Abstract
Cellular eukaryotic mRNAs are capped at their 5′ ends with a 7-methylguanosine nucleotide, a structural feature that has been shown to be important for conferring mRNA stability, stimulating mRNA biogenesis (splicing, poly(A) addition, nucleocytoplasmic transport), and increasing translational efficiency. Whereas yeast mRNAs have no additional modifications to the cap, called cap0, higher eukaryotes are methylated at the 2′-O-ribose of the first or the first and second transcribed nucleotides, called cap1 and cap2, respectively. In the present study, we identify the methyltransferase responsible for cap1 formation in human cells, which we call hMTr1 (also known as FTSJD2 and ISG95). We show in vitro that hMTr1 catalyzes specific methylation of the 2′-O-ribose of the first nucleotide of a capped RNA transcript. Using siRNA-mediated knockdown of hMTr1 in HeLa cells, we demonstrate that hMTr1 is responsible for cap1 formation in vivo.
Keywords: mRNA, RNA, RNA Methylation, RNA Methyltransferase, RNA Modification, CAP1, mRNA cap
Introduction
Eukaryotic cytoplasmic mRNAs are capped at their 5′ end with a 7-methylguanosine (m7GpppN, where N is any nucleotide) (1). This cap is important for mRNA stability and for efficient translation initiation (for reviews, see Refs. 2, 3). The capping of mRNAs is a co-transcriptional process that requires RNA triphosphatase, RNA guanylyltransferase, and RNA methyltransferase activities (4, 5). In higher eukaryotes, the RNA triphosphatase and RNA guanylyltransferase reactions are carried out by a single bifunctional protein, whereas the methyltransferase reaction depends on a separate enzyme. The 5′ cap structure in yeast has no additional modification and is referred to as cap0 (6). However, in higher eukaryotes, the cap structure contains 2′-O-ribose methylations at either the first nucleotide (named cap1) or the first and second nucleotide (cap2) (7, 8) (Fig. 1A). When the first nucleotide of the transcript is an adenosine, the base can also be methylated at the N6 position (9). Viruses that replicate in the cytoplasm, such as vesicular stomatitis virus (10), coronavirus (11), and West Nile virus (12), often encode their own capping enzymes as well as a cap 2′-O-ribose methyltransferase activity.
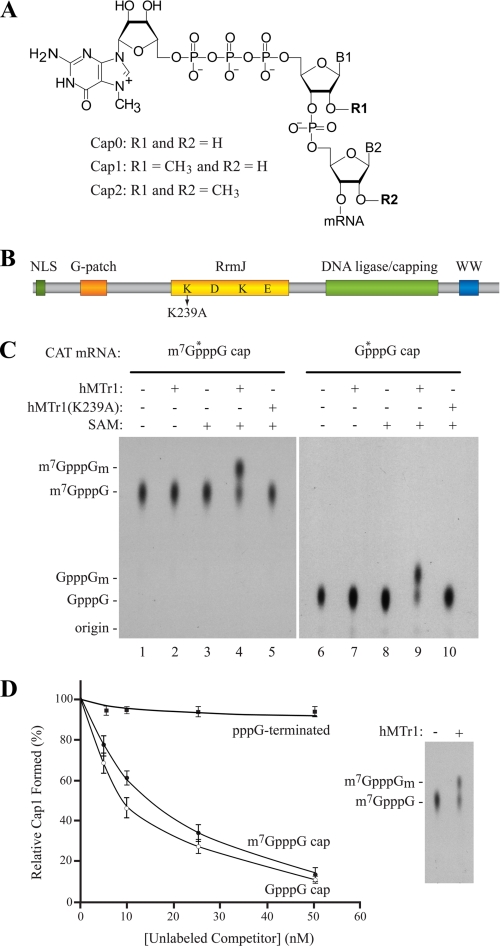
FIGURE 1.
hMTr1 methylates capped mRNA in vitro. A, structure of the 5′-cap in eukaryotic mRNA. B, schematic representation of hMTr1. Predicted domains include a nuclear localization signal (NLS), a G patch RNA interaction domain, am RrmJ homology 2′-O-ribose methyltransferase domain, a putative DNA ligase/capping domain, and a WW protein interaction domain. K239 denotes a point mutation in the conserved KDKE catalytic tetrad. C, hMTr1-methylated mRNAs containing m7GpppG or GpppG cap structures. In vitro transcribed CAT mRNA was capped with [α-32P]GTP and treated with purified recombinant hMTr1 (wild type or K239A mutant) in the presence or absence of 50 μm SAM. Nuclease P1-digested products were analyzed by TLC. m7GpppG- (lanes 1–5) and GpppG- (lanes 6–10) terminated CAT mRNA were used as substrate. *, positions of the radiolabeled phosphate. Unlabeled cap structure analogs were analyzed in parallel, and their positions of migration were detected by UV shadowing and are indicated on the left. D, methylation of RNA by hMTr1 requiring a cap structure. Reactions were performed as in C, with the indicated amounts of unlabeled CAT mRNA competitors. The inset shows that the efficiency of conversion of m7GpppG to m7GpppGm was ∼50% in the absence of competitor.
The kinetoplastids such as Trypanosoma brucei are a special case. Their mRNAs are capped with a spliced leader sequence containing a total of seven methylations, including 2′-O-ribose methylations on the first four nucleotides (cap4) (13–15). The T. brucei 2′-O-ribose methyltransferases responsible for methylation of the first three positions have been identified and characterized (16–20). Knock out of the enzymes required for methylating nucleotides 2 and 3 was shown to decrease translation efficiency by 50% (21). In higher eukaryotes, cap ribose methylation was reported to improve translation during Xenopus oocyte maturation (22).
Although the existence of cap1 and cap2 have been known for >30 years, their role in mRNA biogenesis remains obscure, and the enzymes involved in their formation have not yet been identified in mammals. Cap1 and cap2 methyltransferase activity have been partially purified from HeLa cell extracts, but the responsible enzymes were never identified (23). In the present study, we identify and characterize the enzyme responsible for 2′-O-methylation of the mRNA first transcribed nucleotide. This protein had been cloned previously and is known as FtsJ methyltransferase domain-containing 2 (FTSJD2) and ISG95 (24). Herein, we attribute 2′-O-methyltransferase activity to this gene and demonstrate that it is required for cap1 formation in vivo.
MATERIALS AND METHODS
Cloning, Expression, and Purification of hMTr1
The cDNA clone of KIAA0082 was purchased from Open Biosystems and amplified by PCR using primers: hMTr1-NdeI (5′-GGCAGCCATATGAAGAGGAGAACTGACCCAGAATGC-3′) and hMTr1-XhoI (5′-TCCTTCATCCAGATGCACAGGGCCTAACTCGAGGATCCG-3′). The resulting product was cloned into the NdeI and XhoI restriction sites of pET-15b. His6-hMTr1 expression was induced in Escherichia coli BL21 (DE3/pLysS) with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 30 °C. Bacteria were sonicated in lysis buffer (PBS containing 1% Triton X-100, 50 μg/ml PMSF, and 5 mm imidazole). Bacterial lysate was centrifuged 30 min at 20,000 × g, the supernatant was collected, and the NaCl concentration was adjusted to 750 mm. The supernatant was incubated for 1 h at 4 °C with 1 ml of nickel-nitrilotriacetic acid resin (Qiagen) equilibrated with lysis buffer. After incubation, the flow-through was collected, and the resin was washed with 10 ml of wash buffer (10 mm Tris-HCl, pH 7.8, 500 mm NaCl, 1% Triton X-100, 50 μg/ml PMSF, and 10 mm imidazole), followed by a wash with 10 ml of wash buffer containing 100 mm NaCl. The column was eluted with 0.5-ml fractions of buffer 2 containing 100, 200, 300, and 500 mm imidazole. Fractions shown by SDS-PAGE analysis to contain hMTr1 were further purified on a heparin-Sepharose 6 Fast Flow (GE Healthcare) which had been equilibrated in A100 buffer (20 mm Tris-HCl, pH 7.5, 10% glycerol, 0.1 mm EDTA, 100 mm KCl, and 2 mm DTT). Samples were applied to the column, washed with 2 ml of A100, and eluted with a 100–500 mm NaCl linear gradient. Elutions of 0.5 ml were analyzed by SDS-PAGE, and fractions containing hMTr1 were combined and dialyzed against 20 mm Tris-HCl, pH 7.8, 100 mm NaCl, 15% glycerol and stored at −80 °C.
Preparation of RNA Substrate
CAT4 mRNA was transcribed in vitro from pSP-CAT plasmid linearized with BamHI using SP6 RNA polymerase, essentially as described previously (25). For unlabeled RNA, the use of the dinucleotide m7GpppG or GpppG in the transcription reaction allowed the generation of m7GpppG-terminated or GpppG-terminated mRNA. RNA was internally radiolabeled by including [α-32P]GTP in the transcription reaction mixture. For labeling of the cap structure, in vitro transcribed mRNA was capped and methylated with [α-32P]GTP and SAM using vaccinia guanylyltransferase, as described previously (25). RNA was purified by phenol/CHCl3 extraction, centrifugation through a G50-Sephadex spin column, followed by ethanol precipitation.
Synthesis of Cap Analogs
m7GpppGm was prepared as described earlier (26). GpppGm was obtained by a procedure analogous to that described for GpppG synthesis (27). To this aim 2′-O-methylguanosine was synthesized as previously described (28) and converted to its 5′-monophosphate by the Yoshikawa method (29). Then, the imidazole derivative of 2′-O-methylguanosine-5′-monophosphate was prepared and coupled to GDP (triethylammonium salt obtained from GDP/sodium salt; Sigma). GpppGm was isolated by ion exchange column chromatography and converted to the sodium salt as previously described (27). The product was homogeneous as revealed by HPLC in two systems (>95% purity). HPLC was performed on an Agilent Technologies Series 1200 apparatus. In system 1, Supelcosil LC-SAX1 ion exchange columns (25 cm) were run in a linear gradient of 0.6 m KH2PO4 in buffer A (6 mm KH2PO4 adjusted to pH 4.0 by acetic acid), over 20 min with a flow rate of 1 ml/min and monitored at 260 nm. The retention time for GpppGm was 5.8 min (for comparison, the retention time for GpppG is 5.2 min). In System 2, Supelcosil LC-18-T reverse phase column (25 cm) was run in a linear gradient of methanol from 0 to 20% in 0.1 m triethylammonium acetate, pH 7.0, over 30 min with flow rate of 1 ml/min, monitored at 260 nm. The retention time was 14.1 min (for comparison, the retention time for GpppG is 12.1 min).
In Vitro Methyltransferase Assays
Reactions were performed in 20 μl of 50 mm Tris-HCl, pH 7.9, 2 mm DTT, 50 μm SAM, and RNase inhibitors (NEB) containing 300 pmol of m7G-[32P]ppG-terminated CAT mRNA and 0.1 pmol of recombinant His6-hMTr1 enzyme for 30 min at 30 °C. RNA was extracted with phenol/CHCl3, ethanol-precipitated, and resuspended in 10 μl of 50 mm NaOAc, pH 5.3, containing 2 μg of nuclease P1 (Sigma), and the digestion was performed for 3 h at 37 °C. Samples were analyzed on TLC (PEI-Cellulose F) plates developed with 0.3 m (NH4)2SO4. For tobacco acid pyrophosphatase analysis, RNA pellets were resuspended in 1× tobacco acid pyrophosphatase buffer (Epicenter) containing 1 unit of tobacco acid pyrophosphatase (Epicenter) and incubated at 37 °C for 1 h. TLC plates were developed with 0.45 m (NH4)2SO4. For alkaline hydrolysis, RNA was incubated in 0.4 m KOH for 1 h at 65 °C. Samples were neutralized with HCl and analyzed on a 25% polyacrylamide-7 m urea gel electrophoresis.
Preparation of Cellular Extracts
HeLa cells were transfected with siGENOME SMART pool siRNA (Dharmacon) at a final concentration of 20 nm using Lipofectamine 2000, following the manufacturer's instructions (Invitrogen). Knockdown efficiencies typically averaged 90%. After 48 h, cells were trypsinized, washed in cold PBS, and resuspended in 600 μl of cold lysis buffer (10 mm Hepes-KOH, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 0.5 mm PMSF, 5 μg/ml aprotinin, and 5 μg/ml leupeptin) and incubated on ice for 15 min. Nonidet P-40 was added to a final concentration of 0.5%, samples were vortexed 10 s and centrifuged at 2000 × g for 30 s. The supernatant was collected as the cytoplasmic fraction, and the nuclear pellet was resuspended in 100 μl of nuclear extract buffer (20 mm Hepes-KOH, pH 7.9, 400 mm KCl, 1 mm DTT, 0.5 mm PMSF, 5 μg/ml aprotinin and 5 μg/ml leupeptin) and incubated for 15 min at 4 °C with rotation. The nuclear extract was centrifuged for 5 min at 14,000 × g at 4 °C and the supernatant collected. Extracts were analyzed by Western blotting. Samples were fractionated on a 10% SDS-PAGE, transferred to PVDF membrane (Bio-Rad), and blotted with antibodies to hMTr1 (KIAA0082; Bethyl Laboratory), tubulin (Sigma), and hnRNP A1 (Abcam). Detection was done using Western Lightning Plus ECL (PerkinElmer Life Sciences).
In Vivo RNA Labeling
HeLa cells transfected with siRNA as described above were labeled in 10-cm dishes at a cell confluence of ∼50–70%. Cells were washed twice with PBS, and medium was replaced with methionine-free DMEM (Invitrogen) containing 10% dialyzed serum, 30 μm adenine, 20 μm guanosine, 20 mm sodium formate, and 100 μCi of l-[methyl-3H]methionine (PerkinElmer Life Sciences). After 15 min at 37 °C/5% CO2, 10 μm unlabeled methionine was added to the medium, and labeling continued for 16 h. Total RNA was extracted with TRIzol (Invitrogen) and poly(A)+ mRNAs purified using oligo(dT)-cellulose (GE Healthcare) (30).
For DEAE anion exchange chromatography, 0.7 × 20-cm columns of DEAE (DE52; Whatman) were equilibrated in 50 mm Tris-HCl, pH 7.6, 50 mm NaCl following the manufacturer's instructions (Whatman). Elutions were performed with a linear 50–500 mm NaCl gradient (200 ml total). Fractions of 1.5 ml were collected, and 0.75 ml of each fraction was counted in 10 ml of Universol Mixture (MP Biochemicals) using an LS6500 scintillation counter (Beckman Coulter). Fractions containing the desired RNA fragments were combined and desalted on a DE52 column equilibrated in 10 mm ammonium bicarbonate. Samples were eluted with 1 m ammonium bicarbonate, lyophilized, resuspended in water, and analyzed by TLC.
RESULTS
Identification of a Human Cap1 2′-O-Ribose Methyltransferase
T. brucei cap1 2′-O-ribose methyltransferase (TbMTr1) was first identified following a database search for sequences similar to the E. coli FtsJ/RrmJ 2′-O-ribose methyltransferase (19, 31). In the same database search, the human sequence with the highest similarity was a gene of unknown function, referred to as FtsJ methyltransferase domain-containing 2 (FTSJD2 (KIAA0082)) (Fig. 1B). FTSJD2 is much larger than TbMTr1 (835 amino acids compared with 370 amino acids) and contains a nuclear localization signal, a G patch RNA-binding domain, a putative DNA ligase/mRNA-capping domain, and a WW protein interaction domain (Fig. 1B). We investigated the possibility that FTSJD2 is the human cap1 2′-O-ribose methyltransferase (hMTr1).
We cloned the coding sequence of hMTr1 for expression in E. coli (supplemental Fig. 1A) and tested the purified recombinant protein for cap1 methyltransferase activity in vitro. As substrate, we used an in vitro transcribed CAT mRNA containing a radiolabel in the γ-phosphate of the cap structure. This RNA was incubated with purified hMTr1 and the co-factor SAM. Samples were treated with nuclease P1 to degrade the RNA to single nucleotides, leaving the labeled cap dinucleotide, m7GpppG (supplemental Fig. 1B). Samples were analyzed by TLC to detect the presence of 2′-O-methyl cap (m7GpppGm) (Fig. 1C, left panel). Labeled cap fragment from RNA that had not been treated with hMTr1 or in which the methylation reaction lacked the methyl donor, SAM, yielded a cap structure that co-migrated with the m7GpppG cap analog (Fig. 1C, lanes 1–3). When the RNA was incubated with both recombinant hMTr1 and SAM, the migration of the cap fragment shifted to a position corresponding to that of the synthetic m7GpppGm cap analog (compare lane 4 with lane 1). We also engineered a mutant of hMTr1, K239A, in which we altered one of the residues of the conserved KDKE catalytic tetrad required for methyltransferase activity (Fig. 1B) (31). This mutant completely abolished the methyltransferase activity of hMTr1 (Fig. 1C, compare lane 5 with lane 4).
We investigated whether hMTr1 required a 7-methylguanosine cap structure for activity or could utilize a nonmethylated GpppG structure (Fig. 1C). In the absence of hMTr1 or SAM, the resulting cap fragment from the P1 digestion co-migrated with a synthetic GpppG marker (lanes 6–8). Methylation by hMTr1 and SAM resulted in the appearance of a new spot on the TLC that co-migrated with the GpppGm marker (lane 9). The hMTr1(K239A) mutant showed no activity (lane 10). To test the specificity of this reaction further, we performed in vitro methylation assays with radiolabeled m7GpppG-capped mRNA in the presence of unlabeled competitor mRNAs containing different 5′ termini (Fig. 1D). Unlabeled CAT mRNA containing either a m7GpppG- or GpppG-terminated mRNA competed with labeled substrate, whereas pppG-terminated mRNA failed to compete in this setting (Fig. 1D). These results indicate that hMTr1 has cap1 methyltransferase activity and that this activity does not require 7-methylation of the 5′-terminal guanosine in vitro.
To validate that cap methylation by hMTr1 is on the first transcribed nucleotide of the mRNA, in vitro methylation reactions were performed with m7GpppG or GpppG-terminated mRNAs, and the resulting products were divided into two sets. The first set was digested by nuclease P1 to monitor the efficiency of methylation reaction (Fig. 2A, lanes 1–4). hMTr1 converted cap0 to cap1 with both m7GpppG- and GpppG-terminated mRNAs as substrates (compare lanes 2 and 4 with 1 and 3, respectively). The second set was treated with tobacco acid pyrophosphatase, which cleaves the pyrophosphate bonds of the cap structure, releasing the terminal m7GMP or GMP containing the radiolabeled phosphate moiety (supplementalFig. 1B). TLC analysis revealed that migration of the terminal m7GMP or GMP group was not affected by hMTr1 (Fig. 2A, lanes 5–8). We also performed similar experiments using a CAT mRNA substrate internally labeled with [α-32P]GTP (Fig. 2A, lanes 9 and 10) or [α-32P]ATP (lanes 11 and 12). These results indicate that hMTr1 is unlikely to methylate the 5′-terminal cap structure and internal adenosine or guanosine nucleosides.
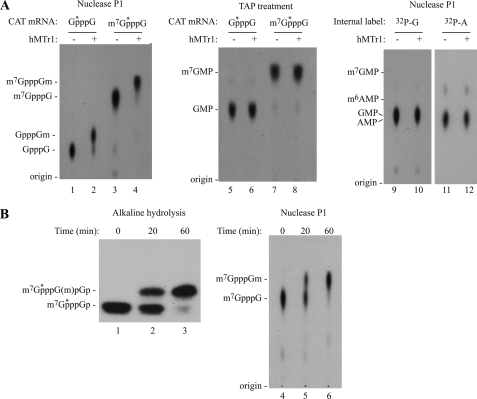
FIGURE 2.
hMTr1 methylates the 2′-O-ribose of the first transcribed nucleotide. A, hMTr1 does not methylate the terminal m7G moiety. Products from in vitro methylation reactions were treated either with nuclease P1 (lanes 1–4) or with tobacco acid pyrophosphatase (TAP; lanes 5–8) to release the terminal [32P]m7GMP or [32P]GMP. Internally labeled CAT mRNA was also used as substrate (lanes 9–12). Products were analyzed by TLC. Migration of unlabeled markers are indicated on the left. B, methylation of the first transcribed nucleotide by hMTr1 occurs on the 2′-O-ribose. Methylation reactions were performed for the indicated time, and products were degraded by complete alkaline hydrolysis and analyzed on a 25% PAGE-urea (lanes 1–3). Digestion of the products by nuclease P1 followed by TLC was performed in parallel (lanes 4–6).
To confirm that methylation by hMTr1 occurs on the 2′-O-ribose position of the first transcribed nucleotide, we performed in vitro methylation reactions on m7GpppG-terminated mRNA and digested the reaction products by alkaline hydrolysis followed by separation on a 25% polyacrylamide-8 m urea gel (Fig. 2B, lanes 1–3 and supplemental Fig. 1B). Alkaline hydrolysis of RNA requires a free 2′-OH to react with the neighboring 3′-phosphate and is therefore blocked by 2′-O-ribose methylation. Complete alkaline hydrolysis of a cap0 mRNA yields m7GpppGp, whereas hydrolysis of a cap1 transcript will yield m7GpppG(m)pGp (7). Treatment of CAT mRNA with hMTr1 for 20 or 60 min resulted in a shift of m7GpppGp by one nucleotide to m7GpppG(m)pGp (Fig. 2B, compare lanes 2 and 3 with lane 1). This indicates that the 2′-O-ribose of the first nucleotide is blocked. We did not see the appearance of any additional bands, indicating that the 2′-O-ribose of neighboring nucleotides is not blocked and that hMTr1 does not convert cap1 to cap2. Nuclease P1 digestion of these methylation reactions followed by thin layer chromatography showed that the appearance of a slower migrating product (lanes 2 and 3) coincides with the appearance of the cap1 dinucleotide (lanes 5 and 6). Taken together, these results indicate that hMTr1 catalyzes methylation of the 2′-O-ribose of the first transcribed nucleotide of a capped mRNA.
Biochemical Characterization of hMTr1 Activity
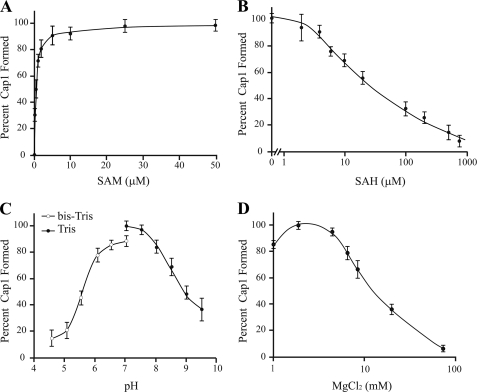
We next characterized the parameters for optimal hMTr1 activity in vitro (Fig. 3). Cap1 formation was optimal at 5 μm SAM, yielding a Kd of 1 μm (Fig. 3A). The activity of hMTr1 was inhibited by concentrations of S-adenosylhomocysteine beyond 2 μm (Fig. 3B). The optimal pH for the methylation reaction was found to be ∼7 (Fig. 3C). Previous studies with the T. brucei cap1 methyltransferase found that it requires magnesium for optimal activity but that it is inhibited above 5 mm (18). We found this also to be the case for hMTr1, which has an optimal activity at 2 mm MgCl2 (Fig. 3D).
FIGURE 3.
Biochemical characterization of hMTr1. A, activity of hMTr1 is dependent on SAM concentration. Reactions were performed with increasing SAM concentrations. Signals corresponding to cap0 and cap1 were quantified and expressed as a percentage of cap1 formed. B, activity of hMTr1 is inhibited by S-adenosylhomocysteine (SAH). Methylation reactions were performed in the presence of 5 μm SAM and increasing concentrations of S-adenosylhomocysteine. C, pH affects hMTr1 activity. Reactions were performed at the indicated pH in either Tris (filled circles) or bis-Tris (open circles). D, magnesium affects hMTr1 methyltransferase activity.
hMTr1 Is Responsible for Cap1 Formation in Vivo
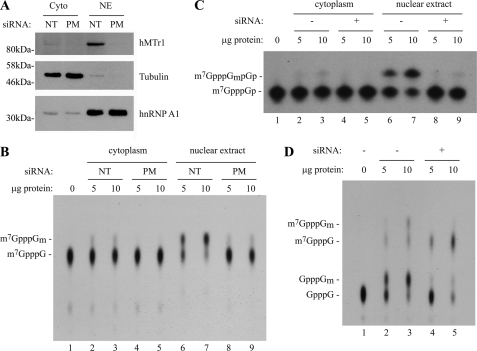
It was important to establish whether hMTr1 is responsible for Cap1 formation in vivo. We first used siRNA to knock down hMTr1 in HeLa cells and examined cap1 formation activity in cellular extracts. It was previously shown that cap1 methyltransferase activity is mostly nuclear (23). To confirm this, we prepared cytoplasmic and nuclear extracts from HeLa cells treated either with siRNAs against hMTr1 (perfect match) or with a nontargeting control siRNA (Fig. 4A). We observed that hMTr1 is present predominantly in the nuclear fraction (Fig. 4A). We next incubated in vitro transcribed m7GpppG-teminated CAT mRNA with cytoplasmic or nuclear extracts and assessed conversion of cap0 to cap1 by TLC separation of nuclease P1-digested products (Fig. 4B). As expected, cap1 methyltransferase activity was localized almost exclusively to the nuclear fraction (compare lanes 6 and 7 with lanes 2 and 3), and knockdown of hMTr1 by siRNA resulted in significant reduction of this activity (compare lanes 8 and 9 with lanes 6 and 7). We confirmed these results by analysis of alkaline hydrolysis of the RNA followed by polyacrylamide-urea gel analysis (Fig. 4C). Treatment of the RNA substrate with nuclear extracts resulted in a blocked 2′-O-ribose characteristic of a cap1 structure, which was abolished upon siRNA knockdown of hMTr1 (Fig. 4C). These results indicate that endogenous hMTr1 from cellular extracts has the same cap1 2′-O-ribose methyltransferase activity as the recombinant hMTr1 used above in our in vitro assays.
FIGURE 4.
Knockdown of hMTr1 in HeLa cells results in loss of cap1 methyltransferase activity. A, Western blots document subcellular fractionation and efficiency of hMTr1 knockdown. HeLa cells were treated with perfect match siRNAs directed against hMTr1 (PM) or with nontargeting control siRNAs (NT) and fractionated into cytoplasmic (Cyto) and nuclear extracts (NE). Western blotting was to detect the presence of hMTr1, tubulin (cytoplasmic), and hnRNP A1 (predominantly nuclear). B, extracts prepared from hMTr1 knockdown cells have reduced 2′-O-methyltransferase activity. Radiolabeled m7GpppG-terminated CAT mRNA was incubated in the presence of HeLa cytoplasmic or nuclear extracts. Formation of m7GpppGm was assessed by nuclease P1 digestion of the reaction products followed by TLC analysis. C, extracts prepared from hMTr1 knockdown cells have reduced 2′-O-methyltransferase activity. Reactions were the same as in B, except the reaction products were analyzed by alkaline hydrolysis and electrophoretic separation of the products on a 25% polyacrylamide-8 m urea gel. D, loss of 2′-O-methylation does not affect conversion of GpppG to m7GpppG. Radiolabeled GpppG-terminated CAT mRNA was incubated in the presence of HeLa nuclear extracts, and reaction products were digested with nuclease P1 and analyzed by TLC.
Upon incubation of GpppG-terminated mRNA substrate with HeLa nuclear extracts, we noted the conversion of GpppG to GpppGm as well as the presence of m7GpppG and m7GpppGm (Fig. 4D, compare lanes 2 and 3 with lane 1). Upon knockdown of hMTr1, we observed conversion of GpppG to m7GpppG but did not see further conversion to m7GpppGm (compare lanes 4 and 5 with lane 1). These results demonstrate that knockdown of hMTr1 abolishes cap1 formation specifically and confirm that this activity is independent of guanosine-7-methyltransferase activity.
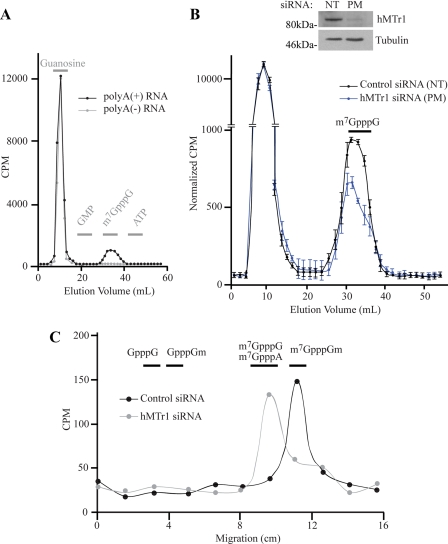
To determine whether hMTr1 is responsible for Cap1 formation on endogenous mRNA templates, we monitored cap1 formation in HeLa cells after in vivo labeling of RNA with l-[methyl-3H]methionine. This should result in the radiolabeling of all methylated nucleotides, internally on bases and on the ribose moieties. Poly(A)+ and poly(A)− RNA were purified from labeled cells and digested with nuclease P1 to generate single nucleotides and cap dinucleotides. Following dephosphorylation of samples, we performed DEAE anion exchange chromatography and observed that the bulk of the radiolabeled material eluted in the nucleoside fraction and corresponds to methylated nucleosides, whereas a second peak co-eluted with the m7GpppG marker (Fig. 5A). When the sample was not treated with phosphatase prior to DEAE chromatography, the first peak of single nucleotides co-migrated with GMP instead of guanosine, as expected for nucleoside monophosphates (data not shown). The second peak was not observed in samples prepared from poly(A)− RNA, consistent with it corresponding to the m7GpppN product specific to poly(A)+ mRNAs (Fig. 5A). In vivo labeling experiments were performed on cells in which hMTr1 had been knocked down by siRNAs or treated with a nontargeting control siRNA (Fig. 5B). RNAi-mediated knockdown of hMTr1 resulted in a decrease of radioactivity eluting at the m7GpppG marker position compared with cells treated with the control siRNA (Fig. 5B). This decrease is consistent with the loss of one methyl group from the m7GpppNm cap structure (for further details, see “Discussion”).
FIGURE 5.
hMTr1 is responsible for Cap1 formation on endogenous mRNAs in vivo. A, HeLa cells were labeled with l-[methyl-3H]methionine. Purified poly(A)+ mRNA and poly(A)− RNAs were treated with nuclease P1 followed by antartic phosphatase and analyzed by DEAE anion exchange chromatography. Gray bars represent the fractions in which unlabeled markers eluted (guanosine, GMP, m7GpppG, and ATP). B, knockdown of hMTr1 decreases incorporation of methyl-3H in the cap structure. HeLa cells were transfected with hMTr1 perfect match siRNA (PM) or a nontargeting control siRNA (NT) before labeling and DEAE analysis. Counts of each samples are normalized to an equivalent amount of total radioactivity. Values are average from three independent experiments. The inset is a Western blot showing the knockdown efficiency of hMTr1. C, knockdown of hMTr1 results in loss of cap1 2′-O-methylation of endogenous mRNAs. Fractions co-eluting with m7GpppG from B were combined and analyzed by TLC. Lanes were cut into 1.5-cm pieces and quantitated by scintillation counting. Horizontal bars denote the positions of various markers.
The fractions eluting at the m7GpppG marker position were pooled, desalted, and analyzed by TLC (Fig. 5C). Fractions from control siRNA-treated cells co-migrated with the m7GpppGm marker (Fig. 5C, black line). In contrast, the majority of the eluent from hMTr1 siRNA-treated cells co-migrated with m7GpppG and m7GpppA markers, with a slight shoulder extending into the m7GpppGm marker position (Fig. 5C). These results confirm that knockdown of hMTr1 causes loss of cap1 2′-O-ribose methylation of endogenous mRNAs in vivo. There was no detectable radioactivity co-migrating with the GpppG marker, indicating that loss of cap1 ribose methylation does not affect guanosine-7-methyltransferase activity.
DISCUSSION
The mRNA cap structure is a determinant of mRNA stability and important for stimulating translation efficiency (2, 3). Many data suggest that cap methylation can be an important control point in regulating gene expression at the level of translation initiation (32). For example, it was recently suggested that c-myc and E2F1 factors both promote N7 cap methylation of mRNAs from their target genes (33). However, the role of ribose methylations associated with cap1 and cap2 structures is not clear.
In the present study we identified and characterized the human cap1 2′-O-ribose methyltransferase, hMTr1. We demonstrate that hMTr1 catalyzes the methylation of the 2′-O-ribose of the first transcribed nucleoside of a capped RNA. The methyltransferase activity of hMTr1 was observed whether the mRNA was capped with a GpppG or m7GpppG moiety and is consistent with similar experiments from previous studies using partially purified cap1 activity from HeLa extracts (23). TbMTr1, the T. brucei homolog of hMTr1, also has no preference for terminal guanosine N7 methylation of the cap structures (18).
It is not yet clear whether cap1 ribose methylation occurs before or after 7-methylation of the 5′-terminal guanosine in vivo. Our own data show that cap1 methylation does not impair 7-methylguanosine formation (Figs. 4D and 5C). The capping enzyme and the guanosine-7-methyltransferase are recruited to the C-terminal domain of RNA polymerase II during transcription, and capping is a co-transcriptional process (34–36). The product encoded by the hMTr1 gene has been previously described (called ISG95), and although its cap1 2′-O-ribose methyltransferase activity for RNA was not investigated, it was noted also to associate with the C-terminal domain of RNA polymerase II (24). Hence, it is plausible that cap1 methylation also occurs co-transcriptionally. In the case of vesicular stomatitis virus, both N-7-methyl and 2′-O-methyl formation are catalyzed by a single multifunctional polymerase protein (L protein) (10, 37), and in that case, ribose methylation precedes and facilitates m7G formation (37). The order of events for higher eukaryotes is not known, but previous reports showed that it need not happen in any specific order (23).
Importantly, we showed that hMTr1 is responsible for cap0 to cap1 conversion in vivo. Knockdown of hMTr1 by siRNA abolished cap1 2′-O-ribose methyltransferase activity in HeLa nuclear extracts but did not affect guanosine-7-methyltransferase activity (Fig. 4D). We used [methyl -3H]methionine labeling of RNA to show that knockdown of hMTr1 eliminates cap1 formation in vivo. In samples prepared from cells treated with hMTr1 siRNA, the relative amount of radioactivity in the cap dinucleotide was 70% of that in the control samples (Fig. 5B).
Cap dinucleotides in mammals contain either 2 or 3 methylations, including the 7-methylguanosine and the 2′-O-methyl of the first transcribed nucleotide. In mammals, the penultimate position can be any of the four bases, but in 46% of cases it is an N-6-methyladenosine (8). Taking into account the presence of this methyl group, then loss of the 2′-O-methyl group will cause a theoretical loss of label in the cap dinucleotide population of ∼40% (1/2.5) of control. We therefore conclude that our observation of a 30% reduction in cap methyl label upon hMTr1 knockdown is consistent with an almost complete loss of cap1 methylation. We observed no cap0 in human poly(A)+ mRNAs in the absence of hMTr1 knockdown (Fig. 5C), which is in agreement with previous reports (7, 8).
The mRNA encoding hMTr1 is up-regulated in human cells during vaccinia virus infection (38) and upon interferon treatment (39). However, up-regulation of this gene during viral infection is transient and relatively modest with a maximum 2-fold increase at 6-h infection with vaccinia. We tested the consequences of hMTr1 knockdown on influenza replication and did not detect any significant effect on viral replication (data not shown). The consequences of an increase in hMTr1 expression levels from these studies are difficult to ascertain because all mammalian mRNAs should contain either cap1 or cap2 after their synthesis (7, 8), so an increase in cap1 methyltransferase activity would not be expected to result in more cap1 methylation. An increase in hMTr1 levels is unlikely to affect viral mRNA processing because vaccinia virus replicates in the cytoplasm and encodes its own cap1 methyltransferase (40). hMTr1 contains a WW domain for protein-protein interactions and can methylate proteins in vitro (24), indicating that additional activities of hMTr1 may exist and be affected by altered expression levels.
The functional role of cap1 and cap2 2′-O-ribose methylation remains elusive. In vitro experiments indicated that cap1 has only a modest effect on ribosome recruitment (41). In Xenopus oocyte, progesterone treatment induced cap1 and cap2 methylation of exogenously injected mRNA that was dependent on polyadenylation (42). The same group showed that progesterone induces cap1 and cap2 methylation on injected c-mos mRNA and that 5′-S-isobutyladenosine, a methyltransferase inhibitor, prevented these methylations and also resulted in decreased expression of endogenous Mos (22). However, 5′-S-isobutyladenosine is a panmethyltransferase inhibitor and thus not very selective in its mode of action (43). In Trypanosoma, gene deletion of the 2′-O-ribose methyltransferases targeting positions 2 and 3 of the cap4 structure, causes a 50% decrease in translation rates (21). In contrast, deletion of TbMTr1, the cap1 methyltransferase in T. brucei, had no effect on protein synthesis, indicating that ribose methylation of the first transcribed nucleotide does not alter translational efficiency in Trypanosoma.
We examined the effect of knockdown of hMTr1 on global protein synthesis using [35S]methionine incorporation, under normal or stress conditions, and did not detect any significant effect (data not shown). The enzyme responsible for cap2 formation in mammals remains to be identified. Unlike cap1 methylation, cap2 formation is thought to be cytoplasmic (23). It is possible that loss of both cap1 and cap2 methylations is required to reveal an effect on mRNA activity. Roughly half of the mRNA population has cap2 structures (7, 8), but the identity of those mRNAs (if there is mRNA bias) is unknown. Our results demonstrate that the absence of cap1 modification does not impart a significant disadvantage to an mRNA ability to translate in vivo. In vivo knockdown of hMTr1 indicates that large reductions in hMTr1 levels do not affect overall cellular viability (data not shown).
The control of mRNA turnover is just as important to gene expression as translational control, and it is possible that cap1 and cap2 methylations could affect mRNA degradation. The Dcp1-Dcp2-decapping enzyme interacts with the body of the mRNA in addition to the 7-methylguanosine moiety (44), but it is not known whether cap ribose methylation affects these interactions. In humans, Dcp2 binds more efficiently to a subset of mRNAs (45), and it would be worthwhile to know whether this correlates with, or can be regulated by, the cap methylation status of those messengers.
The identification and characterization of hMTr1 is a first step in understanding the potential role of this post-transcriptional modification in gene regulation. Genetic models are currently being generated to address the essentiality of hMTr1 in development and differentiation.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Grant MOP-79385 (to J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- CAT
- chloramphenicol acetyltransferase
- bis-Tris
- bis(2-hydroxyethyl)amino-Tris(hydroxymethyl)methane
- SAM
- S-adenosyl-l-methionine.
REFERENCES
- 1.Shatkin A. J. (1976) Cell 9, 645–653 [DOI] [PubMed] [Google Scholar]
- 2.Gu M., Lima C. D. (2005) Curr. Opin. Struct. Biol. 15, 99–106 [DOI] [PubMed] [Google Scholar]
- 3.Shuman S. (2002) Nat. Rev. Mol. Cell. Biol. 3, 619–625 [DOI] [PubMed] [Google Scholar]
- 4.Ensinger M. J., Moss B. (1976) J. Biol. Chem. 251, 5283–5291 [PubMed] [Google Scholar]
- 5.Venkatesan S., Gershowitz A., Moss B. (1980) J. Biol. Chem. 255, 2829–2834 [PubMed] [Google Scholar]
- 6.Sripati C. E., Groner Y., Warner J. R. (1976) J. Biol. Chem. 251, 2898–2904 [PubMed] [Google Scholar]
- 7.Wei C. M., Gershowitz A., Moss B. (1975) Cell 4, 379–386 [DOI] [PubMed] [Google Scholar]
- 8.Furuichi Y., Morgan M., Shatkin A. J., Jelinek W., Salditt-Georgieff M., Darnell J. E. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 1904–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith J. M., Ensinger M. J., Mose B. (1978) J. Biol. Chem. 253, 5033–5039 [PubMed] [Google Scholar]
- 10.Galloway S. E., Richardson P. E., Wertz G. W. (2008) Virology 382, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A. E., Snijder E. J., Canard B. (2008) J. Virol. 82, 8071–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T. S., Zhou Y., Li H., Shi P. Y. (2006) J. Virol. 80, 8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bangs J. D., Crain P. F., Hashizume T., McCloskey J. A., Boothroyd J. C. (1992) J. Biol. Chem. 267, 9805–9815 [PubMed] [Google Scholar]
- 14.Freistadt M. S., Cross G. A., Robertson H. D. (1988) J. Biol. Chem. 263, 15071–15075 [PubMed] [Google Scholar]
- 15.Perry K. L., Watkins K. P., Agabian N. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8190–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arhin G. K., Ullu E., Tschudi C. (2006) Mol. Biochem. Parasitol. 147, 137–139 [DOI] [PubMed] [Google Scholar]
- 17.Hall M. P., Ho C. K. (2006) Nucleic Acids Res. 34, 5594–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mittra B., Zamudio J. R., Bujnicki J. M., Stepinski J., Darzynkiewicz E., Campbell D. A., Sturm N. R. (2008) J. Biol. Chem. 283, 3161–3172 [DOI] [PubMed] [Google Scholar]
- 19.Zamudio J. R., Mittra B., Foldynová-Trantírková S., Zeiner G. M., Lukes J., Bujnicki J. M., Sturm N. R., Campbell D. A. (2007) Mol. Cell. Biol. 27, 6084–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zamudio J. R., Mittra B., Zeiner G. M., Feder M., Bujnicki J. M., Sturm N. R., Campbell D. A. (2006) Eukaryot. Cell 5, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zamudio J. R., Mittra B., Campbell D. A., Sturm N. R. (2009) Mol. Microbiol. 72, 1100–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuge H., Brownlee G. G., Gershon P. D., Richter J. D. (1998) Nucleic Acids Res. 26, 3208–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langberg S. R., Moss B. (1981) J. Biol. Chem. 256, 10054–10060 [PubMed] [Google Scholar]
- 24.Haline-Vaz T., Silva T. C., Zanchin N. I. (2008) Biochem. Biophys. Res. Commun. 372, 719–724 [DOI] [PubMed] [Google Scholar]
- 25.Pelletier J., Sonenberg N. (1985) Mol. Cell. Biol. 5, 3222–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jankowska M., Stepinski J., Stolarski R., Wieczorek Z., Temeriusz A., Haber D., Darzynkiewicz E. (1996) Collect. Czech. Chem. Commun. 81, S197–S202 [Google Scholar]
- 27.Niedzwiecka A., Stepinski J., Antosiewicz J. M., Darzynkiewicz E., Stolarski R. (2007) Methods Enzymol. 430, 209–245 [DOI] [PubMed] [Google Scholar]
- 28.Kusmierek J., Shugar D. (1978) Nucleic Acids Res. Special Publ. 4, S73–S77 [Google Scholar]
- 29.Yoshikawa M., Kato T., Takenishi T. (1967) Tetrahedron Lett. 50, 5065–5068 [DOI] [PubMed] [Google Scholar]
- 30.Aviv H., Leder P. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feder M., Pas J., Wyrwicz L. S., Bujnicki J. M. (2003) Gene 302, 129–138 [DOI] [PubMed] [Google Scholar]
- 32.Schwer B., Saha N., Mao X., Chen H. W., Shuman S. (2000) Genetics 155, 1561–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole M. D., Cowling V. H. (2009) Oncogene 28, 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bentley D. L. (2005) Curr. Opin. Cell Biol. 17, 251–256 [DOI] [PubMed] [Google Scholar]
- 35.Pei Y., Schwer B., Shuman S. (2003) J. Biol. Chem. 278, 7180–7188 [DOI] [PubMed] [Google Scholar]
- 36.Schroeder S. C., Schwer B., Shuman S., Bentley D. (2000) Genes Dev. 14, 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahmeh A. A., Li J., Kranzusch P. J., Whelan S. P. (2009) J. Virol. 83, 11043–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerra S., López-Fernández L. A., Pascual-Montano A., Muñoz M., Harshman K., Esteban M. (2003) J. Virol. 77, 6493–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiss G. K., Carter V. S., He Y., Kwieciszewski B. K., Holzman T., Korth M. J., Lazaro C. A., Fausto N., Bumgarner R. E., Katze M. G. (2003) J. Virol. 77, 6367–6375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnierle B. S., Gershon P. D., Moss B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2897–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthukrishnan S., Moss B., Cooper J. A., Maxwell E. S. (1978) J. Biol. Chem. 253, 1710–1715 [PubMed] [Google Scholar]
- 42.Kuge H., Richter J. D. (1995) EMBO J. 14, 6301–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquemont B., Huppert J. (1977) J. Virol. 22, 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker R., Song H. (2004) Nat. Struct. Mol. Biol. 11, 121–127 [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Song M. G., Kiledjian M. (2008) Mol. Cell. Biol. 28, 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.