Abstract
Bacteriophage endolysins include a group of new antibacterials reluctant to development of resistance. We present here the first structural study of the Cpl-7 endolysin, encoded by pneumococcal bacteriophage Cp-7. It contains an N-terminal catalytic module (CM) belonging to the GH25 family of glycosyl hydrolases and a C-terminal region encompassing three identical repeats of 42 amino acids (CW_7 repeats). These repeats are unrelated to choline-targeting motifs present in other cell wall hydrolases produced by Streptococcus pneumoniae and its bacteriophages, and are responsible for the protein attachment to the cell wall. By combining different biophysical techniques and molecular modeling, a three-dimensional model of the overall protein structure is proposed, consistent with circular dichroism and sequence-based secondary structure prediction, small angle x-ray scattering data, and Cpl-7 hydrodynamic behavior. Cpl-7 is an ∼115-Å long molecule with two well differentiated regions, corresponding to the CM and the cell wall binding region (CWBR), arranged in a lateral disposition. The CM displays the (βα)5β3 barrel topology characteristic of the GH25 family, and the impact of sequence differences with the CM of the Cpl-1 lysozyme in substrate binding is discussed. The CWBR is organized in three tandemly assembled three-helical bundles whose dispositions remind us of a super-helical structure. Its approximate dimensions are 60 × 20 × 20 Å and presents a concave face that might constitute the functional region involved in bacterial surface recognition. The distribution of CW_7 repeats in the sequences deposited in the Entrez Database have been examined, and the results drastically expanded the antimicrobial potential of the Cpl-7 endolysin.
Keywords: Bacteriophage, Biophysics, Circular Dichroism (CD), Computer Modeling, Ultracentrifugation, CW-7 Motif, Cpl-7 Endolysin Structure, SAXS, Streptococcus pneumoniae, Cell Wall Hydrolase
Introduction
Bacteria and bacteriophages produce a variety of enzymes that cleave the peptidoglycan, the cell wall hydrolases (CWHs),3 either to lyse host cells or to re-model the cell wall during growth and division. Many of these enzymes have a modular structure and are composed of catalytic units linked to a number of other modules. The Cpl-7 lysozyme, the lytic enzyme (endolysin) encoded by the pneumococcal bacteriophage Cp-7, consists of a catalytic N-terminal domain fused to a C-terminal region containing three identical tandem repeats of 48 amino acids (the third motif has the last six residues missing), which is essential for activity and unique among CWHs encoded by the pneumococcus and its bacteriophages (1, 2). However, the same type of repeats (termed CW_7 hereafter) has been later identified in other proteins putatively associated with bacterial cell wall degradation activity (3). The catalytic module (CM) of Cpl-7 belongs to the GH25 family of glycosyl hydrolases and shows 85.6% sequence identity with the CM of the Cpl-1 lysozyme, the endolysin encoded by the pneumococcal bacteriophage Cp-1 (4). However, the cell wall binding region (CWBR) of Cpl-1, made of six tandem copies of ∼20 amino acid residues plus a terminal tail, belongs to the family of pneumococcal choline-binding modules (5). Its lytic activity is thus conditioned by the presence of choline moieties in pneumococcal teichoic and lipoteichoic acids and becomes inhibited at high choline concentrations (IC50 = 2 mm) (1, 6). In contrast, Cpl-7 degrades pneumococcal cell walls containing either choline or ethanolamine and retains full activity even in the presence of high concentrations of these amino alcohols (7). This strongly indicates that the C-terminal modules of Cpl-7 and Cpl-1 are responsible for cell wall targeting but recognize different structural elements of the bacterial envelope. Indeed, the loss of the CWBR by controlled proteolytic digestion reduces 4 orders of magnitude the activity of Cpl-7 CM against pneumococcal cell walls containing either choline or ethanolamine (8). The reduced activity of this truncated form is comparable with those reported for the isolated CM of Cpl-1 against both types of substrates and for the full-length form of Cpl-1 when assayed against ethanolamine-containing cell walls, and it reflects the intrinsic activity of Cpl-7 and Cpl-1 CMs without cooperation of their cell wall-binding motifs. The relationship between the CW_7 repeats and the substrate specificity displayed by the Cpl-7 lysozyme was clearly demonstrated by construction of a chimeric protein, LC7, consisting of the CM of the LytA amidase (the major pneumococcal autolysin and a choline-dependent enzyme) fused to the C-terminal region of Cpl-7 (7). This chimeric enzyme exhibited an amidase activity capable of degrading ethanolamine-containing cell walls not inhibited by choline. Furthermore, the CWBR also determined the optimal pH for the catalytic activity of LC7 and the parental enzymes. Interestingly, the acquisition of one or two CW_7-like motifs by the endopeptidase module of the λSa2 endolysin, a bifunctional enzyme containing endopeptidase and β-d-N-acetylglucosaminidase activities, not only increased the activity on several streptococcal strains but was shown to be essential for its activity on staphylococci (3).
The crystallographic structure of Cpl-1, both in its free-state and bound to muropeptide analogues, has been reported (9, 10) and its solution behavior characterized both in the absence and in the presence of choline (11, 12). In contrast, although the CM of Cpl-7 has been recently crystallized (13), there is no available information about the overall structure of the enzyme or its CWBR.
Because of their unique ability to cleave peptidoglycan in a generally species-specific manner, endolysins represent a novel class of antibacterial agents (enzybiotics) and provide a mean of selective and rapid killing of pathogenic bacteria, refractory to resistance development (14, 15). Indeed, the endolysins encoded by the lytic pneumococcal bacteriophages Dp-1 and Cp-1 have been shown to efficiently eradicate pneumococci without affecting the normal microbiota (16–22), a feature related with their targeting of choline moieties.
The presence in the Cpl-7 endolysin of a cell wall-binding motif that may recognize a distinct element of the cell wall opens new possibilities to the potential use of both the wild-type enzyme and chimeric constructions bearing CW_7 repeats as efficient enzybiotics against Streptococcus pneumoniae and other Gram-positive bacterial pathogens. This study reports the first structural characterization of the Cpl-7 lysozyme using low resolution experimental techniques, i.e. CD, analytical ultracentrifugation, and small angle x-ray scattering (SAXS), and three-dimensional modeling. Cpl-7 behaves as a monomer in solution, and its two modules are arranged forming an elongated particle of ≅115 Å long. The CM has the (βα)5β3 barrel characteristic of the GH25 family, whereas the CWBR would include three tandemly assembled, three-helical bundles whose disposition reminds a super-helical structure. The high resolution models proposed here for the isolated modules together with the SAXS-derived model for the overall protein structure provide clues to facilitate the identification of residues and surfaces important for Cpl-7 attachment to the cell wall. Finally, the three-helical bundle formed by the CW_7 repeat seems to be conserved in the related motifs that have been identified, in variable number and position, linked to a variety of catalytic units and cell wall-binding motifs in many putative proteins potentially involved in cell wall metabolism.
EXPERIMENTAL PROCEDURES
Materials
Plasmid pCP700 overexpressing the Cpl-7 lysozyme (7) was constructed by cloning a 2.5-kb BamHI-BclI fragment of pCP70 containing the cpl-7 gene (1) into BamHI-digested pIN-III(lppP-5)-A3 (23), and Cpl-7 lysozyme (38,438 Da; Entrez code, AAA72844) was purified from Escherichia coli RB791 cells harboring pCP700. E. coli cultures were grown at 37 °C in LB medium containing ampicillin (100 μg ml−1) to an absorbance at 600 nm (A600) of ∼0.6. Lactose was then added (2% w/v final concentration) and incubation continued overnight. Cells were harvested at 4 °C by centrifugation in an SLA-3000 rotor (Sorvall) for 20 min at 8,000 rpm, resuspended in 20 mm sodium phosphate buffer, pH 6.9 (Pi buffer), and disrupted in a French pressure cell press. The insoluble fraction was separated by centrifugation (13,000 rpm in an SS-34 rotor (Sorvall) for 20 min), and the supernatant was treated with streptomycin sulfate (0.8% w/v final concentration) to precipitate nucleic acids. The soluble fraction was separated by centrifugation and subjected to a fractioning at increasing concentrations of ammonium sulfate. Proteins precipitated between 20 and 35% (w/v) of salt saturation were resuspended and exhaustively dialyzed against Pi buffer (24 h, 5×, 500 ml), and then loaded onto a DEAE-cellulose column (15 × 2 cm) at 0.5 ml min−1 flow rate. The column was washed with Pi buffer until the initial base line (A280) was recovered. A discontinuous NaCl gradient was then applied using a Pharmacia LKB fast-protein liquid chromatograph (GE Healthcare). After a first gradient from 0 to 0.3 m of salt (60 min), followed by washing with 0.3 m NaCl until base-line recovering, the protein was eluted with a gradient from 0.3 to 0.5 m NaCl (60 min) followed by 30 min of washing with 1 m NaCl. Cpl-7 eluted around 0.46 m salt, and the fractions collected showed a single band in SDS-PAGE (12% acrylamide/bisacrylamide) whose mobility agreed with the protein theoretical mass. Fractions with A280/A260 ratios ≥1.8 were pooled, concentrated, and dialyzed against Pi buffer before being stored at −20 °C. Those with lower absorbance ratios were subjected to an additional gel filtration chromatography step in a SuperoseTM 12 column (GE Healthcare; 30 × 1.5 cm) to remove minor contaminations by nucleic acids. The protein was eluted (0.5 ml min−1) isocratically with Pi buffer, and the collected fractions were stored as above. All purification steps were performed at 4 °C, and reagents (Sigma) were of analytical grade. Protein activity was tested using [methyl-3H]choline-labeled pneumococcal cell walls (7).
Cpl-7 concentration was measured spectrophotometrically using a molar absorption coefficient at 280 nm of 65,700 ± 700 m−1 cm−1, determined according to Gill and von Hippel (24). The N terminus of Cpl-7 was sequenced using an automatic sequencer (model 477A from Applied Biosystems) as described previously (25).
Circular Dichroism
CD spectra (average of four scans) were recorded in the far-UV region using a Jasco-J810 spectropolarimeter (Jasco Corp.) equipped with a Peltier-type cell holder. Measurements were performed using a scan rate of 20 nm min−1, a response time of 4 s, a bandwidth of 1 nm, and protein concentrations of 5.2 and 26 μm (1- and 0.2-mm path length cells, respectively). Buffer contribution was subtracted from the experimental data, and the corrected ellipticity was converted to mean residue ellipticity using an average molecular mass per residue of 112.39. Secondary structure (SS) content was estimated by deconvolution of the experimental curves with CONTIN (26), SELCON (27), and CDNN (28) programs using reference data sets with 17 (CONTIN and SELCOM) and 13 (CDNN) protein spectra. They evaluate four conformations (α-helix, β-sheet, β-turn, and remainder), but CONTIN does not discriminate between parallel and antiparallel β-sheet.
Secondary Structure Prediction
Prediction of SS was performed using Prof (29), PSIPRED (30), and Jpred 3 (31), all of them accessible via the ExPASy website. Final prediction data include only those residues whose secondary structure was predicted with reliability above 86% by at least two of the three methods.
Analytical Ultracentrifugation
Sedimentation velocity measurements were carried out using an Optima XL-A (absorption optics) and an Optima XL-I (interference) analytical ultracentrifuge (Beckman Coulter) for detection of concentration gradients. The experiments were performed in Pi buffer at 45,000 rpm, with 400 μl of protein solution in each cell, using double sector Epon-charcoal centerpieces. Differential sedimentation coefficients, c(s), were calculated by least squares boundary modeling of sedimentation velocity data using the program SEDFIT (32). The SEDNTERP program (33) was used to calculate standard sedimentation coefficients, s20,w, from the experimental values, the effective Stokes radius (RS), and the frictional ratio (f/f0) assuming a specific hydration grade of 0.404 g of water/g of protein. A partial specific volume of 0.722 ml g−1 at 20 °C was calculated from the amino acid composition (33). Equilibrium sedimentation experiments were performed at 20 °C by centrifugation of 80 μl samples at 10,000, 12,000 and 17,000 rpm in the Optima XL-A ultracentrifuge. Conservation of mass in the cell was checked in all the experiments. Data were analyzed following previously reported protocols (34). The theoretical value of the sedimentation coefficient of ab initio and high resolution models proposed for the overall structure were calculated using HYDRO and HYDROPRO (35, 36).
Three-dimensional Modeling of Cpl-7 Modules
A search for homologues of Cpl-7 modules in the nonredundant protein sequence data base at NCBI using PSI-BLAST (37) returned the catalytic module of the Cpl-1 lysozyme (PDB code 1h09) as the best matching structure for the CM of Cpl-7. However, no sequences related to the CWBR with known structure were found. Protein-fold recognition methods were therefore applied using the FUGUE (38), 3D-PSSM (39), and SP4 (40) servers, taking as searching target either the sequence of the whole module or a single CW_7 repeat. Only the sequences identified as the best hits by at least two searching methods showing some relevant residue conservation and a good correlation with the SS of the CWBR were considered as possible templates for the structural model.
Sequence alignments were performed using Clustal_X (41) and corrected, when necessary, by manual adjustment with SEAVIEW (42) using as a guide the predicted SS of the target. The structural models were built automatically using the Swiss-Model server in the alignment mode (43), and their quality was evaluated with PROCHECK (44), VERIFY3D (45), QMEAN (46), and ERRAT (47). Several cycles of model building, realignment, and validation were repeated for the CWBR until no further optimization of the structure was achieved.
The best structural models of each module were inspected visually to identify atomic clashes that were relieved by manually twisting the amino acid side chains using the Sybyl software (Sybyl version 7.2, Tripos Inc.). After addition of all hydrogen atoms, the energy of the optimized structures was minimized with the AMBER force field implemented in the Sybyl package, using 3000 steps of the conjugate gradient minimization, and the quality of the final models was evaluated again as indicated above.
SAXS Measurements
Data collection was performed at the 2.1 station of Daresbury's synchrotron. The camera was set to cover ranges of the scattering vector (defined as reciprocal Bragg spacing, i.e. 2 sinφ/λ) from ∼0.001 to 0.060 Å−1, and data collected ranged from 0.045 to 0.05 Å−1. Absolute values of the scattering vector were obtained by reference to the orders of the 67 nm repeat in wet rat tail collagen. The temperature of the samples was set at 4 °C, and the x-ray scattering profiles were recorded in 60 time frames of 30 s. Prior to SAXS data collection, Cpl-7 samples were centrifuged for 20 min at 50,000 rpm and 4 °C in a Beckman TL-100 (TL100.3 rotor), and the lowest part of the protein solution was discarded to remove possible aggregates.
Data processing was performed using the software package provided by the Collaborative Computational Project for Fiber Diffraction and Solution Scattering. Raw data were normalized by beam intensity and detector response before being processed. Time frames showing radiation damage were removed before averaging.
The Guinier plot of log(I(s)) against s2, where I(s) is the scattered intensity, was used to check the quality of the scattering data and the presence or absence of aggregates or interparticle interferences in solution (48). A first estimation of Rg and the forward scattering intensity (Io) was obtained from the slope and the y-intercept in the range of s values where the Guinier plot fits to a straight line (48). The radius of gyration (Rg), Io, and the pair distance distribution function (p(r)) were then calculated from the experimental scattering data using the package GNOM (49). The maximum dimension of the particle, Dmax, was determined empirically by examining the qualities of the fits of p(r) Fourier transform to the experimental data for a given range of Dmax values. Ab initio bead models of Cpl-7 were constructed with DALAI_GA (50) using SAXS data. Typically, 10 models were generated, superimposed, and aligned in pairs with SUPCOMB (51). The models with lowest average spatial discrepancy were considered to be the most probable, and the most divergent ones were considered as outliers. The selected aligned structures were averaged (DAMAVER) and filtered (DAMFILT) using as cutoff in DAMFILT the 70% of its default value (52).
The molecular envelope of the final model was obtained using the Situs package (53), and the structural models built for the isolated modules were manually docked in this envelope using the USCF-Chimera program (54). The scattering profile and the radius of gyration of Cpl-7 model based in the docked structures were calculated with CRYSOL (55).
RESULTS
CW_7 Repeats in Nature
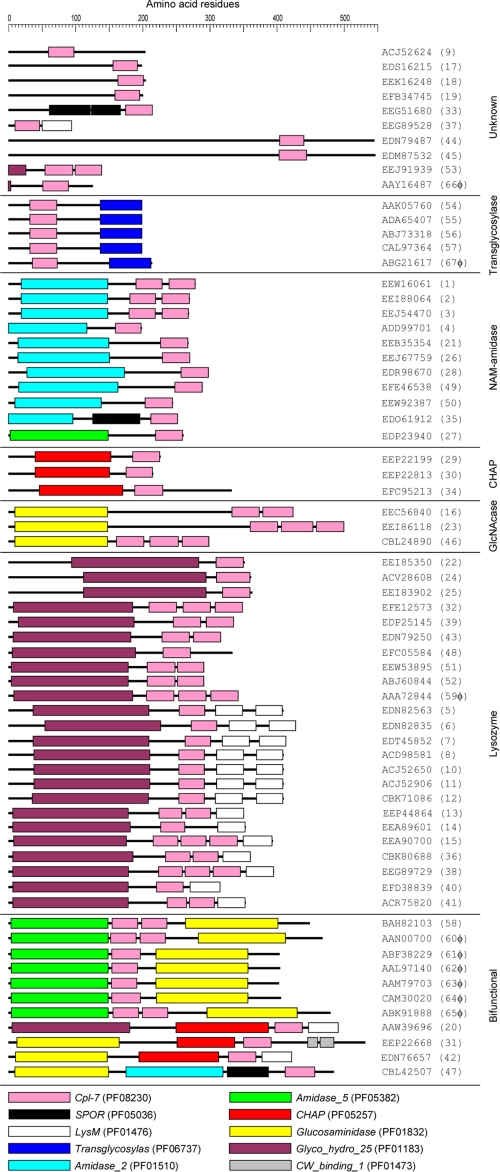
The Cpl-7 lysozyme is the only member known of the pneumococcal CWH family whose activity does not require the presence of phosphocholine residues in teichoic and lipoteichoic acids associated to the bacterial cell wall (2). Instead of the choline-binding repeats that characterize pneumococcal CWHs, the CWBR of Cpl-7 includes three identical repeats of 48 amino acids (the CW_7 repeats), found for the first time in this lysozyme (1), although in the third one the last six residues are missing. However, a search of the Entrez Database (55) showed that the CW_7 motif (Cpl-7 family; Pfam entry PF08230) has significant similarity with 39–42- amino acid long segments present, in variable number (1–3) and position (seven major architectures), in 67 different proteins whose other modules suggest their implication in cell wall metabolism in most cases (last date accessed, April 8, 2010) (Fig. 1). Most bacterial species (30 of 46) belong to the Firmicutes phylum and 21 of them to the order Clostridiales (supplemental Table S1). The majority of species represent normal inhabitants of the microbiota of human gastrointestinal tract (including four members of the phylum Bacteroidetes) (56–58), although some of them may cause different diseases, including endocarditis (e.g. Corynebacterium jeikeium and Granulicatella elegans) and oral (e.g. Bifidobacterium dentium and Parvimonas micra) and skin infections (e.g. Propionibacterium acnes and Streptococcus dysgalactiae subsp. equisimilis). However, some beneficial bacterial species such as certain bifidobacteria also encode CW_7-containing proteins. Besides, Dehalococcoides ethenogenes (phylum Chloroflexi) and Ethanoligenens harbinense (phylum Firmicutes) were isolated from contaminated waters (supplemental Table S1). Nine proteins containing CW_7 motifs are encoded by prophages and lytic phages, including the pneumococcal phage Cp-7. However, when the genomic regions located upstream of the open reading frames encoding CW_7-containing proteins in different bacteria were examined, the frequent presence of genes coding for holins and/or other phage-related proteins strongly suggested that 40 of the 67 CW_7-containing proteins were encoded by prophages (or phage remnants) (supplemental Table S1).
FIGURE 1.
Schematic representation of proteins containing the CW_7 motif. Proteins were ordered by increasing complexity. The accession number of each protein is shown. The modules are color-coded and their designations according to the Pfam data base are shown at the bottom. Additional data are shown in supplemental Table S1.
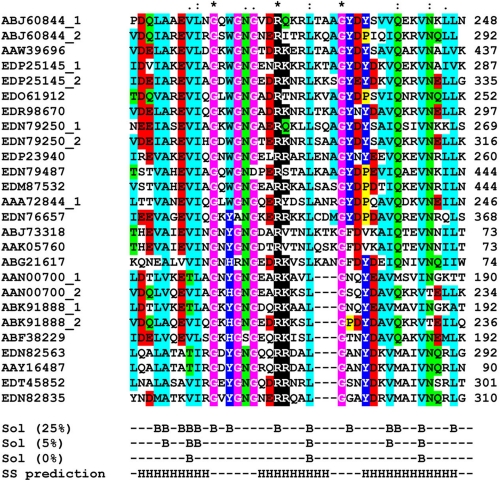
The CW_7 repeats combine with lysozyme modules (Glyco_hydro_25; PF01183), as in Cpl-7, as well as with other modules such as N-acetylmuramoyl-l-alanine amidase (Amidase_2 (PF01510) or Amidase_5 (PF05382)), N-acetylglucosaminidase (Glucosaminidase; PF01832), transglycosylase (Transglycosylase; PF05036), or cysteine, histidine-dependent amidohydrolases/peptidases (CHAP; PF05257) domains. Eleven proteins contain different combinations of CMs (Fig. 1). Moreover, the multiple alignment of a representative sample of the amino acid sequences of CW_7 repeats showed that sequence conservation expands the first 42 residues of the CW_7 motifs of Cpl-7 (Fig. 2). This indicates that the six residues missing in the last repeat of Cpl-7 correspond, in fact, to a linker connecting the CW_7 motifs. Another finding of potential interest was that there are two subfamilies of CW_7 motifs differing by three amino acid residues near the middle of the repeat (Fig. 2). With only three exceptions (i.e. EEA90700 from Collinsela stercoris, EEG 89729 from Coprococcus comes, and EEI88064 from Mobiluncus curtisii), both types of repeats do not coexist in the same protein.
FIGURE 2.
Sequence conservation in the CW_7 motifs. Alignment of representative sequences of the CW_7 motif from Pfam entry PF08230. The Entrez accession numbers are shown. The last amino acid positions are indicated on the right. Asterisks, colons, and dots correspond to strictly conserved residues, conservative, and semi-conservative substitutions (based in BLOSUM substitution matrices), respectively. Color scheme is as follows: pink, glycine; yellow, proline; light blue, small and hydrophobic; dark blue, tyrosine and histidine; green, hydroxyl and neutral polar; red, acidic; and black, basic. Residues with percentages of solvent-accessible surface below 25, 5, and 0%, as well as predicted α-helices (H) along the motif are also indicated.
Cpl-7 Purification
The Cpl-7 lysozyme (predicted molecular mass of 38,438 Da) was purified from extracts of E. coli RB791(pCP700) cells by ion-exchange chromatography in DEAE-cellulose of a crude extract of proteins precipitated between 20 and 35% ammonium sulfate, followed by size-exclusion chromatography on a dextran-agarose column (SuperoseTM 12, GE Healthcare). Typically, up to 35 mg of electrophoretically pure Cpl-7 were obtained from 2 liters of culture. N-terminal sequence determination showed that Met1 was removed under the overexpression and purification conditions used. The specific activity of the purified protein on [3H]choline-labeled pneumococcal cell walls was 1.1 × 105 units mg−1 under optimal conditions (25 mm sodium acetate, pH 5.5, 37 °C), which was slightly lower than the value reported for the Cpl-1 lysozyme at equivalent conditions (6).
Secondary Structure Composition of the Cpl-7 Lysozyme
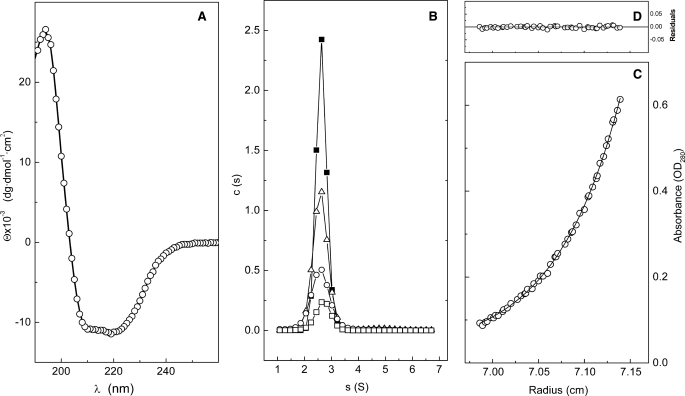
The SS of Cpl-7 was experimentally evaluated using CD. The far-UV region of the CD spectrum presents a maximum at 195 nm and two minima at 209 and 220 nm, characteristic of proteins with a high α-helical content (Fig. 3A). SS composition was calculated by deconvolution of the spectrum using different programs with similar results. The average content was estimated to be 36.8 ± 0.4% α-helix, 17 ± 1% β-sheet (7% antiparallel and 10% parallel), 18 ± 1% turns, and 28 ± 3% nonperiodic structure.
FIGURE 3.
Characterization of Cpl-7 secondary and quaternary structures. A, far-UV CD spectra; symbols are the experimental data and the continuous line represents the theoretical fit of CONTIN. B, distribution of sedimentation coefficients measured at 2.5 μm (open squares), 6.5 μm (circles), 11.7 μm (triangles), and 16.8 μm (black squares). C, sedimentation equilibrium profile at 17,000 rpm showing the fit (continuous line) of the experimental data (circles) to a single species with an average molecular mass of 40.2 ± 0.2 kDa; the residual plot is shown in D. All measurements were performed in Pi buffer at 20 °C.
The SS of the CM was independently estimated using the crystallographic structure of Cpl-1 (PDB code 1h09) as the CMs of both lysozymes show 85.6% identity (Fig. 4A) (1). Its contribution to the whole protein structure was thus estimated to be 14% for β-strands and 16% for α-helices. According to these values and taking into account that the CD estimation for total β-strands was ∼17%, the CWBR would display an all-α-helical fold, contributing around 21% to the total α-helical content of Cpl-7. This value agrees fairly well with the predictions obtained by computational methods. According to them, CWBR residues predicted to be in α-helical conformation represented around 25% of the total sequence, and each repeat would fold into a three α-helical motif containing two clearly amphipathic helices (Fig. 2 and supplemental Fig. S1).
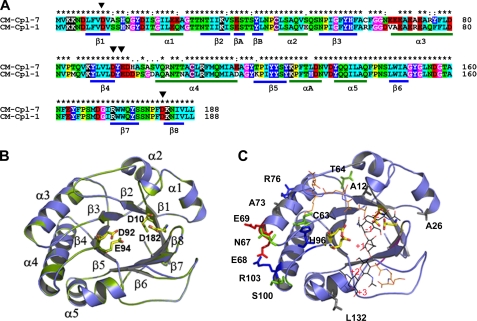
FIGURE 4.
Homology model of the Cpl-7 catalytic module. A, sequence alignments of Cpl-7 and Cpl-1 CMs. Color code is as used in Fig. 2. The position of β-strands and α-helices in the CM of Cpl-1 is shown. Black triangles indicate the catalytic residues (Asp10, Asp92, Glu94, and Asp182). B, superimposition of the template (green) and model (blue) structures. Catalytic residues are in stick representation (Cpl-7, yellow; Cpl-1, green). C, superimposition of the coordinates for the Cpl-1-(2S5P)3 complex (10) with the computational model of Cpl-7 CM. Nonconserved residues together with substitutions relevant for substrate binding are shown in stick representation (red, acidic; blue, basic; green, polar uncharged; and gray, hydrophobic). (2S5P)3 stands for (GlcNAc-MurNAc-(l-Ala-d-isoGln-l-Lys-d-Ala-d-Ala)3); carbon atoms are in black (sugar chain) or orange (peptide stem).
Association State and Hydrodynamic Behavior
Cpl-7 was subjected to sedimentation velocity experiments to analyze the homogeneity of the protein solutions, its association state, and the particle hydrodynamics. The distribution of sedimentation coefficients measured in Pi buffer at 20 °C, using protein concentrations from 2.5 to 16.8 μm, showed that Cpl-7 sediments as a single species with an s20,w0 value of 2.73 ± 0.02 S (Fig. 3B). The same behavior was observed at 4 °C (data not shown) using protein concentrations 1 order of magnitude higher. The molecular weight of the sedimenting species was determined by equilibrium sedimentation experiments. As expected, the equilibrium profiles can be fitted to a single species model (Fig. 3C) with an average molecular mass, Mw(app), of 37 ± 2 kDa (average of 10 experiments), which corresponds to the monomer molecular mass. The effective Stokes radius (RS = 34.6 ± 0.4 Å) and the translational frictional coefficient (f/f0 = 1.52 ± 0.2) were calculated from the experimental data using the SEDNTERP program. The high deviation of f/f0 from the value expected for globular particles (f/f0 ∼1) indicated that Cpl-7 has a highly elongated shape.
Modeling of Cpl-7 Catalytic Module
Protein-Substrate Interactions
The catalytic barrel of Cpl-7 was modeled by homology using as template the CM of Cpl-1 lysozyme. Fig. 4A shows the sequence alignment together with the distribution of SS elements in Cpl-1 structure. The final model (Fig. 4B) displays a good geometry as defined by PROCHECK (98.7% of the residues fall in the most favored regions of the Ramachandran plot), and the energetic evaluation showed comparable scores for the template and the model (Table 1). According to them, the proposed model would represent an excellent approximation of the real structure, as also indicated the full superimposition of the catalytic residues and the r.m.s.d. values (0.07 Å for Cα atoms and 0.43 Å for all atoms). As shown in Fig. 4, A and C, nonconservative substitutions expand primarily along the region included between the C terminus of the loop connecting strand-3 to helix-3 and the beginning of helix-4, including the loop containing the proton donor Glu94. The differences in amino acid composition modify the electrostatic potential of the module surface (data not shown), particularly at the barrel edge, including helices 3 and 4 and also at a locus close to the catalytic cleft (positions 63 and 96) where the catalytic pair Asp92–Glu94 is located. Moreover, the substitution of Arg63 by cysteine in Cpl-7 implies the loss of hydrogen bonding to the side chains of Asp37, Asp95, and His96 (changed to aspartic in Cpl-1), which could also modify the binding locus for the stem peptide of MurNAc unit bound at position −1 (10).
TABLE 1.
Stereochemical and energetic evaluation of the models generated for the CM and the CWBR of Cpl-7
| Protein | PROCHECK summarya |
QMEANb | VERIFY3Dc | ERRATd | |||
|---|---|---|---|---|---|---|---|
| Most favored | Additional allowed | Generously allowed | Disallowed | ||||
| % | % | % | % | ||||
| CM-Cpl-7 | 87.3 | 11.4 | 1.2 | 0.0 | 0.677 | 0.19/0.72 | 94.4 |
| 1h09e | 89.0 | 10.3 | 0.6 | 0.0 | 0.753 | 0.04/0.72 | 91.1 |
| CWBR-M1 | 93.1 | 5.2 | 1.7 | 0.0 | 0.427 | −0.01/0.48 | 90.3 |
| CWBR-M2 | 93.3 | 5.0 | 1.7 | 0.0 | 0.394 | −0.01/0.48 | 93.7 |
| 3cd1 (b)f | 97.0 | 3.0 | 0.0 | 0.0 | 0.623 | 0.24/0.67 | 98.5 |
a Distribution of residues in the Ramachandran plot region is shown.
b Pseudo-energy of the global structure is indicated (46).
c Minimum and maximum values of energy per residue are shown (45).
d Percentage of residues whose structure is under the rejection limit is shown (47).
e Evaluation of the CM structure is shown.
f Protein subunit is indicated in parentheses.
Fig. 4C shows the structural superimposition of the CM model with the coordinates of the complex formed by Cpl-1 and the muropeptide (2S5P)3 (10), a hexasaccharide tri-pentapeptide that expands from positions −2 to +3 of the active site (saccharide units flanking the scissile glycoside bond are assigned as positions −1 and +1). The positions of nonconserved residues suggest that sequence differences between the CMs of Cpl-1 and Cpl-7 would affect substrate recognition by the active site. Major changes would arise from substitutions at positions 12 and 96. The change of Ser12 by alanine would prevent the H-bond formed between the serine oxygen of Cpl-1 and the lactil group of MurNAc at position −1 (10). Moreover, the substitution of Asp96 by histidine also implies the loss of the H-bonds discerned in Cpl-1 between the carboxylate oxygens of Asp96 and the Lys moiety of the peptide bound to the same MurNAc unit. More importantly, it also introduces unfavorable electrostatic interactions with the Lys amino group at neutral and acidic pH values. Finally, the structural changes of the CM derived from the change in the pattern of hydrogen bonds mediated by the residue at position 63 might also affect substrate binding, as indicated above.
According to the model shown in Fig. 4C, it seems unlikely that the glycanic chain bound to the active site might interact with residues forming helices 3 and 4. However, the possibility that this region could interact with adjacent glycopeptidic chains cannot be completely ruled out.
Modeling of Cpl-7 Cell Wall Binding Region
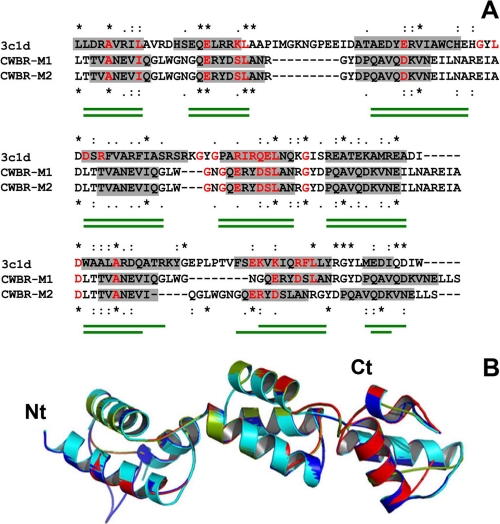
The PSI-Blast search of databases for three-dimensional structures homologous to the CWBR did not identify any protein to be used as a possible template. Protein fold recognition methods were therefore applied using the sequences of either the whole module or a single CW_7 repeat in the search for remotely related structures. The highest scoring results returned as possible templates for the complete module by more than one method are shown in Table 2. RecX (PDB code 3c1d), the first hit of FUGUE and SP4 servers, is a modular protein consisting of three tandem repeats of a three-helix motif (∼50 amino acids each) that can be superimposed despite their low sequence similarity (59). The similarity between RecX and the CWBR was near 46% (≅14% sequence identity), and the alignments returned by FUGUE and SP4 servers showed a good correlation between the distribution of residues involved in α-helices in RecX structure and Cpl-7 SS prediction. In contrast, the structure of PDB code 1ng6, identified by the three servers with much lower scores, consists of two α-helical bundles comprising four- and three-helical motifs, respectively, in tandem disposition, and its compatibility with the secondary structure predicted for the N-terminal half of the CWBR was clearly the worst (data not shown). On the other hand, four proteins were identified as possible templates by at least two servers among the 10 top hits when the sequence of a single CW_7 repeat was used in the search for related structures (Table 3). All of them were α-helical proteins, and the two first-ranked candidates (PDB codes 1ixs and 1gab) display a three-helical bundle topology. Because the confidence levels of the highest scoring results (PDB code 1ixs) were in the order of those obtained for the best full-length candidate (PDB code 3c1d), and the use of a single repeat for modeling the full CWBR structure would entail the additional difficulty of finding the correct orientation of CW_7 repeats within the full structure, the crystal structure of 3c1d (RecX protein) was finally selected as the best possible template. After several steps of alignment-modeling-evaluation, two promising models were finally obtained using as inputs the alignments shown in Fig. 5A, where residues conserved in the RecX family sequences (59) present in the CWBR were also indicated. The two models slightly differ in the length of the α-helices included in the last repeat (Fig. 5A), and their superimposition with the RecX structure is shown in Fig. 5B. Their evaluation was also similar, showing good stereochemical and energetic values (Table 1) and r.m.s.d. for Cα atoms of 3.15 Å (model M1) and 4.83 Å (model M2). For these reasons and taking into account that modeling of the CWBR was based on threading methods, models M1 and M2 will be considered as a single one for discussion. According to them, the CWBR would display an elongated structure with approximated overall dimensions of 60 × 20 × 20 Å and a moderate degree of curvature along the long axis. The side chains of conserved hydrophobic residues (Fig. 2) are tightly packed into the bundle hydrophobic core (Fig. 6A), and their conservation seems to be related to the structural role played in the bundle folding. Moreover, conserved glycines will facilitate the sharp turns between the helices. On the other hand, the fully conserved arginine (Fig. 2), deeply buried at the inside, might contribute to stabilize the bundle structure by making hydrogen bonds with other residues. An equivalent role may play the conserved residues of glutamine and asparagines located at positions 15, 34, and 38 of each repeat (Fig. 2).
TABLE 2.
Structures selected as possible templates for the CWBR of Cpl-7 by more than one method
Only the top 10 hits of each method were considered.
| PDB | FUGUE |
SP4 |
3DPSSM |
Template structure | |||
|---|---|---|---|---|---|---|---|
| Ranking | Score | Ranking | Score | Ranking | Score | ||
| 3c1d (b)a | 1 | 4.35 | 1 | 4.94 | Three assembled 3-helical bundles | ||
| 1ng6 | 9 | 1.95 | 5 | 3.79 | 5 | 6.08 | 4-helical bundle assembled to a 3-helical bundle |
a Protein subunit is indicated in parentheses.
TABLE 3.
Structures selected as possible templates for a CW_ repeat by more than one method
Only the top 10 hits of each method were considered.
| PDBa | FUGUE |
SP4 |
3D-PSSM |
Template structure | |||
|---|---|---|---|---|---|---|---|
| Ranking | Score | Ranking | Score | Ranking | Score | ||
| 1ixs (a) | 2 | 3.11 | 1 | 7.15 | 15 | 34.2 | 3-helical bundle |
| 1gab | 7 | 2.47 | 3 | 24.7 | 3-helical bundle | ||
| 1ytf (b) | 5 | 6.07 | 9 | 30.8 | 2-helix motif | ||
| 1fse (a) | 6 | 2.52 | 10 | 5.54 | 4-helical bundle | ||
a The protein subunit is indicated in parentheses.
FIGURE 5.
Three-dimensional threading model of Cpl-7 CWBR. A, structure-based sequence alignments of the CWBR and the template (PDB code 3c1d; RecX protein). Residues in α-helical conformation according to RecX structure and Cpl-7 SS prediction are highlighted in gray. Strictly conserved residues are marked with asterisks; colons, and dots indicate, respectively, conservative and semi-conservative changes. Residues conserved in RecX family sequences (59) are in red as also are similar residues present in the CWBR sequence. Green traces under the alignments indicate α-helix positions in models 1 (top) and 2 (bottom). B, superimposition of template molecules a (cyan) and b (blue) present in PDB 3c1d structure with the best CWBR structural models (M1, red; M2, green).
FIGURE 6.
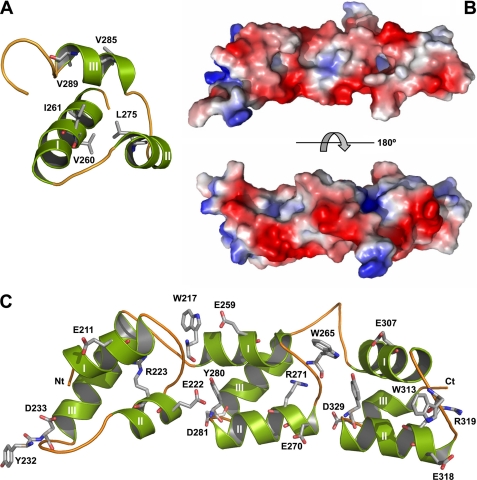
Three-dimensional structure and surface properties of Cpl-7 CWBR. A, three-helical bundle of the second repeat. Side chains of hydrophobic residues conserved in CW_7 family are shown in stick representation (Val260 corresponds to Val8 in the multiple alignment of Fig. 2A). B, molecular surface of the CWBR colored according to its electrostatic potential (top, concave face; bottom, convex face). C, distribution of conserved aromatic and charged residues along the concave face.
Inspection of the model surface showed clear patterns in polarity and charge distribution (Fig. 6B). The convex face is crossed by polar patches due to the solvent-exposed surface of the third helix of each bundle, where neutral and charged residues alternate, and amino acid conservation is rather low. In contrast, fully conserved arginines extend, deep buried, along the central region of the concave face, whose edges are lined by conserved aromatic residues and acidic groups (Fig. 6, B and C). Interestingly, exposed regions involved in functional interactions show a higher degree of amino acid conservation than otherwise expected for surface residues. In addition, both aromatic and polar residues with planar side chains (Asp, Glu, Asn, Gln, Lys, and Arg) are a common signature of glycan-binding sites in carbohydrate recognition motifs (60). Therefore, the presence of both features in the CWBR concave face suggests that it might represent the functional surface involved in the attachment of Cpl-7 to the cell wall.
Modular Organization
SAXS Modeling of the Overall Cpl-7 Structure
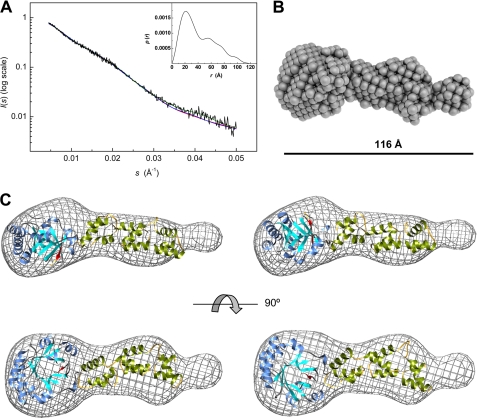
SAXS is a powerful methodology to investigate domain organization in modular proteins, particularly when structural models of the components can be used to analyze the scattering data and to assess the most probable solution (48, 61–62). The modular organization of Cpl-7 in solution was thus investigated using synchrotron x-ray scattering data (Fig. 7) using a protein concentration of 187 μm. First, the quality of the scattering data and the presence or absence of aggregates or interparticle interference was checked from the Guinier plot. The linearity of log(I(s)) against s2 at low s values (supplemental Fig. S2A) indicated the absence of significant aggregates or interparticle effects in solution. The values of Rg (29.8 ± 0.9 Å) and Io were thus obtained using the Guinier approximation. The analysis of the experimental curves with GNOM, which uses the entire scattering profile and which therefore can be more precise, provided an estimation of 30.3 Å for Rg, in good agreement with the Guinier value, and a maximum intraparticle distance, Dmax, of 115 Å (Fig. 7A). A good correlation between the scattering profile and the Fourier transform of P(r) was also observed (supplemental Fig. S2C). These results confirm that Cpl-7 is indeed a rather elongated molecule, as suggested by the sedimentation velocity results. Moreover, the behavior of the P(r) function at extended Dmax (supplemental Fig. S2B) further supported the quality of the scattering profile and the absence of severe interferences from aggregation or interparticle interference (48). The pair-distance distribution function presents three well defined maxima at 22, 54, 74, and 98 Å (Fig. 7A, inset), indicating the presence of distinct structural entities within the molecule, as could be expected from the models derived for the isolated modules. Low resolution bead models of Cpl-7 were reconstructed ab initio using DALAI_GA, and the most probable one is shown in Fig. 7B. The theoretical scattering profile of the model overlaps the experimental one (Fig. 7A, green line), and its Rg = 32.7 ± 0.2 Å, agrees with the experimental one (Table 4). Moreover, its hydrodynamic features correlate well with the experimental data as shown in Table 4. The bead model shows a very elongated molecule (∼116 Å long) with two well differentiated regions (Fig. 7B). One of them resembles a flattened ellipsoid whose dimensions (∼46 × 38 × 30 Å) are in the order of those reported for the CM of Cpl-1 (9). The other one is elongated (∼70 × 30 × 20 Å) as the model proposed for the CWBR and shows a small curvature along the long axis. The assignment was further supported by manual fitting of the high resolution models obtained for the isolated modules into the bead-model surface (Fig. 7C). Two models with similar deviations between the experimental and the theoretical profiles of SAXS were finally selected whose major divergence lies in the relative position of the CM (Fig. 7C). In model a (χ2 = 1.76), the catalytic cavity is oriented toward the same side of the convex face of the CWBR, and it is rotated toward the side of the concave face in model b (χ2 = 1.77). In both models, the region of the β-barrel devoid of α-helices (β6 to β8) seems to be part of the interface with the CWBR. Table 4 summarizes the χ2 values for the fit to the data of the theoretical scattering profiles together with the structural and hydrodynamic parameters estimated by CRYSOL or HYDROPRO for the two models. The theoretical sedimentation coefficient is similar for both models and close to the experimental value, as it also occurred with the Stokes radius and the radius of gyration estimated by HYDROPRO (Table 4), reflecting the compatibility of the proposed models with the hydrodynamic features of Cpl-7.
FIGURE 7.
SAXS-based modeling of Cpl-7 overall structure. A, experimental SAXS profile of Cpl-7 (black line) and theoretical spectra calculated from the ab initio bead model (green line) in B, and the high resolution models in C. The inset shows the pair-distance distribution function, P(r), generated by GNOM with a Dmax of 115 Å (data collected at 4 °C in Pi buffer). B, ab initio bead model of Cpl-7 derived from SAXS data using DALAI_GA (50). C, molecular envelope (grid representation) of Cpl-7 bead model with the best fits of the three-dimensional models built for the CM and the CWBR manually docked inside (the antiparallel β8-strand of the CM is colored in red). Red and blue lines in A are the theoretical SAXS profile derived for models a (left-hand model) and b (right-hand model), respectively, using CRYSOL (54).
TABLE 4.
Comparison of the structural and hydrodynamic parameters of SAXS-based model of Cpl-7 with the experimental values measured by SAXS or sedimentation velocity
| SAXS |
Ab initio model |
High resolution models |
Sedimentation velocity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Guinier | GNOM | DALAI_GA | HYDRO | Model a |
Model b |
||||
| CRYSOL | HYDROPRO | CRYSOL | HYDROPRO | ||||||
| Rg (Å−1) | 29.8 | 30.3 | 32.7 | 33.3 | 27.8 | 31.9 | 27.7 | 30.3 | |
| Dmax (Å−1) | 115 | 116 | 103 | 109 | 103 | 108 | |||
| RS (Å−1) | 32.9 | 32.9 | 32.3 | 34.6 | |||||
| s (S) | 2.87 | 2.87 | 2.93 | 2.73 | |||||
| χ2a | 4.79 × 10−5b | 7.18 × 10−5 | 1.76 | 1.77 | |||||
DISCUSSION
Endolysins are bacteriophage-encoded enzymes produced during the late phase of gene expression in the lytic cycle to degrade peptidoglycan, thereby enabling the virion progeny to be liberated. Their unique ability to cleave peptidoglycan in a generally species-specific manner makes endolysins promising antibacterial agents, capable of killing pathogenic bacteria without affecting the normal microbiota. Moreover, the use of near-species-specific endolysins might help to avoid the resistance that appears with the use of broad range antimicrobials. Species specificity in endolysins produced by pneumococcal bacteriophages relies on the acquisition of additional modules that specifically target the cell wall and increase the lytic activity by several orders of magnitude (2, 6). In the Cpl-7 endolysin, encoded by the pneumococcal bacteriophage Cp-7, the targeting region includes a set of three perfect repeats, the CW_7 motifs, sequentially unrelated with the choline-binding motifs that characterize the cell wall-binding modules of all the other CWHs produced by the pneumococcus and its bacteriophages.
Although Cpl-7 was described in 1990, sequences similar to the CW_7 repeats were not reported until 2002 as being encoded by various prophages of Streptococcus pyogenes and Streptococcus agalactiae (supplemental Table S1). During the last few years, the availability of relatively inexpensive next generation sequencing technologies has permitted the complete genomic sequences of hundreds of genomes of prokaryotic organisms, and today up to 67 different proteins comprising CW_7 motifs have been included in the Entrez Database (last date accessed, April 8, 2010). Although the majority of these proteins are annotated as “hypothetical,” they contain combinations of motifs that permit us to classify them as probable CWHs. Only 10 proteins do not appear to contain any known motif in addition to CW_7 (Fig. 1). To our knowledge, apart from the Cpl-7 lysozyme, only the endolysin encoded by the λSa2 prophage of S. agalactiae strain 2603 V/R has been biochemically characterized so far and has been shown to be a bifunctional enzyme with γ-d-glutaminyl-l-lysine endopeptidase (Amidase_5; PF05382) and β-d-N-acetylglucosaminidase (Glucosaminidase; PF01832) activities (63). Cell wall-degrading activity has also been reported for the LySMP endolysin of the lytic phage SMP infecting Streptococcus suis type 2 strains (64), a close homologue (76% identity) of the λSa2 endolysin. It is worth noting that only in the cpl-7 gene, the long (144 nucleotides) tandem repeat units are identical (1), which suggests that the duplication event occurred very recently, in evolutionary terms.
It is interesting to notice that proteins containing CW_7 repeats appear to be present only in four bacterial phyla (out of 25): Actinobacteria, Bacteroidetes, Chloroflexi, and Firmicutes. Surprisingly, they appear to be absent from bacteria belonging to the Proteobacteria phylum, despite the fact that the NCBI genome data base contains more than 800 proteobacterial genomes. Most of the bacterial species coding for CW_7-containing proteins belong to the Firmicutes phylum and nearly all form part of the human gut (fecal) microbiota where, in healthy adults, 80% of the identified fecal bacteria can be classified into three dominant phyla: Bacteroidetes, Firmicutes, and Actinobacteria, although the ratio of Firmicutes to Bacteroidetes evolves along the different life stages (65). The fecal microbiota is a highly complex and diverse bacterial ecosystem. Prominent clusters include some of the most abundant gut species, such as members of the Bacteroidetes and Eubacterium/Ruminococcus groups and also bifidobacteria (Actinobacteria), Proteobacteria and streptococci/lactobacilli groups (58). The finding that D. ethenogenes (phylum Chloroflexi) and E. harbinense (phylum Firmicutes), two species living in contaminated waters, also code for CW_7-containing proteins may be the result of horizontal gene transfer events. Interestingly, although the root of the tree of life (and the last universal common ancestor) has been suggested to be located within or next to the Chloroflexi (66), evidence suggesting that D. ethenogenes has experimented horizontal transfer events has been provided (67).
As mentioned above, most of the proteins containing CW_7 motifs appear to be related with the presence of prophages (or parts of them). This hypothesis is consistent with the observation that some of these proteins are found only in certain strains of a particular species. For instance, the putative N-acetylmuramoyl-l-alanine amidase EEW16061 (Corynebacterium jeikeium strain ATCC 43734) is absent in the genome of strain K411 (accession number CR931997). Another interesting example is the protein EEJ91939, identified in strains SD2112 and CF48–3A (EEI65431) of Lactobacillus reuteri (supplemental Table S1) but is missing in the genomic sequences of strains DSM 20016 (CP000705), MM2-3 (ACLB01000000), MM4-1A (ACGX01000000), JCM 1112 (AP007281), or 100-23 (ACGW01000000). Phages (either temperate or lytic) may have served as vehicles for horizontal gene transfer among different bacterial species; among other examples, this is strongly supported by the striking similarity found between the prophage φ3396 from S. dysgalactiae subsp. equisimilis and the S. pyogenes φ315.1 temperate phage (68). Furthermore, evidence suggesting that the virulent lactococcal phage 1706 may have originated from a phage infecting Ruminococcus torques and/or Clostridium leptum has been provided (69).
Here, we have reported the first in-depth characterization of the Cpl-7 endolysin. The protein was purified to electrophoretic homogeneity by a simple procedure, including protein fractioning with ammonium sulfate and two chromatographies. Cpl-7 behaves in solution as a monomer, and its hydrolytic activity on pneumococcal cell walls is slightly lower than that of the Cpl-1 endolysin, whose good profile as a potent antimicrobial agent against pneumococci has been already proved (17–19, 22). By combination of SAXS and molecular modeling, a three-dimensional model of the overall protein structure, compatible with the SS inferred from CD and prediction methods, has been proposed. SAXS experiments showed that Cpl-7 adopts in solution an extended configuration, as also indicated by the velocity sedimentation results. Even though SAXS analysis is unable to identify unique structures because of rotational averaging, it is able to rule out incorrect structures, including more globular arrangements of Cpl-7 modules that will have displayed different radii of gyration and maximum inter-particle distances. Moreover, the molecular envelope generated from SAXS data is clearly compatible with the structures derived for the isolated modules of Cpl-7 using molecular modeling, and the high resolution models proposed for the overall protein structure are also compatible with the SAXS spectrum and Cpl-7 hydrodynamic behavior. All this indicates that the proposed models represent a reasonable approximation to the real structure of the protein. Moreover, the fitting of CWBR high resolution models into the SAXS envelope might denote a certain mobility of the last repeat, probably because of the own modular structure displayed by the CWBR of Cpl-7.
The model of the CM displays a very good stereochemistry, and the r.m.s.d. from the template was rather low, which indicates that it constitutes an excellent approximation to the actual module structure. Nonconservative substitutions are primarily localized at the region from helix-3 to helix-4 (Fig. 4C). According to the model inferred for the complex formed by Cpl-1 and (2S5P)3 muropeptide (10), substitutions at positions 12 and 96 would modify the interaction with the substrate as follows: (i) reducing the number of hydrogen bonds formed in the complex with Cpl-7, and (ii) inverting the electrostatic interactions (unfavorable in Cpl-7) between the side chain of the residue at position 96 and the stem peptide of the MurNAc at position −1. Besides, in TIM barrels, the loops at the C terminus of β-strands usually contribute to determine the specific configuration of the active site and the substrate specificity. Therefore, sequence differences in the loop containing the proton donor (residues 93–100) could also contribute to modulate the murolytic activities of Cpl-1 and Cpl-7. A similar effect might also have the drastic change in the protein hydrogen bond network derived from substitution of Arg63 by Cys in Cpl-7 described above. The relevance of the substitutions accumulated on the edge of the CM along helices three and four that strongly modify the surface charge distribution remains unknown. Participation of this region in the CM/CWBR interface seems unlikely considering the best SAXS-based models for the overall Cpl-7 structure, and its distance to the glycanic chain bound to the active site is too large to interact with it (Fig. 4C). However, the possibility that this part of the CM might interact with adjacent glycopeptidic chains, providing additional contacts with the cell wall, presently cannot be discarded.
The stereochemical and energetic profiles of the CWBR model were fairly good. According to the proposed model, the CWBR includes three tandemly assembled three-helical bundles, whose disposition reminds us of a super-helical structure, connected by six amino acid-long linkers. Each bundle contains 42 residues, and the inner core is stabilized by conserved hydrophobic residues, whereas sharp turns between helices are facilitated by the presence of glycine residues highly conserved in all the related sequences so far identified (Fig. 2). The lack of three residues in the middle of certain CW_7 repeats would probably shorten the second helix of the bundle. Although the model may not reveal in detail the module structure, it provides some clues to aid identification of residues and surfaces important for the cell wall attachment. Residues located on the convex face of the CWBR are poorly conserved within the CW_7 family. On the contrary, conserved acidic and aromatic residues are present along the concave face (Fig. 6C), suggesting that this could be the region involved in cell wall attachment, because solvent-exposed areas involved in functional interactions with other macromolecules are subjected to strong evolutionary pressure and therefore show a higher degree of amino acid conservation than otherwise expected for surface residues. Moreover, both aromatic and polar residues with planar side chains, as those conserved in the concave face, are a common signature of glycan-binding sites in carbohydrate recognition motifs (60).
Among the great variety of cell wall-binding motifs acquired by CWHs, only a few have been structurally characterized so far, including SH3_5 (PF08460), LysM (PF01476), SPOR (PF05036), PG_binding_1–3 (clan PGBD; CL0244), and CW_binding_1 (choline-binding repeats; PF01473) domains. They all include relatively short sequences forming different structural motifs. PG_binding motifs show a three-helical bundle fold but, in contrast to the CW_7 motif, the helices have a parallel disposition and are connected by rather long loops. This could explain why they were not identified as potential templates of the CWBR by fold recognition methods and suggests that CW_7 repeats might constitute a new cell wall-binding structural motif. The existence of protein sequences with a single CW_7 repeat evidences the ability of each bundle to fold and recognize its target, as demonstrated by the increase in activity observed upon fusing one CW_7 motif to the endopeptidase module of the λSa2 endolysin (3). However, the common presence (21 of 67) of tandemly arranged repeats suggests a mechanism to improve cell wall recognition, and the model proposed here for the CWBR of Cpl-7 represents a first approximation to the architecture that multiple CW_7 repeats would adopt. It is worth noting that the acquisition of a second CW_7 repeat by the endopeptidase module of the λSa2 endolysin not only enhanced the lytic activity of the truncated protein on different streptococcal species, but it was shown to be essential for its activity on staphylococci (3). Moreover, the acquirement of other cell wall-binding motifs, like LysM and SPOR, by so many CW_7-containing proteins (Fig. 1) could denote alternative ways for tight substrate binding, directing muropeptides into the catalytic cavity, or even modifying the species (or the strain) specificity in certain CWHs.
The CM is linked to the CWBR by an extremely polar segment made of 16 amino acids including six acidic and two lysine residues, which probably adopts an extended conformation and introduces certain intermodular flexibility. The linker sensitivity to the proteolytic action of trypsin (8) also suggests a solvent-exposed disposition according to its polar character. Nevertheless, the mass density at the interface of the module in the SAXS-derived model strongly indicates the existence of a large interacting surface. According to the model, the interface between the Cpl-7 modules would include, as in Cpl-1 (9), the region of the catalytic barrel lacking α-helices, which is fully conserved in both lysozymes. Finally, if the linker would adopt an extended configuration, its estimated length (∼64 Å) does not exclude the possibility of a tail-to-tail disposition of the CM and the CWBR in the protein overall structure. Whatever the case, the relative disposition of Cpl-7 modules within the protein structure diverges from the one adopted in the Cpl-1 lysozyme (9, 11).
The effective lytic spectrum of endolysins is usually constrained by cell wall elements that serve as binding targets and are themselves distributed in species/strain-specific manners. Nevertheless, endolysins often lyse a broader range of species than just the producing organism, and the substrate specificity inherent to the acquirement of the CW_7 motifs by the Cpl-7 endolysin opens appealing perspectives to the use of this lysozyme, either wild-type or in chimeric constructions, to control pathogenic, resistant bacteria. Interestingly, CW_7 repeats are also present in the CWHs encoded by a large variety of Gram-positive pathogens and certain phages of S. agalactiae (a leading cause of bacterial sepsis, pneumonia, and meningitis in neonates) and S. pyogenes (a prevalent human pathogen that has re-emerged as a public health hazard). To that end, the great variety of architectures shown by the CW_7-containing proteins highlights the versatility of this motif to combine with different CMs and cell wall-binding domains in a single polypeptide chain to alter bond and/or substrate specificity while retaining activity.
Supplementary Material
This work was supported by Grants BFU2006-10288, SAF2009-10824, BIO2007-61336, and BFU2009-10052 from Ministerio de Ciencia e Innovación, Grant BIPPED-CM from the Comunidad de Madrid, the CIBER de Enfermedades Respiratorias (CIBERES), an initiative of the ISCIII, Glycodynamics Network Grant FP6-UE MCTN-CT-2005-019561, and European Community beam time Proposal 48086.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- CWH
- cell wall hydrolase
- CM
- catalytic module
- CW_7
- cell wall-binding repeat of Cpl-7
- CWBR
- cell wall-binding region
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- SAXS
- small angle X-ray scattering
- SS
- secondary structure.
REFERENCES
- 1.García P., García J. L., García E., Sánchez-Puelles J. M., López R. (1990) Gene 86, 81–88 [DOI] [PubMed] [Google Scholar]
- 2.García P., García J. L., López R., García E. (2005) in Phages: Their Role in Bacterial Pathogenesis and Biotechnology (Waldor M. K., Friedman D. L., Adhya S. eds) pp. 335–361, American Society for Microbiology, Washington, D. C [Google Scholar]
- 3.Donovan D. M., Foster-Frey J. (2008) FEMS Microbiol. Lett. 287, 22–33 [DOI] [PubMed] [Google Scholar]
- 4.García J. L., García E., Arrarás A., García P., Ronda C., López R. (1987) J. Virol. 61, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López R., García E. (2004) FEMS Microbiol. Rev. 28, 553–580 [DOI] [PubMed] [Google Scholar]
- 6.García P., Paz González M., García E., García J. L., López R. (1999) Mol. Microbiol. 33, 128–138 [DOI] [PubMed] [Google Scholar]
- 7.Diaz E., López R., Garcia J. L. (1991) J. Biol. Chem. 266, 5464–5471 [PubMed] [Google Scholar]
- 8.Sanz J. M., Díaz E., García J. L. (1992) Mol. Microbiol. 6, 921–931 [DOI] [PubMed] [Google Scholar]
- 9.Hermoso J. A., Monterroso B., Albert A., Galán B., Ahrazem O., García P., Martínez-Ripoll M., García J. L., Menéndez M. (2003) Structure 11, 1239–1249 [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Dorado I., Campillo N. E., Monterroso B., Hesek D., Lee M., Páez J. A., García P., Martínez-Ripoll M., García J. L., Mobashery S., Menéndez M., Hermoso J. A. (2007) J. Biol. Chem. 282, 24990–24999 [DOI] [PubMed] [Google Scholar]
- 11.Buey R. M., Monterroso B., Menéndez M., Diakun G., Chacón P., Hermoso J. A., Díaz J. F. (2007) J. Mol. Biol. 365, 411–424 [DOI] [PubMed] [Google Scholar]
- 12.Monterroso B., Sáiz J. L., García P., García J. L., Menéndez M. (2008) J. Biol. Chem. 283, 28618–28628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva-Martin N., Molina R., Angulo I., Mancheño J. M., García P., Hermoso J. A. (2010) Acta Crystallogr. F Struct. Biol. Crystallogr. Commun. 66, 670–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti V. A. (2005) Trends Microbiol. 13, 491–496 [DOI] [PubMed] [Google Scholar]
- 15.Hermoso J. A., García J. L., García P. (2007) Curr. Opin. Microbiol. 10, 461–472 [DOI] [PubMed] [Google Scholar]
- 16.Loeffler J. M., Nelson D., Fischetti V. A. (2001) Science 294, 2170–2172 [DOI] [PubMed] [Google Scholar]
- 17.Jado I., López R., García E., Fenoll A., Casal J., García P. (2003) J. Antimicrob. Chemother. 52, 967–973 [DOI] [PubMed] [Google Scholar]
- 18.Loeffler J. M., Djurkovic S., Fischetti V. A. (2003) Infect. Immun. 71, 6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullers J. A., Karlström A., Iverson A. R., Loeffler J. M., Fischetti V. A. (2007) PLoS Pathog. 3, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez-Cerrato V., García P., Del Prado G., García E., Gracia M., Huelves L., Ponte C., López R., Soriano F. (2007) J. Antimicrob. Chemother. 60, 1159–1162 [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-Cerrato V., García P., Huelves L., García E., Del Prado G., Gracia M., Ponte C., López R., Soriano F. (2007) Antimicrob. Agents Chemother. 51, 3371–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witzenrath M., Schmeck B., Doehn J. M., Tschernig T., Zahlten J., Loeffler J. M., Zemlin M., Müller H., Gutbier B., Schütte H., Hippenstiel S., Fischetti V. A., Suttorp N., Rosseau S. (2009) Crit. Care Med. 37, 642–649 [DOI] [PubMed] [Google Scholar]
- 23.Inouye S., Inouye M. (1985) Nucleic Acids Res. 13, 3101–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 25.Speicher D. W. (1994) Methods 6, 262–273 [Google Scholar]
- 26.Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 27.Sreerama N., Woody R. W. (1993) Anal. Biochem. 209, 32–44 [DOI] [PubMed] [Google Scholar]
- 28.Böhm G., Muhr R., Jaenicke R. (1992) Protein Eng. 5, 191–195 [DOI] [PubMed] [Google Scholar]
- 29.Ouali M., King R. D. (2000) Protein Sci. 9, 1162–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 31.Cole C., Barber J. D., Barton G. J. (2008) Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C. eds) pp. 90–125, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 34.Varea J., Monterroso B., Sáiz J. L., López-Zumel C., García J. L., Laynez J., García P., Menéndez M. (2004) J. Biol. Chem. 279, 43697–43707 [DOI] [PubMed] [Google Scholar]
- 35.Garcia de la Torre J., Navarro S., Lopez Martinez M. C., Diaz F. G., Lopez Cascales J. J. (1994) Biophys. J. 67, 530–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García De La Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi J., Blundell T. L., Mizuguchi K. (2001) J. Mol. Biol. 310, 243–257 [DOI] [PubMed] [Google Scholar]
- 39.Kelley L. A., MacCallum R. M., Sternberg M. J. (2000) J. Mol. Biol. 299, 499–520 [DOI] [PubMed] [Google Scholar]
- 40.Liu S., Zhang C., Liang S., Zhou Y. (2007) Proteins 68, 636–645 [DOI] [PubMed] [Google Scholar]
- 41.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galtier N., Gouy M., Gautier C. (1996) Comput. Appl. Biosci. 12, 543–548 [DOI] [PubMed] [Google Scholar]
- 43.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 44.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 45.Eisenberg D., Lüthy R., Bowie J. U. (1997) Methods Enzymol. 277, 396–404 [DOI] [PubMed] [Google Scholar]
- 46.Benkert P., Tosatto S. C., Schomburg D. (2008) Proteins Struct. Funct. Bioinformat. 71, 261–277 [DOI] [PubMed] [Google Scholar]
- 47.Colovos C., Yeates T. O. (1993) Protein Sci. 2, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacques D. A., Trewhella J. (2010) Protein Sci. 19, 642–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 50.Chacón P., Morán F., Díaz J. F., Pantos E., Andreu J. M. (1998) Biophys. J. 74, 2760–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 52.Wriggers W., Chacón P. (2001) J. Appl. Crystallogr. 34, 773–776 [Google Scholar]
- 53.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 54.Svergun D. I., Barberato C., Koch M. H. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 55.Sayers E. W., Barrett T., Benson D. A., Bolton E., Bryant S. H., Canese K., Chetvernin V., Church D. M., Dicuccio M., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Lu Z., Madden T. L., Madej T., Maglott D. R., Marchler-Bauer A., Miller V., Mizrachi I., Ostell J., Panchenko A., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Slotta D., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Wang Y., John Wilbur W., Yaschenko E., Ye J. (2010) Nucleic Acids Res. 38, D5–D16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turroni F., Ribbera A., Foroni E., van Sinderen D., Ventura M. (2008) Antonie Van Leeuwenhoek 94, 35–50 [DOI] [PubMed] [Google Scholar]
- 57.Mahowald M. A., Rey F. E., Seedorf H., Turnbaugh P. J., Fulton R. S., Wollam A., Shah N., Wang C., Magrini V., Wilson R. K., Cantarel B. L., Coutinho P. M., Henrissat B., Crock L. W., Russell A., Verberkmoes N. C., Hettich R. L., Gordon J. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J. M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S. D., Wang J. (2010) Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragone S., Maman J. D., Furnham N., Pellegrini L. (2008) EMBO J. 27, 2259–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solís D., Menéndez M., Romero A., Jiménez-Barbero J. (2009) in The Sugar Code. Fundamentals of Glycoscience (Gabius H. J. ed) pp. 233–245, Wiley-Blackwell, Mörlenbach, Germany [Google Scholar]
- 61.Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 62.Buey R. M., Chacón P., Andreu J. M., Díaz J. F. (2009) Lect. Notes Phys. 776, 245–263 [Google Scholar]
- 63.Pritchard D. G., Dong S., Kirk M. C., Cartee R. T., Baker J. R. (2007) Appl. Environ. Microbiol. 73, 7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y., Sun J. H., Lu C. P. (2009) Curr. Microbiol. 58, 609–615 [DOI] [PubMed] [Google Scholar]
- 65.Mariat D., Firmesse O., Levenez F., Guimarães V., Sokol H., Doré J., Corthier G., Furet J. P. (2009) BMC Microbiol. 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valas R. E., Bourne P. E. (2009) Biol. Direct. 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Regeard C., Maillard J., Dufraigne C., Deschavanne P., Holliger C. (2005) Appl. Environ. Microbiol. 71, 2955–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davies M. R., McMillan D. J., Van Domselaar G. H., Jones M. K., Sriprakash K. S. (2007) J. Bacteriol. 189, 2646–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garneau J. E., Tremblay D. M., Moineau S. (2008) Virology 373, 298–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.