Abstract
IL-17, the hallmark cytokine of the Th17 population, mediates immunity to extracellular pathogens and promotes autoimmune immunopathology. The signaling mechanisms triggered by the IL-17 receptor (IL-17RA) and related receptors are strikingly different from other cytokine subclasses. Namely, IL-17Rs contain a conserved SEF/IL-17R (SEFIR) subdomain that engages Act1, leading to activation of TRAF6, NF-κB, and other events. Although the SEFIR is critical for signaling, the molecular details of the functional subdomains within IL-17RA remain poorly characterized. Here, we provide a detailed structure-function analysis delineating the C-terminal boundary of the SEFIR-containing region of IL-17RA. We show that functionality of this domain requires a large extension to the previously identified SEFIR motif. In contrast to the SEFIR, this extension is not conserved among IL-17R family members. Surprisingly, Act1 recruitment is not sufficient for downstream signaling activation, whereas ubiquitination of TRAF6 correlates tightly with functional receptors. We further demonstrate that IL-17RA exhibits signaling properties that are nonredundant with other IL-17R family members. Finally, we report that IL-17 signals synergistically with lymphotoxin-α3, using the same signaling motifs within IL-17RA. These studies provide new insight into the structure-function relationships of IL-17RA and reveal distinct signaling differences among IL-17R family members.
Keywords: Cytokine, Immunology, Inflammation, Interleukin, Receptor Structure-Function, IL-17, SEFIR
Introduction
Recently, the Th17 subset of CD4+ T helper cells was discovered to play an essential role in promoting inflammatory reactions in both autoimmune disease and defense against infection with extracellular microbes. IL-17 (also called IL-17A) is the hallmark cytokine secreted by Th17 cells, which also produce IL-17F, IL-22, and IL-21. A similar profile of cytokines is made by certain subsets of γδ-T cells, NK (natural killer), NKT, and LTi (lymphoid tissue inducer) cells (reviewed in Refs. 1 and 2).
Whereas most T helper cell-derived cytokines activate JAK-STAT-dependent signal transduction, the IL-17 family mediates signaling via pathways more typical of innate immune effectors, such as IL-1 and Toll-like receptor (TLR)4 ligands (reviewed in Ref. 3). Specifically, IL-17 activates the NF-κB, C/EBP, and AP-1 transcription factors, which collectively initiate transcription and expression of proinflammatory proteins, such as IL-6, CXC chemokines, and lipocalin-2/24p3 (4, 5). The receptor for IL-17 is composed of two subunits, IL-17RA and IL-17RC (6). Although the expression profiles of these receptors are surprisingly different (7, 8), both are co-expressed on epithelial cells and fibroblasts, where they are required for IL-17- as well as IL-17F-dependent signal transduction (6, 8, 9). IL-17 also signals cooperatively with other cytokines, particularly TNFα, with which it mediates potent synergy through a variety of mechanisms (3).
In terms of signal transduction, the cytoplasmic tails of the IL-17R family are distinct in sequence from other cytokine receptor families and encode a conserved motif called a SEF/IL-17 receptor (SEFIR) domain (10). The SEFIR bears some homology to Toll/IL-IR (TIR) domains, the key functional subdomains used by the Toll/IL-1 family receptors (11). A SEFIR domain is also found in Act1/CIKS, an essential adaptor downstream of IL-17R family members that is required for activation of NF-κB and other signals (12–16). Thus, the SEFIR is a protein-protein interaction domain used by IL-17R-dependent signaling cascades.
In studies that mapped functional domains within the IL-17RA cytoplasmic tail, we previously reported that the SEFIR alone is not sufficient to mediate IL-17 signaling to NF-κB or NF-κB-dependent genes. In that study, we identified a short additional motif located at the C-terminal end of the SEFIR that is required for IL-17-dependent activation of the NF-κB, C/EBP, and MAPK pathways (17). This motif was identified on the basis of homology to a substructure of TIR domains known as the BB-loop (18) and was accordingly called a TIR-like loop (TILL). A single point mutation within the TILL renders IL-17RA non-functional, and internal deletions of the SEFIR or the TILL abrogated signaling (17). However, it was not clear from this work whether the SEFIR/TILL encompasses the entire signaling unit or whether additional sequences beyond the TILL might be involved in IL-17RA-mediated signaling. It was also unclear whether simply binding to Act1 was sufficient to trigger downstream signaling. Interestingly, there is no obvious TILL domain in other IL-17R family members (3), so we questioned whether IL-17RA is unique or if SEFIR domains of other IL-17R family members might be functionally interchangeable.
Here, we report a detailed structure-function analysis delineating the C-terminal boundary of the functional SEFIR-containing region in IL-17RA. We show that this region requires a large extension to the previously identified SEFIR/TILL motif to promote expression of IL-17 target genes. Surprisingly, based on these mutants, binding of Act1 to IL-17RA is not sufficient for signaling because certain mutants that bind Act1 nonetheless fail to promote downstream signaling. Also surprising is that the SEFIR extension is not conserved among other IL-17R family members. We further demonstrate unique properties of IL-17RA compared with other IL-17 family receptors by means of chimeric receptors. Finally, we show that IL-17 signals synergistically with lymphotoxin-α3 (LTα3), an event that is dependent on the same SEFIR extension within IL-17RA. These studies provide new insight into structure-function relationships of IL-17RA and reveal distinct signaling activities among IL-17R family receptors.
MATERIALS AND METHODS
Cell Lines and Cytokines
IL-17RA−/− fibroblasts were created from tail biopsies of IL-17RA−/− mice (from Amgen) and immortalized with SV40 T antigen. Cell lines expressing IL-17RA mutants were created by stable transfection and selection in zeocin as described (17). IL-17RA−/−, ST2, and HEK293T cells were cultured in α-minimum Eagle's medium (Sigma) with 10% FBS (Gemini Bioproducts). Transfections were performed with CaPO4, FuGENE6 (Roche Applied Science), or an Amaxa Nucleofector system (Lonza, Gaithersburg, MD). IL-17 and TNFα were from Peptrotech (Rocky Hill, NJ) or R&D Systems (Minneapolis, MN). LTα3 was from Alexis Biochemicals (San Diego, CA).
Plasmids
Murine IL-17RB (NM_019583), IL-17RC, and IL-17RD (NM_134437) cDNAs were generated by RT-PCR of mRNA from primary kidney cells. IL-17RA deletions were created by PCR and fused to a C-terminal HA tag, as described (17). Chimeras were created by overlapping PCR. Constructs were subcloned in the pcDNA3.1-Zeo vector (Invitrogen). Act1 was generated by RT-PCR from ST2 cells and fused at the N terminus with Myc. HA-tagged TLR2 was kindly provided by Dr. S. Sarkar (University of Pittsburgh), and HA-TLR4 was generously provided Dr. R. Tapping (University of Illinois, Chicago, IL).
Antibodies, Flow Cytometry, ELISA, and Luciferase Assays
Abs to TRAF6 (sc-7221) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), IL-17RA (AF448) (R&D Systems, Minneapolis, MN), and Myc (9B11) (Cell Signaling, Beverly, MA) were used for Western blotting. IL-17RA monoclonal antibodies (clone M751) used in FACS were generously provided by Amgen. Anti-IgG-phycoerythrin Abs were from BD Biosciences or eBioscience. Flow cytometry was performed on a FACSCalibur flow cytometer (BD Biosystems) and analyzed with Cellquest software. IL-6 ELISA kits were from eBioscience or R&D Systems. Luciferase assays were performed on a Turner Biosystems Veritas Microplate Luminometer and Promega Dual Reporter Assay System (21). Statistical analysis was performed with GraphPad Prism (version 4).
Immunoprecipitations and Real-time RT-PCR
HEK293T cells were plated 16–18 h before transfection and stimulated with 200 ng/ml IL-17 and/or 2 ng/ml TNFα 15 min prior to lysis. Lysates were prepared in 1% Nonidet P-40 buffer as described (17), and immunoprecipitations were performed with 5 μg of α-IL-17RA Abs and Protein G beads (Roche Applied Science). For real-time RT-PCR, cells were stimulated with 2 ng/ml TNFα and/or 200 ng/ml IL-17. Total RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA) with on-column DNase digestion. cDNA was synthesized using a SuperScript first strand synthesis system (Invitrogen). Real-time PCR was performed in triplicate, as described previously (21).
RESULTS
Delineating the SEFIR-containing IL-17RA Signaling Domain
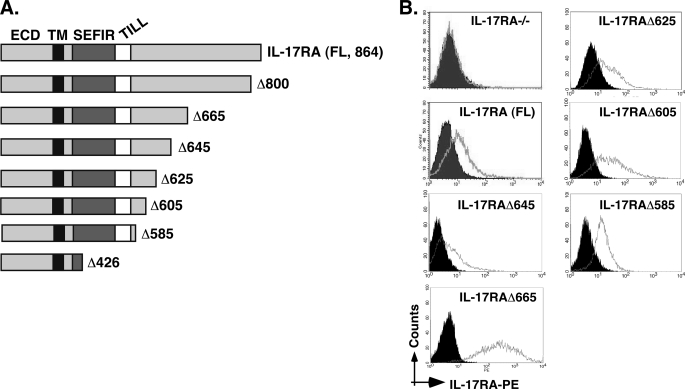
We previously demonstrated that the SEFIR and TILL domains are essential for IL-17RA to activate NF-κB, C/EBP, and ERK and expression of IL-6, 24p3 (lipocalin 2), C/EBPδ, and CXCL5 (17). However, it was not clear from those studies whether the SEFIR plus TILL (ending at residue 553) was sufficient for signal transduction (Fig. 1A). Because an IL-17RA construct deleted at residue 665 retained functionality (measured by NF-κB activation and induction of downstream genes), it was apparent that the end of the IL-17RA signaling domain lies between residues 554 and 665.
FIGURE 1.
IL-17RA deletion series to evaluate IL-17RA structure-function relationships. A, schematic diagram of the IL-17RA truncation mutants used in this study. ECD, extracellular domain; TM, transmembrane domain. SEFIR and TILL domains are indicated. B, expression of mutant IL-17 receptors. Stable cell lines expressing each receptor were created in the IL-17RA−/− background, and staining for IL-17RA was performed with anti-IL-17RA Abs followed by anti-IgG-phycoerythrin.
To delineate the C-terminal boundary of the SEFIR-containing domain more precisely, we created a series of IL-17RA deletions and expressed them stably in IL-17RA−/− fibroblast cells. Similar to what we routinely observe for IL-17RA mutants, these receptors expressed well on the surface, indicating no obvious impairments in protein folding or transport caused by deletions of the cytoplasmic tail (Fig. 1B). As we have also seen before, the IL-17RAΔ665 deletion was expressed more robustly than most other receptors, which correlated with a somewhat stronger signaling capability. This observation is not surprising because the IL-17RAΔ665 mutant lacks a known signaling inhibitory domain (see “Discussion”) (19).
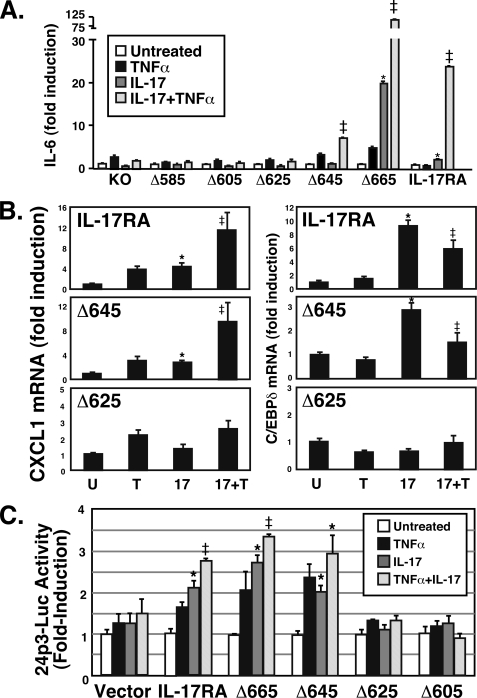
Cells expressing the panel of IL-17RA deletion mutants were treated with IL-17 in the presence or absence of low dose TNFα (with which IL-17 mediates potent synergy (20)), and IL-6 was evaluated by ELISA (Fig. 2A). As noted previously, IL-17 signaling alone is weak, and only modest signals were observed even with the full-length (FL) IL-17RA construct. However, strong IL-6 secretion was seen after treatment of cells with IL-17 plus TNFα in cells expressing IL-17RA or IL-17RAΔ665 (17, 19). Similarly, the IL-17RAΔ645 mutant was also able to activate IL-6, although the signal was reproducibly weaker than with IL-17RA or IL-17RAΔ665. In contrast, the panel of deletions from IL-17RAΔ585 to IL-17RAΔ625 failed to induce detectable expression of IL-6 (Fig. 2A, summarized in Table 1). Several independent stable cell lines for each mutant were screened, with similar results (data not shown).
FIGURE 2.
The C-terminal boundary of the IL-17RA SEFIR-containing signaling domain lies between amino acid residues 645 and 625. A, induction of IL-6 by IL-17RA deletion mutants. IL-17RA−/− cells stably reconstituted with the indicated IL-17RA mutant series were stimulated with IL-17 (200 ng/ml) and/or suboptimal doses of TNFα (2 ng/ml) for 24 h, and IL-6 secretion was assessed by ELISA of conditioned supernatants, in triplicate. Data are normalized to the untreated sample. *, p < 0.05 compared with untreated samples; ‡, p < 0.05 compared with TNF-treated samples. B, induction of IL-17-dependent gene expression by IL-17RA deletion mutants. IL-17RA−/− cells stably reconstituted with the indicated IL-17RA mutants were stimulated with IL-17 (200 ng/ml) and/or suboptimal doses of TNFα (2 ng/ml) for 8 h, and expression of CXCL1 and C/EBPδ was evaluated by real-time RT-PCR, normalized to an internal GAPDH control. -Fold induction over the untreated samples is shown. *, p < 0.05 compared with untreated samples; ‡, p < 0.05 compared with TNF-treated samples. C, induction of the 24p3 promoter by IL-17RA deletion mutants. IL-17RA−/− cells were transiently transfected with the 24p3-luciferase reporter construct (21) and stimulated with IL-17 and/or TNFα as in A, and lysates were analyzed for luciferase activity, normalized to an internal Renilla-luc control. Data are expressed as -fold induction over untreated samples. *, p < 0.05 compared with untreated samples; ‡, p < 0.05 compared with TNF-treated samples. Error bars, S.D.
TABLE 1.
Summary of IL-17RA deletion mutant phenotypes
Shown is a summary of signal transduction mediated by IL-17RA deletion series. ND, not determined. +, competent for signaling; −, incompetent for signaling.
| IL-6 secretion | 24p3-luciferase | Act1 recruitment | TRAF6 ubiquitination | C/EBPδ mRNA | IκBζ mRNA | CXCL1 mRNA | |
|---|---|---|---|---|---|---|---|
| IL-17RA | + | + | + | +/− | + | + | + |
| Δ800 | + | + | ND | ND | ND | ND | ND |
| Δ665 | + | + | + | + | + | + | + |
| Δ645 | + | + | + | + | + | + | + |
| Δ625 | − | − | + | − | − | − | − |
| Δ605 | − | − | − | − | ND | ND | ND |
| Δ585 | − | − | − | ND | ND | ND | − |
| Δ426 | − | − | − | ND | ND | ND | ND |
To confirm these findings in additional IL-17-dependent assays, we evaluated the ability of the most salient mutants to induce several representative IL-17 target genes by real time RT-PCR (Fig. 2B) (20). Full-length IL-17RA and IL-17RAΔ645 induced transcription of CXCL1 (KC or Groα), whereas the IL-17RAΔ625 mutant reproducibly failed to do so. Cooperative induction of CXCL1 by IL-17 plus TNFα was also impaired in the IL-17RAΔ625 mutant but not in cells expressing the FL or IL-17RAΔ645 constructs. Consistently, IL-17-mediated induction of C/EBPδ mRNA was impaired in the IL-17RAΔ625 mutant but not the IL-17RAΔ645 or FL constructs, although induction of C/EBPδ was not cooperatively induced by IL-17 plus TNFα in this background. We also saw a similar pattern of IL-17-dependent induction I-κBζ (MAIL or NFkBiz) in FL and IL-17Δ645-expressing cells but not in IL-17RAΔ625-expressing cells (Table 1).
We then transiently transfected the panel of IL-17RA mutants into IL-17RA−/− fibroblasts together with an IL-17-dependent reporter consisting of the 24p3/lipocalin 2 promoter linked to luciferase (21). IL-17RA, IL-17RAΔ665, and IL-17RAΔ645 activated expression of the reporter in response to IL-17 alone or in combination with TNFα (Fig. 2C), whereas IL-17RAΔ625 and IL-17RAΔ605 did not. Collectively, these experiments demonstrate that the C-terminal border of IL-17RA lies between residues 645 and 625, which is considerably larger than would be predicted on the basis of homology among IL-17R family members (see Fig. 4A) (10). We term the entire domain downstream of the SEFIR a SEFIR extension (SEFEX) domain.
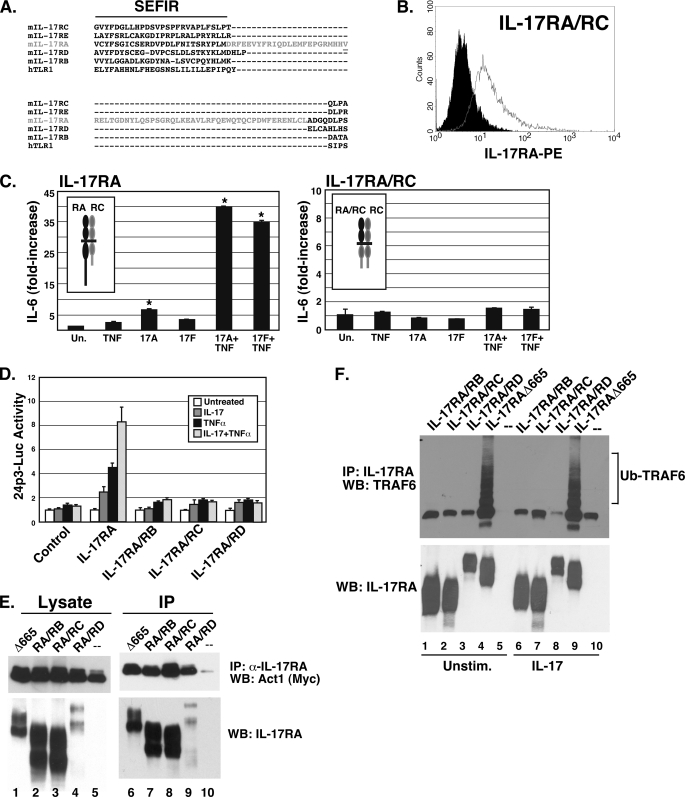
FIGURE 4.
Distinct and unique functions of the IL-17RA SEFIR domain. A, alignment of the extended SEFIR domain of IL-17RA family members. ClustalW2 alignment (42) of murine IL-17RA through IL-17RE cytoplasmic tails is depicted. The TILL domain is highlighted in red, and the crucial Val553 residue (required for signal transduction by IL-17RA) (17) is underlined. B, IL-17RA/RC chimeric receptor expression. IL-17RA−/− cells stably transfected with the IL-17RA/RC chimera were stained with anti-IL-17RA Abs and analyzed by flow cytometry. C, replacing the IL-17RA cytoplasmic tail with that of IL-17RC fails to rescue IL-17 induction of IL-6. The IL-17RA/RC-expressing cell line shown in B was treated with IL-17A or IL-17F (200 ng/ml) and/or TNFα (2 ng/ml) for 24 h, and IL-6 in the supernatant was evaluated by ELISA in triplicate. Very similar results were obtained with two other independently derived cell lines (not shown). D, replacing the IL-17RA cytoplasmic tail with that of other IL-17 receptor family members fails to rescue IL-17-induction of the 24p3 promoter. IL-17RA−/− cells were transiently transfected with IL-17RA or the indicated chimeric receptors together with the 24p3-luciferase reporter, and luciferase activity was assessed in triplicate. E, Act1 associates with multiple IL-17R cytoplasmic tails. HEK293T cells were transfected with IL-17RAΔ665 or the chimeric receptors together with Myc-Act1. Lysates were immunoprecipitated with anti-IL-17RA Abs. Lysates (lanes 1–5) and immunoprecipitates (lanes 6–10) were immunoblotted with Abs to Myc (top) or IL-17RA (bottom). Data are representative of at least two experiments. F, ubiquitination of TRAF6 is unique to IL-17RA. HEK293T cells were transiently transfected with the indicated receptors and TRAF6, cells were treated with or without IL-17 for 15 min, and lysates were immunoprecipitated with Abs to IL-17RA. IP samples were separated by SDS-PAGE and immunoblotted (WB) with Abs to TRAF6 (top) or IL-17RA (bottom). Error bars, S.E.
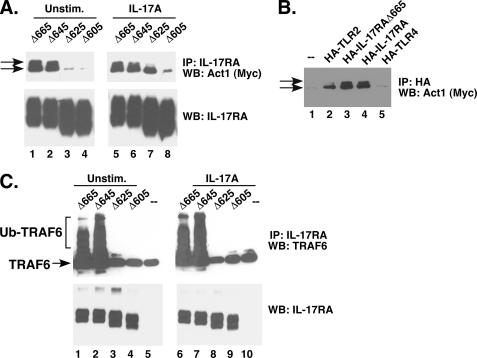
The adaptor/E3 ubiquitin ligase Act1 is required for IL-17-induced activation of NF-κB and expression of all of the genes assayed above (12, 13, 22). We therefore assessed the ability of each mutant to associate with Act1. HEK293T cells were transiently transfected with IL-17RA deletions together with Myc-tagged Act1. Full-length IL-17RA generally does not express efficiently in overexpression studies5; therefore, we used the IL-17RAΔ665 mutant as a positive control. Cells were treated with IL-17 for 10 min, lysed, and immunoprecipitated with Abs to IL-17RA. IP samples were then immunoblotted to detect Act1 or IL-17RA. As expected (Fig. 3A), Act1 associated strongly with the functional IL-17RA mutants (namely IL-17RAΔ665 and IL-17RAΔ645). We also saw ligand-dependent association with a larger migrating form of Act1 (top arrow), which we have shown in other studies to be phosphorylated (23). Unexpectedly, one of the non-functional deletions (IL-17RAΔ625) bound strongly to Act1 in the stimulated condition (lane 7), but the larger Act1 isoform was not pulled down with this mutant. However, deletions extending beyond this residue (IL-17RAΔ605) did not bind well to Act1 regardless of whether cells were treated with IL-17. We reproduced this finding by co-IP with a CFP-tagged Act1 and a Myc-tagged IL-17RA (data not shown) (23). To verify that Act1 association was specific to IL-17RA, control co-IP experiments were performed with related receptors, TLR2 and TLR4. As shown (Fig. 3B), we observed weak association of Act1 with TLR2 and TLR4, which is not surprising given the homology between SEFIR and TIR domains. Notably, however, only the smaller isoform of Act1 associated with TLR2 and TLR4, consistent with the idea that the phosphorylated version of Act1 represents the functional isoform. Together, these data show that sequences extending far beyond the SEFIR are needed to permit IL-17RA downstream signaling. However, because Act1 binding does not correlate with IL-17 signaling, recruitment of Act1 (or at least with its smaller isoform) is not in itself sufficient to trigger distal signal transduction.
FIGURE 3.
Activation of TRAF6 but not Act1 requires an intact SEFEX domain. A, association of IL-17RA deletion mutants with Act1. HEK293T cells were transiently transfected with the indicated IL-17RA deletion mutants and Myc-tagged Act1, and lysates were immunoprecipitated with Abs to IL-17RA. IP samples from untreated cells (lanes 1–4) or cells treated with IL-17 for 10 min (lanes 5–8) were separated by SDS-PAGE and immunoblotted (WB) with Abs to Myc (top) or IL-17RA (bottom). The arrows indicate two isoforms of Act1, one of which we have shown to be phosphorylated (23). Note that all lanes were derived from the same gel. Data are representative of at least three experiments. B, Act1 associates preferentially with IL-17RA. HEK293T cells were transiently transfected with HA-tagged receptors, as indicated. Lysates were subjected to IP with anti-HA Abs and immunoblotted with Abs to Myc. C, ubiquitination of TRAF6 by IL-17RA deletion mutants. HEK293T cells were transiently transfected with the indicated IL-17RA deletion mutants and TRAF6, cells were stimulated with or without IL-17 for 15 min, and lysates were immunoprecipitated with Abs to IL-17RA. IP samples were separated by SDS-PAGE and immunoblotted with Abs to TRAF6 (top) or IL-17RA (bottom). Larger ubiquitinated TRAF6 isoforms are indicated. Data are representative of at least three experiments.
Act1 is an E3 ubiquitin ligase that targets TRAF6 (22). We therefore evaluated TRAF6 ubiquitination following stimulation of the mutant receptor series. HEK293T cells were transfected with IL-17RA constructs together with TRAF6, and the appearance of slower migrating, ubiquitinated TRAF6 isoforms was evaluated by immunoblotting (Fig. 3C). There was a dramatic appearance of ubiquitinated TRAF6 in cells expressing the functional receptors (IL-17RAΔ665 and -Δ645) but not the defective receptors (IL-17RAΔ625 and -Δ605). Therefore, in contrast to Act1, activation of TRAF6 correlates tightly with functional receptors.
The IL-17RA Signaling Motif Is Functionally Distinct from Other IL-17R Family Members
The SEFIR is found in all 5 IL-17R family members and is required for signaling downstream of IL-17RC, IL-17RB, and IL-17RA (10, 16, 23). However, the extended domain downstream of the SEFIR is not conserved among these receptors (Fig. 4A). This raises the question of whether other IL-17 family receptor SEFIR domains mediate unique signaling events. Consequently, to determine whether other IL-17R cytoplasmic tails can substitute for IL-17RA, we created a panel of chimeric receptors composed of the extracellular domain and transmembrane domain of IL-17RA fused to the cytoplasmic tails of IL-17RB, IL-17RC, and IL-17RD. All receptor constructs expressed well when transfected into HEK293T cells, although the IL-17RA/RD chimeric receptor was present at somewhat lower surface levels than the other receptors (data not shown). Because IL-17RA partners with IL-17RC to mediate IL-17-dependent signaling, we also created stable cell lines expressing the IL-17RA/RC chimera in the IL-17RA−/− fibroblast background (Fig. 4B). Three independent clones expressing the IL-17RA/RC chimera failed to respond to either IL-17A or IL-17F as measured by IL-6 secretion (representative data shown in Fig. 4C). To evaluate the ability of the chimeras to activate the IL-17-dependent 24p3-luciferase reporter, cells were transiently transfected with IL-17RA/RC, IL-17RA/RB, and IL-17RA/RD, but no detectable IL-17-dependent signaling was observed (Fig. 4D). Accordingly, we conclude that there are unique signals mediated by IL-17RA, which cannot be replaced by other SEFIR-containing receptors.
To determine whether the IL-17R chimeras can recruit Act1 or activate TRAF6, HEK293T cells were transfected with the chimeras and Myc-tagged Act1. The chimeric receptors were immunoprecipitated with Abs to the extracellular domain of IL-17RA. Act1 was pulled down with all the receptors at levels indistinguishable from wild type IL-17RA (Fig. 4E). This finding confirms the IL-17RA deletion analysis (Fig. 3A) showing that, although recruitment of Act1 to the receptor complex is necessary for IL-17-dependent signaling, it apparently is not sufficient to transduce an effective downstream signal. Moreover, when the chimeras were transfected with TRAF6, only IL-17RA led to ubiquitination (Fig. 4F). This is also parallel to the deletion analysis in Fig. 3, demonstrating that TRAF6 activation occurs only with functional receptors.
IL-17 Synergizes with Lymphotoxin, Which Requires the Extended SEFIR Motif in IL-17RA
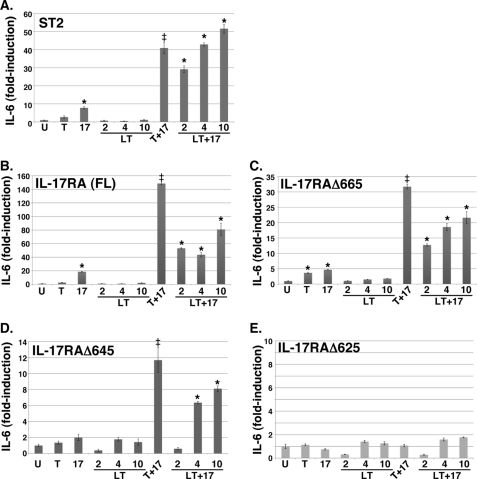
IL-17 acts most potently in combination with other inflammatory receptors (20, 24, 25). Best characterized is its synergy with TNFα, but IL-17 has also been shown to signal cooperatively with IL-22, IL-1β, LPS, and most recently with BAFF (reviewed in Ref. 26). Lymphotoxin (LT) (formerly called TNF-β) is another member of the TNF superfamily. The functional forms of LT are a secreted homotrimer of LTα (LTα3) or membrane-associated LTα paired with LTβ in trimeric configurations. LTα3 binds to the same receptor and activates a similar inflammatory pathway as TNFα (reviewed in Ref. 27), and LTα blockade ameliorates IL-17-dependent disease models, such as collagen-induced arthritis (28). Moreover, surface-associated LTα is found at high levels on Th17 cells (28). Therefore, we questioned whether LTα3 might also signal synergistically with IL-17. Due to difficulty obtaining recombinant mouse LTα3 commercially, we used human LTα3, which interacts with both TNFR1 and TNFR2 (29). As shown, treatment of ST2 stromal cells with IL-17 and LTα3 induced IL-6 expression far more potently than either cytokine alone (Fig. 5A). We next evaluated the panel of IL-17RA mutants in combination with LTα3 and found that the C-terminal end of the functional activation domain was located between residues 645 and 625 (Fig. 5, B–E). This finding indicates that the mechanisms of synergy between IL-17 with either TNFα or LTα3 appear to be the same and further extends the range of IL-17 influence within the inflammatory milieu.
FIGURE 5.
IL-17 synergizes with LTα3, which requires the extended SEFIR domain. A, IL-17 and LTα synergize in a stromal cell line. ST2 stromal cells were treated with IL-17 (200 ng/ml), TNFα (2 ng/ml), and/or LTα3 (2, 4, or 10 ng/ml, as indicated) for 24 h, and supernatants were evaluated for IL-6 in triplicate by ELISA. *, p < 0.05 compared with untreated samples; ‡, p < 0.05 compared with TNF-treated samples. B–E, synergy of IL-17 and LTα3 requires residues in the IL-17RA cytoplasmic tail through amino acid 645. IL-17RA−/− fibroblasts reconstituted with IL-17RA (full-length) or the indicated IL-17RA deletion mutants were treated for 8 or 24 h (not shown) with IL-17, TNFα, and LTα3, and supernatants were evaluated for IL-6 in triplicate as in A.
DISCUSSION
The IL-17 receptor family has long been intriguing in that it is notably different in structure from other classic cytokine subfamilies (3, 30, 31). IL-17RA, the first of the family to be cloned (32), has an unusually large cytoplasmic tail yet encodes few recognizable motifs that might lend insight into its modes of signal transduction. The discovery of the SEFIR and its homology with members of the TLR/IL-1R family (10) led to the prediction that the IL-17 cytokines might function akin to IL-1 and TLR ligands. This was consistent with observations that IL-17-induced genes are associated with innate immune responses and are regulated by proinflammatory transcription factors, such as NF-κB and C/EBP (5, 20, 32–34). The discoveries that IL-17 activates TRAF6 and Act1 rather than the JAK-STAT pathway were also consistent with this view (12, 13, 35). Thus, a general picture of IL-17R signaling has emerged in which the SEFIR domain recruits Act1 via a homotypic SEFIR-SEFIR motif, leading to ubiquitination of TRAF6 and activation of the classical NF-κB pathway, ultimately stimulating proinflammatory gene expression.
The molecular nature of the IL-17R cytoplasmic tail is poorly defined. Deletion of SEFIR domains within IL-17RA or Act1 abrogates the IL-17 response (13, 17). Despite homology between SEFIR and TIR domains, IL-17RA signals independently of TLR signaling intermediates, such as MyD88, TRIF, IRAK1, and IRAK4 (12, 17). We previously reported that a short extension to the IL-17RA SEFIR termed a TILL was required for signaling (17). However, we did not delineate the C-terminal boundary of the SEFIR/TILL-containing region, although modeling studies predicted that the domain was similar in size to a TIR (data not shown). Here we show that a surprisingly long non-conserved region in IL-17RA extending at least 92 amino acids beyond the SEFIR/TILL is necessary for IL-17-mediated signaling, which we term a SEFEX domain (Fig. 2 and Table 1).
We consistently observed that the IL-17RAΔ645 mutant usually signaled weakly compared with IL-17RAΔ665 and FL, which was consistent in several independent clones (data not shown). We speculate that this truncation may lie very close to the C terminus of the functional subdomain and therefore renders the receptor less stable than the IL-17RAΔ665. This idea is consistent with its somewhat reduced surface expression (Fig. 1).
Although no structural information regarding this region is yet available, we predict that the SEFIR and SEFEX regions comprise a composite motif rather than separate substructures. If true, this domain is much larger than a TIR and probably will have a different three-dimensional architecture. Similar to IL-17RA, we recently found that an extended region of IL-17RC downstream of the SEFIR is also needed for IL-17-dependent signaling, although the IL-17RC SEFEX does not bear detectable homology to the IL-17RA SEFEX motif (23, 36). Therefore, whereas SEFIR domains are required for IL-17-mediated signal transduction, sequences distal to the SEFIR also provide critical functions, which we speculate confer unique signaling properties to each subunit.
Because SEFIRs are conserved in the IL-17R family, we asked whether they are functionally interchangeable by replacing the cytoplasmic tail of IL-17RA with that of other IL-17R family members. Replacing IL-17RA with other SEFIR domains did not rescue signaling (Fig. 4). Furthermore, replacing just the IL-17RA SEFIR with the IL-17RC SEFIR while retaining IL-17RA distal sequences also failed to restore function,6 supporting the notion that the overall structure of the IL-17RA SEFIR/SEFEX domain is distinct from the rest of the family.
The IL-17RA cytoplasmic tail is by far the largest in the IL-17R family. We have shown previously that the distal ∼200 amino acids of the receptor comprise a second functional domain involved in regulation of C/EBPβ (17, 19). Like the SEFEX, the C/EBPβ activation domain is absent in other IL-17R family members, and almost nothing is known about its structure. Our prior studies showed that the C/EBPβ activation domain exerts an inhibitory effect on signaling, which is manifested in consistently enhanced signaling by the IL-17RAΔ665 deletion compared with full-length IL-17RA (19).
IL-17RA has been shown to participate in other receptor complexes. For example, IL-17RA pairs with IL-17RB to make up the IL-17E·IL-25R complex (37). IL-17RA−/− mice are deficient in IL-25 signaling, and IL-17RB also recruits Act1 (15, 16). It is not clear whether the same extended sequences of IL-17RA are required for responses to IL-25. IL-17RA has also been suggested to partner with IL-17RD (SEF) (38), although the ligand that binds to this pairing is unknown.
Act1 is critical for signaling downstream of IL-17RA and IL-17RC (39). In the case of IL-17RA, deletions that lack Act1 binding capacity also fail to mediate IL-17-dependent signaling (Fig. 3 and Table 1). However, one non-signaling deletion was still able to recruit at least one isoform of Act1 efficiently (IL-17RAΔ625), demonstrating that Act1 recruitment is not enough to drive downstream signaling (Fig. 3A).
To probe this issue further, we employed a chimeric receptor system to show that Act1 in fact binds to many members of the IL-17R family. An advantage of this chimeric system is that it enables us to directly compare the binding capacity of each receptor to Act1 because the extracellular domain (where the immunoprecipitating Ab binds) is held constant. This experiment highlights the unexpected observation that simply recruiting Act1 is not sufficient for mediating signal transduction because none of the chimeric receptors were able to mediate IL-17-dependent signaling (Fig. 4). It is not completely clear which isoform(s) of Act1 are pulled down with each chimeric receptor, which is an interesting issue that will be addressed in ongoing studies. Rather, ubiquitination of TRAF6 appears to be the critical event necessary for downstream signaling to NF-κB, and apparently recruitment of Act1 alone cannot lead to modification of TRAF6. Moreover, even recruitment of Act1 to the IL-17RAΔ625 mutant is not enough to drive ubiquitination of TRAF6 (Fig. 3, A and C); therefore, additional as yet unknown components must be necessary.
A hallmark feature of IL-17 is that it synergizes potently with other cytokines (40). This is probably an accurate reflection of the inflammatory environment, where many cytokines participate in fine tuning the magnitude and duration of immune responses. Because TNFα and LTα3 share a common receptor, and the LTα monomer is highly expressed on Th17 cells (28), we reasoned that IL-17 was likely to exhibit a similar synergy with this cytokine, which was indeed the case (Fig. 5). Although TNFα and LTα3 have overlapping functions due to shared receptor usage, they also exhibit divergence in vivo. For example, LTα−/− mice lack lymph nodes, which is not true of TNFα−/− or IL-17−/− mice (27). Whereas IL-17 is considered to mediate immunity to extracellular pathogens, LT plays an important role in anti-viral immunity (27). However, there is mounting evidence that IL-17 contributes to anti-viral responses (26), and thus it is conceivable that this might be mediated in cooperation with LT. In addition, membrane-bound LT from RORγt+ cells (which includes all IL-17-producing cells) is required for host defense against intestinal infection with Citrobacter rodentium (41). Intriguingly, blockade of LT reduces disease symptoms in several models dependent on Th17 cells, including collagen-induced arthritis, experimental autoimmune encephalomyelitis, and delayed type hypersensitivity (28). Given the connection of IL-17, TNFα, and LT to rheumatoid arthritis pathogenesis, it is not surprising that IL-17 cooperates with both cytokines.
In summary, we have delineated several important new features of IL-17RA-mediated signal transduction. We identified the C-terminal boundary of the SEFIR-containing motif and show that it includes an unexpectedly large extension. We have also shown that the cytoplasmic domains of IL-17R subfamily members are not interchangeable and that IL-17 signals through the SEFIR/SEFEX motif in cooperation with LTα3.
Acknowledgments
We thank Dr. Joel Tocker of Amgen (Seattle, WA) for anti-IL-17RA Abs and IL-17RA−/− mice from which cell lines were derived. We thank Dr. L. Garrett-Sinha (State University of New York, Buffalo, NY) for primary kidney mRNA, Drs. R. Tapping (University of Illinois, Chicago, IL), and S. Sarkar (University of Pittsburgh) for TLR2 and TLR4 constructs, and we thank L. Kane and C. Coyne (University of Pittsburgh) for valuable suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant AR054389 (to S. L. G.).
R. M. Onishi, S. J. Park, A. Maitra, and S. L. Gaffen, unpublished observations.
S. J. Park, unpublished data.
- TLR
- Toll-like receptor
- SEFIR
- SEF/IL-17R
- C/EBP
- CCAAT/enhancer-binding protein
- FL
- full-length
- LT
- lymphotoxin
- SEFEX
- SEFIR extension
- TILL
- TIR-like loop
- Ab
- antibody
- IP
- immunoprecipitation
- TIR
- Toll/IL-IR.
REFERENCES
- 1.McGeachy M. J., Cua D. J. (2008) Immunity 28, 445–453 [DOI] [PubMed] [Google Scholar]
- 2.Korn T., Bettelli E., Oukka M., Kuchroo V. K. (2009) Annu. Rev. Immunol. 27, 485–517 [DOI] [PubMed] [Google Scholar]
- 3.Gaffen S. L. (2009) Nat. Rev. Immunol. 9, 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen F., Gaffen S. L. (2008) Cytokine 41, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen F., Hu Z., Goswami J., Gaffen S. L. (2006) J. Biol. Chem. 281, 24138–24148 [DOI] [PubMed] [Google Scholar]
- 6.Toy D., Kugler D., Wolfson M., Vanden Bos T., Gurgel J., Derry J., Tocker J., Peschon J. J. (2006) J. Immunol. 177, 36–39 [DOI] [PubMed] [Google Scholar]
- 7.Ishigame H., Kakuta S., Nagai T., Kadoki M., Nambu A., Komiyama Y., Fujikado N., Tanahashi Y., Akitsu A., Kotaki H., Sudo K., Nakae S., Sasakawa C., Iwakura Y. (2009) Immunity 30, 108–119 [DOI] [PubMed] [Google Scholar]
- 8.Kuestner R. E., Taft D. W., Haran A., Brandt C. S., Brender T., Lum K., Harder B., Okada S., Ostrander C. D., Kreindler J. L., Aujla S. J., Reardon B., Moore M., Shea P., Schreckhise R., Bukowski T. R., Presnell S., Guerra-Lewis P., Parrish-Novak J., Ellsworth J. L., Jaspers S., Lewis K. E., Appleby M., Kolls J. K., Rixon M., West J. W., Gao Z., Levin S. D. (2007) J. Immunol. 179, 5462–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y., Valdez P. A., Danilenko D. M., Hu Y., Sa S. M., Gong Q., Abbas A. R., Modrusan Z., Ghilardi N., de Sauvage F. J., Ouyang W. (2008) Nat. Med. 14, 282–289 [DOI] [PubMed] [Google Scholar]
- 10.Novatchkova M., Leibbrandt A., Werzowa J., Neubüser A., Eisenhaber F. (2003) Trends Biochem. Sci. 28, 226–229 [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki A., Medzhitov R. (2004) Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 12.Chang S. H., Park H., Dong C. (2006) J. Biol. Chem. 281, 35603–35607 [DOI] [PubMed] [Google Scholar]
- 13.Qian Y., Liu C., Hartupee J., Altuntas C. Z., Gulen M. F., Jane-Wit D., Xiao J., Lu Y., Giltiay N., Liu J., Kordula T., Zhang Q. W., Vallance B., Swaidani S., Aronica M., Tuohy V. K., Hamilton T., Li X. (2007) Nature Immunology 8, 247–256 [DOI] [PubMed] [Google Scholar]
- 14.Li X. (2008) Cytokine 41, 105–113 [DOI] [PubMed] [Google Scholar]
- 15.Swaidani S., Bulek K., Kang Z., Liu C., Lu Y., Yin W., Aronica M., Li X. (2009) J. Immunol. 182, 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claudio E., Sønder S. U., Saret S., Carvalho G., Ramalingam T. R., Wynn T. A., Chariot A., Garcia-Perganeda A., Leonardi A., Paun A., Chen A., Ren N. Y., Wang H., Siebenlist U. (2009) J. Immunol. 182, 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maitra A., Shen F., Hanel W., Mossman K., Tocker J., Swart D., Gaffen S. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7506–7511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., Tong L. (2000) Nature 408, 111–115 [DOI] [PubMed] [Google Scholar]
- 19.Shen F., Li N., Gade P., Kalvakolanu D. V., Weibley T., Doble B., Woodgett J. R., Wood T. D., Gaffen S. L. (2009) Sci. Signal. 2, ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruddy M. J., Wong G. C., Liu X. K., Yamamoto H., Kasayama S., Kirkwood K. L., Gaffen S. L. (2004) J. Biol. Chem. 279, 2559–2567 [DOI] [PubMed] [Google Scholar]
- 21.Shen F., Ruddy M. J., Plamondon P., Gaffen S. L. (2005) J. Leukoc. Biol. 77, 388–399 [DOI] [PubMed] [Google Scholar]
- 22.Liu C., Qian W., Qian Y., Giltiay N. V., Lu Y., Swaidani S., Misra S., Deng L., Chen Z. J., Li X. (2009) Sci. Signal. 2, ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho A. W., Shen F., Conti H. R., Patel N., Childs E. E., Peterson A. C., Hernández-Santos N., Kolls J. K., Kane L. P., Ouyang W., Gaffen S. L. (2010) J. Immunol. 185, 1063–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruddy M. J., Shen F., Smith J. B., Sharma A., Gaffen S. L. (2004) J. Leukoc. Biol. 76, 135–144 [DOI] [PubMed] [Google Scholar]
- 25.Patel D. N., King C. A., Bailey S. R., Holt J. W., Venkatachalam K., Agrawal A., Valente A. J., Chandrasekar B. (2007) J. Biol. Chem. 282, 27229–27238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onishi R. M., Gaffen S. L. (2010) Immunology 129, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tracey D., Klareskog L., Sasso E. H., Salfeld J. G., Tak P. P. (2008) Pharmacol. Ther. 117, 244–279 [DOI] [PubMed] [Google Scholar]
- 28.Chiang E. Y., Kolumam G. A., Yu X., Francesco M., Ivelja S., Peng I., Gribling P., Shu J., Lee W. P., Refino C. J., Balazs M., Paler-Martinez A., Nguyen A., Young J., Barck K. H., Carano R. A., Ferrando R., Diehl L., Chatterjea D., Grogan J. L. (2009) Nat. Med. 15, 766–773 [DOI] [PubMed] [Google Scholar]
- 29.Bossen C., Ingold K., Tardivel A., Bodmer J. L., Gaide O., Hertig S., Ambrose C., Tschopp J., Schneider P. (2006) J. Biol. Chem. 281, 13964–13971 [DOI] [PubMed] [Google Scholar]
- 30.Aggarwal S., Gurney A. L. (2002) J. Leukoc. Biol. 71, 1–8 [PubMed] [Google Scholar]
- 31.Ely L. K., Fischer S., Garcia K. C. (2009) Nat. Immunol. 10, 1245–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Z., Fanslow W. C., Seldin M. F., Rousseau A. M., Painter S. L., Comeau M. R., Cohen J. I., Spriggs M. K. (1995) Immunity 3, 811–821 [DOI] [PubMed] [Google Scholar]
- 33.Fossiez F., Djossou O., Chomarat P., Flores-Romo L., Ait-Yahia S., Maat C., Pin J. J., Garrone P., Garcia E., Saeland S., Blanchard D., Gaillard C., Das Mahapatra B., Rouvier E., Golstein P., Banchereau J., Lebecque S. (1996) J. Exp. Med. 183, 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. (2005) Nat. Immunol. 6, 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwandner R., Yamaguchi K., Cao Z. (2000) J. Exp. Med. 191, 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho A. W., Gaffen S. L. (2010) Semin. Immunopathol. 32, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rickel E. A., Siegel L. A., Yoon B. R., Rottman J. B., Kugler D. G., Swart D. A., Anders P. M., Tocker J. E., Comeau M. R., Budelsky A. L. (2008) J. Immunol. 181, 4299–4310 [DOI] [PubMed] [Google Scholar]
- 38.Rong Z., Wang A., Li Z., Ren Y., Cheng L., Li Y., Wang Y., Ren F., Zhang X., Hu J., Chang Z. (2009) Cell Res. 19, 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindén A. (2007) Sci. STKE 2007, re4. [DOI] [PubMed] [Google Scholar]
- 40.Miossec P. (2003) Arthritis Rheum. 48, 594–601 [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Koroleva E. P., Kruglov A. A., Kuprash D. V., Nedospasov S. A., Fu Y. X., Tumanov A. V. (2010) Immunity 32, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]