Abstract
Members of the K+ channel-interacting protein (KChIP) family bind the distal N termini of members of the Shal subfamily of voltage-gated K+ channel (Kv4) pore-forming (α) subunits to generate rapidly activating, rapidly inactivating neuronal A-type (IA) and cardiac transient outward (Ito) currents. In heterologous cells, KChIP co-expression increases cell surface expression of Kv4 α subunits and Kv4 current densities, findings interpreted to suggest that Kv4·KChIP complex formation enhances forward trafficking of channels (from the endoplasmic reticulum or the Golgi complex) to the surface membrane. The results of experiments here, however, demonstrate that KChIP2 increases cell surface Kv4.2 protein expression (∼40-fold) by an order of magnitude more than the increase in total protein (∼2-fold) or in current densities (∼3-fold), suggesting that mechanisms at the cell surface regulate the functional expression of Kv4.2 channels. Additional experiments demonstrated that KChIP2 decreases the turnover rate of cell surface Kv4.2 protein by inhibiting endocytosis and/or promoting recycling. Unexpectedly, the experiments here also revealed that Kv4.2·KChIP2 complex formation stabilizes not only (total and cell surface) Kv4.2 but also KChIP2 protein expression. This reciprocal protein stabilization and Kv4·KChIP2 complex formation are lost with deletion of the distal (10 amino acids) Kv4.2 N terminus. Taken together, these observations demonstrate that KChIP2 differentially regulates total and cell surface Kv4.2 protein expression and Kv4 current densities.
Keywords: Cell Surface Protein, Endocytosis, Potassium Channels, Protein Assembly, Protein Stability, KChIP, Kv4
Introduction
Members of the Shal subfamily of voltage-gated K+ (Kv) channel pore-forming (α) subunits encode rapidly activating and inactivating Kv channels that also recover rapidly from inactivation and are important in the generation of IA channels in neurons (1–4) and Ito channels in cardiac myocytes (5, 6). Accumulating evidence suggests that functional Kv4 channels reflect the assembly of Kv4 α subunits with one or more Kv channel accessory subunits and other regulatory proteins that influence channel cell surface expression and/or biophysical properties (7). The K+ channel-interacting proteins (KChIP),4 members of the Neuronal Calcium Sensor superfamily (8, 9), for example, are cytosolic accessory subunits that were initially identified in a yeast two-hybrid screen using the N terminus of Kv4.2 as bait (10).
Heterologous co-expression with accessory KChIP subunits increases Kv4.2 current densities, as well as altering the time- and voltage-dependent properties of currents (10–14). Truncation of the first 40 amino acids in the Kv4.2 N terminus results in the loss of KChIP-mediated current modulation but a paradoxical increase in Kv4.2 current densities (11, 14). Progressive truncation of the N terminus (up to 40 amino acids) was reported to result in progressively greater increases in current densities, although it was not determined whether the observed increase reflected increased total and/or cell surface Kv4.2 protein expression. Previous mutagenesis studies have been interpreted as suggesting that the major binding site for KChIP2 on the Kv4.2 N terminus is between residues 11 and 23 (15). Structural analysis of the N terminus of Kv4.3, crystallized in complex with the core region (conserved across all family members) of KChIP1 (16, 17), however, revealed that KChIPs bind the distal 20 N-terminal residues of Kv4 α subunits in a hydrophobic binding pocket.
The KChIP-mediated increases in Kv4.2 current densities have been ascribed to increased trafficking of channels from the endoplasmic reticulum (ER) to the surface membrane (10, 11, 18), although a precise motif that regulates ER retention has yet to be identified. The amino acid motif Arg-Xaa-Arg (RXR) has been shown to play a role in ER retention of inwardly rectifying K+ channels (Kir) (19). For example, ATP-sensitive K+ channels (KATP) formed by the co-assembly of Kir6 α subunits and sulfonylurea receptor accessory subunits are retained in the ER when either subunit is expressed alone. Subunit co-assembly, however, masks RXR retention motifs, promoting forward trafficking to the cell surface of channel complexes (20). Although the Kv4.2 N terminus contains (at positions 35–37) an RKR sequence, previous studies suggest that this sequence does not function as an ER retention motif (18). It has also been suggested that Kv4.2 alone traffics out of the ER but fails to progress beyond the Golgi complex in the absence of KChIPs (21). Rather than or in addition to effects on trafficking, Kv4 protein stabilization resulting from complex formation with KChIPs may play a role in increasing functional expression of Kv4-encoded channels. Functional cell surface expression of Kv1-encoded channels, for example, appears to be mediated by stabilization of assembled Kv α and accessory Kvβ subunit proteins rather than masking of ER retention motifs (22, 23). Indeed, stabilization of Kv4.2 protein with KChIP2 co-expression has been suggested (18).
It has also been reported that ventricular myocytes isolated from mice with a targeted genetic disruption of either Kv4.2 (Kv4.2−/−) or KChIP2 (KChIP2−/−) lack Kv4.2 encoded (Ito,f) currents (6, 24). Additionally, although Kcnip2 (KChIP2) transcript was not decreased, there was a dramatic decrease of KChIP2 protein expression in Kv4.2−/− ventricles (6). Expression of KChIP2 and KChIP3 proteins was also decreased in Kv4.2−/− brain (3, 25). Taken together, these results reveal that both of the Kv4 and KChIP subunits are critical for the generation of native Kv4-encoded currents and suggest that Kv channel protein stability may be a key mechanism regulating functional Kv channel expression in native cells. The goal of the experiments presented here was to explore these hypotheses directly, combining biochemical and electrophysiological approaches to examine Kv4.2 and KChIP2 protein expression and functional Kv4.2 currents in parallel.
EXPERIMENTAL PROCEDURES
Molecular Modeling of Kv4.2/KChIP2 Interaction
Schematic representation of the molecular interactions between the N terminus of Kv4.2 and KChIP2 were generated using the protein structure modeling program Modeler v9.2 (Accelrys Inc., San Diego, CA) (26) and the published crystal structure of the N terminus of Kv4.3 and KChIP1 (Protein Data Bank code 2NZ0) (17) substituting the Kv4.2 and KChIP2 sequences for the (homologous) sequences in Kv4.3 and KChIP1, respectively.
Plasmid Construct Generation
The coding sequence for the red fluorescent protein tdTomato (27) (a gift from R.Y. Tsien) was cloned in the multiple cloning site of the pBK-CMV phagemid vector (Stratagene, La Jolla, CA) between the NheI and NotI restriction sites to generate pBK-CMV.tdTomato. The AdEGI vector (28) (a gift from D. C. Johns) is a bi-cistronic adenoviral shuttle vector expressing enhanced GFP and a second open reading frame separated by an internal ribosomal entry site in a single transcript driven by the ecdysone promoter. NsiI and NotI restrictions sites were introduced into AdEGI at base pair 468 (prior to the ecdysone promoter) and 2498 (following enhanced GFP) by site-directed mutagenesis using the QuikChange XL kit (Stratagene) with the following primers: NsiI, 5′-tttattattatagtcaggatctcggatgcatattaagtgcattgttctcgat, and 3′-atcgagaacaatgcacttaatatgcatccgagatcctgactataataataaa; and NotI, 5′-gagctgtacaagtgcggccgctagtactccggt and 3′-accggagtactagcggccgcacttgtacagctc. This construct was then digested with NsiI and NotI restriction endonucleases to excise the ecdysone promoter region together with enhanced GFP, and the CMV promoter together with tdTomato were subcloned into the NsiI and NotI sites to generate the vector AdloxCRI.
The coding sequence for Kcnd2 (NM_019697), encoding mouse Kv4.2, was cloned into AdloxCRI at the SacI and EcoRI restriction sites to generate the vector AdloxCRI.mKv4.2. The coding sequence for KCNIP2 (NM_173192), encoding human KChIP2, was cloned into the pCMV-Script vector (Stratagene) at the NheI and NotI restriction sites to generate the vector pCMV-Script.hKChIP2.
Kv4.2 N-terminal truncation constructs were generated in which amino acids 2–10 (Kv4.2Δ10N), 2–23 (Kv4.2Δ23N), 2–30 (Kv4.2Δ30N), or 2–40 (Kv4.2Δ40N) were deleted by PCR cloning of truncated Kv4.2 sequence from AdloxCRI.mKv4.2 with 5′ SacI and 3′ EcoRI restriction sites using the following 5′ primers: attaagagctcatgtttgccagggcagccgccat (Kv4.2Δ10N), actaagagctcatgtcggggcctatgcctgc (Kv4.2Δ23N), actacgagctcatgccaagacaggagagaaaaagg (Kv4.2Δ30N), and actacgagctcatggctctgatagtgctgaa (Kv4.2Δ40N) and the 3′ primer gccgtgaattcttac-aaagcagacaccctg in all cases. Kv4.2 truncation constructs were cloned into AdloxCRI between the SacI and EcoRI restriction sites.
Two Kv4.2 point mutant constructs were generated in which amino acids 35RKR37 were mutated to all alanines (Kv4.2AAA) or all lysines (Kv4.2KKK) by site-directed mutagenesis of AdloxCRI.mKv4.2 using the QuikChange XL kit (Stratagene) with the following primers: Kv4.2AAA, 5′-gcctgcgcccccaagacaggaggcagcagcgactcaggacgctctgatagtg and 3′-cactatcagagcgtcctgagtcgctgctgcctcctgtcttgggggcgcaggc; and Kv4.2KKK, 5′-cccccaagacaggagaaaaaaaagactcaggacgctctg and 3′-cagagcgtcctgagtcttttttttctcctgtcttggggg. All of the constructs were sequenced to ensure that no unintentional mutations were introduced by PCR.
Cell Culture and Transient Transfections
HEK 293 cells, obtained from the American Tissue Culture Collection, were maintained in DMEM (Invitrogen) supplemented with 5% horse serum (Invitrogen), 5% heat-inactivated FCS (Invitrogen), and 1 unit/ml penicillin-streptomycin (Invitrogen). The cells were passaged at confluence every 3–4 days by brief trypsinization (29).
For experiments, the plasmids were mixed with Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen) and incubated at room temperature for ∼30 min prior to addition to the cultures. For electrophysiology, the cells were transfected in 35-mm dishes using 2 μl of Lipofectamine mixed with 0.5 μg of total plasmids. For biochemistry, the cells were transfected in 25-cm2 flasks using 4 μl of Lipofectamine mixed with 4 μg of total plasmids. In both cases, a 1:1 ratio of the Kv4.2 construct to pCMV-Script.hKChIP2 (in presence of KChIP2) or to Adlox.CRI (in absence of KChIP2) was used. Approximately 8 h later, the plasmid-containing medium was removed and replaced with the normal cell culture medium (see above). Electrophysiological recordings and biochemical studies were performed 24–36 h after transfections.
Electrophysiology
Whole cell recordings were obtained at room temperature (22–24 °C) within 36 h of transfections using an Axopatch-1D amplifier (Axon Instruments, Sunnyvale, CA) interfaced to a Dell (model, Precision 340) personal computer using a Digidata 1322A A/D converter (Axon Instruments). Voltage-clamp paradigms were controlled, and data were collected using Clampex 9.2 (pClamp 9; Axon Instruments). The data were acquired at 100 kHz, and current signals were filtered on-line at 5 kHz prior to digitization and storage. Recording pipettes contained: 115 mm KCl, 20 mm KOH, 10 mm EGTA, 10 mm HEPES, and 5 mm glucose (pH 7.2; 300–310 mOsm). Pipette resistances were 1.5–3.0 MΩ when filled with the recording solution. The bath solution contained 140 mm NaCl, 4 mm KCl, 2 mm MgCl2, 1 mm CaCl2, 10 mm HEPES, and 5 mm glucose (pH 7.4; 300–310 mOsm). All of the reagents used to make the recording solutions come from Sigma.
After establishing the whole cell configuration, ±10-mV steps from a holding potential (HP) of −70 mV were applied to allow measurements of whole cell membrane capacitances and input resistances. Whole cell membrane capacitances and series resistances were routinely compensated (≥85%) electronically. Only data obtained from cells with input resistances ≥300 mΩ were analyzed. Leak currents were always <200 pA and were not corrected. Voltage-gated Kv4-encoded K+ currents were evoked in response to 450-ms depolarizing voltage steps to potentials between −60 and +40 mV from an HP of −70 mV; voltage steps were presented in 10-mV increments at 10-s intervals. To determine the time course of recovery of the currents from steady-state inactivation, a three-pulse protocol was used. From a HP of −70 mV, the cells were first depolarized to +30 mV for 450 ms (conditioning pulse), subsequently hyperpolarized to −70 mV for various times ranging from 5 ms to 5 s to allow for recovery, and finally depolarized to +30 mV (test pulse) to assess the extent of recovery.
Analysis of electrophysiological data were completed using Clampfit 9.2 (Axon). Whole cell membrane capacitances were calculated by integrating the capacitive transients elicited during +10-mV voltage steps from the HP. Kv4-encoded currents (at each test potential) were measured as the difference between the maximal outward current amplitudes and the currents remaining at 450 ms. The decay phases of the outward K+ currents evoked during 450-ms depolarizing voltage steps were fitted to the sum of two exponentials using the following equation: y(t) = A1exp(−t/τ1) + A2exp(−t/τ2) + B, where t is time, A1 and A2 are the amplitudes, and τ1 and τ2 are the time constants of decay of the inactivating K+ current components. The mean ± S.E. decay time constants (τdecay) reported here were obtained from recordings at +30 mV. To quantify the rates of recovery from steady-state inactivation, the amplitudes of Kv4.2-encoded currents evoked during each of the test pulses were measured and normalized to the amplitudes of currents measured during the first (conditioning) pulse. The normalized recovery data were plotted against the interpulse interval and were fitted with single exponential functions.
All data are presented as the means ± S.E. The statistical significance of observed differences between groups of cells was evaluated using a one-way Student's t test; the p values are presented in the text, and statistical significance was set at the p < 0.05 level.
Western Blot Analysis
Using previously described methods (30), Western blot analyses were performed on total protein lysates prepared from HEK 293 cells transfected with the Kv4 and KChIP2 plasmids described above. All of the reagents were from Sigma unless otherwise noted. Transfected cells were scraped from 25-cm2 tissue culture flasks and briefly vortexed in 500 μl of ice-cold lysis buffer: PBS (136 mmol/liter NaCl, 2.6 mmol/liter KCl, 10 mmol/liter Na2HPO4, 1.7 mmol/liter KH2PO4, pH 7.4) containing protease inhibitor mixture tablet (Roche) and Triton X-100 (1%). After a 15-min incubation with slow rotation at 4 °C, the insoluble fraction was removed by centrifugation at 3000 rpm for 10 min at 4 °C. Total protein in each sample was quantified using the BCA protein assay kit (Pierce). For Western blots, 20 μg of protein was loaded into each lane of the SDS-PAGE gels. Following fractionation, proteins were transferred to PVDF membranes (Bio-Rad) and blocked with 5% nonfat dry milk in PBS + 0.1% Tween 20. Following blocking PVDF membranes were incubated with the mouse monoclonal anti-Kv4.2 or anti-KChIP2 antibody (developed by and obtained from the University of California, Davis/National Institutes of Health NeuroMab facility, supported by National Institutes of Health Grant U24NS050606 and maintained by the University of California at Davis) at 4 °C overnight. To ensure equal protein loading of lanes, the membranes were also probed with mouse monoclonal anti-transferrin receptor (Invitrogen) antibody at room temperature for 1 h.
After washing, the membranes were incubated with a rabbit anti-mouse horseradish peroxidase-conjugated secondary antibody (Bethyl Laboratories, Montgomery, TX) followed by SuperSignal West Dura Extended Duration substrate (Pierce). The signals were detected using a Molecular Imager Chemidoc XRS system running Quantity One software version 4.6 (Bio-Rad). The intensities of the wild type and mutant Kv4.2 protein bands were determined using volume analysis from the Quantity One software, and channel subunit protein intensities were normalized to transferrin receptor and wild type Kv4.2 intensities on the same blot. The results are expressed as the means ± S.E. from six to nine experiments. Statistical differences between conditions were assessed using the Student's t test or Wilcoxon Signed Rank test when appropriate.
Immunoprecipitation of Kv4.2 Channel Subunits
Cell lysates from HEK 293 cells co-transfected with KChIP2 and one of the Kv4.2 constructs described above were used for immunoprecipitations with 5 μg of an anti-Kv4.2 rabbit polyclonal antibody (RbαKv4.2; Sigma). Parallel control experiments were completed using the same amount (5 μg) of nonspecific rabbit immunoglobulin G (RbIgG; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Prior to immunoprecipitations, antibodies were bound to 12.5 μl of protein A-magnetic beads (Invitrogen). Protein samples and antibody-coupled beads were mixed for 2 h at 4 °C. Magnetic beads were then collected and washed rapidly four times with ice-cold lysis buffer, and isolated protein complexes were eluted from the beads in 1× SDS sample buffer. Purified cell surface proteins were then analyzed by Western blot as described above. The results are representative of four experiments.
Cell Surface Biotinylation and Endocytosis Assays
Surface biotinylation of transfected HEK 293 cells was completed as described previously (30). Briefly, transfected cells were incubated in PBS at 4 °C to inhibit membrane protein internalization, followed by incubation with 0.25 mg/ml Sulfo-NHS-SS-Biotin (Pierce) in PBS for 30 min on ice. The biotinylation reaction was quenched with Tris-saline solution (10 mmol/liter Tris, pH 7.4, 120 mmol/liter NaCl), and the cells were collected. Detergent-soluble lysates were prepared, and biotinylated cell surface proteins were affinity-purified using NeutrAvidin-conjugated agarose beads (Pierce). Purified cell surface proteins were then subjected to Western blot analyses as described above.
In endocytosis assays, the cells were first biotinylated (pulse) and washed with Tris-saline solution as described above. The cells were then returned to culture medium (chase) at 37 °C for varying times (0, 15, 30, and 60 min). At the end of each chase time, the cells were rinsed with ice-cold PBS and incubated with the impermeant reducing agent sodium 2-mercaptoethane-sulfonate (100 mm, in 50 mm Tris, pH 8.6, 100 mm NaCl, 2.5 mm CaCl2) at 4 °C for 15 min to remove the biotin on proteins remaining at the cell surface. This procedure was repeated twice, and the cells were then incubated with 5 mg/ml iodoacetamide in PBS at 4 °C for 15 min to modify free SH groups. Detergent-soluble cell lysates were prepared, and biotinylated proteins were purified using NeutrAvidin-conjugated agarose beads. For each chase time, nonreduced samples were used to estimate the degradation of biotinylated proteins or spontaneous debiotinylation. Reduced samples at 0 min (t0 min, reduced) were used to evaluate background (usually ≤10%). The results are expressed as the percentages of biotinylated surface proteins that were endocytosed at each time point, i.e. percentage of fraction endocytosed (at x min), calculated as (timex min, reduced − time0 min, reduced)/timex min, nonreduced × 100.
RESULTS
Kv4.2 N-terminal Truncation Partially Mimics the Effects of KChIP2 on K+ Currents
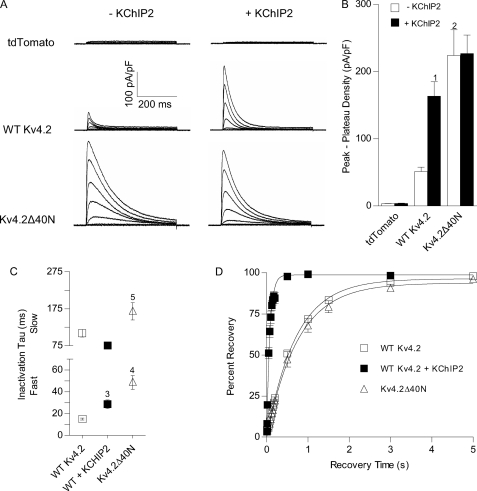
As illustrated in Fig. 1, heterologous expression of WT Kv4.2 resulted in rapidly activating, rapidly inactivating voltage-gated outward K+ currents. Co-expression of KChIP2 significantly (p < 0.001) increased Kv4.2 current (defined as the peak minus the plateau (450 ms) current) densities, consistent with previous findings (10). On average, Kv4.2 current densities were increased ∼3-fold with KChIP2 co-expression (Fig. 1B). Importantly, the unitary conductance of Kv4 channels was not affected by KChIP co-expression (31), and the observed augmentation in Kv4.2-encoded currents has been attributed to increased protein expression (10, 11, 18, 31).
FIGURE 1.
Kv4.2 N-terminal truncation partially mimics the effects of KChIP2 co-expression on Kv4.2 currents. Whole cell voltage-gated outward K+ currents were recorded from HEK 293 cells transiently transfected with cDNA constructs encoding tdTomato plus WT Kv4.2 or Kv4.2Δ40N (in which residues 2–40 were deleted), in the absence and in the presence of KChIP2. Whole cell K+ currents were recorded in response to depolarizing voltage steps between −60 and +40 mV (in 10-mV increments) from a HP of −70 mV. Representative records are shown in A, and the mean ± S.E. (n = 4–23 cells) (peak-plateau) K+ current densities (at −10 mV) are presented in B. Only endogenous, non-voltage-gated K+ currents were evident in HEK 293 cells expressing tdTomato (in the absence and presence of KChIP2). Rapidly activating and inactivating K+ currents were routinely observed in cells expressing WT Kv4.2, and Kv4.2 current densities were increased significantly (1, p < 0.001) with KChIP2 co-expression (B). Transient K+ current densities were also significantly (2, p < 0.001) increased in cells expressing Kv4.2Δ40N. In contrast to WT Kv4.2, however, Kv4.2Δ40N current densities (B) were not increased with co-expression of KChIP2. Additionally, transient K+ current densities were not significantly different between cells co-expressing WT Kv4.2 and KChIP2 and cells co-expressing Kv4.2Δ40N and KChIP2. C, mean ± S.E. (n = 6–23 cells) time constants of slow (τSlow) and fast (τFast) inactivation of Kv4.2 currents, measured at +30 mV, are plotted. Co-expression of WT Kv4.2 with KChIP2 and N-terminal truncation of Kv4.2 both resulted in slowing of macroscopic current inactivation (A). Only the fast component of inactivation of WT Kv4.2 currents, however, was increased significantly (3, p < 0.05) with co-expression of KChIP2, whereas both the fast (4, p < 0.001) and slow (5, p < 0.05) inactivation time constants were increased significantly for Kv4.2Δ40N (compared with WT Kv4.2) currents. The mean ± S.E. (n = 5–7 cells) time course of recovery from inactivation is plotted in D. Recovery was significantly (p < 0.01) faster in cells co-expressing KChIP2 compared with cells expressing WT Kv4.2 alone. In contrast, the time course of recovery of Kv4.2Δ40N currents is slow.
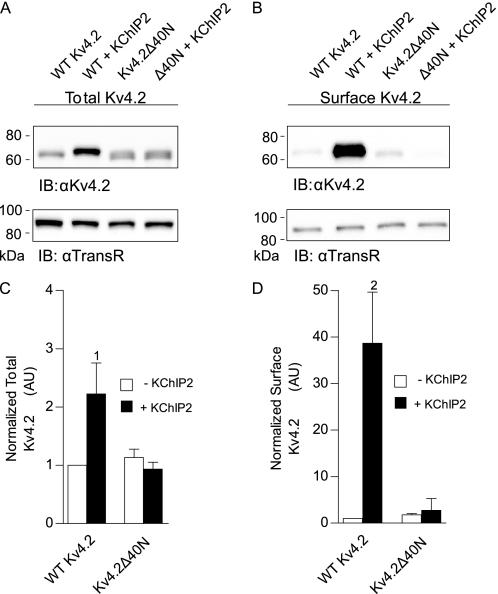
As also reported previously (10), the kinetic properties of the Kv4.2 currents were also affected by KChIP2 co-expression. In heterologous cells, Kv4.2 inactivation is best described by the sum of two exponentials that differ by approximately an order of magnitude (32). In cells expressing WT Kv4.2 in the absence of KChIP2, the rapidly inactivating component, with a time constant (τFast) of 15 ± 1 ms (at +30 mV), contributed 82 ± 1% to the peak Kv4.2 current, with the minor (17 ± 1%) component inactivating more slowly, with a time constant (τSlow) of 109 ± 10 ms (Fig. 1C). KChIP2 co-expression resulted in significant (p < 0.05) slowing (τFast = 29 ± 4 ms) of the rapid component of Kv4.2 current inactivation and a decrease (to 50 ± 6%) in the fraction of total current that inactivated rapidly. Consistent with previous findings (10), however, the kinetics of Kv4.2 recovery from steady-state inactivation (Fig. 1D) were significantly (p < 0.01) faster with KChIP2 co-expression. Parallel biochemical experiments demonstrated that KChIP2 co-expression increased total Kv4.2 protein expression 2–3-fold (Fig. 2A) compared with cells expressing Kv4.2 alone. As reported previously (18), the increase in total Kv4.2 protein in the lysate reflected, at least in part, increased Kv4.2 solubility (data not shown). In addition, however, KChIP2 co-expression resulted in a marked (>40-fold) increase in cell surface Kv4.2 protein expression (Fig. 2B) compared with cells expressing Kv4.2 alone.
FIGURE 2.
KChIP2 co-expression increases total and cell surface Kv4.2 protein expression. Representative Western blots of fractionated total (A) and cell surface (B) proteins prepared from HEK 293 cells transiently transfected with cDNA constructs encoding WT Kv4.2 or Kv4.2Δ40N in the absence and in the presence of KChIP2. The intensities of the Kv4.2 bands were measured and normalized, first to the expression of endogenous transferrin receptor (TransR) in the same lane and subsequently to WT Kv4.2 in the same blot. The mean ± S.E. (n = 3–12 blots) total (A) and cell surface (B) Kv4.2 protein expression levels in cells expressing WT Kv4.2 or Kv4.2Δ40N in the presence and in the absence of KChIP2 are plotted. Co-expression of WT Kv4.2 with KChIP2 significantly increased total (1, p < 0.05) and cell surface (2, p < 0.01) Kv4.2 protein expression. In contrast to the marked differences in current densities (Fig. 1B), however, total (C) and cell surface (D) Kv4.2Δ40N protein expression were not measurably higher than WT Kv4.2 in the absence of KChIP2. IB, immunoblot.
Similar to the current augmentation seen with KChIP2 (Fig. 1, A and Bs), truncation of the first 40 amino acid residues of Kv4.2 (Kv4.2Δ40N) resulted in significantly (p < 0.001) higher Kv4.2 current densities than in cells expressing WT Kv4.2 alone (Fig. 1, A and B). In cells expressing Kv4.2Δ40N, K+ current densities, on average, were 4.4-fold higher than in cells expressing WT K4.2. In contrast to cells expressing WT Kv4.2, however, co-expression with KChIP2 did not measurably affect Kv4.2Δ40N current densities (Fig. 1B). Analyses of the decay phases of the currents also revealed subtle, yet significant, differences in the properties of the currents in Kv4.2Δ40N-expressing cells compared with WT Kv4.2 + KChIP2-expressing cells. Both components of inactivation, for example, were slowed with N-terminal deletion, and 26 ± 6% of the peak Kv4.2Δ40N current inactivated rapidly. Similar to the lack of effect on current densities, co-expression with KChIP2 also did not measurably affect the fast or slow components of Kv4.2Δ40N current inactivation (data not shown). In addition, N-terminal deletion did not measurably affect the kinetics of Kv4.2 recovery from steady-state inactivation (Fig. 1D). Surprisingly, despite the marked increases in K+ current densities (Fig. 1B), neither the total (Fig. 2A) nor the cell surface (Fig. 2B) expression of the Kv4.2Δ40N protein was measurably different from WT Kv4.2 (expressed alone, i.e. in the absence of KChIP2).
Molecular Dissection of Kv4.2 N-terminal Interactions
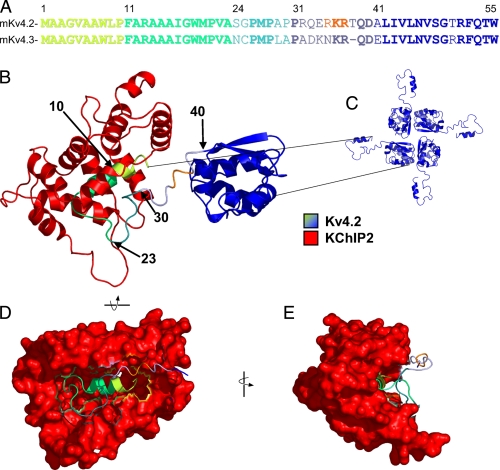
To guide further investigation into the molecular determinants that mediate Kv4.2/KChIP2 interactions, the predicted structure of the N terminus of Kv4.2, together with KChIP2, was modeled (26) based on the published structure of Kv4.3 N terminus·KChIP1 complex (16, 17). Sequence homology was high between Kv4.2 and Kv4.3 in the distal N terminus (Fig. 3A), as well as between KChIP2 and KChIP1 (alignment not shown) in the crystallized C termini (10). Not surprisingly, therefore, the modeling-predicted interactions between KChIP2 and the N terminus of Kv4.2 (Fig. 3B) very closely matched the interactions between KChIP1 and Kv4.3 revealed in the structural data (16, 17). Specifically, the distal N terminus of Kv4.2 (∼20 residues) was buried in a hydrophobic binding pocket of KChIP2. Additionally, the N terminus of Kv4.2 exited this pocket but remained in close proximity to KChIP2 through the first 30 residues (Fig. 3B).
FIGURE 3.
Predicted KChIP2-Kv4.2 N-terminal interaction. Sequence alignment revealed substantial homology in the distal N termini of mouse Kv4.2 and Kv4.3 (A). The structure of the Kv4.2 N terminus was predicted, therefore, from the reported crystal structure of the Kv4.3 N terminus in complex with KChIP1 (16, 17). The KChIP2 sequence replaced the highly homologous KChIP1, and the structure of the N terminus of Kv4.2 together with KChIP2 (B) was predicted using comparative protein modeling by satisfaction of spatial restraints (26). A schematic illustrating the assembly of Kv4 α subunits into functional tetrameric channels is shown in C. Amino acid residues 10, 23, 30, and 40 in the Kv4.2 N terminus are indicated. The structure depicted in B is rotated first vertically by 90° (D) and subsequently horizontally by 90° (E), and the surface of the KChIP2 molecule is rendered to illustrate the KChIP2 binding pocket encapsulating the N terminus of Kv4.2. The predicted structure of the Kv4.2 N terminus·KChIP2 complex suggests that KChIP2 interacts closely with the first 30 residues of Kv4.2.
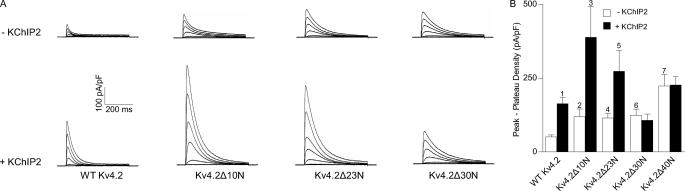
To explore further the critical domains in the Kv4.2 N terminus regulating interactions with KChIP2, additional Kv4.2 N-terminal constructs were generated in which residues 2–10 (Kv4.2Δ10N), 2–23 (Kv4.2Δ23N), or 2–30 (Kv4.2Δ30N) were removed. Whole cell K+ currents were recorded from HEK 293 cells transiently transfected with tdTomato and each of the mutant constructs, Kv4.2Δ10N, Kv4.2Δ23N, and Kv4.2Δ30N, in the absence or presence of KChIP2. As illustrated in Fig. 4, whole cell Kv4.2 current densities in cells expressing Kv4.2Δ10N were significantly (p < 0.05) higher (∼2.3-fold) than in cells transfected with WT Kv4.2. Similar to cells expressing WT Kv4.2, co-expression of KChIP2 significantly (p < 0.01) increased Kv4.2Δ10N current densities (Fig. 4B), and the magnitude (3.3-fold) of the effect was similar to that observed for WT Kv4.2 (Fig. 4B). Outward current densities were also increased significantly (p < 0.005) in cells expressing Kv4.2Δ23N (Fig. 4A), and the magnitude of the increase (2.3-fold) was similar to Kv4.2Δ10N-expressing cells. In addition, co-expression of KChIP2 again resulted in a significant (p < 0.05) increase in mean current density (Fig. 4B). The magnitude of the increase (2.4-fold) in current densities, however, was less than with WT Kv4.2 (3.3-fold) or Kv4.2Δ10N (3.3-fold), revealing that truncation of the first 23 residues (but not the first 10 residues) of Kv4.2 attenuates KChIP2-mediated effects on current densities (see “Discussion”).
FIGURE 4.
Differential effects of Kv4.2 N-terminal truncations on K+ current densities and modulation by KChIP2. Whole cell outward K+ currents were recorded as described in the legend to Fig. 1 from HEK 293 cells transiently transfected with cDNA constructs encoding tdTomato together with WT Kv4.2 or Kv4.2 in which N-terminal residues 2–10 (Kv4.2Δ10N), 2–23 (Kv4.2Δ23N), or 2–30 (Kv4.2Δ30N) were deleted in the absence and in the presence of KChIP2. Representative records are shown in A, and the mean ± S.E. Kv4.2 current densities (n = 6–23 cells) measured at −10 mV are plotted in B. The mean ± S.E. Kv4.2 current densities from cells expressing Kv4.2Δ40N in the absence and in the presence of KChIP2 are repeated from Fig. 1B for comparison. Whole cell K+ current amplitudes/densities in cells expressing Kv4.2Δ10N (2, p < 0.05), Kv4.2Δ23N (4, p < 0.005), or Kv4.2Δ30N (6, p < 0.01) were significantly higher than in WT Kv4.2 expressing cells. Additionally, K+ current densities in cells expressing Kv4.2Δ40N were higher (7, p < 0.05) than in cells expressing Kv4.2Δ10N, Kv4.2Δ23N, or Kv4.2Δ30N. Co-expression with KChIP2 resulted in 2–3-fold increases in K+ current densities in cells expressing WT Kv4.2 (1, p < 0.001), Kv4.2Δ10N (3, p < 0.01), or Kv4.2Δ23N (5, p < 0.05). In contrast, co-expression with KChIP2 did not significantly increase current densities in cells expressing Kv4.2Δ30N or Kv4.2Δ40N.
Similar to Kv4.2Δ10N and Kv4.2Δ23N, peak Kv4.2 current densities were significantly (p < 0.01) higher in cells expressing Kv4.2Δ30N (alone) compared with cells expressing WT Kv4.2 (Fig. 4B). Co-expression with KChIP2, however, did not measurably affect current densities in cells expressing Kv4.2Δ30N. The kinetic properties of the currents (supplemental Fig. S1) produced by the various Kv4.2 N-terminal truncation mutants were similar to those of Kv4.2Δ40N (Fig. 1). Interestingly, the magnitude of the current enhancement (2.4-fold) in Kv4.2Δ10N-, Kv4.2Δ23N-, and Kv4.2Δ30N-expressing cells (Fig. 4B) was significantly (p < 0.05) smaller than the 4.4-fold increase observed in cells expressing Kv4.2Δ40N (Fig. 1B), suggesting that residues between positions 30 and 40 function to suppress Kv4.2 current densities.
In support of this hypothesis, further studies revealed that expression of Kv4.2AAA, in which neutral alanines were substituted for three positively charged residues (RKR) at positions 35–37 (Fig. 3), resulted in significantly (p < 0.05) higher current densities than WT Kv4.2 (supplemental Fig. 2B). In contrast, expression of Kv4.2KKK in which the arginines at positions 35 and 37 were mutated to lysines resulted in peak current densities that were not measurably different from WT Kv4.2 (supplemental Fig. 2B), suggesting that charged residues in this Δ30-Δ40 region play a role in limiting Kv4.2 currents (see “Discussion”).
Distinct Effects of N-terminal Truncations on (Total and Cell Surface) Kv4.2 Protein Expression
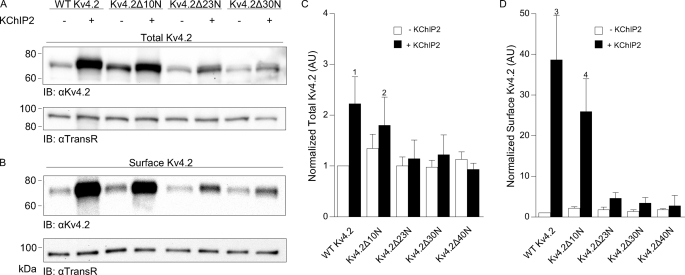
Western blots of fractionated proteins from HEK 293 cells transfected with WT Kv4.2 or one of the N-terminal truncation mutants in the absence and in the presence of KChIP2 are illustrated in Fig. 5. Similar to the results obtained with Kv4.2Δ40N (Fig. 2), the total and cell surface protein expression levels of the other (Kv4.2Δ10N, Kv4.2Δ23N, and Kv4.2Δ30N) truncation mutants were not significantly higher than WT Kv4.2 in the absence of KChIP2 (Fig. 5, A and B). These results suggest that the observed increases in functional current densities in cells expressing the Kv4.2 N-terminal truncation mutants in the absence of KChIP2 therefore do not result from increased total or cell surface Kv4.2 protein expression (see “Discussion”).
FIGURE 5.
KChIP2-mediated increases in total and cell surface Kv4.2 protein expression are lost with progressive truncations of the Kv4.2 N terminus. Representative Western blots of fractionated total (A) and cell surface (B) proteins from HEK 293 cells transiently transfected with cDNA constructs encoding WT Kv4.2, Kv4.2Δ10N, Kv4.2Δ23N, or Kv4.2Δ30N in the absence (−) and in the presence (+) of KChIP2. The intensities of the Kv4.2 protein bands were measured and normalized first to the expression of endogenous transferrin receptor (TransR) and subsequently to WT Kv4.2 in the same blot. The mean ± S.E. (n = 6–9 blots) total (C) and cell surface (D) Kv4.2 protein expression levels in cells expressing the various Kv4.2 truncation constructs in the presence and absence of KChIP2 are plotted; in addition, WT Kv4.2 and Kv4.2Δ40N data from Fig. 2 are replotted here for comparison. Although K+ current densities were higher in Kv4.2Δ10N-expressing cells than in WT Kv4.2-expressing cells in the absence of KChIP2 (Fig. 4), neither total (C) nor cell surface (D) protein expression levels were significantly different from WT Kv4.2. Similar to WT Kv4.2 (1, p < 0.05; and 3, p < 0.01), however, co-expression of KChIP2 with Kv4.2Δ10N significantly increased total (2, p < 0.05) (C) and cell surface (D) (4, p < 0.05) protein expression. Further truncation of the N terminus of Kv4.2 also did not result in significant increases in total or cell surface Kv4.2 protein expression in the absence of KChIP2. In addition, although KChIP2 co-expression appeared to modestly increase total and surface protein levels in Kv4.2Δ23N- or Kv4.2Δ30N-expressing cells (A), these differences did not reach statistical significance. IB, immunoblot.
As with WT Kv4.2, KChIP2 co-expression significantly (p < 0.05) increased total Kv4.2Δ10N protein expression. For WT Kv4.2, the increase in total Kv4.2 protein with KChIP2 co-expression was modest (∼2-fold), whereas the increase in surface Kv4.2 protein expression was ∼40-fold. Truncation of the first 10 residues in Kv4.2 partially attenuated this effect, with surface Kv4.2 protein expression increased by ∼25-fold in Kv4.2Δ10N-expressing cells (Fig. 5B). In contrast, KChIP2 co-expression did not significantly increase total or surface Kv4.2 protein expression in Kv4.2Δ23N- or Kv4.2Δ30N-expressing cells (Fig. 5, A and B).
Kv4.2·KChIP2 Complex Formation Results in Mutual Protein Stabilization
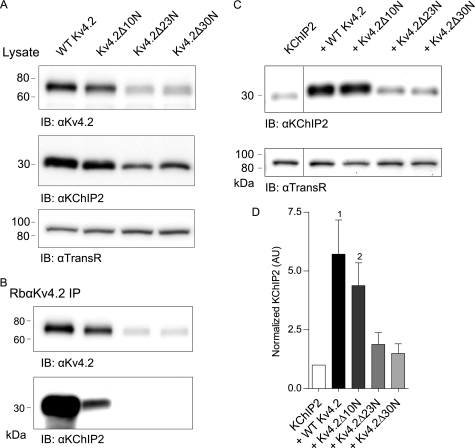
Using an antibody targeting the C terminus of Kv4.2, subsequent experiments were focused on examining the interaction between KChIP2 and the various Kv4.2 mutants illustrated in Figs. 4 and 5. As illustrated in Fig. 6B, KChIP2 was co-immunoprecipitated with the anti-K4.2 antibody when co-expressed with WT Kv4.2 and, to a much lesser extent, with Kv4.2Δ10N (Fig. 6B). In contrast, KChIP2 was not co-immunoprecipitated with either Kv4.2Δ23N or Kv4.2Δ30N (Fig. 6B), although both mutant Kv4.2 proteins were efficiently immunoprecipitated with the anti-Kv4.2 antibody (Fig. 6A). Consistent with the biochemical studies described above, there was a marked decrease in Kv4.2 protein expression as the Kv4.2 N terminus was truncated and the interaction with KChIP2 was lost. In addition, the relative amount of KChIP2 that was recovered following co-immunoprecipitation with WT Kv4.2 was several orders of magnitude greater than with Kv4.2Δ10N, suggesting that the first 10 amino acids of the Kv4.2 N terminus, which are predicted to be located in the KChIP2 hydrophobic binding pocket (Fig. 3B), impart the greatest relative binding affinity (see “Discussion”).
FIGURE 6.
Kv4.2-KChIP2 co-expression also results in reciprocal increases in KChIP2. Lysates from HEK 293 cells co-transfected with KChIP2 and Kv4.2 or one of the Kv4.2 N-terminal truncation mutants were immunoprecipitated (IP) with a rabbit anti-Kv4.2 antibody directed against the C terminus of Kv4.2. Representative Western blots of the cell lysates before immunoprecipitation (A) and of the immunoprecipitated proteins (B) are illustrated. Although the Kv4.2 protein was recovered in all cases, KChIP2 was only detectable in samples immunoprecipitated from cells expressing WT Kv4.2 or Kv4.2Δ10N; KChIP2 was not detected in immunoprecipitates from cells expressing Kv4.2Δ23N or Kv4.2Δ30N. In addition, Western blot analyses (C) of fractionated proteins from HEK 293 cells transiently transfected with KChIP2 and WT Kv4.2, Kv4.2Δ10N, Kv4.2Δ23N, or Kv4.2Δ30N revealed that KChIP2 protein expression was markedly increased in cells transfected with WT Kv4.2 or Kv4.2Δ10N, whereas co-transfection with Kv4.2Δ23N or Kv4.2Δ30N did not result in increased KChIP2. The intensities of the KChIP2 bands were measured and normalized first to the expression of the transferrin receptor (TransR) in the same lane and subsequently to KChIP2 in cells expressing KChIP2 alone in the same blot. D, mean ± S.E. (n = 5) KChIP2 protein expression levels in cells expressing KChIP2 with WT Kv4.2 (1, p < 0.05) or Kv4.2Δ10N (2, p < 0.05) were significantly higher than in cells expressing KChIP2 alone, whereas KChIP2 levels in Kv4.2Δ23N- and Kv4.2Δ30N-expressing cells were not significantly different from cells expressing KChIP2 alone. (The vertical line in C marks the deletion of a lane in the blot from cells expressing WT Kv4.2 alone.) IB, immunoblot.
These biochemical experiments also revealed that although the same amount of KChIP2 cDNA was used in the transfections with each of the Kv4.2 constructs, there were marked differences in KChIP2 protein expression levels evident in the Western blots (Fig. 6A). In fact, there were parallel decreases in KChIP2 and Kv4.2 protein expression with the truncation of Kv4.2 (Fig. 6A). Co-expression with WT Kv4.2 or Kv4.2Δ10N significantly (p < 0.05) increased KChIP2 protein expression (Fig. 6C) ∼6- and ∼5-fold, respectively (Fig. 6D). In contrast, expression of the KChIP2 protein was not measurably affected by the co-expression of either Kv4.2Δ23N or Kv4.2Δ30N (Fig. 6D).
KChIP2 Decreases the Turnover Rate of Cell Surface Kv4.2
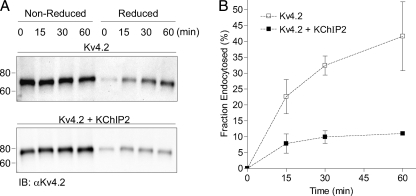
The dramatic increase in the surface expression of the Kv4.2 protein with co-expression of KChIP2 (Fig. 2) suggested the possibility that KChIP2 acts at the cell surface, specifically, to stabilize Kv4.2 protein expression. This hypothesis was explored in pulse-chase assays in which surface protein was biotinylated (pulse) followed by incubation at 37 °C for various times up to 1 h (chase) to allow for internalization of surface labeled proteins. Following incubation, biotin was removed from proteins remaining at the surface by treating cells with the impermeant reducing agent sodium 2-mercaptoethanesulfonate (MESNA). The cell lysates were then prepared from control (nonreduced) and MESNA-treated (reduced) cells. As illustrated in Fig. 7, Western blot analyses of the biotinylated (internalized) protein fraction from (reduced) cells exposed to MESNA revealed that co-expression with KChIP2 slowed the turnover rate of cell surface Kv4.2 compared with Kv4.2 expressed alone. When expressed alone, Kv4.2 was endocytosed at a moderate rate with ∼60% of the initial labeled protein remaining at the cell surface following 1 h of incubation at 37 °C. Co-expression of KChIP2 slowed the rate of Kv4.2 endocytosis markedly, with ∼90% of the initial labeled protein remaining at the cell surface following 1 h of incubation at 37 °C. In agreement with previous studies (33), parallel control experiments revealed that the rate of the surface transmembrane control, transferrin receptor, was rapid, showing complete endocytosis of labeled protein after 15 min (data not shown).
FIGURE 7.
KChIP2 decreases the turnover rate of cell surface Kv4.2. Approximately 24 h following transfection with Kv4.2 and KChIP2, cell surface proteins were biotinylated as described under “Experimental Procedures” and subsequently incubated at 37 °C for varying times. At each time indicated, the cells were treated with the reducing agent MESNA to remove the remaining surface biotin. The cell lysates were then prepared from control (nonreduced) and MESNA-treated (reduced) cells, and biotinylated proteins were isolated by streptavidin pulldown. The fraction (of Kv4.2) endocytosed (at x min) was calculated as (Kv4.2x min, reduced − Kv4.20 min, reduced)/Kv4.2x min, nonreduced × 100. Representative Western blots are illustrated in A, and the mean ± S.E. (2–4 blots) time course of protein internalization in the absence and in the presence of KChIP2 is plotted in B. Western blot analyses (A) of the biotinylated (internalized) protein fraction from (reduced) cells revealed that co-expression with KChIP2 slowed the rate of Kv4.2 endocytosis compared with Kv4.2 expressed alone. IB, immunoblot.
DISCUSSION
Although previous studies have provided important insights into the molecular determinants of KChIP-mediated effects on Kv4 currents (11, 15, 18, 34, 35), the links between channel subunit protein expression/stability and functional Kv4-encoded currents have not been clearly defined. Biochemical and electrophysiological approaches were combined in the studies here to examine total and cell surface Kv4.2 protein expression and functional Kv4.2 current densities directly. The results of these parallel analyses have revealed that KChIP2 co-expression increases total and cell surface Kv4.2 protein expression and Kv4.2 current densities to dramatically different extents. Previous studies have suggested that KChIP2 plays a critical role in controlling the solubility (18) and the trafficking of Kv4.2 out of the ER and Golgi complex (18, 21). Additional experiments presented here revealed that KChIP2 also promotes Kv4.2 cell surface expression by prolonging retention of Kv4.2·KChIP2 channel complexes at the cell surface. Unexpectedly, the experiments here also revealed that the interaction (binding) of the Kv4.2 and KChIP2 subunits and the formation of Kv4.2·KChIP2 complexes result in mutual, reciprocal stabilization of both proteins.
Kv4 N-terminal Truncations Increase Kv4 Currents without Increasing Kv4.2 Protein Expression
Consistent with previous studies (11, 14), truncation of the Kv4.2 N terminus (Kv4.2Δ40N) increased Kv4.2 current densities, partially mimicking the effects of KChIP2 co-expression. One interpretation of the observed increases in current densities is that KChIP2 promotes functional Kv4.2 (current) expression by masking an ER retention signal in the Kv4.2 N terminus and allowing forward trafficking of Kv4.2·KChIP2 complexes (11). Previous studies have suggested that KChIP2 promotes forward trafficking of Kv4.2 from the ER or the Golgi complex (18, 21). The experiments here, however, revealed that although Kv4 current densities were increased, deletion of 40 amino acids in the distal Kv4.2 N terminus (Kv4.2Δ40N) did not result in increased total or cell surface Kv4.2 protein expression. The observed augmentation of currents with N-terminal truncation therefore likely results from changes in channel gating as suggested previously (36). This hypothesis is supported by the observation that deletion of the distal 40 amino acid residues results in slowing of current inactivation in Kv4.2Δ40N-expressing cells. Further experiments revealed that more modest deletions of the Kv4.2 N terminus (in Kv4.2Δ10N, Kv4.2Δ23N, and Kv4.2Δ30N) also did not result in increased total or cell surface Kv4.2 protein expression. The observed increases in current densities with N-terminal truncation therefore are not consistent with the removal of an N-terminal ER retention motif and increased forward trafficking of assembled Kv4 channels. Rather, the results demonstrate direct inhibitory effects of the distal N terminus on Kv4.2 current densities.
Unexpectedly, the analyses here also revealed the presence of two separable domains in the Kv4.2 N terminus. Deletion of the distal 10 residues (Kv4.2Δ10N) resulted in increased (2.3-fold) Kv4.2 current densities (in the absence of KChIP2), and similar increases were observed with the Kv4.2Δ23N and Kv4.2Δ30N mutants. The structural modeling (Fig. 3) suggests that KChIP2 binds up to approximately the first 20 residues of the N terminus of Kv4.2 in a hydrophobic binding pocket. The electrophysiological data suggest the interesting possibility that the distal 10 residues (1–10) in Kv4.2 inhibit Kv4.2 currents and that the physical sequestration of these Kv4.2 N-terminal residues (by KChIP2) is one mechanism by which KChIP2 increases Kv4.2 current densities. Deletion of the first 10 amino acids also reduced (by an order of magnitude) the amount of KChIP2 protein co-immunoprecipitating with the Kv4.2 antibody (Fig. 6), revealing the critical role for these residues in controlling binding to KChIP2. This conclusion appears to be in conflict with previously published results (11) suggesting a critical role for amino acids 11–23, but not amino acids 1–10. These early studies, however, were performed on in vitro translated Kv4.2N termini, rather than on full-length proteins expressed in intact cells, and this may underlie the difference in experimental results.
A further increase in Kv4.2 current densities was observed with deletion of amino acid residues 31–40 (Kv4.2Δ40N). The increase in current densities in cells expressing Kv4.2Δ40N, compared with cells expressing Kv4.2Δ30N (or the smaller truncations, Kv4.2Δ10N and Kv4.2Δ23N), suggests that residues in this region (residues 30–40) independently inhibit Kv4.2 currents. Consistent with this hypothesis, neutralization of the positive charges at residues 35–37 within this region (in the absence of N-terminal truncation), in the Kv4.2AAA mutant, also resulted in increased Kv4.2 current densities (compared with WT Kv4.2). In contrast, conservative mutations in this region (Kv4.2KKK), which maintained the positive charge, resulted in Kv4 current densities similar to WT Kv4.2. These observations suggest that the three (RKR) positively charged residues form a second site that inhibits functional Kv4.2 currents. Importantly, and consistent with the results of the N-terminal truncation mutants, neutralization of the positively charged RXR sequence, although increasing K+ currents, did not result in increased total or cell surface Kv4.2 protein expression (supplemental Fig. 1C).
KChIP2-mediated Increases in Cell Surface Kv4.2 Protein Expression Are Not Paralleled by Increased Kv4.2 Current Densities
The increase in the surface expression of Kv4.2 channels with KChIP2 co-expression has been interpreted as suggesting that KChIP2 promotes proper folding and forward trafficking of Kv4.2 (10, 11, 18, 21). The experiments here, however, revealed that KChIP2 co-expression also stabilizes Kv4.2 protein expression. In addition, the results of the co-immunoprecipitation experiments (Fig. 6) demonstrate that the elimination of the interaction (binding) between Kv4.2 and KChIP2 results in the loss of KChIP2-mediated stabilization of the Kv4.2 protein. Interestingly, the biochemical experiments also revealed that co-expression with KChIP2 differentially effects total and cell surface Kv4.2 expression. Although the effects of KChIP2 on total Kv4.2 protein expression were modest (∼2-fold increase), cell surface Kv4.2 protein expression was increased dramatically (∼40-fold). This KChIP2-mediated increase in cell surface Kv4.2 protein is also greater (by an order of magnitude) than the observed increases in Kv4.2 current densities. These results demonstrate that Kv4.2 current expression is separable from the Kv4.2 cell surface protein expression and suggest the exciting possibility that a pool of electrically silent Kv4.2 channels exist at the cell surface that might be recruited to respond to perturbations in cellular activity. In addition, these results suggest that additional mechanisms are required to shift cell surface WT Kv4.2 channel complexes into a functional state. Additionally, pulse-chase experiments, designed to monitor the rate of cell surface protein endocytosis, revealed that retention of Kv4.2 channel complexes at the cell surface is an additional mechanism by which KChIP2 promotes the surface expression of Kv4.2.
Interestingly, the detailed comparison of Kv4.2 protein expression levels and functional current densities also revealed a disparity in Kv4.2Δ23N-expressing cells. KChIP2 co-expression did not increase total or cell surface Kv4.2 protein levels in Kv4.2Δ23N-expressing cells, and KChIP2 did not co-immunoprecipitate with Kv4.2Δ23N. In contrast, KChIP2-mediated increases in Kv4.2 current densities were observed in Kv4.2Δ23N-expressing (but not in Kv4.2Δ30N-expressing) cells. Taken together, these results suggest that residues 11–23 contribute to KChIP2-mediated total and cell surface Kv4.2 protein stability and that amino acid residues 23–30 contribute to a low affinity interaction that results in increased Kv4.2 current densities without affecting Kv4.2 protein stability.
Kv4 Co-expression Reciprocally Stabilizes KChIP2 Protein Expression
The results presented here also revealed that co-expression with Kv4.2 results in increased KChIP2 protein expression. The increase in KChIP2 protein expression was also lost with Kv4.2 N-terminal deletions, which eliminate the interactions between Kv4.2 and KChIP2, suggesting that the formation of the Kv4.2·KChIP2 complex results in mutual co-stabilization of the two (Kv4.2 and KChIP2) subunit proteins. Consistent with this interpretation, KChIP2 protein expression was shown to be reduced dramatically in Kv4.2−/− ventricles (6), and both KChIP2 and KChIP3 are reduced in the brains of Kv4.2−/− mice (3, 25). Interestingly, previous studies suggest that accessory Kvβ subunits stabilize assembled Kv1 α subunits early in protein biosynthesis and subsequently chaperone assembled channel complexes from the ER to the cell surface to increase the functional cell surface expression of Kv1-encoded channels (22, 23). In addition, it was recently reported that the cytosolic accessory subunit Sleepless regulates the functional expression of Shaker Kv channels in Drosophila through mutual stabilization of the two (Sleepless and Shaker) subunits (37). Taken together, these results suggest that co-stabilization of protein complexes may be a generalized mechanism regulating Kv channel expression in native cells.
Supplementary Material
Acknowledgments
We thank Aaron Norris, Dr. Scott Marrus, and Dr. Yarimar Carrasquillo for many valuable discussions and for comments on this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL034161 (to J. M. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- KChIP
- K+ channel-interacting protein
- ER
- endoplasmic reticulum
- HP
- holding potential
- MESNA
- sodium-2-mercaptoethane sulfonate.
REFERENCES
- 1.Chen X., Yuan L. L., Zhao C., Birnbaum S. G., Frick A., Jung W. E., Schwarz T. L., Sweatt J. D., Johnston D. (2006) J. Neurosci. 26, 12143–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serôdio P., Kentros C., Rudy B. (1994) J. Neurophysiol. 72, 1516–1529 [DOI] [PubMed] [Google Scholar]
- 3.Nerbonne J. M., Gerber B. R., Norris A., Burkhalter A. (2008) J. Physiol. 586, 1565–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norris A. J., Nerbonne J. M. (2010) J. Neurosci. 30, 5092–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon J. E., Shi W., Wang H. S., McDonald C., Yu H., Wymore R. S., Cohen I. S., McKinnon D. (1996) Circ. Res. 79, 659–668 [DOI] [PubMed] [Google Scholar]
- 6.Guo W., Jung W. E., Marionneau C., Aimond F., Xu H., Yamada K. A., Schwarz T. L., Demolombe S., Nerbonne J. M. (2005) Circ. Res. 97, 1342–1350 [DOI] [PubMed] [Google Scholar]
- 7.Nerbonne J. M., Kass R. S. (2005) Physiol. Rev. 85, 1205–1253 [DOI] [PubMed] [Google Scholar]
- 8.Burgoyne R. D., Weiss J. L. (2001) Biochem. J. 353, 1–12 [PMC free article] [PubMed] [Google Scholar]
- 9.Braunewell K. H., Gundelfinger E. D. (1999) Cell Tissue Res. 295, 1–12 [DOI] [PubMed] [Google Scholar]
- 10.An W. F., Bowlby M. R., Betty M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. (2000) Nature 403, 553–556 [DOI] [PubMed] [Google Scholar]
- 11.Bähring R., Dannenberg J., Peters H. C., Leicher T., Pongs O., Isbrandt D. (2001) J. Biol. Chem. 276, 23888–23894 [DOI] [PubMed] [Google Scholar]
- 12.Li H., Guo W., Mellor R. L., Nerbonne J. M. (2005) J. Mol. Cell Cardiol. 39, 121–132 [DOI] [PubMed] [Google Scholar]
- 13.Schrader L. A., Birnbaum S. G., Nadin B. M., Ren Y., Bui D., Anderson A. E., Sweatt J. D. (2006) Am. J. Physiol. Cell Physiol. 290, C852–C861 [DOI] [PubMed] [Google Scholar]
- 14.Salvador-Recatalà V., Gallin W. J., Abbruzzese J., Ruben P. C., Spencer A. N. (2006) J. Exp. Biol. 209, 731–747 [DOI] [PubMed] [Google Scholar]
- 15.Callsen B., Isbrandt D., Sauter K., Hartmann L. S., Pongs O., Bähring R. (2005) J. Physiol. 568, 397–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pioletti M., Findeisen F., Hura G. L., Minor D. L., Jr. (2006) Nat. Struct. Mol. Biol. 13, 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. (2007) Nat. Neurosci. 10, 32–39 [DOI] [PubMed] [Google Scholar]
- 18.Shibata R., Misonou H., Campomanes C. R., Anderson A. E., Schrader L. A., Doliveira L. C., Carroll K. I., Sweatt J. D., Rhodes K. J., Trimmer J. S. (2003) J. Biol. Chem. 278, 36445–36454 [DOI] [PubMed] [Google Scholar]
- 19.Ma D., Jan L. Y. (2002) Curr. Opin. Neurobiol. 12, 287–292 [DOI] [PubMed] [Google Scholar]
- 20.Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. (1999) Neuron 22, 537–548 [DOI] [PubMed] [Google Scholar]
- 21.Hasdemir B., Fitzgerald D. J., Prior I. A., Tepikin A. V., Burgoyne R. D. (2005) J. Cell Biol. 171, 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manganas L. N., Trimmer J. S. (2000) J. Biol. Chem. 275, 29685–29693 [DOI] [PubMed] [Google Scholar]
- 23.Shi G., Nakahira K., Hammond S., Rhodes K. J., Schechter L. E., Trimmer J. S. (1996) Neuron 16, 843–852 [DOI] [PubMed] [Google Scholar]
- 24.Kuo H. C., Cheng C. F., Clark R. B., Lin J. J., Lin J. L., Hoshijima M., Nguyêñ-Trân V. T., Gu Y., Ikeda Y., Chu P. H., Ross J., Giles W. R., Chien K. R. (2001) Cell 107, 801–813 [DOI] [PubMed] [Google Scholar]
- 25.Menegola M., Trimmer J. S. (2006) J. Neurosci. 26, 12137–12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 27.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 28.Johns D. C., Marx R., Mains R. E., O'Rourke B., Marbán E. (1999) J. Neurosci. 19, 1691–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelan M. C. (2007) Basic Techniques in Cell Tissue Culture, Current Protocols in Cell Biology, Chapter 1, Unit 1.1, Wiley Interscience, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 30.Marionneau C., Brunet S., Flagg T. P., Pilgram T. K., Demolombe S., Nerbonne J. M. (2008) Circ. Res. 102, 1406–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck E. J., Bowlby M., An W. F., Rhodes K. J., Covarrubias M. (2002) J. Physiol. 538, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X. R., Wulf A., Schwarz M., Isbrandt D., Pongs O. (1999) Receptors Channels 6, 387–400 [PubMed] [Google Scholar]
- 33.van Renswoude J., Bridges K. R., Harford J. B., Klausner R. D. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 6186–6190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han W., Nattel S., Noguchi T., Shrier A. (2006) J. Biol. Chem. 281, 27134–27144 [DOI] [PubMed] [Google Scholar]
- 35.Hatano N., Ohya S., Imaizumi Y. (2002) Pfluegers Arch. Eur. J. Physiol. 444, 80–88 [DOI] [PubMed] [Google Scholar]
- 36.Bähring R., Boland L. M., Varghese A., Gebauer M., Pongs O. (2001) J. Physiol. 535, 65–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M. N., Joiner W. J., Dean T., Yue Z., Smith C. J., Chen D., Hoshi T., Sehgal A., Koh K. (2010) Nat. Neurosci. 13, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.