Abstract
PA1b (pea albumin 1, subunit b) is a small and compact 37-amino acid protein, isolated from pea seeds (Pisum sativum), that adopts a cystine knot fold. It acts as a potent insecticidal agent against major pests in stored crops and vegetables, making it a promising bioinsecticide. Here, we investigate the influence of individual residues on the structure and bioactivity of PA1b. A collection of 13 PA1b mutants was successfully chemically synthesized in which the residues involved in the definition of PA1b amphiphilic and electrostatic characteristics were individually replaced with an alanine. The three-dimensional structure of PA1b was outstandingly tolerant of modifications. Remarkably, receptor binding and insecticidal activities were both dependent on common well defined clusters of residues located on one single face of the toxin, with Phe-10, Arg-21, Ile-23, and Leu-27 being key residues of the binding interaction. The inactivity of the mutants is clearly due to a change in the nature of the side chain rather than to a side effect, such as misfolding or degradation of the peptide, in the insect digestive tract. We have shown that a hydrophobic patch is the putative site of the interaction of PA1b with its binding site. Overall, the mutagenesis data provide major insights into the functional elements responsible for PA1b entomotoxic properties and give some clues toward a better understanding of the PA1b mode of action.
Keywords: Disulfide, Insect, Mutant, NMR, Peptide Chemical Synthesis, Plant, Protein Structure, Biopesticides, Knottin, Structure/Function
Introduction
The recent isolation of peptides that are toxic for insect pests in stored vegetables and crops has enlarged the possibilities for cereal grain protection (1, 2). These peptides could be a valuable alternative to chemical pesticides, reducing concern about the toxicity of remainders. PA1b (pea albumin 1, subunit b), extracted from pea seeds (3), induces mortality in several pests, including cereal weevils (Sitophilus sp.), which are the major pests of cereals (1, 3, 4). Moreover, PA1b has also been described as being involved in signal transduction, under the name leginsulin (5) or aglycin (6).
PA1b is a cysteine-rich peptide consisting of 37 amino acids. Some sequence variations have been characterized in the PA1b homologues (3, 4, 7, 8). Except for these homologues, the sequence of this protein does not share any similarity with those of other known toxins. The three-dimensional structure, resolved from the extracted peptide (9), shows that PA1b adopts a typical knottin fold with a triple-stranded antiparallel β-sheet and three buried interlocked disulfide bonds, a signature of the inhibitory cystine knot family (10, 11). The ring embedded in the structure, through which the third disulfide bond threads, is composed of eight residues. Such a topological arrangement of the disulfides confers high stability to PA1b, which has proven to be stable to attack by numerous proteases, as well as to pH and temperature changes (1, 12). Moreover, it exposes hydrophobic residues at the molecular surface that are well conserved residues among PA1b homologues. The significant similarities with the cystine knot toxin ACTX-Hi-OB4219, from an Australian spider (13) in terms of amphiphilic character and electrostatic surface potentials, as well as sequence similarities in presumably functionally relevant loops, may be indicative of a neighboring mode of action, possibly as an ion channel blocker (9).
A screening of up to 90 cereal weevil strains for their susceptibility to PA1b has revealed that four strains of the Sitophilus oryzae species are fully resistant to the toxin and that a single gene is responsible for this resistance (14). A proteinaceous saturable and reversible binding site for PA1b has been identified in the microsomes of S. oryzae, with a specific affinity in the nanomolar range (15). The binding site was present in all susceptible species and strains within the insect orders, including Coleoptera, Diptera, and Hemiptera, and in Sf9 cells but was not detectable in extracts from the four resistant S. oryzae strains (12, 15). This wide distribution might indicate a conservation of the protein-binding site among insects. It has also been shown that PA1b is internalized into the Sf9 insect cell after binding, and this binding site-mediated entry of the peptide would be a mechanism for detoxification (16).
A better understanding of the entomotoxin structure-activity relationships would help reveal the mechanism(s) mediating PA1b bioactivity. Hence, in the present study, we analyzed a collection of chemically synthesized alanine mutants for structural and functional perturbations relative to native PA1b and determined their activities against weevils and Sf9 insect cells. This approach provided significant insights into the functional elements responsible for the entomotoxic properties of PA1b.
EXPERIMENTAL PROCEDURES
Synthesis and Purification of Peptides
The primary sequence of the synthetic PA1b (ASCNGVCSPFEMPPCGTSACRCIPVGLVIGYCRNPSG) is identical to the sequence of the 3741 Da (average molecular mass) isoform purified from pea seeds (15, 16). Alanine mutants were synthesized and folded following the optimized procedure described for the production of synthetic PA1b, using solid-phase peptide methods and the Fmoc/tBu (N-(9-fluorenyl)methoxycarbonyl/tert-butyl) strategy (17). The synthetic mutants were purified by semipreparative RP-HPLC.2 The purity of the peptides was assessed using RP-HPLC and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. The peptide concentrations were determined using UV spectrophotometry at 274 nm (ϵTyr: 1420 m−1 cm−1). The concentration of the Y31A variant was determined by measuring its RP-HPLC peak area at 210 nm, referring to known quantities of pure peptides used as standards.
NMR Spectroscopy and Structure Calculation
Various amounts of the synthetic peptides were dissolved in 330 μl of a mixture of trifluoroethanol and H2O, in a 1:1 ratio (v/v) and 0.02% NaN3, leading to protein concentrations ranging from 0.5 to 3 mm. The final pH of the solutions, adjusted with minute increments of 1 n HCl, was 4.7. 1H-NMR spectra were recorded on a 600-MHz Varian INOVA NMR spectrometer equipped with a z axis field gradient unit. NOESY (18), clean total correlation spectroscopy (19), and DQF-COSY (20) experiments were performed at 293 K. All spectra were referenced to internal 2,2-dimethyl-2-silapentanesulfonic acid at 0 ppm. The data were processed using NMRPipe/NMRDraw (21) and analyzed within NmrView (22).
The three-dimensional structures of the inactive mutants F10A and R21A and of the active mutant R33A were determined with the support of ARIA 1.1 (23). Because the disulfide pairing was already known, covalent bonds were built between the sulfur atoms of the paired cysteines. For the F10A and R21A mutants, dihedral angle restraints derived from Talos (24) were applied to ϕ, ψ backbone angles. The calculations were initiated using the default parameters of ARIA and a first set of easily assigned NOEs. At the end of each run, the new assignments proposed by ARIA were checked manually and introduced (or not) into the subsequent calculation. This iterative process was repeated until the assignment of the NOESY map was complete. In the last run, 100 structures were calculated using the final list of NOE-derived distance restraints, of which 10 structures, in very close agreement with all the experimental data and the standard covalent geometry, were considered as being characteristic of the peptide structure.
Biological Tests
The affinity of synthetic PA1b and its mutants for the PA1b-binding site was determined by ligand binding using 125I-toxin (15). Toxicity assays were performed both on the susceptible WAA42 and on the resistant ISOR3 strains of S. oryzae. The insects used for bioassays were adults aged 2–3 weeks, collected from experimental 1-week cohorts and deposited in batches of 30 individuals (for each WAA42 and ISOR3 strain) on food pellets incorporating the tested synthetic PA1b in a whole wheat-based diet (1). The concentration of PA1b mutant varied from 400 to 4000 μg/g of food. Bioactivity was evaluated by scoring daily insect survival during the first 2 weeks of contact with the test food (27.5 °C, 70% room humidity) and by standard survival analysis. The data were analyzed using the free software Simfit, allowing the LT50 values (lethal time 50%, or median life duration) to be calculated. A toxicity assay was also performed on cultured Sf9 insect cells using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide protocol (16).
Peptide Degradation Assays
Enzymatic digests were identical to those described previously (12), using the enzymes trypsin, chymotrypsin, proteinase K, Pronase E, and papain (Sigma).
RESULTS
Synthesis and Structural Characterization of PA1b Mutants
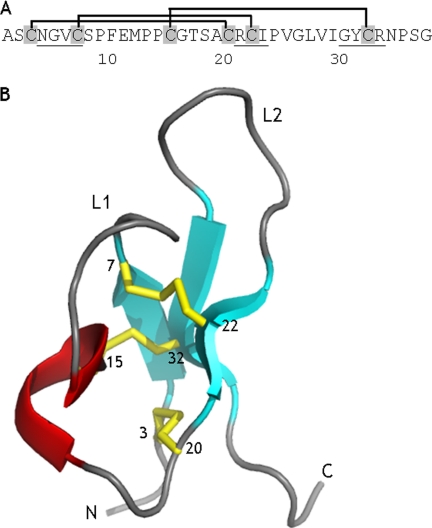
A former sequence analysis of several PA1b isoforms from different origins showed that two regions are well conserved: the N-terminal part of loop L1, spanning residues 7–11 (CSPFE), and loop L2 24–29, where the overall hydrophobicity of the residues is conserved (3) (Fig. 1). We decided to chemically synthesize a collection of PA1b mutants, with mutated residues mainly located in these two loops and/or with involvement in the definition of the PA1b amphiphilic and electrostatic characteristics. Thus, mutated residues were either in or surrounding loop L1 (Val-6, Ser-8, Phe-10, Glu-11, and Met-12) and loop L2 (Ile-23, Val-25, Leu-27, Val-28, Ile-29, and Tyr-31), and the three charged residues were mutated (Glu-11 in loop L1, Arg-21 and Arg-33). Regarding the crucial role of the three-dimensional structure in toxin activity and their very high level of conservation among PA1b isoforms, the residues that are major determinants of this structure (the six cysteines, the five prolines, and the five glycines) were not mutated.
FIGURE 1.
Sequence and three-dimensional structure of PA1b. A, primary structure of PA1b. The network of disulfide bonds is included. Residues found to be part of the triple-stranded antiparallel β-sheet in the solution structure of the pea-extracted peptide are underlined. B, ribbon representation of PA1b three-dimensional structure. The cystine knot motif is characterized by three disulfide bonds, shown as yellow sticks, and by the triple-stranded antiparallel β-sheet (cyan arrows). The numbers of the cysteine residues and the two loops L1 and L2 are also indicated.
Synthetic PA1b and its 13 alanine mutants (Table 1) were achieved by solid phase peptide synthesis. After cleavage from the resin, the reduced peptides were engaged in the in vitro oxidative folding step following an optimized procedure, and the oxidized form was purified by HPLC (17). The oxidative folding yields of the alanine mutants (about 60% of folded peptide) were similar to that of the synthetic native PA1b. The mutants were checked by mass spectrometry. All of them have molecular masses corresponding to the theoretical masses of the mutants harboring the three disulfide bridges (supplemental Table S1).
TABLE 1.
Affinity to the PA1b binding site and insecticidal activities of PA1b mutants
ND, not determined.
| Mutant | Kia | LT50b |
LD50c | ||
|---|---|---|---|---|---|
| (400 μg/g) | (1000 μg/g) | (4000 μg/g) | |||
| nm | days | μm | |||
| WT | 8 ± 1 | 8.2 ± 0.6 | 4.8 ± 0.3 | ND | 0.48 ± 0.12 |
| V6A | 8 ± 2 | 7.3 ± 0.7 | 5.3 ± 0.3 | 4.7 ± 0.3 | 1.81 ± 0.14 |
| S8A | 102 ± 9 | 11.3 ± 0.9 | 8.4 ± 1.0 | ND | 4.60 ± 1.70 |
| F10A | >5000 | >14 | >14 | >14 | >50 |
| E11A | 13 ± 3 | 13.7 ± 1.1 | ND | ND | 1.09 ± 0.52 |
| M12A | 10 ± 1 | 7.2 ± 0.6 | ND | ND | 0.62 ± 0.38 |
| R21A | >4093 | >14 | >14 | 12.3 ± 0.9 | >50 |
| I23A | 1640 ± 123 | >14 | >14 | >14 | >50 |
| V25A | 28 ± 3 | 7.8 ± 0.7 | 6.3 ± 0.4 | ND | 0.53 ± 0.25 |
| L27A | 420 ± 53 | >14 | >14 | >14 | >50 |
| V28A | 72 ± 7 | 8.5 ± 0.9 | 6.2 ± 0.8 | ND | 3.60 ± 0.90 |
| I29A | 79 ± 12 | 10.2 ± 0.9 | 7.9 ± 0.9 | ND | 5.58 ± 1.37 |
| Y31A | 7 ± 1 | 8.5 ± 0.7 | 6.9 ± 0.8 | ND | 1.13 ± 0.36 |
| R33A | 58 ± 17 | 6.5 ± 0.3 | 5.8 ± 0.4 | 4.2 ± 0.3 | 0.45 ± 0.24 |
a The Ki of PA1b and its mutants was determined by ligand binding using 125I-PA1b, according to Gressent et al. 15.
b LT50 deduced from toxicity assays performed on the susceptible WAA42 strain and the resistant ISOR3 strain for concentrations of PA1b mutants varying from 400 to 4000 μg/g of food. LT50 values on the resistant ISOR3 strain are not indicated and are all greater than 14 days.
c LD50 calculated from biological assays performed on cultured Sf9 cells using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide protocol, according to Rahioui et al. 16.
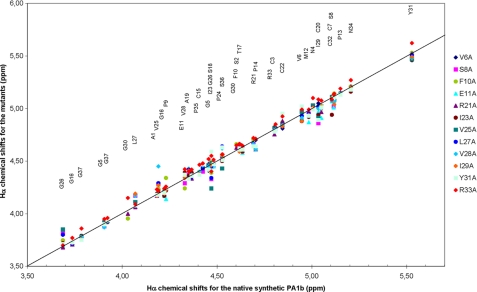
Next, fold validation of the alanine mutants with respect to the native synthetic PA1b, in similar experimental conditions, was processed by NMR. The classical set of the COSY, total correlation spectroscopy, and NOESY homonuclear spectra was acquired from all mutants, except for mutant M12A, which was insufficiently concentrated to obtain usable spectra and whose correct folding was assessed according to the one-dimensional 1H-NMR spectrum only. The α-proton chemical shifts of peptides are sensitive indicators of the backbone dihedral angles and constitute quick and reliable indicators of the secondary structure (25). The α-proton chemical shifts were determined for most residues of the mutants, except for some glycines and for Pro-14 and Cys-15. Fig. 2 shows that the alanine mutants exhibit no significant α-proton chemical shift perturbation when compared with native PA1b. Chemical shift deviations, with respect to the native peptide, concern the residues neighboring the mutations, the largest deviations (>0.15 ppm) being observed for Val-25 in mutant V28A, Gly-26 in mutant V25A, and Ile-29 in mutant Y31A. Altogether, this chemical shift analysis strongly supports the fact that no major structural change is generally induced by alanine substitution and that all the mutants adopt the same knottin fold as native PA1b.
FIGURE 2.
NMR chemical shifts of the α-H signals of PA1b alanine mutants when compared with those of the wild-type peptide.
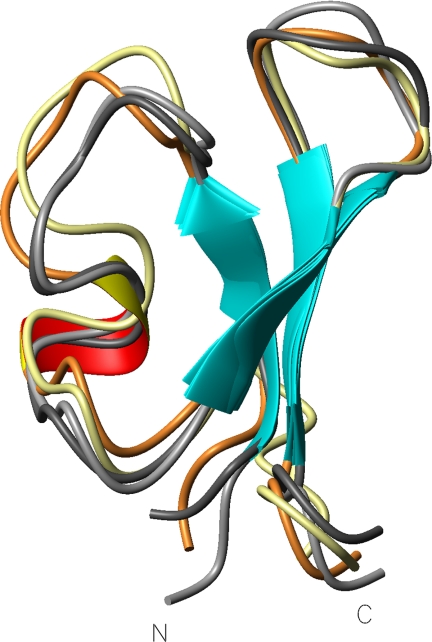
Finally, three alanine mutants were selected for a full analysis of their structure. The three-dimensional structures of F10A, R21A, and R33A were determined by simulated annealing, using distance restraints derived from the two-dimensional NMR spectra. ARIA calculations gave ensembles of 10 structures with backbone root mean square deviations of 0.82, 0.98, and 0.78 Å for F10A, R21A, and R33A, respectively (supplemental Table S2). The structures of the native and mutant PA1bs are shown in Fig. 3, and these confirm that the overall fold of the mutants does not substantially differ from that of the native PA1b.
FIGURE 3.
Comparison of the three-dimensional structure of three alanine mutants with that of native PA1b. Shown is a superposition of the ribbon representations of PA1b (light gray), F10A (yellow), R21A (orange), and R33A (dark gray), using the mean structure of each NMR ensemble fitted onto the triple-stranded β-sheet.
Bioactivity of Alanine Mutants
The synthesized mutants were next examined in biological and biochemical assays to determine the residues implicated in the peptide entomotoxic activity. The activity of the mutants was assessed by measuring their affinities for the PA1b-binding site, their LD50 (lethal dose, 50%) on cultured Sf9 insect cells, and their LT50 (lethal time, 50%), both on the susceptible WAA42 strain and on the resistant ISOR3 strain of S. oryzae. The activities of the alanine mutants are reported in Table 1, where it is clear that substitution of a specific set of individual residues leads to the loss of bioactivity. Indeed, substitution of any residue in the third β-sheet strand of PA1b (Arg-21 and Ile-23) was observed to significantly reduce, or even abolish, the binding and toxic abilities of the peptide, as did mutations of Phe-10 and Leu-27 in loops L1 and L2, respectively. Mutation of Ser-8, Val-28, or Ile-29 surrounding the four residues critical to the bioactivity of PA1b resulted in a decrease, but not a loss, of the binding and toxic abilities. In addition, mutation of Val-6, Glu-11, Met-12, Val-25, Tyr-31, or Arg-33, which are not located around the four critical residues on the molecular surface, led to biological properties similar to those of the wild-type toxin. It is important to note that no PA1b mutant exhibited toxicity against the resistant ISOR3 strain of S. oryzae, confirming that PA1b-like toxicity was observed in the active mutants. The tests performed on Sf9 cultured cells confirmed the results obtained by binding affinity and toxicity assays on cereal weevils.
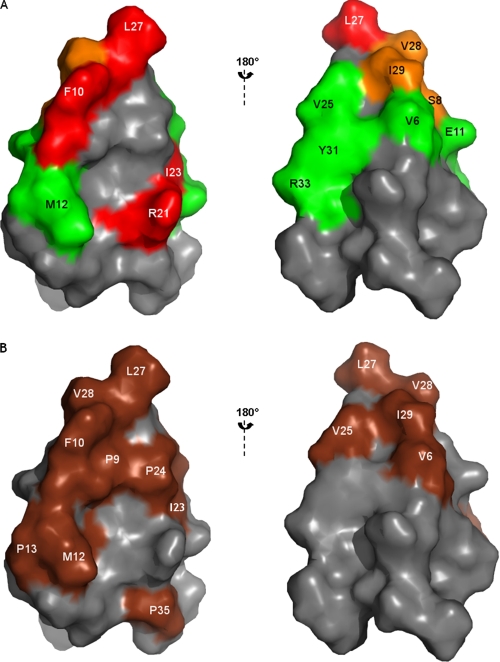
As shown in Fig. 4A, mapping of the bioassay results onto the three-dimensional structure of native PA1b highlights the fact that all the residues critical for entomotoxic activity are localized on one face of the peptide. Moreover, except for Arg-21, they are all hydrophobic (Phe-10, Ile-23, and Leu-27). This result clearly demonstrates the importance of the surface-exposed hydrophobic patch of PA1b as regards its bioactivity (Fig. 4B).
FIGURE 4.
Surface representation showing the localization of residues of PA1b replaced by alanine and the hydrophobic residues of PA1b. The figures on the right are rotated 180° relative to those on the left. A, mutated residues giving the same bioactivity as PA1b are shown in green, those giving intermediate activity are colored in orange, and the mutated residues leading to a loss of the insecticidal activity of PA1B are indicated in red. B, the hydrophobic residues of PA1b are highlighted in brown.
Mutant Resistance to Proteases
The alanine mutants of PA1b were assessed for their resistance to proteolysis. All 13 mutants were completely resistant to enzymatic digestion by the two serine endoproteinases trypsin and chymotrypsin, by the cysteine endoproteinase K, and by papain, and they were hydrolyzed by Pronase E, as was native PA1b (supplemental Table S3). On the other hand, the control non-folded reduced mutants were always sensitive to degradation by the four proteases tested in this assay.
DISCUSSION
In this study, we synthesized a rational collection of PA1b single point alanine mutants and determined their biological activities in terms of binding properties and cellular and insecticidal toxicity. A collection of 13 mutants was selected from previous bioinformatic (3) and structural (9) studies, which had revealed the importance of residues located in or around loops L1 and L2 in terms of sequence conservation and/or definition of the PA1b amphiphilic and electrostatic characteristics. The mutants were generated using the optimized synthesis procedure developed for the chemical production of native PA1b (17). All the mutants were efficiently folded, as judged by RP-HPLC, with a yield similar to that of native PA1b. Bearing in mind the difficulties encountered during the oxidative step of native PA1b, the mutants were carefully characterized using mass spectrometry and NMR. These structural analyses showed that mutant and native PA1bs exhibit the same structures. The cystine knot framework can accommodate sequence variations and maintain the well defined three-dimensional structure of the wild-type PA1b. These results, together with the resistance shown by all the mutants to protease degradation, clearly demonstrate that the resilience of the scaffold can be retained and indicate that the cystine knot alone is a structural feature sufficient to confer an extremely high degree of stability. Such findings have already been reported for other cystine knot proteins, such as insecticidal plant cyclotides (26).
Given the conservation of the overall fold in the PA1b mutants and their resistance to degradation, it appears likely that the inactivity of some mutants cannot be ascribed to improper folding or to protease degradation in the insect digestive tract. Conversely, a major role in the bioactivity of PA1b should be assigned to the residue side chains. The biological data obtained on mutants have shown that four residues, namely Phe-10, Arg-21, Ile-23, and Leu-27, are of outstanding importance both for binding and for toxicity. When a PA1b mutant conserved a binding activity, its insecticidal ability was also maintained, and vice versa. These results show that the binding ability is directly correlated to the in vivo entomotoxicity of PA1b. Moreover, the acute responsiveness of the bioactivity to the loss of critical residues clustered on a single face of PA1b (Fig. 4A) is consistent with a specific recognition process that would be mediated via the membrane protein-based receptor sensitive to PA1b, found in various insect species (12, 15).
An isoform of PA1b, extracted from soybean and named leginsulin, has been shown to be a ligand for the 43-kDa protein in legumes that controls cell proliferation and differentiation (27). Leginsulin shares almost 60% of sequence identity and slightly more than 80% of similarity with PA1b (Table 2). In terms of three-dimensional structure, when considering the average structures of the ensembles of NMR-derived models, the root mean square deviation between the backbone heavy atom coordinates of PA1b and leginsulin is very low: 1.5 Å (Table 2). However, despite these sequence and structure similarities, the residues involved in the binding of PA1b and of leginsulin (27) to their respective membrane targets are different and do not align (Table 2). Their only common feature is hydrophobicity concerning PA1b, except for Arg-21. Finally, the topological arrangement of each set of critical residues, located on opposite faces of the molecules, suggests a different receptor binding mode (supplemental Fig. S1).
TABLE 2.
Sequence alignment of PA1b with leginsulin
Amino acids critical for the binding activities of PA1b and leginsulin to their respective membrane receptors, as determined by alanine scanning (see Ref. 20) for leginsulin), are highlighted in grey.
To date, PA1b isoforms constitute a unique family of peptides that may show three different biological activities, mediated by specific protein-based receptors. (i) In plants, PA1b behaves as a hormone-like peptide stimulating the protein kinase activity of the 43-kDa protein to which it binds (27)/ (ii) In mammals, PA1b can modulate the blood glucose concentration by interacting with voltage-dependent anion channel 1 (VDAC-1) on the pancreatic cell membrane (28). iii) In insects, PA1b acts as a potent insecticidal agent via another membrane protein-based receptor. Hence, concerning PA1b entomotoxic activity, our findings suggest an original mode of action of PA1b as a cystine knot insecticidal peptide. As far as we know, only a few cystine knot proteins have an insecticidal activity. They include insecticidal plant cyclotides (29–31) and a knottin, Amaranthus α-amylase inhibitor, found in the seeds of Amaranthus hypochondriacus (32). The insecticidal activity of Amaranthus α-amylase inhibitor relies on a specific inhibition of the α-amylase of insect larvae, such as Tribolium castaneum. Concerning cyclotides, it has been postulated that their bioactivities would be mediated purely, or at least mainly, through membrane interactions (33). Until now, PA1b is the only insecticidal knottin acting via a membrane protein-based receptor. The purification and identification of this PA1b-binding partner will, consequently, be of major importance in understanding the mode of action of this entomotoxic peptide because that seems to be the key to deciphering PA1b toxicity.
CONCLUSION
Our mutagenesis strategy based on the rational synthesis of PA1b mutants has allowed us to clearly identify the residues critical for PA1b bioactivity (Phe-10, Ile-23, Arg-21, and Leu-27). An analysis of the structural properties of the mutants revealed the extraordinary stability of the cystine knot fold. The crucial role played by these mainly hydrophobic residues, in both binding and insecticidal activities, established the major involvement of the membrane protein-based receptor of PA1b in its mode of action. Such observations pave the way for the design of more potent toxins. Moreover, the identification of the PA1b receptor and a better understanding of the relationship between these two partners would lead to the validation of the binding site of PA1b as a new target for the development of bioinsecticides, with PA1b as the lead compound.
Supplementary Material
Acknowledgment
We thank Christophe Chambon from the mass spectrometry platform at the Meat Research Unit (INRA, Clermont-Ferrand, France) for the majority of the mass analyses.
This work was supported in part by INSA-Lyon and Region Centre (PA1bbio) grants.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. S1.
- RP-HPLC
- reverse phase-HPLC.
REFERENCES
- 1.Delobel B., Grenier A. M., Gueguen J., Ferrasson E., Mbaiguinam M. (July28, 2004) Patent Cooperation Treaty Patent WO99/58695 [Google Scholar]
- 2.Taylor W. G., Fields P. G., Elder J. L. (2004) J. Agric. Food Chem. 52, 7491–7498 [DOI] [PubMed] [Google Scholar]
- 3.Louis S., Delobel B., Gressent F., Duport G., Diol O., Rahioui I., Charles H., Rahbé Y. (2007) Phytochemistry. 68, 521–535 [DOI] [PubMed] [Google Scholar]
- 4.Louis S., Delobel B., Gressent F., Rahioui I., Quillien L., Rahbé Y. (2004) Plant Sci. 167, 705–714 [Google Scholar]
- 5.Yamazaki T., Takaoka M., Katoh E., Hanada K., Sakita M., Sakata K., Nishiuchi Y., Hirano H. (2003) Eur. J. Biochem. 270, 1269–1276 [DOI] [PubMed] [Google Scholar]
- 6.Dun X. P., Wang J. H., Chen L., Lu J., Li F. F., Zhao Y. Y., Cederlund E., Bryzgalova G., Efendic S., Jörnvall H., Chen Z. W., Bergman T. (2007) FEBS J. 274, 751–759 [DOI] [PubMed] [Google Scholar]
- 7.Higgins T. J., Chandler P. M., Randall P. J., Spencer D., Beach L. R., Blagrove R. J., Kortt A. A., Inglis A. S. (1986) J. Biol. Chem. 261, 11124–11130 [PubMed] [Google Scholar]
- 8.Taylor W. G., Sutherland D. H., Olson D. J., Ross A. R., Fields P. G. (2004) J. Agric. Food Chem. 52, 7499–7506 [DOI] [PubMed] [Google Scholar]
- 9.Jouvensal L., Quillien L., Ferrasson E., Rahbé Y., Guéguen J., Vovelle F. (2003) Biochemistry 42, 11915–11923 [DOI] [PubMed] [Google Scholar]
- 10.Craik D. J., Daly N. L., Waine C. (2001) Toxicon. 39, 43–60 [DOI] [PubMed] [Google Scholar]
- 11.Norton R. S., Pallaghy P. K. (1998) Toxicon. 36, 1573–1583 [DOI] [PubMed] [Google Scholar]
- 12.Gressent F., Duport G., Rahioui I., Pauchet Y., Bolland P., Specty O., Rahbe Y. (2007) J. Insect Sci. 7, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosengren K. J., Wilson D., Daly N. L., Alewood P. F., Craik D. J. (2002) Biochemistry 41, 3294–3301 [DOI] [PubMed] [Google Scholar]
- 14.Grenier A. M., Mbaiguinam M., Delobel B. (1997) Heredity 79, 15–23 [Google Scholar]
- 15.Gressent F., Rahioui I., Rahbé Y. (2003) Eur. J. Biochem. 270, 2429–2435 [DOI] [PubMed] [Google Scholar]
- 16.Rahioui I., Laugier C., Balmand S., Da Silva P., Rahbe Y., Gressent F. (2007) Biochimie 89, 1539–1543 [DOI] [PubMed] [Google Scholar]
- 17.Da Silva P., Strzepa A., Jouvensal L., Rahioui I., Gressent F., Delmas A. F. (2009) Biopolymers 92, 436–444 [DOI] [PubMed] [Google Scholar]
- 18.Kumar A., Ernst R. R., Wüthrich K. (1980) Biochem. Biophys. Res. Commun. 95, 1–6 [DOI] [PubMed] [Google Scholar]
- 19.Griesinger C., Otting G., Wuethrich K., Ernst R. R. (1988) J. Am. Chem. Soc. 110, 7870–7872 [Google Scholar]
- 20.Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. (1983) Biochem. Biophys. Res. Commun. 117, 479–485 [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 22.Johnson B. A., Blevins R. A. (1994) J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 23.Linge J. P., O'Donoghue S. I., Nilges M. (2001) Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 24.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 25.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 26.Simonsen S. M., Sando L., Rosengren K. J., Wang C. K., Colgrave M. L., Daly N. L., Craik D. J. (2008) J. Biol. Chem. 283, 9805–9813 [DOI] [PubMed] [Google Scholar]
- 27.Hanada K., Nishiuchi Y., Hirano H. (2003) Eur. J. Biochem. 270, 2583–2592 [DOI] [PubMed] [Google Scholar]
- 28.Dun X. P., Li F. F., Wang J. H., Chen Z. W. (2008) Peptides 29, 891–897 [DOI] [PubMed] [Google Scholar]
- 29.Barbeta B. L., Marshall A. T., Gillon A. D., Craik D. J., Anderson M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennings C. V., Rosengren K. J., Daly N. L., Plan M., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. (2005) Biochemistry 44, 851–860 [DOI] [PubMed] [Google Scholar]
- 32.Chagolla-Lopez A., Blanco-Labra A., Patthy A., Sánchez R., Pongor S. (1994) J. Biol. Chem. 269, 23675–23680 [PubMed] [Google Scholar]
- 33.Craik D. J., Mylne J. S., Daly N. L. (2010) Cell. Mol. Life Sci. 67, 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.