Abstract
The future clinical use of embryonic stem cell (ESC)-based hepatocyte replacement therapy depends on the development of an efficient procedure for differentiation of hepatocytes from ESCs. Here we report that a high density of human ESC-derived fibroblast-like cells (hESdFs) supported the efficient generation of hepatocyte-like cells with functional and mature hepatic phenotypes from primate ESCs and human induced pluripotent stem cells. Molecular and immunocytochemistry analyses revealed that hESdFs caused a rapid loss of pluripotency and induced a sequential endoderm-to-hepatocyte differentiation in the central area of ESC colonies. Knockdown experiments demonstrated that pluripotent stem cells were directed toward endodermal and hepatic lineages by FGF2 and activin A secreted from hESdFs. Furthermore, we found that the central region of ESC colonies was essential for the hepatic endoderm-specific differentiation, because its removal caused a complete disruption of endodermal differentiation. In conclusion, we describe a novel in vitro differentiation model and show that hESdF-secreted factors act in concert with regional features of ESC colonies to induce robust hepatic endoderm differentiation in primate pluripotent stem cells.
Keywords: Cell Differentiation, Embryonic Stem Cell, Hepatocyte, Induced Pluripotent Stem Cell (iPSC), Liver
Introduction
Patients with end stage and genetic liver diseases have been shown to benefit from liver cell replacement therapy (1). However, one of the major factors currently limiting the utilization of cellular replacement in the general population with severe liver disease is the lack of consistent sources of transplantable hepatocytes. Ideally, a proliferative stem cell population with robust capacity to generate hepatic cells in vitro will overcome the shortage of transplantable hepatic cells. Over the past several years, pluripotent embryonic stem cells (ESCs)3 have been successfully derived from nonhuman primates (2–4) and human embryos (5, 6). The ability of these cells to proliferate indefinitely and to develop into virtually any cell or tissue type, including hepatocytes, makes them likely candidates for cell-based treatment of human diseases.
Over the past few decades, studies in vertebrate models have described several signaling pathways critical for the embryonic development of hepatocytes (7, 8). For example, it has been shown that selective FGFs can substitute for the cardiac mesoderm to activate the expression of hepatic genes in the endoderm (9). In addition to the FGF pathway, bone morphogenic factors (BMPs), which are highly expressed in the septum transversum mesenchyme, are capable of inducing early hepatic fate independently in mice (10). Subsequently, Wnt signaling plays an important role in enhancing the growth of the liver bud (7, 11). Other inductive signals, such as activin (12, 13) and Sonic hedgehog (14, 15), also contribute to endoderm or early liver development. Terminal maturation of hepatocytes requires additional signals, such as hepatocyte growth factor (HGF) (16, 17) and oncostatin M (17, 18).
The future clinical use of ESC-based hepatocyte replacement therapy depends on the development of an efficient procedure for differentiation of hepatocytes from ESCs. To achieve this goal, researchers have successfully exploited the molecular signals for liver development outlined above to direct the development of mouse (19–21) and subsequently monkey (22, 23) and human (24–30) ESCs into hepatocyte-like cells (HLCs). Additionally, a few studies have demonstrated that mouse or monkey ESCs can be coaxed to generate HLCs in the presence of murine hepatocytes (31, 32) or human liver nonparenchymal cells (33).
Recent demonstrations of the generation of induced pluripotent stem cells (iPSCs) with defined transcription factors (34, 35) that allowed the derivation of patient- and disease-specific pluripotent stem cells (36, 37) without using human embryos have heightened interest in the in vitro hepatic differentiation potential of iPSCs (38–40). The demonstration of the hepatic differentiation capability of iPSCs suggests their possible application for in vitro personalized pharmacogenetics, toxicology, and metabolism studies and future in vivo transplantation trails.
In this study, we explored the potential of our previously established human ESC-derived fibroblast-like cells (hESdFs) (41) for derivation of hepatocytes from pluripotent stem cells. We found that exposure of monkey and human ESCs and human iPSCs to a high density of hESdFs induced robust hepatic endoderm differentiation in a regionally specific manner in ESC/iPSC colonies. Additionally, we elaborate the possible mechanisms responsible for such hepatic induction.
EXPERIMENTAL PROCEDURES
Cell Culture and Differentiation
Mitomycin C-inactivated mouse embryonic fibroblasts (MEFs) and hESdFs were generated as described previously (41). The monkey (ORMES-5 and ORMES-6) and human ESCs (NTU1, NTU2, and NTU3) (3) were grown on MEF feeders (2 × 104 cells/cm2) in DMEM/F-12 medium plus 20% knock-out serum replacement (Invitrogen) and basic FGF (4 ng/ml; Sigma-Aldrich). To induce differentiation, ESCs and iPSCs were grown on a high density of hESdFs (1 × 104 cells/cm2) in the ESC medium.
Immunocytochemistry (ICC)
ICC assay was performed as described previously (41). The antibodies used are listed in supplemental Table S1.
RNA Isolation, RT-PCR, and QRT-PCR
Assays were performed as described previously (41, 42). All of the primers used are listed in supplemental Table S2. All of the QRT-PCRs were performed in triplicate.
Quantitative Measurement of Albumin and Urea
Albumin and urea levels in the cell culture media were measured with ALB-G kits (Denka Seiken, Japan) or the ELISA kit for human albumin (Assaypro) and urea liquid kits (Sentinel Diagnostics), respectively. All of the steps were performed according to the manufacturers' instructions. Colorimetric determinations were made using Toshiba 200FR (Toshiba).
ELISA
Haptoglobin, activin A, and FGF2 levels in hESdF-conditioned media were determined by ELISA kits for human haptoglobin (Alpco Diagnostics, Salem, NH), activin A (R & D, Minneapolis, MN), and FGF2 (Sigma) according to the manufacturers' instructions.
Periodic Acid-Schiff Staining
The cells were fixed in 4% paraformaldehyde, treated with 0.5% periodic acid for 10 min, washed three times with distilled water, and treated with Schiff's buffer for 20 min. Afterward, the cells were washed with distilled water and stained with hematoxylin.
Ethoxyresorufin-O-deethylase
Ethoxyresorufin-O-deethylase assays were performed as described previously (43). The detailed procedure is included in the supplemental “Materials and Methods”.
Microarray Analysis and Data Processing
Ten μg of total RNA purified by TRIzol (Invitrogen) was used for cDNA synthesis and to generate biotin-labeled cRNA probe, which was hybridized to an Affymetrix Rhesus Macaque genome array (Affymetrix, Santa Clara, CA) by the Affymetrix Gene Expression Service Laboratory at Academia Sinica in Taiwan. The chips were scanned with an Affymetrix GeneChip Scanner 7G, and GeneSpring X software (Agilent, Santa Clara, CA) was used for data mining. Raw data were normalized by robust multichip average, and the weakly expressed signals (<20% of total samples) were excluded. The filtered genes were clustered by Pearson centered complete clustering. The average log intensities of the biological replicates were normalized to the average log intensity of ESCs to obtain the changes in expression between different conditions. Genes highly expressed in the liver, pancreas, or lung were identified with reference to UniGene transcriptome cluster annotation (National Center for Biotechnology Information). The raw microarray data are available through the Gene Expression Omnibus (GEO, GSE19964).
Statistical Analysis
All of the in vitro results were derived from triplicate experiments if not otherwise indicated. The results are presented as the means ± S.D. Grubb's test was used to detect possible outliers. Student's t test was used to examine the significance of differences between group means, and differences with p values less than 0.05 were considered significant.
RESULTS
hESdFs Induced and Promoted Regional Differentiation in Monkey and Human ESC Colonies
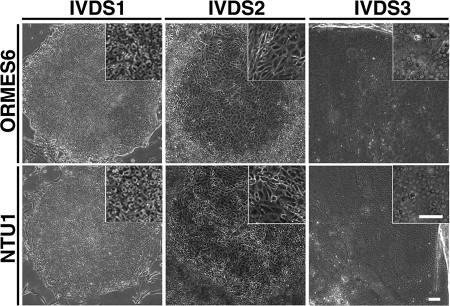
To see whether hESdFs could support the undifferentiated status of multiple primate ESC lines, we evaluated the growth pattern of monkey ORMES-5 and ORMES-6 (3) and human NTU1, NTU2, and NTU3 ESCs (44) on mitotically inactivated hESdFs. Surprisingly, we found that hESdFs induced the formation of a small flattened area with differentiated cells in the central area of ESC colonies of all examined ESC lines after prolonged co-culture without splitting. Subsequently, the early flattened area expanded, resulting in the formation of differentiated foci containing HLCs (Fig. 1). Moreover, we found that a higher density of hESdFs (5 × 104 cells/cm2) further enhanced the differentiation and eventually led to the formation of differentiated foci containing HLCs in the majority of human and monkey ESC colonies. In contrast, only a few ESC colonies were able to develop into HLC foci in the MEF co-culture group. We divided the differentiation process into three stages according to the morphological changes seen in the differentiated cells along the course of focus formation and expansion. The first 5-day period (days 1–5) after the seeding of ESCs on hESdFs was defined as in vitro differentiation stage (IVDS) 1. The colonies growing at this stage normally contained cells exhibiting characteristic ESC morphology, occasionally with a small differentiated area (Fig. 1). The following 5-day period (days 6–10) was defined as IVDS2. At this stage an extended flattened area with spiky differentiated cells started to emerge in the central region (Fig. 1). At approximately day 15 (IVDS3), differentiated cells exhibiting typical hepatocyte morphological features, such as compact cell-cell contacts, polygonal shapes, and distinctive round nuclei (Fig. 1), started to emerge. These eventually formed a homogenous hepatic-like cell population in a fully expanded focus surrounded by a bulge of compact piled up cells. The hepatic foci could be further maintained in serum-free culture for up to 1–2 months (supplemental Fig. S1).
FIGURE 1.
Stages of hESdF induced regional differentiation in human and monkey ESC colonies. The in vitro differentiation stages (IVDS1, IVDS2, and IVDS3) of ESC colonies were defined by the temporal morphological change of human or monkey ESCs after replating onto hESdF feeders. IVDS1 colonies (days 1–5) resembled ESC colonies; IVDS2 colonies (days 6–10) contained central foci filled with flattened and spiky cells. IVDS3 colonies (days 10–15) contained hepatocyte-like cells. Scale bar, 50 μm.
hESdF-primed Regional Differentiation Recapitulated Hepatic Endoderm Differentiation in Primate ESCs
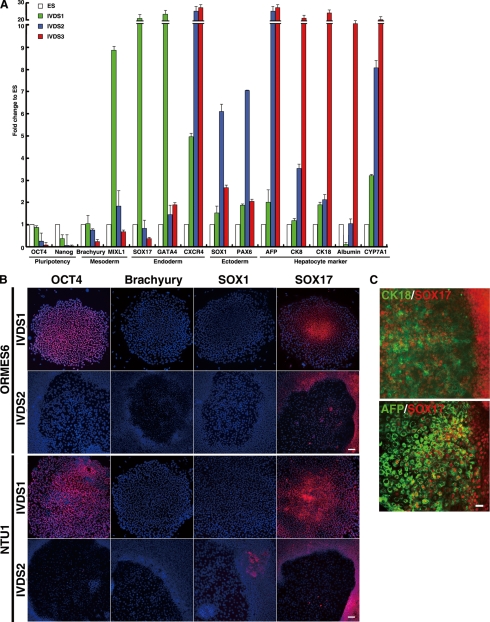
To confirm that the differentiated monkey ESCs in the foci did develop into endoderm and then hepatocytes in the hESdF-primed culture, we performed QRT-PCR analysis on the cells at various differentiation stages with primers specific to genes of three embryonic germ layers and hepatic lineages (Fig. 2A). The expression levels of pluripotency genes, Nanog and Oct4, gradually decreased at IVDS1 and IVDS2 and eventually disappeared at IVDS3 (Fig. 2A). In parallel, the expression levels of mesoendodermal (MIXL1) and early endodermal marker genes (SOX17 and GATA4) were highly up-regulated at IVDS1 (Fig. 2A), suggesting the induction of endodermal fate. Although the early endodermal marker genes were gradually down-regulated during IVDS2, expression of the definitive endodermal gene CXCR4 was up-regulated through IVDS2 and IVDS3 (Fig. 2A). Moreover, the expression of hepatocyte genes, AFP, CK8, and CK18, and mature hepatic marker genes, Albumin and CYP7A1, started at IVDS1–2 and peaked at IVDS3 (Fig. 2A), indicating a transition from endodermal to hepatic differentiation. In contrast, the expression levels of ectodermal and mesodermal genes were only transiently up-regulated or suppressed in the course of differentiation (Fig. 2A). To confirm whether hESdFs promoted homogenous hepatic endoderm differentiation among the differentiating ESC colonies, we randomly picked up individual colonies of monkey (ORMES6) and human ESCs (NTU1) at various differentiation stages and subjected them to QRT-PCR analysis with primers specific for genes of endoderm and hepatic lineages. We found that the relative endodermal and hepatic gene expression level did not vary significantly among individual colonies (supplemental Fig. S2), indicating that the differentiated ESC colonies exhibited a similar trend of differentiation under hESdF co-culture.
FIGURE 2.
hESdFs promote a sequential endodermal-to-hepatic differentiation in human and monkey ESCs. A, QRT-PCR analysis of IVDS 1, 2, and 3 for pluripotency-related markers (Oct4 and Nanog), mesodermal markers (Brachyury MIXL1), endodermal markers (SOX17, GATA4, and CXCR4), ectodermal markers (SOX1 and PAX 6), and hepatocyte markers (AFP, CK8, CK18, albumin, and CYP7A1). B, the expression of Oct4 (pluripotency-related), early ectodermal marker SOX1, early mesodermal Brachyury, and early endodermal SOX17 in differentiating human and monkey ESC colonies of IVDS 1 and 2 were analyzed by ICC. C, co-expression of endodermal marker SOX 17 with hepatocyte marker CK18 or AFP is as indicated. IVDS2 foci derived from monkey ESC were stained for SOX17 (red) and CK18 (green) or AFP (green). Scale bar, 25 μm.
To further dissect lineage information along the course of differentiation, we performed ICC analysis on monkey and human ESC colonies at different differentiation stages. We found that cells in the central area of ESC colonies expressed a high level of SOX17 at IVDS1 (Fig. 2B). Subsequently at IVDS2, expression of SOX17 and down-regulation of OCT4 were mainly detected in the peripheral ring region of the foci area of ESC colonies (Fig. 2B), whereas cells in the foci expressed SOX17 sporadically and weakly (Fig. 2B). These results suggest that most endoderm-like cells in the foci might have already differentiated into early hepatic cells at IVDS2. Double ICC for SOX17 and the hepatocyte markers AFP or CK18 demonstrated that CK18 and AFP were expressed in the majority of cells in the IVD2 foci, with sporadic cells co-expressing both SOX17 and CK18 or AFP (Fig. 2C). However, the expression of CK18 or AFP was completely absent from the SOX17-expressing cells surrounding the foci (Fig. 2C). Consistent with the RT-PCR results, the differentiation of both mesodermal and ectodermal lineages was mostly suppressed in the presence of hESdFs because Brachyury expression was absent in all stages, and only a few occasional SOX1-expressing cells were found outside the foci (Fig. 2B).
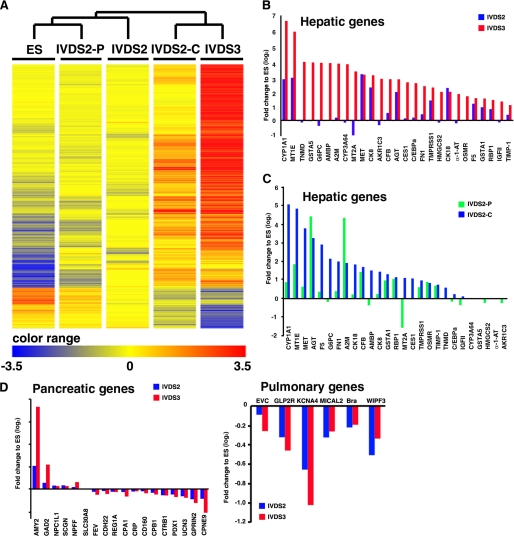
To elucidate the spatial-temporal gene expression pattern in the differentiating ESC colonies, we looked at RNA expression from ORMES-6 ESC, cells at IVDS2 and IVDS3, and cells in the central foci (IVDS2-C) and peripheral area (IVDS2-P) of ESC colonies at IVDS2, by cDNA microarray analysis. The global gene expression analysis revealed a dramatic increase in the expression of genes highly expressed in the hepatic lineage at IVDS2 and IVDS3 (Fig. 3, A and B). Hierarchical clustering analysis showed that the IVDS2-C expression pattern was similar to that at IVDS3, whereas the IVDS2-P pattern was closer to ESCs, indicating differences in the fate of cells in the central and peripheral regions of IVDS2 colonies (Fig. 3, A and C). To ascertain whether hESdF co-culture can induce nonhepatic endoderm, we compared the expression patterns of subsets of genes highly expressed in the pancreatic or pulmonary endoderm in cells from IVDS2 and IVDS3; we found that the majority of these genes were down-regulated at IVDS2 and IVDS3 (Fig. 3D). Taken together, these results demonstrate that hESdFs induced a stepwise endodermal-to-hepatic differentiation of ESCs along with morphological change in the ESC colonies emanating from the central region.
FIGURE 3.
Microarray analysis of the spatial and temporal global gene expression pattern in differentiated monkey ES induced by hESdF co-culturing. A, hierarchical clustering of ESCs and cells of IVDS2, IVDS2-P, IVDS2-C, and IVDS3 according to genes that were highly differentially expressed (>10-fold change) between ESCs and IVDS3 differentiated cells as highlighted by heat map. B and C, differential expression (>2-fold change) of genes known to be highly expressed in the liver by UniGene transcriptome cluster annotation were used to analyze the expression biases of cells of IVDS2 and IVDS3 (B) or IVDS2-P and IVDS2-C (C). D, genes known to be highly expressed in the pancreas and lung by UniGene transcriptome cluster annotation were used to analyze the expression biases of cells of IVDS2 and IVDS3 (B).
Primate ESCs Gave Rise to Differentiated Cells Exhibiting Mature Hepatic Phenotypes in hESdF Co-culture
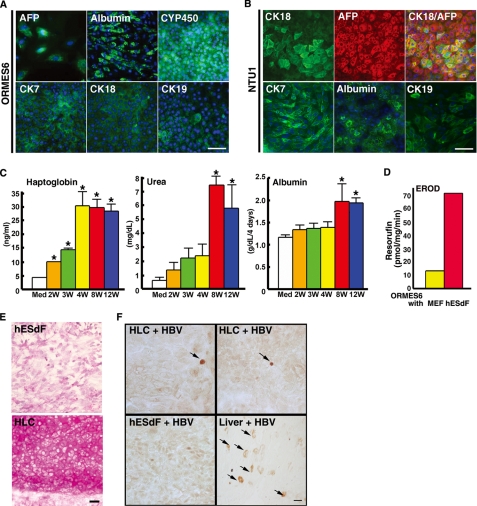
To determine whether HLCs derived from our hESdF culture system displayed mature characteristics of hepatic lineages, we analyzed the expression patterns of early hepatocyte marker genes AFP and HNF4A, mature hepatocyte marker genes, such as Albumin, CK8, CK18, α-1AT, TOD2, and CYP3A4, and cholangiocyte markers CK19 and AQP1 in the HLCs collected from IVDS3 by RT-PCR. All of these genes were highly expressed in the HLCs from both monkey and human ESCs (NTU1 and 3) co-cultured with hESdF derived either from H9 (see Fig. 5A and supplemental Fig. S3A) or NTU1 (supplemental Fig. S3B). QRT-PCR assay further demonstrated that albumin expression was dramatically increased at IVDS3 in both human and monkey ESCs (supplemental Fig. S4A). Moreover, hESCs had increased expression of cholangiocyte markers CK19 and AQP1 from IVDS2 to IVDS3, and monkey ESCs had a higher expression of CK19 at IVDS2 (supplemental Fig. S4A). ICC analysis also showed that the majority of the primate ESC-derived HLCs expressed key hepatocyte proteins, such as AFP, CK7, CK18 (Fig. 4, A and B, and supplemental Fig. S3), and albumin (Fig. 4, A and B, and supplemental Fig. S3). However, only a small subpopulation of the HLCs expressed cholangiocyte marker, CK19 (supplemental Fig. S4B). These results demonstrate that hESdFs derived from different parental ESC lines were able to promote hepatic differentiation in most populations in multiple primate ESC lines.
FIGURE 5.
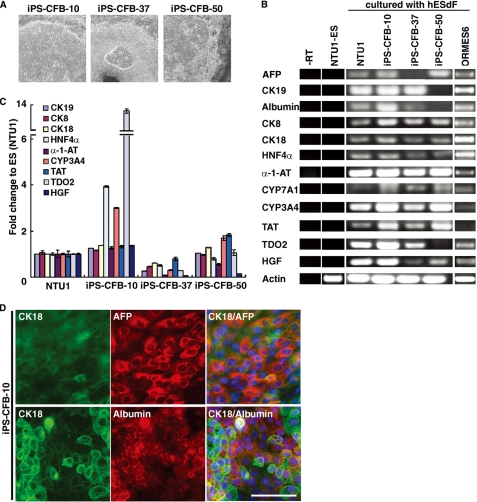
Derivation of human iPSCs and characterization of iPSC-derived HLCs under hESdF co-culture conditions. A, phase contrast images of three human iPSC lines (cell lines 10, 37, and 50), which showed characteristic hepatocyte-like morphology after 3 weeks co-culture with feeder hESdFs. B, RT-PCR analysis of hepatic markers (CK8, CK18, CK19, AFP, Albumin, HNF4α, α-1-AT, CYP3A4, CYP7A1, TAT, TDO2, and HGF) expression in the HLCs derived from human ESCs (NTU1) and monkey ESCs (ORMES6) and three different iPSC clones, and in control ESCs (NTU1) without differentiation. C, QRT-PCR analysis of total RNA isolated from HLCs derived from iPSC-CFB-10, -37, and -50 clones for CK8, CK18, CK19, HNF4α, α-1-AT, CYP3A4, TAT, TDO2, and HGF expression. For each sample, relative expression levels were normalized to corresponding levels in human ESCs (NTU1). The data correspond to the averages and standard deviations of triplicate experiments. D, ICC analysis of hepatocyte markers in human iPSC-derived HLCs. HLCs derived from iPSC-CFB-10 clone at day 15 of differentiation were double-stained with anti-human CK18 (green) and AFP (red) or albumin (red) as indicated. The nuclei were stained with DAPI (blue). Scale bar, 25 μm.
FIGURE 4.
The HLCs derived from primate ESCs co-cultured with hESdFs exhibited mature and functional phenotypes of hepatocytes. A and B, ICC analysis of hepatocyte marker expression in human (NTU1) and monkey (ORMES-6) ESC-derived HLCs showed that the majority of the HLCs were positively stained by the indicated hepatic markers. The nuclei were stained with DAPI (blue). Scale bar, 25 μm. C, temporal production of haptoglobin, albumin, and urea hepatocyte proteins during differentiation and maturation of monkey ESC-derived HLCs under hESdF co-culture conditions. The production of haptoglobin, albumin, and urea in the medium of differentiated cells increased in parallel with differentiation duration (Med, medium; 2W, 2 weeks; 4W, 4 weeks; 8W, 8 weeks; 12W, 12 weeks). The data correspond to the averages and S.D. of triplicate experiments, except the albumin data, which were the averages of four independent experiments. Significant differences between each time point and media are labeled (*, p < 0.05). D, ethoxyresorufin-O-deethylase assay showed that differentiated cells had a higher activity than control cells. E, periodic acid-Schiff staining showed glycogen storage in HLCs (lower panel), whereas the hESdF feeder cells were negative. F, immunoperoxidase staining showed positive nuclear staining of HBcAg (arrows, upper panels) in monkey ESC-derived HLCs infected with serum from patients with HBV infection for 3 days. The feeder cells (left lower panel) were negative. An HBV-infected liver section (right lower panel), stained for both cytoplasmic and nuclear (arrows); HBcAg was the positive control. Scale bar, 25 μm.
Next, we investigated the functional properties of the ESC-derived HLCs. Conditioned media previously incubated with monkey HLCs for 2, 3, 4, 8, and 12 weeks were collected for hepatocyte functional tests, including ureagenesis and haptoglobin and albumin synthesis. These monkey HLCs exhibited urea, haptoglobin, and albumin synthesis that significantly increased in parallel with the duration of hepatic differentiation (Fig. 4C), indicating enhanced hepatic maturation over the course of prolonged culture. In addition to monkey HLCs, we also tested the albumin synthesis ability of HLCs differentiated from human ESCs (NTU1) using ELISA and found that albumin secretion was significantly increased after co-culture with hESdFs for 4 or 8 weeks compared with that without differentiation (supplemental Fig. S5). The increase of albumin in both human and monkey HLCs was comparable with the increase using the three-step differentiation protocol (supplemental Fig. S6). These HLCs also exhibited other characteristics of hepatocytes, including activation of cytochrome P-450 activity (Fig. 4D) and glycogen storage (Fig. 4E), as demonstrated by an ethoxyresorufin-O-deethylase assay and by periodic acid-Schiff staining, respectively. Remarkably, we found that a small number of monkey ESC-derived HLCs could be infected by human hepatitis B virus (HBV), as shown by the positive nuclear staining for HBV core antigen (Fig. 4F). These results demonstrate that this hESdF co-culture system did support the production of mature and functional hepatocytes from both human and monkey ESCs.
hESdFs Promote Hepatic Differentiation in Human iPSC Cells
To determine whether human iPSCs are equally as amenable to induction by hESdFs as ESCs, we generated iPSCs by reprogramming human foreskin fibroblasts with methods described previously (34). After various in vitro and in vivo characterizations, three iPSC clones (iPSC-CFB 10, 37, and 50) (supplemental Fig. S7) were selected for experiments. The iPSC colonies treated with hESdFs formed hepatic foci after 10 days of differentiation, although clone 37 was less efficient than in the other two clones (Fig. 5A). RT-PCR showed that the iPSC derived HLCs from IVD3 expressed hepatic genes, such as α-1-AT, CYP3A4, TAT, and TDO2 (Fig. 5B). QRT-PCR analysis showed that the expression levels of hepatic genes varied among the HLCs of different iPSC clones (Fig. 5C), indicating that different iPSC clones responded differently to the stimulation from hESdFs. Finally, ICC analysis further indicated that the majority of the HLCs from all three lines expressed CK18 and AFP, but only HLCs from iPSC-CFB-10 expressed abundant albumin (>70% cells; Fig. 5D).
hESdFs Produced Factors Supporting Hepatic Differentiation of Primate ESCs
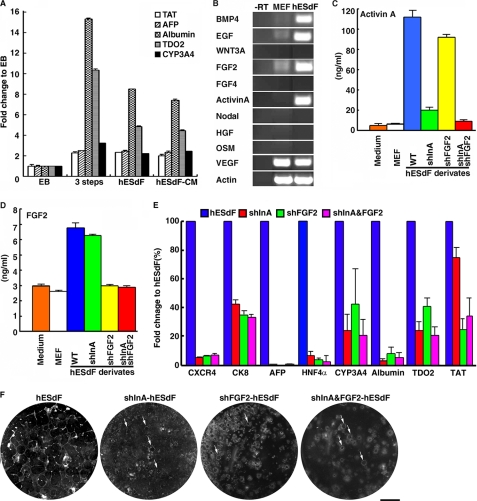
Next, we reasoned that such hepatic differentiation of ESCs/iPSCs might result from factors produced by hESdFs. Thus, we investigated whether hESdF-conditioned medium (hESdF-CM) alone could induce ESC differentiation. We found that >90% of the monkey ESCs colonies formed hepatocyte foci in hESdF-CM. Moreover, QRT-PCR revealed that hESdF-CM supported efficient hepatocyte differentiation of ESCs, because the expression levels of hepatic marker genes in the HLCs (at IVDS3) induced by hESdF-CM, hESdF co-culture and the three-step mixture condition (27) were similar (Fig. 6A). To identify factors that may be responsible for the hepatocyte differentiation, we analyzed the expression of a panel of growth factors/cytokines that have been reported to promote endodermal differentiation of ESCs by RT-PCR of hESdFs and MEFs (Fig. 6B). We found that hESdFs expressed BMP4, EGF, FGF2, activin A, and VEGF at high levels. We next tested whether inhibition of these factors could block the hESdF-induced differentiation. Accordingly, we knocked down FGF2 and/or activin A in hESdFs by lentiviruses stably expressing short hairpin RNAs against human FGF2 and/or activin A transcripts and grew ORMES-6 ESCs on the modified hESdFs (Fig. 6, C and D). Under these conditions, the typical morphological change and hepatocyte focus formation induced by hESdFs were significantly suppressed at IVDS3 (Fig. 6F). Furthermore, FGF2 and/or activin A knockdown in hESdFs also reduced the expression level of endoderm and hepatocyte marker genes in the differentiating ESCs at IVDS3 (Fig. 6E). Taken together, these experiments indicate that factors produced by hESdFs were directly responsible for hepatic development in primate ESCs.
FIGURE 6.
hESdF secreted factors promoting hepatic differentiation in pluripotent stem cells. A, QRT-PCR analysis of hepatic markers TAT, AFP, albumin, TDO2, and CYP3A4 in ESCs cultured under EB-based differentiation conditions (29), three-step culture conditions, and hESdF co-culture conditions and in hESdF-CM (27). The cells were harvested for analysis 10 days post-differentiation. Gene expression levels are presented as the relative change of the 2−ΔΔCt value relative to that of day 10 EB-based differentiation conditions. The data correspond to the averages and standard deviations of triplicate experiments. B, RT-PCR analysis of BMP4, EGF, FGF2, WNT3A, FGF2, FGF4, activin A, Nodal, HGF, oncostatin M (OSM), and VEGF in hESdFs. Actin was used as a loading control, and a reaction without reverse transcriptase (−RT) was applied to rule out genomic DNA contamination. C and D, ELISAs for activin A (C) and FGF2 protein (D) expression in conditioned media derived from MEFs, hESdFs, and hESdFs transduced with short hairpin RNA specific to INHBA (shInA) and/or FGF2 (shFGF2). E, QRT-PCR analysis for hepatic markers in monkey ESCs co-cultured with activin A and/or FGF2 knockdown hESdFs for 15 days. Gene expression levels relative to those in hESdF co-cultured cells are shown. F, morphology of monkey ESC colonies co-cultured with hESdFs and shInA and/or shFGF2 hESdFs for 15 days. The number and size of hepatic foci were dramatically reduced in shFGF2 and/or shInA hESdF group(s). Small hepatic foci (arrows) were occasionally found in the ESC colonies cultured in the modified hESdFs.
The Central Region of Colonies Is Constitutionally Essential for Hepatocyte Differentiation
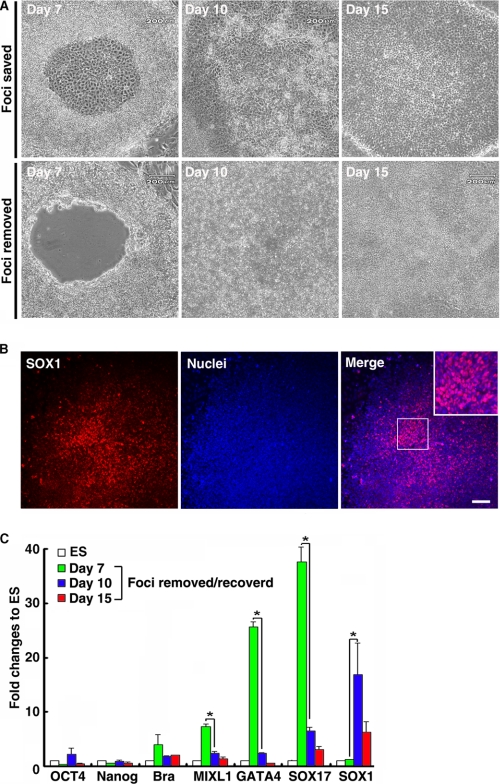
In our system, hepatocyte differentiation always followed focus formation in the central region of ESC colonies. To explore how the central foci formation affects the subsequent hepatic differentiation, we manually removed the central foci area (composed of ∼20–40% of the monkey ESC colonies) at IVDS2 and investigated the growth pattern of the excised colonies (Fig. 7A). The empty central region recovered after 3 days by replacement of cells from the surrounding region (Fig. 7A). Interestingly, further co-culturing the recovered colonies with hESdF failed to regenerate the hepatocyte foci (Fig. 7A) and instead resulted in emergence of SOX1-expressing neural rosettes (Fig. 7B). Furthermore, QRT-PCR analysis (Fig. 7C) showed that SOX1 was up-regulated in the regenerated cells, but the expression levels of endodermal and mesodermal marker genes were dramatically reduced compared with their counterparts in the colonies without excision (Fig. 7C). These results indicated that removal of the central area of ESC colonies in hESdF co-culture conditions likely disrupted the progress of hepatic endoderm differentiation and converted the differentiation direction from endoderm to ectoderm and therefore highlight the critical role of central region cells for hepatic endoderm differentiation.
FIGURE 7.
Removal of the central foci of monkey ESC colonies from IVDS2 disrupted hepatic endoderm differentiation under hESdF-primed culture conditions. A, the differentiated cells in the central area of ESC colonies at early IVDS2 (day 7) were excised and removed with a glass micropipette. The central area of an ESC colony was repopulated by cells grown from peripheral areas 3 days (day 10) after cell removal. B, ICC for SOX1 on ESC colonies 3 days after cell removal, showing that the central area of ESC colonies was repopulated with SOX1-expressing cells. Scale bar, 25 μm. C, QRT-PCR analysis of total RNA isolated from foci removed IVDS2 (day 7), regenerated cells at 3 (day 10) and 8 days (day 15) after central area removal using primers specific for Oct4, Nanog, Brachyury, MIXL1, GATA4, SOX17, and SOX1. The data correspond to the means and standard deviations of triplicate experiments. *, p < 0.05 by Student's t test.
DISCUSSION
Several protocols (22–30) have been published that claim to effectively coax human or monkey ESCs to differentiate into hepatocytes. The majority of the protocols described involve an initial differentiation step of embryoid body (EB) formation (22, 23, 25, 28, 29), and others include direct differentiation of ESCs on MEF (24, 30) or Matrigel-coated plates (26, 27). Subsequent strategies mostly involve multiple culture steps with sequential supplements of small molecules and recombinant growth factors to the culture medium. In this study, we demonstrate that the hESdF co-culture system is at least as efficient for hepatocyte differentiation as the previously published protocols (22–30). EB formation has been the most popular method exploited to differentiate ESCs into many cell types including hepatocytes; however, the uncontrolled nature of EB differentiation has made this approach less likely to generate scalable specific cell types with satisfactory purity. Monolayer-adherent culture conditions, on the other hand, have been used successfully to direct primate ESC differentiation into various cell types including hepatocytes (26, 27). In this study, the efficient hepatocyte differentiation induced by the hESdF co-culture system required ESC/iPSCs to be cultured as a monolayer adhering to the culture surface. The monolayer-adherent culture helped to maintain the structural integrity of the two-dimensional ESC colonies and therefore possibly provided a foundation for establishing an unknown feature in the central region of ESC/iPSC cell colonies that is susceptible to differentiation signal(s) secreted by hESdFs.
We showed that FGF2 and activin A secreted by hESdFs played critical roles in mediating the hepatic endoderm differentiation in primate pluripotent stem cells. This finding is well supported by results of previous reports, indicating that activin A and FGF2 have positive effects in promoting ESCs to differentiate into endoderms (28, 33). Unlike most protocols involving supplementation of extraneous factors (28, 33), it is not possible to manipulate the expression level of the factors secreted by hESdFs along the course of differentiation.
However, we still observed efficient hepatocyte differentiation with the presence of mature hepatocytes. Therefore, the persistent presence of factors such as activin A and FGF2, which are important for early stages of differentiation, may not affect the subsequent differentiation and maturation of hepatocytes. Additionally, other growth factors secreted by hESdFs (Fig. 5B), such as BMP4, EGF, and VEGF, which have been shown to be required for hepatic specification of mouse ESC-derived endoderm (21, 45) and to enhance hepatocyte specification of ESCs, respectively, were very likely responsible for the hepatic specification, differentiation, and maturation following the endodermal differentiation mediated by hESdF secreted FGF2 and activin A (21). Finally, the nonparenchymal cells generated along the hepatic lineages differentiation may also contribute to the differentiation and maturation process by secreting growth factors, because HGF expression was normally detected in hepatic foci after 3 weeks of differentiation (Fig. 5B). Taken together, these results strongly suggest that a combination of hESdF and differentiated ESC secreted factors might be responsible for the hepatocyte differentiation and maturation in our system.
In the hESdF co-culture system, the central regions of ESC colonies always transformed into endoderm-like foci, a step that preceded hepatocyte differentiation. Removal of central foci disrupted the progress of hepatic endoderm differentiation and resulted in the acquisition of ectodermal fate by the regenerated cells. We can only speculate about why the central region of the ESC colonies appears so important for hepatic differentiation. It seems unlikely that the hESdF feeders created a growth factor/cytokine signal gradient along the central to peripheral axis of the ESC colonies, because the differentiation foci could still be readily induced in hESdF-conditioned medium alone.
Alternatively, there might be heterogeneous populations of pluripotent cells in ESC colonies that are assembled in specific topological compartments in the two-dimensional ESC/iPSC colonies; perhaps only those allocated to the central region of ESC/iPSC colonies could properly respond to hESdF-secreted factors to form hepatic foci. This hypothesis is consistent with a recent observation showing that subpopulations of ESCs with distinct tissue-specific fates can be selected from pluripotent cultures (46) and was supported by our microarray analysis comparing the gene expression patterns of the central and rim regions of the ESC colonies (Fig. 3). Future identification of the molecular mechanism by which the distinct hepatocyte-promoting capability of the central region of ESC colonies is established may lead to more efficient control of hepatic differentiation.
Consistent with a previous study (40), we found that different iPSC lines have different capabilities to generate hepatic lineages in response to differentiation stimuli (e.g. hESdF treatment), even though that they are all regarded as fully reprogrammed iPSC lines according to currently used standards (supplemental Fig. S7) for iPSC characterization. These discrepancies may be attributed to currently unknown factors that modulate hepatocyte differentiation, which were somehow defective in some of the iPSC lines. It will be of interest to further explore the mechanisms that constitute the discrepancies among different iPSC clones.
In summary, we have developed a simple method that allows scalable hepatocyte generation from primate pluripotent stem cells. Furthermore, we provide compelling evidence indicating that hESdF-secreted factors are responsible for the topology-dependent differentiation of endodermal cells to hepatocytes in ESC/iPSC colonies. These findings provide opportunities to further explore the mechanisms underpinning hepatic lineage differentiation and may be useful for future pharmacotoxicology and transplantation applications.
Supplementary Material
Acknowledgments
We thank Drs. Chun-Jen Liu and Yung-Ming Jeng for sera and liver sections, respectively, from HBV-infected patients. We thank the National RNAi Core Facility (supported by National Science Council Grant 97-3112-B-001-016) for lentiviral shRNA clones and the Affymetrix Gene Expression Service Lab of Academia Sinica for microarray assays.
This work was supported by Summit Project II Intramural Grant 5202402020-0 from Academia Sinica (to H.-C. K.), Stem Cell Priority Grants 97-3111-B-001-009 and 98-2811-B-001-022 (to H.-C. K.), and Grant 97-2321-B-002-020-MY3 (to H.-P. H.) from the National Science Council of Taiwan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1 and S2, and Figs. S1–S7.
- ESC
- embryonic stem cell
- hESdF
- human ESC-derived fibroblast-like cell
- HLC
- hepatocyte-like cell
- iPSC
- induced pluripotent stem cell
- BMP
- bone morphogenic factor
- HGF
- hepatocyte growth factor
- MEF
- mouse embryonic fibroblast
- ICC
- immunocytochemistry
- QRT-PCR
- quantitative RT-PCR
- IVDS
- in vitro differentiation stage
- HBV
- hepatitis B virus
- hESdF-CM
- hESdF-conditioned medium
- EB
- embryoid body.
REFERENCES
- 1.Akhter J., Johnson L. A., Gunasegaram A., Riordan S. M., Morris D. L. (2007) Surgeon 5, 155–164 [DOI] [PubMed] [Google Scholar]
- 2.Thomson J. A., Kalishman J., Golos T. G., Durning M., Harris C. P., Becker R. A., Hearn J. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitalipov S., Kuo H. C., Byrne J., Clepper L., Meisner L., Johnson J., Zeier R., Wolf D. (2006) Stem Cells 24, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 4.Suemori H., Tada T., Torii R., Hosoi Y., Kobayashi K., Imahie H., Kondo Y., Iritani A., Nakatsuji N. (2001) Dev. Dyn. 222, 273–279 [DOI] [PubMed] [Google Scholar]
- 5.Reubinoff B. E., Pera M. F., Fong C. Y., Trounson A., Bongso A. (2000) Nat. Biotechnol. 18, 399–404 [DOI] [PubMed] [Google Scholar]
- 6.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 7.Zaret K. S. (2008) Nat. Rev. Genet. 9, 329–340 [DOI] [PubMed] [Google Scholar]
- 8.Zhao R., Duncan S. A. (2005) Hepatology 41, 956–967 [DOI] [PubMed] [Google Scholar]
- 9.Jung J., Zheng M., Goldfarb M., Zaret K. S. (1999) Science 284, 1998–2003 [DOI] [PubMed] [Google Scholar]
- 10.Rossi J. M., Dunn N. R., Hogan B. L., Zaret K. S. (2001) Genes Dev. 15, 1998–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monga S. P., Pediaditakis P., Mule K., Stolz D. B., Michalopoulos G. K. (2001) Hepatology 33, 1098–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber H., Holewa B., Jones E. A., Ryffel G. U. (1996) Development 122, 1975–1984 [DOI] [PubMed] [Google Scholar]
- 13.D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E., Baetge E. E. (2005) Nat. Biotechnol. 23, 1534–1541 [DOI] [PubMed] [Google Scholar]
- 14.Gualdi R., Bossard P., Zheng M., Hamada Y., Coleman J. R., Zaret K. S. (1996) Genes Dev. 10, 1670–1682 [DOI] [PubMed] [Google Scholar]
- 15.Deutsch G., Jung J., Zheng M., Lóra J., Zaret K. S. (2001) Development 128, 871–881 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt C., Bladt F., Goedecke S., Brinkmann V., Zschiesche W., Sharpe M., Gherardi E., Birchmeier C. (1995) Nature 373, 699–702 [DOI] [PubMed] [Google Scholar]
- 17.Kamiya A., Kinoshita T., Miyajima A. (2001) FEBS Lett. 492, 90–94 [DOI] [PubMed] [Google Scholar]
- 18.Kamiya A., Kinoshita T., Ito Y., Matsui T., Morikawa Y., Senba E., Nakashima K., Taga T., Yoshida K., Kishimoto T., Miyajima A. (1999) EMBO J. 18, 2127–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamazaki T., Iiboshi Y., Oka M., Papst P. J., Meacham A. M., Zon L. I., Terada N. (2001) FEBS Lett. 497, 15–19 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto H., Quinn G., Asari A., Yamanokuchi H., Teratani T., Terada M., Ochiya T. (2003) Hepatology 37, 983–993 [DOI] [PubMed] [Google Scholar]
- 21.Gouon-Evans V., Boussemart L., Gadue P., Nierhoff D., Koehler C. I., Kubo A., Shafritz D. A., Keller G. (2006) Nat. Biotechnol. 24, 1402–1411 [DOI] [PubMed] [Google Scholar]
- 22.Tsukada H., Takada T., Shiomi H., Torii R., Tani T. (2006) In Vitro Cell Dev. Biol. Anim. 42, 83–88 [DOI] [PubMed] [Google Scholar]
- 23.Momose Y., Matsunaga T., Murai K., Takezawa T., Ohmori S. (2009) Biol. Pharm. Bull. 32, 619–626 [DOI] [PubMed] [Google Scholar]
- 24.Cai J., Zhao Y., Liu Y., Ye F., Song Z., Qin H., Meng S., Chen Y., Zhou R., Song X., Guo Y., Ding M., Deng H. (2007) Hepatology 45, 1229–1239 [DOI] [PubMed] [Google Scholar]
- 25.Duan Y., Catana A., Meng Y., Yamamoto N., He S., Gupta S., Gambhir S. S., Zern M. A. (2007) Stem Cells 25, 3058–3068 [DOI] [PubMed] [Google Scholar]
- 26.Hay D. C., Fletcher J., Payne C., Terrace J. D., Gallagher R. C., Snoeys J., Black J. R., Wojtacha D., Samuel K., Hannoun Z., Pryde A., Filippi C., Currie I. S., Forbes S. J., Ross J. A., Newsome P. N., Iredale J. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12301–12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hay D. C., Zhao D., Fletcher J., Hewitt Z. A., McLean D., Urruticoechea-Uriguen A., Black J. R., Elcombe C., Ross J. A., Wolf R., Cui W. (2008) Stem Cells 26, 894–902 [DOI] [PubMed] [Google Scholar]
- 28.Basma H., Soto-Gutiérrez A., Yannam G. R., Liu L., Ito R., Yamamoto T., Ellis E., Carson S. D., Sato S., Chen Y., Muirhead D., Navarro-Alvarez N., Wong R. J., Roy-Chowdhury J., Platt J. L., Mercer D. F., Miller J. D., Strom S. C., Kobayashi N., Fox I. J. (2009) Gastroenterology 136, 990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiao E., Elazar M., Xing Y., Xiong A., Kmet M., Millan M. T., Glenn J. S., Wong W. H., Baker J. (2008) Stem Cells 26, 2032–2041 [DOI] [PubMed] [Google Scholar]
- 30.Agarwal S., Holton K. L., Lanza R. (2008) Stem Cells 26, 1117–1127 [DOI] [PubMed] [Google Scholar]
- 31.Saito K., Yoshikawa M., Ouji Y., Moriya K., Nishiofuku M., Ueda S., Hayashi N., Ishizaka S., Fukui H. (2006) World J. Gastroenterol. 12, 6818–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukumitsu K., Ishii T., Yasuchika K., Amagai Y., Saitoh M., Kawamoto T., Kawase E., Suemori H., Nakatsuji N., Ikai I., Uemoto S. (2009) Tissue Eng. Part A 12, 3847–3856 [DOI] [PubMed] [Google Scholar]
- 33.Soto-Gutiérrez A., Navarro-Alvarez N., Zhao D., Rivas-Carrillo J. D., Lebkowski J., Tanaka N., Fox I. J., Kobayashi N. (2007) Nat. Protoc. 2, 347–356 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 35.Lowry W. E., Richter L., Yachechko R., Pyle A. D., Tchieu J., Sridharan R., Clark A. T., Plath K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2883–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park I. H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M. W., Cowan C., Hochedlinger K., Daley G. Q. (2008) Cell 134, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha K., Jaenisch R. (2009) Cell Stem Cell 5, 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Z., Cai J., Liu Y., Zhao D., Yong J., Duo S., Song X., Guo Y., Zhao Y., Qin H., Yin X., Wu C., Che J., Lu S., Ding M., Deng H. (2009) Cell Res. 19, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 39.Sullivan G. J., Hay D. C., Park I. H., Fletcher J., Hannoun Z., Payne C. M., Dalgetty D., Black J. R., Ross J. A., Samuel K., Wang G., Daley G. Q., Lee J. H., Church G. M., Forbes S. J., Iredale J. P., Wilmut I. (2010) Hepatology 51, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Si-Tayeb K., Noto F. K., Nagaoka M., Li J., Battle M. A., Duris C., North P. E., Dalton S., Duncan S. A. (2010) Hepatology 51, 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H. F., Chuang C. Y., Shieh Y. K., Chang H. W., Ho H. N., Kuo H. C. (2009) Hum. Reprod. 24, 1114–1125 [DOI] [PubMed] [Google Scholar]
- 42.Chen H. F., Kuo H. C., Chen W., Wu F. C., Yang Y. S., Ho H. N. (2009) Hum. Reprod. 24, 71–80 [DOI] [PubMed] [Google Scholar]
- 43.Wortelboer H. M., de Kruif C. A., de Boer W. I., van Iersel A. A., Falke H. E., Blaauboer B. J. (1987) Mol. Toxicol. 1, 373–381 [PubMed] [Google Scholar]
- 44.Chen H. F., Kuo H. C., Chien C. L., Shun C. T., Yao Y. L., Ip P. L., Chuang C. Y., Wang C. C., Yang Y. S., Ho H. N. (2007) Hum. Reprod. 22, 567–577 [DOI] [PubMed] [Google Scholar]
- 45.Runge D. M., Runge D., Dorko K., Pisarov L. A., Leckel K., Kostrubsky V. E., Thomas D., Strom S. C., Michalopoulos G. K. (1999) J. Hepatol. 30, 265–274 [DOI] [PubMed] [Google Scholar]
- 46.King F. W., Ritner C., Liszewski W., Kwan H. C., Pedersen A., Leavitt A. D., Bernstein H. S. (2009) Stem Cells Dev. 18, 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.