., Davies J. E., Huang Z., Tunnacliffe A., Rubinsztein D. C. (2007) J. Biol. Chem. 282, 5641–5652 [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto K., Seki T., Adachi N., Takahashi T., Tanaka S., Hide I., Saito N., Sakai N. (2010) Genes Cells 15, 425–438 [DOI] [PubMed] [Google Scholar]
- 34.Crowe J. H. (2007) Adv. Exp. Med. Biol. 594, 143–158 [DOI] [PubMed] [Google Scholar]
- 35.Davies J. E., Sarkar S., Rubinsztein D. C. (2006) Hum. Mol. Genet. 15, 23–31 [DOI] [PubMed] [Google Scholar]
- 36.Attanasio F., Cascio C., Fisichella S., Nicoletti V. G., Pignataro B., Savarino A., Rizzarelli E. (2007) Biochem. Biophys. Res. Commun. 354, 899–905 [DOI] [PubMed] [Google Scholar]
- 37.Agorogiannis E. I., Agorogiannis G. I., Papadimitriou A., Hadjigeorgiou G. M. (2004) Neuropathol. Appl. Neurobiol. 30, 215–224 [DOI] [PubMed] [Google Scholar]
- 38.Glabe C. G. (2006) Neurobiol. Aging 27, 570–575 [DOI] [PubMed] [Google Scholar]
- 39.Lins R. D., Pereira C. S., Hünenberger P. H. (2004) Proteins 55, 177–186 [DOI] [PubMed] [Google Scholar]
- 40.Richards A. B., Krakowka S., Dexter L. B., Schmid H., Wolterbeek A. P., Waalkens-Berendsen D. H., Shigoyuki A., Kurimoto M. (2002) Food Chem. Toxicol. 40, 871–898 [DOI] [PubMed] [Google Scholar]
- 41.Conlin L. K., Nelson H. C. (2007) Mol. Cell. Biol. 27, 1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver A. E., Jamil K., Crowe J. H., Tablin F. (2004) Cell Preserv. Technol. 2, 35–49 [Google Scholar]
- 43.Elliott G. D., Liu X. H., Cusick J. L., Menze M., Vincent J., Witt T., Hand S., Toner M. (2006) Cryobiology 52, 114–127 [DOI] [PubMed] [Google Scholar]
- 44.Verbeek D. S., Knight M. A., Harmison G. G., Fischbeck K. H., Howell B. W. (2005) Brain 128, 436–442 [DOI] [PubMed] [Google Scholar]
- 45.Lin D., Shanks D., Prakash O., Takemoto D. J. (2007) Exp. Eye. Res. 85, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verbeek D. S., Goedhart J., Bruinsma L., Sinke R. J., Reits E. A. (2008) J. Cell. Sci. 121, 2339–2349 [DOI] [PubMed] [Google Scholar]
- 47.Asai H., Hirano M., Shimada K., Kiriyama T., Furiya Y., Ikeda M., Iwamoto T., Mori T., Nishinaka K., Konishi N., Udaka F., Ueno S. (2009) Hum. Mol. Genet. 18, 3533–3543 [DOI] [PubMed] [Google Scholar]
- 48.Adachi N., Kobayashi T., Takahashi H., Kawasaki T., Shirai Y., Ueyama T., Matsuda T., Seki T., Sakai N., Saito N. (2008) J. Biol. Chem. 283, 19854–19863 [DOI] [PubMed] [Google Scholar]
- 49.Lin D., Takemoto D. J. (2007) Biochem. Biophys. Res. Commun. 362, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.