Abstract
m-calpain plays a critical role in cell migration enabling rear de-adhesion of adherent cells by cleaving structural components of the adhesion plaques. Growth factors and chemokines regulate keratinocyte, fibroblast, and endothelial cell migration by modulating m-calpain activity. Growth factor receptors activate m-calpain secondary to phosphorylation on serine 50 by ERK. Concurrently, activated m-calpain is localized to its inner membrane milieu by binding to phosphatidylinositol 4,5-bisphosphate (PIP2). Opposing this, CXCR3 ligands inhibit cell migration by blocking m-calpain activity secondary to a PKA-mediated phosphorylation in the C2-like domain. The failure of m-calpain activation in the absence of PIP2 points to a key regulatory role, although whether this PIP2-mediated membrane localization is regulatory for m-calpain activity or merely serves as a docking site for ERK phosphorylation is uncertain. Herein, we report the effects of two CXCR3 ligands, CXCL11/IP-9/I-TAC and CXCL10/IP-10, on the EGF- and VEGF-induced redistribution of m-calpain in human fibroblasts and endothelial cells. The two chemokines block the tail retraction and, thus, the migration within minutes, preventing and reverting growth factor-induced relocalization of m-calpain to the plasma membrane of the cells. PKA phosphorylation of m-calpain blocks the binding of the protease to PIP2. Unexpectedly, we found that this was due to membrane anchorage itself and not merely serine 50 phosphorylation, as the farnesylation-induced anchorage of m-calpain triggers a strong activation of this protease, leading notably to an increased cell death. Moreover, the ERK and PKA phosphorylations have no effect on this membrane-anchored m-calpain. However, the presence of PIP2 is still required for the activation of the anchored m-calpain. In conclusion, we describe a novel mechanism of m-calpain activation by interaction with the plasma membrane and PIP2 specifically, this phosphoinositide acting as a cofactor for the enzyme. The phosphorylation of m-calpain by ERK and PKA by growth factors and chemokines, respectively, act in cells to regulate the enzyme only indirectly by controlling its redistribution.
Keywords: Calpain, Cell Migration, Cell Motility, Chemokines, Growth Factors, Phosphorylation Enzymes, Phosphotyrosine Receptor, CXCR3, Wound Healing
Introduction
Calpains are intracellular cysteine proteases involved in numerous physiological and pathological phenomena, such as embryo development and tumor invasion (1). Among the 15 members of the calpain family, the two ubiquitous calpains, calpain 1 and calpain 2, are the best described. They form with the calpain small subunit 1 (calpain S1) two heterodimers, μ-calpain for calpain 1 and m-calpain for calpain 2, that are strongly involved in the regulation of cell motility. Cell migration was previously described as a four-stage process: cell membrane protrusion, adhesion to the substrate, contraction of the cell body, and finally, release of the adhesion contacts at the rear of the cell (2–4). Cell migration is, thus, governed by a succession of adhesion and de-adhesion steps, and a balance between these two processes is required for an optimal cell movement (5, 6). Previous studies have shown that the ubiquitous calpains regulate these adhesion and de-adhesion steps. Indeed, μ-calpain is involved in the formation of the adhesion complexes at the front of the migrating cell, notably by regulating the activity of Rho GTPases (7, 8) and promoting adhesion turnover (9). At the rear end of the cell, m-calpain allows cell detachment by cleaving proteins constituent of the adhesion complexes, such as the focal adhesion kinase, talin, vinculin, and α-actinin (10–12). The role played by m-calpain is crucial as the de-adhesion of the cell rear was shown to be rate-limiting in both hapto- and chemokinetic motility (13, 14).
Cell migration is critical for skin wound healing, a complex process that consists of a succession of highly orchestrated and regulated events (15). The first stages of wound healing (inflammatory and regenerative phases) require the in-migration of keratinocytes, fibroblasts, and endothelial cells to repopulate the wound and create new blood vessels (16–18). The growth factors EGF, VEGF, and PDGF control these first steps by stimulating the migration of these cells (19–21). This massive influx of cells leads to a hypercellular wound bed containing a provisional and immature matrix synthesized by the migrating fibroblasts. The maturation of the wound occurs during the final stage, the resolving phase. Different signals are required to stop the proliferation and the migration of the cells and to induce the contraction of the matrix by the fibroblasts (22). Previous studies have highlighted the crucial role played by CXCR3 ligands in this “stop healing” process (9, 23, 25, 26). Indeed, the ELR-negative CXC chemokines IP-9 (CXCL11 or I-TAC) and IP-10 (CXCL10) inhibit the growth factor-induced motility of fibroblasts and endothelial cells and also induce the apoptosis of these supernumerary cells (27). These chemokines thereby induce the contraction of the matrix (28) and restore the physiologic paucicellularity, leading to the formation of a strong and organized dermis.
As ubiquitous calpains are centrally involved in cell migration, their regulation is crucial during wound healing. The regulation of μ-calpain is well known, and the calcium influxes are mainly responsible for the activation of this enzyme (9). On the opposite, the regulation of m-calpain remains unclear. The calcium concentrations required for its activation in vitro are supraphysiological and not consistent with cell survival (estimated between 400 and 800 μm (1)). Recent studies have highlighted the roles played by phosphorylations in m-calpain regulation particularly during wound healing process. The growth factors secreted during the first steps of wound closure promote cell migration by activating m-calpain. EGF and other classical growth factors stimulate m-calpain activity by inducing the enzyme phosphorylation on the serine 50 residue via the ERK/MAPK pathway (29, 30). The question remained as to why m-calpain leads to detachment primarily at the rear. It was recently determined that asymmetric phosphoinositide distribution directs this polarity (31). EGF induces the activation of PLCγ1 at the front of the cells, leading to the degradation of phosphatidylinositol 4,5-bisphosphate (PIP2)2 and, thus, to the formation of a gradient toward the rear (32). The phosphorylation of m-calpain by ERK allows the interaction of the domain III of this enzyme with PIP2. m-calpain is, thus, relocalized to the rear of the cells, more particularly at the membrane, close to the adhesion complexes. Interestingly, in the absence of PIP2, growth factors cannot activate m-calpain in living cells, although whether this is due to failure to be ERK phosphorylated is still uncertain.
The CXCR3 ligands IP-9 and IP-10 secreted during the resolving phase of wound healing block the growth factor-induced migration by inhibiting m-calpain activity (23–25). This inhibition is due to the phosphorylation of the protease by PKA on the serine 369 (33). By inhibiting the growth factor-induced m-calpain activity, IP-9 and IP-10 prevent cell retraction, leading to the contraction and the maturation of the wound matrix (34). Although the serine 369 phosphorylation is modeled to “freeze” m-calpain into a conformation that prevents the active cleft from cleaving proteins, the dominance of this inhibitory phosphorylation and its effects on m-calpain localization are unknown. Moreover, the importance of the localization of the enzyme at the membrane and the role played by PIP2 (simple docking site or a cofactor) in the m-calpain activation process are still not completely understood.
In this study we asked whether the effects of the CXCR3 ligands on migration were immediate and dominant over EGF and VEGF and whether these chemokines have any effect on the redistribution of m-calpain induced by these growth factors. We observed that the inhibition of the growth factor-induced migration by IP-9 and IP-10 is immediate and dominant over EGF and VEGF effects. Moreover, the redistribution of m-calpain at the rear and at the cell membrane induced by these growth factors is prevented and even reverted by these two chemokines. These effects are not due to a change in PIP2 localization but to the PKA-mediated inhibition of the interaction between m-calpain and this phosphoinositide. This led us to query the role of the membrane localization of m-calpain in its activation process; we bypassed the PIP2-dependent membrane localization by directly anchoring the enzyme at the plasma membrane of the cells. Farnesylation-induced anchorage of m-calpain at the plasma membrane induced a very strong activation of the enzyme. This anchored m-calpain does not require the ERK phosphorylation to be activated and is resistant to PKA phosphorylation. Interestingly, this anchored m-calpain still requires the presence of PIP2 to be activated. Taken together, our results clearly show that m-calpain activity is mainly regulated by its relocalization at the plasma membrane and by its binding to PIP2, whereas the ERK and PKA phosphorylations appear to only indirectly regulate the enzyme activity in living cells by controlling its redistribution. This represents a quantal and unexpected advance of our understanding of molecular control of m-calpain activation.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Dulbecco's modified Eagle's medium (DMEM), α-minimum essential medium, and fetal bovine serum (FBS) were purchased from Cellgro. The MCDB131 medium was purchased from Invitrogen. The growth factors EGF and VEGF were purchased from BD Biosciences and Sigma, respectively, and the chemokines IP-9 and IP-10 were from PeproTech Inc. The primary antibodies used for the immunoblots were purchased from Santa Cruz Biotechnology for m-calpain and GFP, from Sigma for actin, GAPDH, and vinculin, and from Cell Signaling Technology for PLCγ1 (total and phosphorylated on Tyr-783). The HRP-conjugated secondary antibodies were purchased from BIOSOURCE. Concerning the immunofluorescent staining, the primary antibodies were purchased from Calbiochem (m-calpain) and Echelon (phosphatidylinositol 4,5-bisphosphate), and the secondary antibodies were from Invitrogen. The anti-GFP antibody used for immunoprecipitation was also purchased from Invitrogen. TRITC-phalloidin and DAPI were purchased from Sigma. The purified PKA and PKI (PKA inhibitor) were purchased from Promega, and purified m-calpain was from Calbiochem. The phosphoinositides and the active human ERK1 were purchased from Sigma. Finally, the MicroSpin columns S-200 HR were purchased from GE Healthcare.
Cell Culture
Human fibroblasts Hs68, human microvascular endothelial cells, and murine fibroblasts NR6WT were used. The Hs68 fibroblasts were grown in DMEM supplemented with 10% FBS. The murine fibroblasts were grown in α-minimum essential medium plus 7.5% of FBS, and human endothelial cells were grown in MCDB131 supplemented with 10% of FBS. For the different experiments, cells were incubated overnight in quiescent media (0.5% dialyzed FBS (dFBS) for Hs68, 0.1% dFBS for human microvascular endothelial and NR6 WT cells). As cell migration and EGF responsiveness decreases with cell age (35), all cells were used as early passage (<10).
Cell Treatments
EGF was used at 1 nm for human fibroblasts and at 10 nm for human endothelial cells. VEGF used to treat the endothelial cells was added at 200 ng/ml. The CXCR3 ligands IP-9 and IP-10 were used at 15 and 200 ng/ml for fibroblasts and endothelial cells, respectively. Insulin, used to activate farnesyltransferase, was used at 100 nm for 20 or 120 min (36).
In Vitro Wound Healing Assays
Confluent cells were incubated overnight in quiescent medium. The monolayer was then scraped with a pipette tip to create an acellular area. Pictures of the created wounds were taken. The cells were then treated with EGF or VEGF alone or in combination with IP-9 or IP-10. For combination treatments, the chemokines were added 4 h before or after the growth factors. After an overnight incubation (18 h; before proliferation), pictures of the wound were taken. The wound closure was estimated with MetaMorph software (Molecular Devices) by measuring the area of the wounds before and after incubation.
Continuous Cell Tracking Experiments
For cell tracking experiments, human fibroblasts and endothelial cells, incubated in quiescent media overnight, were visualized every 4 min. EGF was first added to the media. IP-9 or IP-10 was added 40 min later for fibroblasts and 75 min later for endothelial cells. The velocity of the cells was determined with MetaMorph software.
Cell Footprint Isolation
The apical (dorsal) aspect of cells was removed by a modification of a method described previously (37). Quiescent cells plated on collagen-coated coverslips were treated overnight with EGF, IP-9, or IP-10 alone or in combination. For combination treatments, IP-9 and IP-10 were added 4 h before or after EGF. After the overnight incubation, the cells were washed with PBS and with MES-buffered saline (MBS; 20 mm MES ((pH 5.5), 135 mm NaCl, 0.5 mm CaCl2, 1 mm MgCl2). The cells were then coated with a 1% solution of cationic colloidal silica (silica prepared as a 30% stock colloid as described previously (38)). After washing with MBS, the cells were coated with 1% polyacrylic acid in MBS (stock 25% aqueous polyacrylic acid solution, 100,000 average molecular weight; Sigma). The cells were washed with MBS and then swelled in hypotonic lysis buffer (2.5 mm imidazole (pH 7.0) supplemented with protease inhibitors (1:100 dilution, protease mixture set V, Calbiochem)) for 30 min. Cells were then unroofed by squirting the monolayer with lysis buffer through a 5-ml syringe fitted with a blunted, flattened 18-gauge needle. The degree of unroofing was monitored by observing cells with an inverted phase-contrast microscope. The isolated footprints were then used to visualize m-calpain by immunostaining or to quantify m-calpain by immunoblot. For immunostaining, the footprints were washed once using MBS and fixed using a 2% paraformaldehyde solution prepared in PBS. For immunoblot, the footprints were washed using PBS, and the proteins were extracted with 1× SDS sample loading buffer.
Immunofluorescent Staining
The cells and the footprints were washed using PBS and fixed with a 2% paraformaldehyde solution (in PBS). The cells were permeabilized with a 0.1% Triton-X solution for 5 min. The cells or the footprints were then blocked with non-immune goat serum (5% in PBS). After washing, the cells or the footprints were incubated for 2 h with the primary antibodies (against calpain 2 subunit, phosphatidylinositol 4,5-bisphosphate, vinculin, or calpain S1) diluted at 1:100. Then they were washed and incubated with the secondary antibodies (Alexa Fluor 488 or 594, 1:500 dilution, 1 h). Actin filaments were stained by incubating the cells or the footprints for 40 min with TRITC-phalloidin. DAPI was used to stain the cell nuclei. The coverslips were mounted using Gelvatol and observed using a fluorescent microscope. For the quantification of the fluorescence of the rear and of the front parts of the cells, the pictures were analyzed using MetaMorph software (20 cells were analyzed for each condition).
Generation of a Phosphomimetic Variant of m-calpain (S369E)
To mimic PKA phosphorylation of m-calpain, the serine 369 was replaced by a glutamic acid. The plasmid encoding the GFP-WT-m-calpain (described previously (33)) was mutated using a PCR-based mutagenesis kit (Stratagene) and primers that encoded the mutation (5′-GAA CTG GAG GCG GGG CGA GAC CGC GGG AGG TTG C-3′ and 5′-GCA ACC TCC CGC GGT CTC GCC CCG CCT CCA GTT C-3′). The generated mutant referred to as GFP-S369E-m-calpain was confirmed by DNA sequencing.
Construction of the CAAX- and SAAX-calpain 2 Plasmids
The K-Ras farnesylation sequence CAAX (and the negative control SAAX) was added to the gene encoding the human calpain 2 subunit by three successive PCR. The plasmid encoding the human WT-calpain 2 was used as a template. The calpain 2 gene containing the CAAX and SAAX sequences was inserted in the plasmid pEGFP-C1 (obtained from the Prof. Chang Lu) using HindIII restriction enzyme (New England Biolabs). The mutation of the serine 50 to alanine was performed using a PCR-based mutagenesis kit (Stratagene) and primers that encoded the mutation (5′-TCC AGG ACC CGG CCT TCC CGG CCA T-3′ and 5′-ATG GCC GGG AAG GCC GGG TCC TGG A-3′). The generated mutants referred to as CAAX-calpain 2 S50A and SAAX-calpain 2 S50A were confirmed by DNA sequencing.
Transfection of the NR6WT Fibroblasts
NR6WT fibroblasts seeded in antibiotic-free medium were transfected with the different plasmids using Lipofectamine 2000 (Invitrogen). The cells transfected with the plasmids encoding the WT-, the ST369AA-, and the S369E-m-calpain were incubated overnight in the presence of 15 μm MDL28170 (Calbiochem) to reduce cell death due to calpain overexpression. For the expression of the CAAX- and SAAX-calpain 2, the cells were transfected at the same time with the plasmids and a mix of two siRNAs directed against the murine calpain 2 (IDT DNA). The cells transfected with the plasmid encoding the CAAX-calpain 2 were incubated overnight in the presence of 5 μm calpain inhibitor MDL28170 (Calbiochem, EMD Biosciences) to reduce the cell death due to the very strong activity of this mutated m-calpain. After the overnight incubation, the cells expressing CAAX- and SAAX-calpain 2 were washed with PBS and treated for 20 or 120 min with insulin.
Immunoprecipitation of the GFP m-calpains
The transfected cells were washed and treated for 30 min with EGF with and without IP-10 to induce the phosphorylations of m-calpain. IP-10 was added at the same time as EGF. After the incubation, the proteins were extracted using radioimmune precipitation assay buffer plus protease inhibitors, centrifuged, and incubated overnight with an anti-GFP antibody (0.5 μg, Invitrogen). The proteins were then incubated with protein A-Sepharose beads. The beads were washed with radioimmune precipitation assay buffer, and the GFP-m-calpains were eluted using Gentle Ag/Ab elution buffer (Pierce). The purified GFP-m-calpains were then used for PIP2 binding assay (using liposomes).
In Vitro Phosphorylation of m-calpain by ERK1 and PKA
Purified m-calpain (from porcine kidney; Calbiochem) was phosphorylated in vitro using PKA (Promega) and/or active ERK1 (Sigma). For ERK1 phosphorylation, 2.5 μg of m-calpain were incubated with 0.1 μg of active ERK1 in the following buffer: 20 mm MOPS, pH 7.2, 25 mm β-glycerolphosphate, 1 mm sodium orthovanadate, 1 mm DTT, 100 μm ATP, and 15 mm MgCl2. After 30 min of incubation at 30 °C, m-calpain was used for a PIP2 binding assay or for in vitro phosphorylation by PKA. For PKA phosphorylation, m-calpain (2.5 μg) was incubated with 25 units of PKA for 2 h in the following buffer: 50 mm Tris-HCl (pH 7.5), 7 mm MgCl2, 1 mm DTT, 0.06% CHAPS, 50 μm ATP. Samples were also treated with PKA and PKI (2 mm) as negative control. After a 2-h incubation at 30 °C, m-calpain was used for PIP2 binding assay (using liposomes).
m-calpain Binding to Phospholipid-containing Liposomes
Preparation of the liposomes and assessment of m-calpain-phospholipid binding were performed as previously reported (31, 39). Briefly, liposomes were prepared from a crude bovine brain extract of phosphoinositides that contains 20–40% di- and triphosphoinositide (a minimum of 5 to 10% of each). Binding of m-calpain (in vitro phosphorylated m-calpain or GFP-m-calpain purified by immunoprecipitation) to lipid vesicles was assayed by spin column size exclusion chromatography on MicroSpin S200 HR columns (GE Healthcare). The amount of protein in elute was detected by SDS-gel electrophoresis followed by immunoblotting. The quantification of the band density was carried out with ImageJ software.
In Vitro Calpain Activity Assay
The GFP m-calpains, immunoprecipitated and eluted in non-denaturing conditions, were desalted using the Zeba Desalt Spin Columns purchased from Pierce Thermo Scientific. The activity of the desalted samples was assessed using the calpain activity assay kit from BioVision. The fluorescence was quantified using a Spectra Fluor plate reader (Tecan).
Cell Survival Assay
The NR6WT fibroblasts were transfected with the empty plasmid (GFP) or with the plasmids encoding the CAAX/SAAX-calpain 2. For the transfection with CAAX-calpain 2 or CAAX-calpain 2 S50A, the cells were incubated overnight with 5 μm MDL28170 to protect them from the strong activity of these m-calpain mutants. After the overnight incubation, the transfected cells were washed several times with PBS to remove the calpain inhibitor used for the CAAX-calpain 2. They were then treated with 100 nm insulin for 20 min. The cells were washed to remove the insulin and incubated in regular medium. Pictures of three different fields were taken at the beginning of the experiment and after 3 and 6 h. The GFP cells were then counted for each time point, and cell survival was expressed as a percentage of the cells counted at the beginning of the experiment.
PIP2 Depletion
To deplete PIP2, the NR6WT cells, transfected with the empty plasmid or with the plasmids encoding the CAAX- or SAAX-calpain 2, were treated with medium containing butanol-1 (1.5%) for 45 min (31). Butanol-2 was used as a negative control. Insulin (100 nm) was added 20 min before the end of the incubation to induce the activation of the farnesyltransferase. After the treatments, calpain activity was assessed by observing vinculin degradation or by using t-Boc-LM-CMAC (Calbiochem). The cells remained viable during the short duration of this treatment (31).
t-Butoxycarbonyl (Boc) Assay
To quantify calpain activity, cells were incubated for 20 min in the presence of 25 mm t-Boc-LM-CMAC. After incubation, the cells were washed three times with PBS, and fluorescence was quantified using a plate reader (SpectraFluor, Tecan).
Statistical Analysis
All comparisons were performed with Student's t test with minimum significance deemed to be p < 0.05.
RESULTS
CXCR3 Ligands Override Growth Factor-induced Migration of Fibroblasts and Endothelial Cells
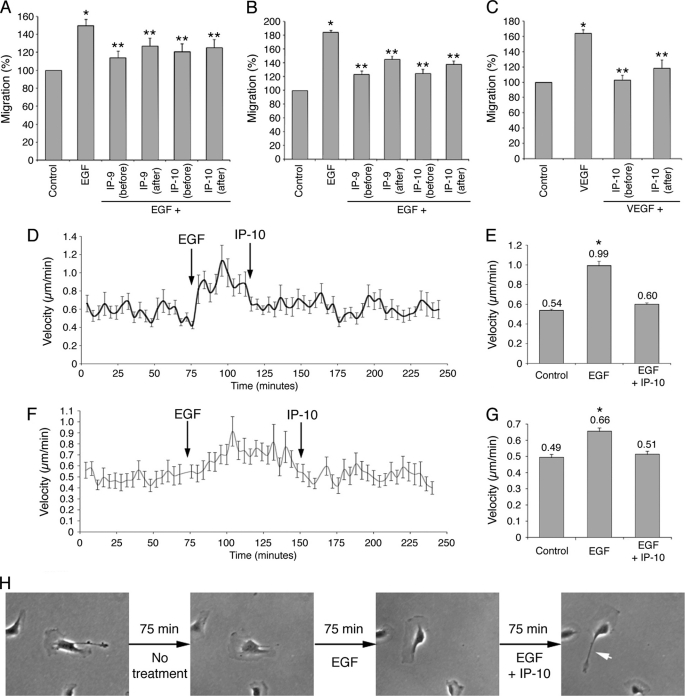
Previous studies have shown that the CXCR3 ligands IP-9/CXCL11 and IP-10/CXCL10 inhibit the migration of fibroblasts and endothelial cells induced by EGF and VEGF, respectively (23, 40). However, little is known concerning the efficiency, rapidity, and dominance of the migration inhibition; for instance, could these ligands stop ongoing locomotion. To have a better understanding of this phenomenon, the migration of human fibroblasts (Hs68) and human dermal microvascular endothelial cells treated with EGF or VEGF was quantified by wound healing assays in the presence of IP-9 or IP-10. These two chemokines were added to the media 4 h before (pretreatment) or after the growth factors. The results showed that the stimulation of fibroblast and endothelial cell migration observed after EGF treatment was almost totally inhibited when the cells were pretreated with IP-9 and IP-10 (Fig. 1, A and B). A similar decrement in in vitro “wound” closure was also observed when the two CXCR3 ligands were added 4 h after EGF, although not to the full extent as when added before EGF. This non-significant difference in the extent of inhibition was deemed as possibly due to 4 h of treatment with EGF alone. In both situations no differences were observed between IP-9 and IP-10, as would expected as they act via the common CXCR3. Similar data were obtained with endothelial cells treated with VEGF and IP-10 (Fig. 1C). These results show that the effects of IP-9 and IP-10 are likely dominant over EGF and VEGF stimulation of migration and disrupt ongoing events rather than preventing subsequent activation by growth factors. To probe this postulate, live cells were tracked. The velocities of untreated fibroblasts and endothelial cells were quantified before the addition of EGF and subsequently IP-10. The results demonstrate that the fibroblast migration is stimulated within minutes by EGF. In the same manner, IP-10 reduces the motility back toward pre-EGF speeds within minutes (Fig. 1D). The velocity of the cells, increased from 0.54 to 0.99 μm/min by EGF, was decreased to 0.6 μm/min by IP-10 (Fig. 1E). Similar results were observed using endothelial cells (Fig. 1, F and G). Observing the migration of these cells shows that CXCR3-mediated inhibition of the migration is due to the inability to retract the cell tail (Fig. 1H and supplemental Movie 1). The extension of the lamellipodia seems unaffected by CXCR3 ligands. Taken together, these results show that the effects of IP-9 and IP-10 on cell migration and retraction are immediate and dominant over EGF and VEGF in both human fibroblasts and endothelial cells.
FIGURE 1.
Inhibition of growth factor-mediated migration by IP-9 and IP-10. Wound healing assays were carried out to study the effects of CXCR3 ligands on the growth factor-mediated migration of human fibroblasts (Hs68, A) and endothelial cells (B and C). The cells were grown until 80–100% confluence. After 24 h of quiescence, the cells were scraped using a pipette tip to create a wound and treated with EGF (A and B) or VEGF (C). The chemokines IP-9 and IP-10 were added 4 h before or after the growth factors. The wound closure was quantified using Metamorph software by comparing the wound areas before and after the overnight treatments. The results were expressed as percentage (100% for untreated cells). Experiments were repeated three times, and the error scale bars show S.D. * and **, significantly different from untreated cells and cells treated with growth factors, respectively (p < 0.05). The effects of IP-10 on EGF-mediated migration were also studied by cell-tracking assays with fibroblasts (D and E) and endothelial cells (F, G, and H). Pictures of the untreated cells were taken every 4 min for 4 h. EGF was added to the quiescent medium after 75 min. IP-10 was then added to the medium 75 min after EGF for endothelial cells and 40 min after EGF for fibroblasts. The velocities of at least 50 migrating cells were determined for each time point using Metamorph software (D and F). The average velocities of the cells were also calculated for each treatment (E and G). The error scale bars show S.E. Pictures illustrating the effects of EGF and IP-10 on an endothelial cell are shown in H. The white arrow shows the rear of the cell, impossible to retract after IP-10 treatment.
CXCR3 Ligands Prevent EGF-induced Relocalization of m-calpain in Fibroblasts and Endothelial Cells
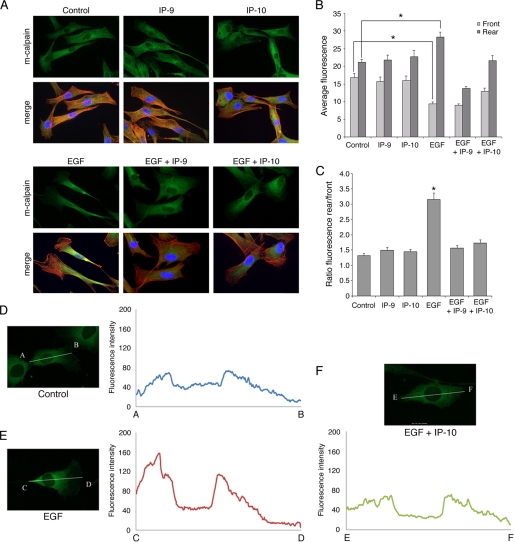
As m-calpain-mediated rear detachment is critical to EGF-induced cell migration (6, 33, 41) and CXCR3 signaling blocks this (Fig. 1F and supplemental Movie 1), we sought to determine whether the growth factor-induced redistribution of m-calpain toward the cell body and rear was affected. We localized m-calpain in human Hs68 fibroblasts treated overnight with diluent or IP-9, IP-10, and/or EGF alone or in combination. For combination treatments, IP-9 and IP-10 were added 4 h before EGF. As shown in Fig. 2A, m-calpain was localized randomly in the cytoplasm of untreated fibroblasts. The same localization was observed when the cells were treated with CXCR3 ligands. EGF exposure strongly redirected this protease into the central body and tail of the cells with almost no m-calpain detectable in the lamellipodia as previously reported for murine fibroblasts (31). Pretreating the cells with IP-9 or IP-10 before EGF prevented the re-localization of m-calpain, resulting in a picture similar to untreated or CXCR3 ligand-treated cells. To quantify the m-calpain distribution, fluorescence intensity was measured in the front part and in the rear part of the cells (Fig. 2, B and C). The ratio between the fluorescence of the rear and the fluorescence of the front was about 1.5 in untreated fibroblasts and in cells treated with only IP-9 or IP-10. EGF exposure resulted in this ratio doubling to more than 3 with m-calpain enriched in the cell rear. This redistribution was prevented by pre-exposure of the cells to CXCR3 ligands before EGF. These effects of EGF and CXCR3 ligands on m-calpain localization were noted more graphically by tracing m-calpain density from the tail to the tip of the lamellipod in one cell (Fig. 2, D–F).
FIGURE 2.
Localization of m-calpain in EGF-stimulated fibroblasts pretreated with IP-9 and IP-10. Hs68 fibroblasts were grown on coverslips coated with collagen. After 24 h in a quiescent medium, the cells were treated using EGF (1 nm), IP-9, and/or IP-10 (15 ng/ml). IP-9 and IP-10 were added 4 h before EGF. After an overnight incubation, the cells were fixed using 2% paraformaldehyde, permeabilized with 0.1% Triton X, and stained for m-calpain (green) and F-actin using phalloidin (red). Nuclei are stained in blue using DAPI. Pictures showing m-calpain are on the top rows, and the merge is on the bottom rows (A). The pictures shown are representative for three independent experiments. The pictures obtained were then analyzed using Metamorph. The average fluorescence was quantified in the front and in the rear of the cells exposed to the different treatments (B). The ratios between the average fluorescence of the rear and the average fluorescent of the front were also calculated (C). The fluorescence intensities were also measured using the line-scan tool of Metamorph software (D, E, and F). The results are expressed as the means ± S.E. The experiments were repeated 3 times, and 20 cells were analyzed for each treatment in each independent experiment. *, significantly different from the results obtained with untreated cells (p < 0.01).
Like fibroblasts, endothelial cell migration is critical for wound healing. Previous studies also have shown that the CXCR3 ligands block the angiogenic factor-induced migration of these cells (23). There is a critical distinction as CXCR3 ligands activate μ-calpain (calpain 1) in these cells in addition to blocking m-calpain activation (42, 43). Thus, the mode by which growth factor-induced migration is diminished needs to be discerned. The localization of m-calpain was, thus, determined using the same approaches as with fibroblasts. The results obtained with endothelial cells treated with EGF were very similar to those obtained with human fibroblasts. The EGF treatment induced the redistribution of m-calpain toward the rear, whereas IP-9 or IP-10, added 4 h before EGF and remaining during the entire EGF exposure, totally blocked this redistribution (supplemental Fig. S1A). The quantification of the fluorescence confirmed these observations (supplemental Fig. S1, B and C). The same experiments were done with VEGF as it was reported previously that this growth factor up-regulates the migration of the endothelial cells, whereas IP-10 inhibits their motility (23). The results were very similar to those obtained for EGF (supplemental Fig. S2). The effects of CXCR3 ligands on the growth factor-induced redistribution of m-calpain are, thus, not specific to EGF or to the cell types used in this study.
Taken together, these results show that both EGF and VEGF induce a change in m-calpain localization toward the cell rear in polarized and presumably motile human fibroblasts and endothelial cells. CXCR3 ligands do not alter m-calpain localization in and of themselves but do prevent growth factor-induced redistribution. This redistribution of m-calpain is not based on excessive degradation as total cell m-calpain levels remain similar at this extended time point under all conditions (see Fig. 4C and supplemental Fig. S4C).
FIGURE 4.
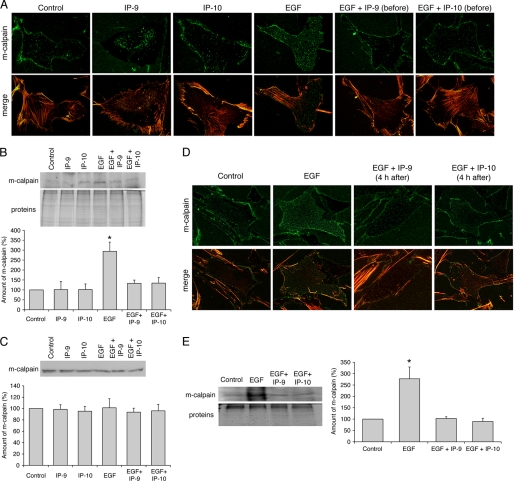
Localization of m-calpain in the footprints of Hs68 fibroblasts. Fibroblasts were grown on coverslips coated with collagen. After 24 h in a quiescent medium, the cells were treated using EGF (1 nm) and IP-9 or IP-10 (15 ng/ml). IP-9 and IP-10 were added 4 h before (A, B, and C) or after (D and E) EGF. After an overnight incubation, the cells were unroofed. The isolated footprints were fixed using 2% paraformaldehyde and stained for m-calpain (green) and actin (TRITC-phalloidin) (A and D) or scraped to extract the proteins. Immunoblots were carried out with these proteins, and m-calpain was stained (B and E). Immunoblots were also carried out with proteins extracted from whole cells treated with the same treatments used for the footprints (C). The immunoblots, representative of three different experiments, were quantified using ImageJ. The amounts of m-calpain were expressed as the percentage of the corresponding control ± S.D. *, significantly different from the results obtained for the control (p < 0.05).
CXCR3 Ligands Reverse the EGF-induced Redistribution of m-calpain
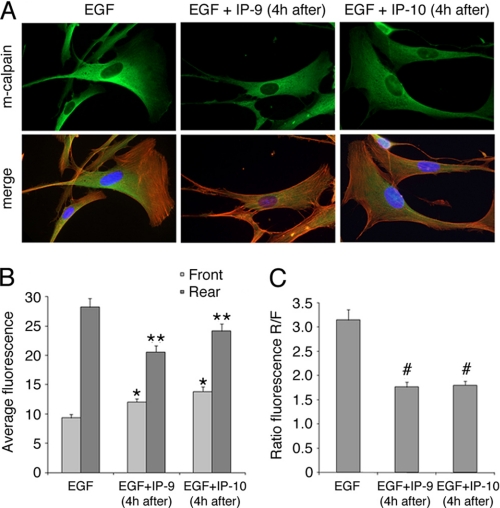
CXCR3 ligands present themselves late in the regenerative phase at a time that growth factors are already present (44). Thus, we sought to determine whether CXCR3 ligands are also capable of reversing the effects of EGF on m-calpain distribution. The fact that CXCR3 ligands were dominant inhibitors of growth factor-induced motility, via m-calpain inhibition, did not implicate the molecular basis of the near immediate cessation of migration (Fig. 1). Human fibroblasts were treated with EGF and IP-9 or IP-10 as described above with the exception that the two chemokines were added 4 h after the growth factor. As previously observed, the fibroblasts treated with EGF presented lamellipodia containing almost no m-calpain, whereas the protease was concentrated toward the rear (Fig. 3A). The morphologies of the cells treated with EGF alone or in combination with IP-9 or IP-10 were very similar; the cells treated with both EGF and the CXCR3 ligands had the same elongated shape with prominent lamellipodia as those treated with EGF alone. The addition of IP-9 and IP-10 4 h after EGF induced major changes in the localization of m-calpain in comparison to the fibroblasts treated with EGF alone. The enzyme was localized throughout the cytoplasm, even in the lamellipodia, and the major part of m-calpain was not localized in the rear part of the cells (Fig. 3A). These observations were confirmed by the quantification of the fluorescence. IP-9 or IP-10 treatment eliminated the front-rear asymmetry of m-calpain noted in cells treated with EGF alone (Fig. 3B), with the ratios of fluorescence reduced from 3.2 to 1.7 (Fig. 3C). These experiments were repeated in the same conditions with endothelial cells treated with EGF or VEGF with the results being very similar to those obtained with fibroblasts. The EGF-induced redistribution of m-calpain at the rear of the cell was reverted by IP-9 and IP-10, and the same phenomenon was observed for VEGF (supplemental Figs. S2 and S3). However, some differences were observed between the fibroblasts and the endothelial cells. Indeed, the rear parts of the endothelial cells treated with EGF and the chemokines were very spread out, perhaps because of a lack of retraction (supplemental Fig. S3A). Moreover, in the endothelial cells, m-calpain was concentrated in the perinuclear cytoplasm after the addition of IP-9 and IP-10. Even if the lamellipodia were devoid of m-calpain, this protease was almost absent from the rear of these cells (supplemental Fig. S3A). However, the quantifications of the fluorescence in the endothelial cells gave results very similar to those obtained with fibroblasts (supplemental Figs. S2, B and C, and S3, B and C). Taken together these results clearly show that the two CXCR3 ligands are able to not only prevent but also to reverse the redistribution of m-calpain induced by growth factors in fibroblasts and in endothelial cells.
FIGURE 3.
Localization of m-calpain in fibroblasts treated with IP-9 and IP-10 4 h after the addition of EGF. Hs68 fibroblasts were grown on coverslips coated with collagen. After 24 h in a quiescent medium, the cells were treated using EGF (1 nm). Four hours after this first treatment, IP-9 and IP-10 were added to the medium (15 ng/ml). After an overnight incubation, the cells were fixed using 2% paraformaldehyde, permeabilized with 0.1% Triton X, and stained for m-calpain (green) and F-actin using phalloidin (red). Nuclei were stained in blue using DAPI. The pictures obtained with the different treatments (A) were analyzed using Metamorph, and the average fluorescence was quantified in the front and in the rear of the cells (B). The ratios between the rear and the front were also calculated (C). The results are expressed as the means ± S.E. The experiments were repeated three times, and 20 cells were analyzed for each treatment in each independent experiment. *, **, #, significantly different from the results obtained for the cells treated with EGF alone (p < 0.01).
CXCR3 Ligands Block the EGF-induced Localization of m-calpain to the Plasma Membrane
Previous studies have shown that the growth factor-induced redistribution of m-calpain toward the rear of fibroblasts is due to the m-calpain binding to PIP2 (31), with EGF treatment resulting in loss of PIP2 in the lamellipodia secondary to PLCγ1 hydrolysis (32, 45). Thus, we questioned whether IP-9 and IP-10 prevented this interaction of m-calpain with PIP2. We probed the localization of m-calpain to membrane footprints of human fibroblasts treated with diluent (control) or IP-9, IP-10, and/or EGF alone or in combination. For combination treatments, IP-9 and IP-10 were added 4 h before EGF. The amount of m-calpain localized to the plasma membrane was unchanged in the fibroblasts treated with IP-9 or IP-10 alone (Fig. 4A). As expected (31), EGF treatment increases the amount of this protease localized to the membrane in both cell types. However, the increase of m-calpain induced by EGF was prevented when the cells were pretreated with IP-9 or IP-10. Indeed, the amount of m-calpain in the footprints of pretreated fibroblasts was similar to the control, as validated by immunoblotting of footprint-associated m-calpain (Fig. 4B). These effects of EGF, IP-9, and IP-10 were not due to changes in the levels of cellular m-calpain (Fig. 4C). To determine whether the CXCR3 ligands were also able to reverse the effects of EGF, the same experiments were performed with fibroblasts treated with IP-9 and IP-10 4 h after the addition of EGF. As shown in Fig. 4D, the amount of m-calpain localized at the membrane of the fibroblasts treated with EGF and IP-9 or IP-10 was similar to the control. m-calpain, translocated to the membrane by the EGF stimulation, was thus removed from the membrane by IP-9 and IP-10. These observations were confirmed by immunoblots (Fig. 4E). The same experiments were performed with endothelial cells in the same conditions and gave very similar results (supplemental Fig. S4). Taken together these results clearly show that CXCR3 ligands are able to prevent and also revert the EGF-induced translocation of m-calpain to the membrane of human fibroblasts and endothelial cells.
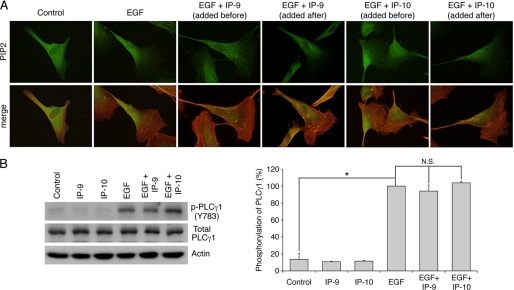
CXCR3 Ligands Do Not Affect EGF-induced Distribution of PIP2 and Activation of PLCγ1
As previously stated, the growth factor-induced redistribution of m-calpain at the rear and at the membrane of the cells is due to the binding of the enzyme to PIP2. The altered localization of m-calpain could, thus, be secondary to a redistribution of PIP2 induced by the CXCR3 ligands. We, therefore, determined whether PIP2 distribution was altered by IP-9 or IP-10. PIP2 was visualized by antibody detection in human fibroblasts treated with EGF and IP-9 or IP-10 (added 4 h before or after the growth factor). The growth factor and chemokine concentrations used were the same than those used for the previous experiments. The results show that PIP2 was localized randomly in untreated fibroblasts (Fig. 5). The addition of EGF induced the concentration of PIP2 at the rear of the cells. However, and in contrast to the results observed for m-calpain, IP-9, added before or after EGF, had no effects on the localization of PIP2 induced by the growth factor. Similarly, the EGF-induced localization of PIP2 at the rear of the cell remains unchanged in the presence of IP-10. The same results were obtained with endothelial cells treated in the same conditions (supplemental Fig. S5). Previous studies have shown that the asymmetric localization of PIP2 in the cells is due to the depletion of this phosphoinositide at the front of the cell by PLCγ1. This enzyme is phosphorylated and, thus, activated after EGF treatment. The CXCR3 ligands had no effect on the phosphorylation of PLCγ1 induced by EGF (Fig. 5B). These chemokines are, thus, unable to block the EGF-mediated activation of PLCγ1. This result confirms the data obtained by immunolocalization of PIP2.
FIGURE 5.
Effects of IP-9 and IP-10 on EGF-mediated localization of PIP2 (A) and EGF-induced activation of PLCγ1 (B). For PIP2 immunolocalization, Hs68 fibroblasts were grown on coverslips coated with collagen. After 24 h in quiescent media, the cells were treated using EGF (1 nm). IP-9 and IP-10 were added to the media 4 h before or after the EGF treatment (15 ng/ml). After an overnight incubation, the cells were fixed using 2% paraformaldehyde, permeabilized with 0.1% Triton X, and stained for PIP2 (green) and F-actin using phalloidin (red). The pictures of the stained fibroblasts are representative for three independent experiments (A). For PLCγ1 phosphorylation, the cells were incubated 24 h in a quiescent medium. The chemokines IP-9 and IP-10 were added to the medium at a final concentration of 15 ng/ml. After 4 h of incubation, EGF was added to the medium at 10 nm for 15 min. The cells were then lysed, and the extracted proteins were loaded on gel. Immunodetection was carried out using antibodies against actin, total PLCγ1, and phosphorylated PLCγ1 (Tyr-783). The immunoblots presented in B are representative of three different experiments. The amount of phosphorylated PLCγ1, quantified using ImageJ, were expressed as the percentage ± S.D. (100% for EGF). *, significantly different from the results obtained for the control (p < 0.05). N.S., not significant.
Taken together, these results showed that the effects of the CXCR3 ligands on EGF-induced localization of m-calpain are not due to changes in the localization of PIP2. IP-9 and IP-10 could be able to block the interaction between m-calpain and PIP2 allowed by EGF, thus preventing and also reversing the redistribution of m-calpain observed in the presence of EGF.
PKA Phosphorylation of m-calpain Induced by CXCR3 Ligands Blocks the Interaction between m-calpain and PIP2
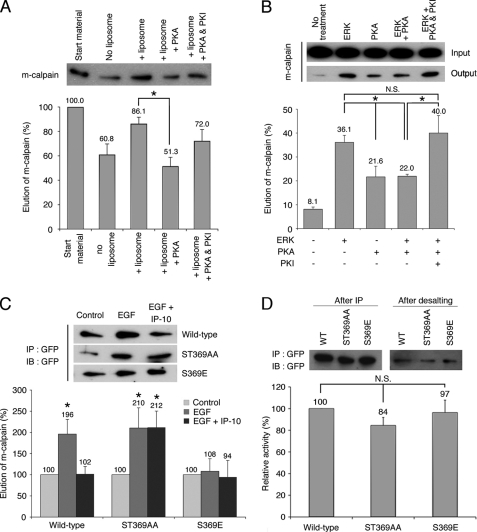
As previously stated, CXCR3 ligands inhibit m-calpain activity by inducing the phosphorylation of m-calpain by PKA (33). To parse the effects of this phosphorylation of m-calpain ability to bind PIP2, we performed a protein-liposome assay with purified m-calpain treated in vitro with PKA. As shown in Fig. 6A, the in vitro phosphorylation of m-calpain by PKA strongly reduced the binding of m-calpain to PIP2-containing liposomes. Indeed the elution of m-calpain (due to the binding of m-calpain to the liposomes) was reduced from 86 to 51% after PKA phosphorylation. This inhibition of m-calpain binding to PIP2 was abrogated in the presence of the PKA specific inhibitor PKI. In vivo the CXCR3 ligands intervene after the growth factors; it is, thus, important to know if the stimulation of the interaction between m-calpain and PIP2 induced by ERK can be blocked by the PKA phosphorylation of m-calpain. To this aim, the ability of m-calpain to bind PIP2 was studied after the successive phosphorylations of the protease by ERK1 and PKA. As shown in Fig. 6B, the strong stimulation of the binding of m-calpain to PIP2 induced by ERK1 phosphorylation is limited by PKA. Indeed the phosphorylation of m-calpain by PKA reduces the ERK-stimulated elution of m-calpain from 36.1 to 22%. This 1.6-fold decrease is prevented by the addition of PKI. The phosphorylation of m-calpain by PKA is, thus, able to limit the effects of the ERK phosphorylation. To confirm these data obtained in vitro, the same protein-liposome assays were performed on isolated m-calpain expressed in cell. Fibroblasts were transfected with GFP-WT-m-calpain or the PKA-resistant GFP-ST369AA-m-calpain and treated with EGF alone or with IP-10. The addition of EGF induced a very strong increase of the ability of m-calpain to bind PIP2, as previously reported (31). This increase was almost totally inhibited by the addition of IP-10 at the same time as EGF (Fig. 6C, wild type). IP-10 was, thus, able to block the EGF-induced interaction of m-calpain with PIP2. This effect of IP-10 was not observed with the mutated m-calpain resistant to PKA phosphorylation (Fig. 6C, ST369AA). To confirm the role of negative phosphorylation at the PKA site, we generated a phosphomimetic S369E m-calpain variant. As shown in Fig. 6C, the mutation mimicking the PKA phosphorylation of m-calpain was able to totally block the effects of EGF on the m-calpain ability to bind PIP2. The effects of IP-10 on the interaction of m-calpain with PIP2 are, therefore, clearly mediated by PKA. These data are correlated to those obtained in vitro (see Fig. 8, A and B). Taken together, these results show that the CXCR3 ligands block the EGF-stimulated binding of m-calpain to PIP2 by inducing the PKA phosphorylation of this protease on the serine 369.
FIGURE 6.
Effects of m-calpain phosphorylations on its interaction with PIP2in vitro (A and B) and in vivo (C) and on its activity (D). The binding of m-calpain to PIP2 was studied by protein-liposome assay with MicroSpin columns. When m-calpain interacts with the PIP2-containing liposomes, the amount of eluted enzyme is increased. For in vitro experiments (A and B), purified m-calpain was phosphorylated in vitro by PKA only or successively by ERK1 and PKA. For the phosphorylation by PKA only (A), purified m-calpain was incubated for 2 h with PKA with and without PKI in PKA phosphorylation buffer. m-calpain was then used for the protein-liposome assay. The experiment was also realized without PKA and without liposome as control. The amount of eluted m-calpain was visualized by immunoblot. For the successive phosphorylations of m-calpain by ERK1 and PKA (B), purified m-calpain was incubated 30 min with or without active ERK1 in ERK phosphorylation buffer. Then, PKA phosphorylation buffer containing PKA with and without PKI was added to the samples. The samples were then used for liposome binding assay. The amount of eluted m-calpain was visualized by immunoblot and compared with the input. For in vivo experiments (C), NR6WT fibroblasts, transfected with GFP-WT-m-calpain, GFP-ST369AA-m-calpain, or GFP-S369E-m-calpain, were treated with EGF to induce the phosphorylation of m-calpain by ERK or with EGF and IP-10 to also induce PKA-mediated phosphorylation. After a 30-min incubation, the proteins were extracted, and GFP-m-calpain was immunoprecipitated (IP). The enzyme was then used for a protein-liposome assay. The amount of eluted m-calpain was visualized by immunoblot (IB). For in vitro calpain activity assay (D), the GFP-m-calpains were immunoprecipitated in non-denaturing conditions. The eluted samples were desalted, and the activity was measured in vitro using a kit. The results are presented as percentage of the activity of the wild type m-calpain. ImageJ software was used to quantify the immunoblots. The results are presented as the mean ± S.D.
FIGURE 8.
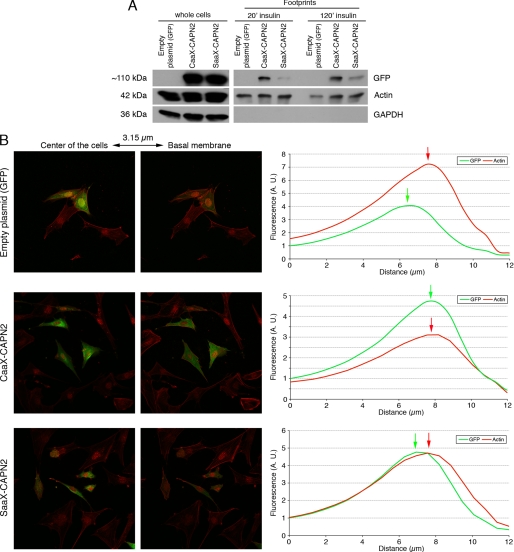
Localization of the CAAX/SAAX-calpain 2 at the membrane of the murine fibroblasts. The correct anchorage of the CAAX-calpain 2 was verified by extracting the proteins of the footprints (A) and by confocal microscopy (B). The NR6WT fibroblasts were transfected with the different plasmids and the siRNA mix. After an overnight incubation (in the presence of 5 μm MDL28170 for CAAX), the cells were washed and treated with 100 nm insulin for 20 min. For footprints isolation, the cells were also treated for 120 min. The cells were then unroofed to extract the footprints (A) or fixed, permeabilized, and stained for actin fibers with rhodamine-phalloidin (B). The proteins extracted from the footprints were loaded on SDS-PAGE, and the membranes were stained for GFP, actin (loading control), and GAPDH (to control the absence of contamination of the footprints by cytosolic proteins). For the confocal microscopy, the cells were observed, and pictures were taken every 0.63 μm. The distances presented on the x axis correspond to the distance from the top of the cells. The fluorescence of each stack was quantified using Metamorph software. A.U., absorbance units.
PKA Phosphorylation of m-calpain Does Not Directly Inhibit the Enzyme Activity
Previous works have clearly shown that the PKA phosphorylation of m-calpain induces the inhibition of this enzyme. However, the mechanism of this inhibition is not known, and we do not know if the phosphorylation is directly responsible for the inhibition or if it blocks an activation step. We have, therefore, assessed the in vitro activity of the S369E m-calpain. This mutant form as well as the wild type m-calpain and the ST369AA mutant, was expressed in fibroblasts and purified by non-denaturing immunoprecipitation. The activity was quantified using in vitro calpain activity assay. As shown in Fig. 6D, this mutant m-calpain mimicking the PKA phosphorylation presents the same in vitro activity as the wild type and ST369AA forms of m-calpain. The fact that this enzyme can still be activated shows that the PKA phosphorylation of m-calpain does not block directly the activity. This result is correlated with the data obtained for m-calpain localization. The PKA phosphorylation would inhibit m-calpain by inducing its removal from the membrane. The membrane localization of m-calpain would, thus, be responsible for the enzyme activation.
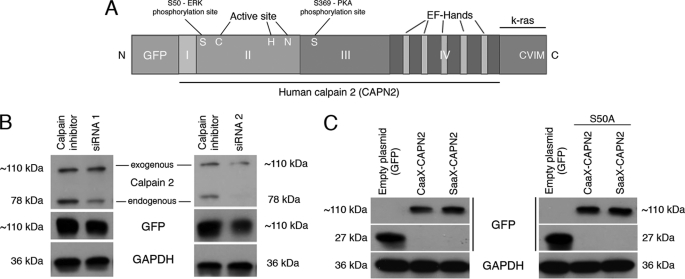
Anchorage of m-calpain at the Plasma Membrane Induces Its Activation
Our results and those obtained in previous studies seem to show that the localization of m-calpain at the plasma membrane would be responsible for the enzyme activation. To test this hypothesis and to determine the role of PIP2 in the activation process of m-calpain (cofactor or docking site), we have studied the effects of the anchorage of m-calpain at the membrane on its activity. In this aim we have created a mutant calpain 2 subunit including the farnesylation sequence (CAAX) of K-Ras at the C terminus of the protease (and GFP for identification and tracking at the N terminus; Fig. 7A). The farnesylation sequence of K-Ras contains the four amino acids CVIM, leading to the farnesylation of the protein on the cysteine, and a polylysine domain allowing the localization at the membrane without palmitoylation and passage through the Golgi (contrary to H- and N-Ras (46)). A SAAX mutant (the cysteine of CAAX was replaced by a serine) was also created and used as a negative control. To express the human CAAX- and SAAX-calpain 2 in murine fibroblasts, we have down-regulated the endogenous expression of m-calpain using two siRNA (Fig. 7B). As shown in Fig. 7C, the CAAX- and SAAX-calpain 2 were correctly expressed in the cells. To verify the correct anchorage of the CAAX-calpain 2 at the plasma membrane, we extracted the footprints of the transfected cells. The cells were treated for 20 or 120 min with insulin before the extraction to activate the farnesyltransferase enzyme required for the farnesylation of the mutant protein (36). As shown in Fig. 8A, the CAAX-calpain 2 is correctly localized at the plasma membrane of the cells treated with insulin. The SAAX-calpain 2, expressed at the same level as CAAX-calpain 2 in the whole cells, is not present at the membrane of the cells treated with insulin for 20 min. However, the amount of SAAX observed in the footprints of the cells is slightly increased after 2 h of insulin treatment but is still weaker than the amount of CAAX. This redistribution of the SAAX-calpain 2 at the membrane after 2 h is probably due to the phosphorylation of the protein by ERK. The anchorage of the CAAX-calpain 2 at the plasma membrane was confirmed by confocal microscopy. Although the GFP and the SAAX-calpain 2 were mainly localized in the cell cytoplasm and were absent at the basal membrane, the CAAX-calpain 2 was localized at the membrane, colocalizing with the actin fibers (Fig. 8B). The fluorescence was quantified, and the data showed that the peaks of fluorescence observed for GFP and actin are very close for CAAX-calpain 2, whereas they are separated for GFP and SAAX-calpain 2. The same phenomenon was observed with cells stained for the adhesion plaque component and calpain substrate vinculin rather than actin fibers (supplemental Fig. S7). Moreover, the fluorescence observed for actin and vinculin was decreased for CAAX m-calpain in comparison to GFP and SAAX m-calpain (Fig. 8B and supplemental Fig. S7). This could be due to an increased degradation of vinculin and to the disorganization of the actin cytoskeleton.
FIGURE 7.
Structure and expression of the CAAX- and SAAX-calpain 2. The structure of the CAAX-calpain 2 is represented in A. The expression of the endogenous murine m-calpain was knocked down using siRNA directed against the 3′-UTR of the gene before the expression of the mutated calpain 2 (B). To be sure that the siRNA was not blocking the expression of the human calpain 2, the cells were transfected with the plasmid encoding the GFP-calpain 2 (human) with or without the siRNA. The cells transfected without siRNA were treated with calpain inhibitor to reduce the mortality due to calpain overexpression. GAPDH was using as loading control (B). The mutant forms of calpain 2 (CAAX- and SAAX-calpain 2, CAAX- and SAAX-calpain 2 S50A) were transfected in murine fibroblasts as the same time than the siRNA mix (C). The correct expression of the mutants was observed by immunoblots against GFP. GAPDH was used as a loading control.
As m-calpain is a heterodimer constituted by calpain 2 and by the small subunit calpain S1, we have verified that the mutation of the larger calpain 2 subunit has no effect on the interaction between this and calpain S1. As shown in supplemental Fig. S8A, the CAAX- and SAAX-calpain 2 constructs strongly colocalized with calpain S1. Moreover, the calpain S1 also colocalized with the CAAX-calpain 2 anchored at the membrane. This interaction of CAAX-/SAAX-calpain 2 with calpain S1 was confirmed by co-immunoprecipitation (supplemental Fig. S8B). It is likely that this interaction is correct at the molecular level as the SAAX-containing construct is activated or suppressed by EGF and CXCR3 ligands indistinguishable from the endogenous m-calpain.
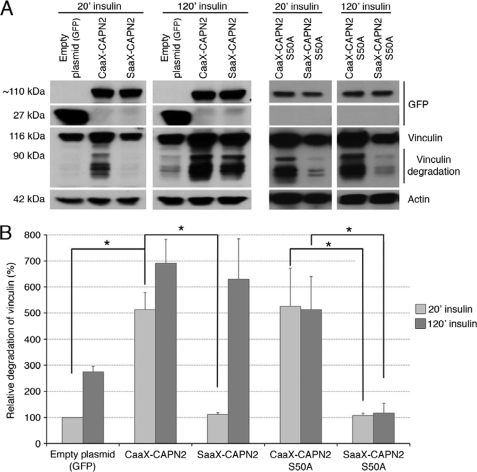
As the CAAX-calpain 2 is correctly anchored at the membrane of the cells and as its interaction with calpain S1 is not disrupted, we have assessed the effects of this anchorage on the enzyme activity. To this aim, the degradation of vinculin, a well known calpain substrate (47), was observed in cells transfected with the empty plasmid (GFP) or the plasmids encoding CAAX- and SAAX-calpain 2 and treated for 20 or 120 min with insulin. As shown in Fig. 9A, no degradation of vinculin was observed in the cells expressing GFP or SAAX-calpain 2 after 20 min of insulin treatment. On the contrary, an important degradation of vinculin was observed in the cells expressing the CAAX-calpain 2, attesting to a strong calpain activity. Indeed, the degradation is five times more in the cells expressing CAAX-calpain 2 than in the cells expressing equal amounts of the control GFP and SAAX-calpain 2 plasmids (Fig. 9B). When the cells were treated with insulin for 120 min, the degradation of vinculin was increased in the cells expressing CAAX-calpain 2 and also SAAX-calpain 2, probably due to ERK phosphorylation. The anchorage of calpain 2, therefore, induces a strong activation of the enzyme.
FIGURE 9.
Degradation of vinculin in the cells expressing the different mutants of m-calpain. The activity of the different mutants of m-calpain was estimated by quantifying the degradation of a well known calpain substrate, the vinculin. The NR6WT fibroblasts were transfected with the different plasmids and the siRNA mix. After an overnight incubation (in the presence of 5 μm MDL28170 for CAAX and CAAX S50A), the cells were washed and treated with 100 nm insulin for 20 or 120 min. The cells were washed and lysed, and the extracted proteins were loaded on a SDS-PAGE gel. The membranes were stained for GFP, vinculin, and actin (loading control). The band corresponding to the full-length vinculin was observed at 116 kDa. The bands observed below correspond to the degradation of the protein. The immunoblots (presented in A) were quantified using ImageJ. The degradation of vinculin is presented in B as a percentage (100% for the cells transfected with the empty plasmid and treated with insulin for 20 min). The results are presented as the mean ± S.D. *, significantly different (p < 0.05).
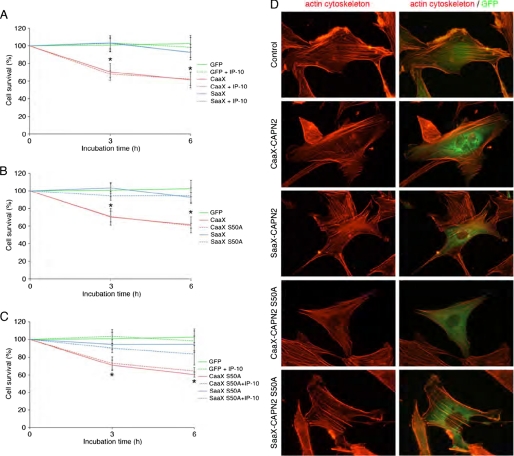
A strong increase of calpain activity can lead to the disorganization of the actin cytoskeleton and to cell death. We have, thus, quantified the survival of the cells expressing GFP alone, CAAX-calpain 2, and SAAX-calpain 2 and observed the organization of the actin cytoskeleton in these cells. As shown in Fig. 10A, the expression of CAAX-calpain 2 induces a significant increase of the cell death (40% of dead cells after 6 h), whereas the expression of SAAX-calpain 2 has no effect on cell survival. The strong activity induced by the anchorage of calpain 2 leads, therefore, to the death of the cells. To know if the anchored m-calpain could be inhibited by the PKA phosphorylation of the serine 369, we have repeated the same experiment in the presence of IP-10. No significant difference was observed between the cells treated with or without IP-10 (Fig. 10A), showing that the activity induced by the anchorage of m-calpain at the membrane is resistant to PKA phosphorylation. The observation of the cytoskeleton of the cells expressing GFP, CAAX, or SAAX-calpain 2 shows that the expression of CAAX-calpain 2 induces a decrease in the number of actin fibers (Fig. 10D). Even if the cytoskeleton is not completely disorganized, the fibers are less numerous and concentrated in the periphery of the CAAX-expressing cells.
FIGURE 10.
Effects of the expression of the different mutant forms of m-calpain on cell survival (A–C) and on actin cytoskeleton organization (D). For cell survival, the NR6WT fibroblasts were transfected with the different plasmids and the siRNA mix. After an overnight incubation (in the presence of 5 nm MDL28170 for CAAX and CAAX S50A), the cells were washed and treated with 100 nm insulin for 20 min. The cells were then washed and incubated in the presence or in the absence of 15 ng/ml IP-10 for 0, 3, and 6 h. Pictures of three different fields were taken at each time point, and the GFP expressing cells were counted. The results were expressed as percentage of the cells counted at the beginning of the experiment. The results are presented as the mean ± S.D. *, significantly different from the results obtained for GFP (p < 0.05). For the observation of the actin cytoskeleton (D), the transfected cells treated with insulin were fixed, permeabilized, and stained for actin fibers using rhodamine-phalloidin.
The Membrane-anchored Calpain 2 Does Not Require ERK Phosphorylation for Its Activation
Activation of calpain 2 upon membrane anchoring could be due to localization for ERK phosphorylation independent of growth factors. To determine whether the anchored calpain 2 still requires ERK phosphorylation to be active, we have mutated the CAAX- and SAAX-calpain 2 by replacing the serine 50 by an alanine. As shown in Fig. 7C, right panel, the CAAX- and SAAX-calpain 2 S50A are correctly expressed in the cells. The anchorage of the CAAX-calpain 2 S50A was confirmed by confocal microscopy like for CAAX- and SAAX-calpain 2 (supplemental Figs. S6 and S7). The activity of the mutants was assessed as above. The degradation of vinculin and, therefore, the calpain activity, is strongly increased in the CAAX-calpain 2 S50A after 20 min of insulin treatment (Fig. 9, A, right panels, and B). On the contrary, the expression of the SAAX-calpain 2 S50A does not increase the degradation of vinculin. The treatment of the cells with insulin for 120 min has no effect on the degradation of vinculin for both CAAX- and SAAX-calpain 2 S50A. The increased activity observed for the SAAX-calpain 2 but not the SAAX-calpain 2 S50A observed after 120 min of insulin treatment was, thus, due to the phosphorylation of the enzyme by ERK.
The effects of the mutation S50A on the cell survival and the actin cytoskeleton were also observed. The results show that the mutation S50A has no effect on the cell death induced by the expression of CAAX-calpain 2. Indeed no significant difference was observed between the cells expressing CAAX-calpain 2 and those expressing CAAX-calpain 2 S50A (Fig. 10B). As for SAAX-calpain 2, no cell death was observed when the cells express SAAX-calpain 2 S50A. Moreover, the IP-10-induced PKA phosphorylation of CAAX- and SAAX-calpain 2 S50A has no effect on the cell survival (Fig. 10C). Concerning the actin cytoskeleton organization, the results obtained for CAAX- and SAAX-calpain 2 S50A are very similar to those obtained for the non-mutated CAAX- and SAAX-calpain 2. Taken together, these results show that by anchoring calpain 2 within the plasma membrane milieu, it can be activated independently of ERK phosphorylation.
PIP2 Is Required for the Activation of the Membrane-anchored m-calpain
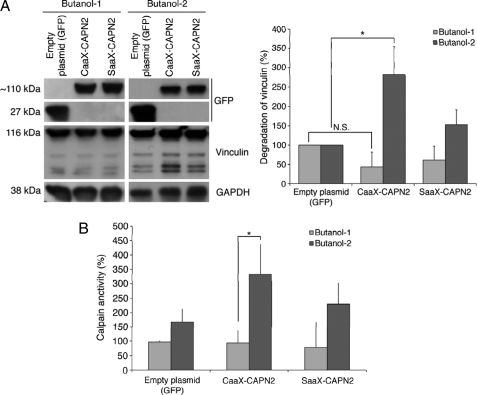
The phosphoinositide PIP2 was previously shown to be required for the activation for m-calpain (31); however, its exact mechanistic role during the activation process remains unknown. PIP2 could simply be a docking site, allowing m-calpain localization at the membrane for subsequent activation, or it could be a docking site and a cofactor (48). We bypassed the PIP2-dependent localization of m-calpain at the membrane by anchoring the enzyme and found this enzyme to be active. To explore the role of PIP2 as a cofactor, we quantified the activity of the CAAX- and SAAX-calpain 2 in the absence of PIP2 using butanol-1 to deplete the phosphoinositide and butanol-2 as a control. The results presented in Fig. 11 clearly show that the CAAX-calpain 2 is still strongly activated after treatment with butanol-2; however, the depletion of PIP2 due to the butanol-1 treatment blocks the enzyme activation. Indeed the activity of the CAAX-calpain 2 after butanol-1 treatment is not significantly different from GFP and SAAX (Fig. 11, A and B). These data show that the binding of m-calpain to PIP2 is still required for the enzyme activation even after membrane anchorage. PIP2 is, thus, a docking site and a cofactor of m-calpain.
FIGURE 11.
Effects of the depletion of PIP2 on the activity of CAAX- and SAAX-calpain 2. The activity of CAAX- and SAAX-calpains was assessed by observing vinculin degradation (A) or by using t-Boc-LM-CMAC (B) after treatments with butanol-1 or butanol-2. To deplete PIP2, the transfected NR6WT cells were treated for 45 min with butanol-1 (1.5%). Butanol-2 was used as a negative control. Insulin (100 nm) was added to the medium for the last 20 min of the treatment to induce the activation of farnesyltransferase. The results of the quantification are expressed as percentage (100% for cells transfected with the empty plasmid). *, significantly different (p < 0.05).
DISCUSSION
The ubiquitous calpains are intimately involved in cell migration both under physiological conditions (during wound closure) and pathological conditions (during tumor invasion). For this reason, understanding the regulation of these enzymes is critical. For μ-calpain, previous studies have highlighted the role played by calcium influxes in its activation. Concerning m-calpain, the regulation process remains uncertain. The amount of calcium required for the in vitro activation of this enzyme is clearly not compatible with cell survival. Recent studies have proved that m-calpain activity is regulated by phosphorylation, particularly during wound healing. Indeed, the phosphorylation of the serine 50, induced by growth factors and mediated by the ERK/MAPK pathway, was shown to trigger m-calpain activation (29). On the contrary, the PKA phosphorylation of the serine 369 induced by the CXCR3 ligands inhibits the enzyme activity, blocking in this way the growth factor-induced migration (33). The effects of the EGF-induced phosphorylation of m-calpain were described recently. The ERK phosphorylation promotes the binding of m-calpain to PIP2 and, therefore, its localization at the rear and at the plasma membrane of the cells, close to its substrates (31). To the contrary, the inhibition mechanism induced by the CXCR3 ligands remains unknown. The PKA phosphorylation at the interface of domains 3 and 4 has been modeled to result in a salt bridge that “locks” the molecule in an inactive conformation. However, domain 3 is also the C2-like domain in m-calpain that has weak PIP2 binding affinity. Altering domain 3 flexibility would likely further reduce the ability to complex with PIP2. Moreover, the removal of m-calpain from the membrane, where its substrates are located, would be a very efficient way to block cell retraction and subsequently cell migration.
Therefore, our first aim was to identify the effects of the CXCR3 ligands on the growth factor-induced relocalization of m-calpain at the membrane. Moreover, it was unknown whether IP-9 and IP-10 were dominant over the growth factors and, thus, if their effects are immediate. Our data show that the EGF-induced migration of fibroblasts is strongly inhibited by the chemokines within minutes after their addition (Fig. 1, A, D, and E). Importantly, similar results were obtained with endothelial cells treated with EGF or VEGF, showing that this phenomenon is not specific for fibroblasts and EGF (Fig. 1, B, C, E, and F). These data prove that the CXCR3 chemokines are able to override the growth factor-induced stimulation of cell migration and that these inhibitory effects are almost immediate. This inhibition of cell motility is at least in part due to preventing cell retraction, as observed by cell tracking within minutes after the addition of the chemokines (Fig. 1H and supplemental Movie 1). This result agrees with previous studies showing that IP-9 and IP-10 are able to inhibit m-calpain, which is critically involved in cell retraction by cleaving several components of the adhesion complexes (11). Moreover, we found that CXCR3 ligands not only prevented growth factor-induced m-calpain relocalization in fibroblasts (Fig. 2) but also reverted membrane localization when added after the growth factors (Fig. 3). Similar results were obtained with endothelial cells treated with EGF or with the angiogenic growth factor VEGF, showing that the effects of CXCR3 chemokines are generalizable (supplemental Figs. S1–S3). It is also interesting to note that the chemokines have no effect on the shape of the growth factor-stimulated cells. Indeed, the cell polarity remained with lamellipodia still present at the front of the cells (Fig. 3). By inhibiting the localization of m-calpain at the rear of the cells without blocking the formation of the lamellipodia, the CXCR3 ligands inhibit cell retraction (and, thus, productive locomotion), whereas still allowing for matrix contraction required for dermal maturation. This would not be possible if both cell retraction and lamellipodia formation were blocked (28). Relocalization of m-calpain to the membrane in the cell body and rear results from increased binding of m-calpain to PIP2 (31). Our results show that the CXCR3 chemokines prevent and even revert this EGF-induced localization of m-calpain at the plasma membrane of both fibroblasts and endothelial cells (Fig. 4 and supplemental Fig. S4). The chemokines are, thus, able to separate m-calpain from its substrates, preventing the cleavage of the adhesion complexes. This phenomenon is not due to a modification of the PIP2 asymmetry that determines cell front from rear (Fig. 5A). In agreement with this, the chemokines have no effect on the activation of PLCγ1 by EGF, which is responsible for the PIP2 asymmetric relocation away from the lamellipodia (Fig. 5B). It was, thus, interesting to study the effects of the CXCR3 ligands on the ability of m-calpain to bind PIP2. Our liposome binding assays showed that the phosphorylation of m-calpain by PKA reduced the interaction between m-calpain and PIP2 even when m-calpain was previously phosphorylated by ERK1 (Fig. 6, A and B). Moreover, the treatment of the cells with IP-10 reduced the EGF-stimulated ability of m-calpain to bind PIP2. This effect is clearly due to the PKA phosphorylation of m-calpain on the serine 369, as IP-10 has no effect on the PKA-resistant mutant form of m-calpain (ST369AA). It is also possible to prevent the effects of EGF by mimicking this phosphorylation with the S369E-m-calpain (Fig. 6C). By blocking the interaction between PIP2 and m-calpain, the PKA phosphorylation of m-calpain is, thus, clearly responsible for the effects of the CXCR3 ligands on m-calpain redistribution at the rear and at the plasma membrane of the cells.
Our results and those obtained in previous studies seem to show that the PIP2-dependent localization of m-calpain at the membrane could be responsible for its activation and that the phosphorylations would regulate m-calpain activity only indirectly. Second, we have, thus, studied the effects of the anchorage of m-calpain at the membrane on its activity. To this aim, we have created a mutant calpain 2 containing the K-Ras farnesylation sequence (CAAX, Fig. 7A). The CAAX-calpain 2 was correctly anchored at the plasma membrane, whereas the negative control SAAX-calpain 2 was mainly cytoplasmic and totally absent from the membrane (Fig. 8, A and B, and supplemental Fig. S7). This artificial and forced localization of calpain 2 at the cell membrane triggered a very strong activation of the enzyme and subsequently the degradation of its substrates (Fig. 9). As an important control, the SAAX-calpain 2 presents no increased activity when the cells are treated with insulin for only 20 min. The activity of the SAAX-calpain 2 is increased only after a 120-min insulin treatment because of ERK phosphorylation. The increased activity observed with CAAX-calpain 2 induced the death of the cells expressing this mutant protease and disorganized the actin cytoskeleton (Fig. 10, A and D), probably by degrading the protein of the adhesion complexes. The activity of the anchored calpain 2 is independent of the ERK phosphorylation. Indeed, the mutation of the serine 50 to alanine has no effect on the activity of the CAAX-calpain 2 and on the subsequent cell death (Figs. 9 and 10B). On the contrary, the mutation S50A blocks the insulin-induced activation of the SAAX-calpain 2. In the same manner the CAAX-calpain 2 is resistant to the PKA phosphorylation. Indeed the addition of IP-10 has no effect on the cell death resulting from CAAX-calpain 2 expression. Very similar results were also obtained when the cells expressing CAAX-calpain 2 S50A were treated with IP-10. However, the activation of the CAAX-calpain 2 still requires the presence of PIP2 (Fig. 11). This phosphoinositide was previously shown to be crucial for the activation of m-calpain (31); however, it was unclear if it was only a docking site necessary for the membrane localization or also a cofactor allowing m-calpain activity. Our data show that even anchored at the membrane and, thus, not relying on PIP2 binding to be localized at the membrane, m-calpain still requires the presence of PIP2 to be activated. These very interesting results show clearly that PIP2 acts a cofactor of m-calpain, crucial for the enzyme activation. The hypothesis of PIP2 being the final activating factor is intellectually satisfying as the charged head group of PIP2 may subsume the electrostatic structural rearrangement directed by calcium at supraphysiological levels (24).
On the surface these findings and the resulting model of the Ser-50 and Ser-369 phosphorylations on m-calpain controlling activation only indirectly due to regulation of localization within the plasma membrane milieu of PIP2 sites would appear to conflict with earlier reports from our own studies (29, 49). Although we do not have a structural basis for this seeming discrepancy, the main reasons for divergent findings are likely the difference between activation in cells and activation of bacterially produced calpains. As m-calpain is multiply, although substoichiometrically phosphorylated on numerous serines and threonine in cells (1), these other post-translational modifications absent in bacterially expressed m-calpain may further constrain this potent and irreversible signaling protease (7). Intriguingly, we were not able to structurally model the Ser-50 phosphorylation as this is in a highly disordered domain, suggesting that phosphorylation allows activation by either removing steric hindrance to the active cleft or, more likely, based on the observed data, that phosphorylation serves to help bind the m-calpain to its membrane cofactor PIP2. Still, the molecular structural basis of this activation is speculative at present and awaits experimentation beyond the scope of the present missive.
In conclusion, our results clearly show that the localization of m-calpain at the plasma membrane and its binding to PIP2 are the keystone of its activation process. The ERK phosphorylation of m-calpain induces the enzyme localization at the membrane by promoting the binding between m-calpain on PIP2 leading to its activation. On the contrary, PKA phosphorylation, induced by CXCR3 ligands, blocks the binding to PIP2 and, thus, induces the removal of m-calpain from the membrane and its inactivation. The phosphorylations of m-calpain have no direct effect on the enzyme activity. They control indirectly the activation process by controlling the redistribution of the protease, therefore allowing, preventing, or reverting its activation.
Supplementary Material
Acknowledgments
We thank members of the Wells and Roy laboratories and Stephen Chirieleison for helpful comments and technical assistance. We also thank Prof. Chang Lu of Purdue University for providing the plasmid pEGFP-C1.
This work was supported, in whole or in part, by National Institutes of Health NIGMS grant GM069668.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Movie 1.
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PKI
- cAMP-dependent protein kinase inhibitor
- PLCγ1
- phospholipase C gamma 1
- VEGF
- vascular endothelial growth factor
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 2.Lauffenburger D. A., Horwitz A. F. (1996) Cell 84, 359–369 [DOI] [PubMed] [Google Scholar]
- 3.Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 4.Sheetz M. P., Felsenfeld D., Galbraith C. G., Choquet D. (1999) Biochem. Soc. Symp. 65, 233–243 [PubMed] [Google Scholar]
- 5.Cox E. A., Huttenlocher A. (1998) Microsc. Res. Tech. 43, 412–419 [DOI] [PubMed] [Google Scholar]
- 6.Hautaniemi S., Kharait S., Iwabu A., Wells A., Lauffenburger D. A. (2005) Bioinformatics 21, 2027–2035 [DOI] [PubMed] [Google Scholar]
- 7.Glading A., Lauffenburger D. A., Wells A. (2002) Trends Cell Biol. 12, 46–54 [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni S., Saido T. C., Suzuki K., Fox J. E. (1999) J. Biol. Chem. 274, 21265–21275 [DOI] [PubMed] [Google Scholar]
- 9.Satish L., Blair H. C., Glading A., Wells A. (2005) Mol. Cell. Biol. 25, 1922–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooray P., Yuan Y., Schoenwaelder S. M., Mitchell C. A., Salem H. H., Jackson S. P. (1996) Biochem. J. 318, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., Huttenlocher A. (2004) Nat. Cell Biol. 6, 977–983 [DOI] [PubMed] [Google Scholar]
- 12.Palecek S. P., Huttenlocher A., Horwitz A. F., Lauffenburger D. A. (1998) J. Cell Sci. 111, 929–940 [DOI] [PubMed] [Google Scholar]
- 13.Glading A., Chang P., Lauffenburger D. A., Wells A. (2000) J. Biol. Chem. 275, 2390–2398 [DOI] [PubMed] [Google Scholar]
- 14.Huttenlocher A., Ginsberg M. H., Horwitz A. F. (1996) J. Cell Biol. 134, 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broughton G., 2nd, Janis J. E., Attinger C. E. (2006) Plast. Reconstr. Surg. 117, Suppl. 7, 1e-S–32e-S [DOI] [PubMed] [Google Scholar]
- 16.Diegelmann R. F., Evans M. C. (2004) Front Biosci. 9, 283–289 [DOI] [PubMed] [Google Scholar]
- 17.Li J., Zhang Y. P., Kirsner R. S. (2003) Microsc. Res. Tech. 60, 107–114 [DOI] [PubMed] [Google Scholar]
- 18.Raja, Sivamani K., Garcia M. S., Isseroff R. R. (2007) Front Biosci. 12, 2849–2868 [DOI] [PubMed] [Google Scholar]
- 19.Imanishi J., Kamiyama K., Iguchi I., Kita M., Sotozono C., Kinoshita S. (2000) Prog. Retin. Eye Res. 19, 113–129 [DOI] [PubMed] [Google Scholar]
- 20.Kunimoto B. T. (1999) Ostomy. Wound Manage. 45, 56–64 [PubMed] [Google Scholar]
- 21.Wells A., Gupta K., Chang P., Swindle S., Glading A., Shiraha H. (1998) Microsc. Res. Tech. 43, 395–411 [DOI] [PubMed] [Google Scholar]
- 22.Nedelec B., Ghahary A., Scott P. G., Tredget E. E. (2000) Hand Clin. 16, 289–302 [PubMed] [Google Scholar]
- 23.Bodnar R. J., Yates C. C., Wells A. (2006) Circ. Res. 98, 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moldoveanu T., Hosfield C. M., Jia Z., Elce J. S., Davies P. L. (2001) Biochim. Biophys. Acta 1545, 245–254 [DOI] [PubMed] [Google Scholar]
- 25.Yates C. C., Whaley D., Y.-Chen A., Kulesekaran P., Hebda P. A., Wells A. (2008) Am. J. Pathol. 173, 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yates C. C., Whaley D., Hooda S., Hebda P. A., Bodnar R. J., Wells A. (2009) Wound Repair Regen. 17, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao H., Yi X. M., Wells A. (2008) Wound Repair Regen. 16, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen F. D., Asnes C. F., Chang P., Elson E. L., Lauffenburger D. A., Wells A. (2002) Wound Repair Regen. 10, 67–76 [DOI] [PubMed] [Google Scholar]
- 29.Glading A., Bodnar R. J., Reynolds I. J., Shiraha H., Satish L., Potter D. A., Blair H. C., Wells A. (2004) Mol. Cell. Biol. 24, 2499–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glading A., Uberall F., Keyse S. M., Lauffenburger D. A., Wells A. (2001) J. Biol. Chem. 276, 23341–23348 [DOI] [PubMed] [Google Scholar]
- 31.Shao H., Chou J., Baty C. J., Burke N. A., Watkins S. C., Stolz D. B., Wells A. (2006) Mol. Cell. Biol. 26, 5481–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou J., Stolz D. B., Burke N. A., Watkins S. C., Wells A. (2002) Int. J. Biochem. Cell Biol. 34, 776–790 [DOI] [PubMed] [Google Scholar]
- 33.Shiraha H., Glading A., Chou J., Jia Z., Wells A. (2002) Mol. Cell. Biol. 22, 2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith K. D., Wells A., Lauffenburger D. A. (2006) Exp. Cell Res. 312, 1970–1982 [DOI] [PubMed] [Google Scholar]
- 35.Shiraha H., Gupta K., Drabik K., Wells A. (2000) J. Biol. Chem. 275, 19343–19351 [DOI] [PubMed] [Google Scholar]
- 36.Goalstone M. L., Draznin B. (1996) J. Biol. Chem. 271, 27585–27589 [DOI] [PubMed] [Google Scholar]
- 37.Stolz D. B., Jacobson B. S. (1992) J. Cell Sci. 103, 39–51 [DOI] [PubMed] [Google Scholar]
- 38.Chaney L. K., Jacobson B. S. (1983) J. Biol. Chem. 258, 10062–10072 [PubMed] [Google Scholar]
- 39.Tompa P., Emori Y., Sorimachi H., Suzuki K., Friedrich P. (2001) Biochem. Biophys. Res. Commun. 280, 1333–1339 [DOI] [PubMed] [Google Scholar]
- 40.Shiraha H., Glading A., Gupta K., Wells A. (1999) J. Cell Biol. 146, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kharait S., Hautaniemi S., Wu S., Iwabu A., Lauffenburger D. A., Wells A. (2007) BMC Syst. Biol. 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodnar R. J., Yates C. C., Rodgers M. E., Du X., Wells A. (2009) J. Cell Sci. 122, 2064–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satish L., Yager D., Wells A. (2003) J. Invest. Dermatol. 120, 1110–1117 [DOI] [PubMed] [Google Scholar]
- 44.Satish L., Babu M., Tran K. T., Hebda P. A., Wells A. (2004) Wound Repair Regen. 12, 183–192 [DOI] [PubMed] [Google Scholar]
- 45.Chou J., Burke N. A., Iwabu A., Watkins S. C., Wells A. (2003) Exp. Cell Res. 287, 47–56 [DOI] [PubMed] [Google Scholar]
- 46.Hancock J. F. (2003) Nat. Rev. Mol. Cell Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 47.Serrano K., Devine D. V. (2004) Cell Motil. Cytoskeleton 58, 242–252 [DOI] [PubMed] [Google Scholar]
- 48.Pontremoli S., Melloni E., Sparatore B., Salamino F., Michetti M., Sacco O., Horecker B. L. (1985) Biochem. Biophys. Res. Commun. 129, 389–395 [DOI] [PubMed] [Google Scholar]
- 49.Smith S. D., Jia Z., Huynh K. K., Wells A., Elce J. S. (2003) FEBS Lett. 542, 115–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.