Abstract
Several protein-protein interactions within the SARS-CoV proteome have been identified, one of them being between non-structural proteins nsp10 and nsp16. In this work, we have mapped key residues on the nsp10 surface involved in this interaction. Alanine-scanning mutagenesis, bioinformatics, and molecular modeling were used to identify several “hot spots,” such as Val42, Met44, Ala71, Lys93, Gly94, and Tyr96, forming a continuous protein-protein surface of about 830 Å2, bearing very conserved amino acids among coronaviruses. Because nsp16 carries RNA cap 2′-O-methyltransferase (2′O-MTase) activity only in the presence of its interacting partner nsp10 (Bouvet, M., Debarnot, C., Imbert, I., Selisko, B., Snijder, E. J., Canard, B., and Decroly, E. (2010) PLoS Pathog. 6, e1000863), functional consequences of mutations on this surface were evaluated biochemically. Most changes that disrupted the nsp10-nsp16 interaction without structural perturbations were shown to abrogate stimulation of nsp16 RNA cap 2′O-MTase activity. More strikingly, the Y96A mutation abrogates stimulation of nsp16 2′O-MTase activity, whereas Y96F overstimulates it. Thus, the nsp10-nsp16 interface may represent an attractive target for antivirals against human and animal pathogenic coronaviruses.
Keywords: Positive Strand RNA Viruses, Protein-Protein Interactions, RNA Methyltransferase, Viral Replication, Viral Transcription, nsp10-nsp16 Interaction
Introduction
Coronaviruses (CoVs),7 classified into the family Coronaviridae in the order Nidovirales, possess a viral RNA genome that is among the largest known (2). They include important pathogens of livestock, wild and companion animals, and humans, such as the severe acute respiratory syndrome CoV (SARS-CoV) (3–5). They are mainly etiological agents of respiratory and enteric diseases, exemplified by the worldwide pandemic of SARS-CoV spreading in 2003 from Asia, with a final number of cases around 8,000 and a 10% mortality.
The genome of SARS-CoV contains a single-stranded plus-sense RNA of ∼29.7 kb (2). At the molecular level, CoVs employ a variety of unusual strategies to accomplish a complex program of gene expression (5). Coronavirus replication requires the synthesis of both genomic and multiple subgenomic RNA species and the assembly of progeny virions by a pathway that is unique among enveloped RNA viruses (5–7). Fourteen open reading frames (ORFs) have been identified, of which 12 are located in the 3′-end of the genome. The other two ORFs (1a and 1b), which are located in the 5′-proximal two-thirds of the genome, encode two large polyproteins translated directly from genomic RNA. ORF 1b is expressed by a −1 ribosomal frameshifting at the end of pp1a, extending its coding sequence and thus generating the pp1ab polyprotein (6). These two polyproteins are cleaved into 16 functional viral replicase proteins called nsp1 to -16 (for non-structural proteins 1–16). Those nsps form the membrane-bound replication-transcription complex, which is localized to a network of endoplasmic reticulum-derived membranes in the infected cell (8, 9). Bioinformatics, structural biology, (reverse) genetics, and biochemical studies have contributed to the characterization of CoV nsps (10). Many enzymatic activities have been documented for products of ORF 1a and ORF 1b. Among these are the proteolytic activities endowed by nsp3 and nsp5, required to mature pp1a and pp1ab polyproteins into individual non-structural proteins (11, 12); the RNA-dependent RNA polymerase by nsp12, and a putative RNA primase, nsp8, involved in replication and transcription (13–15); several RNA modification enzymes, such as nsp14, an exoribonuclease, nsp15, an endoribonuclease, and nsp13, a helicase/RNA triphosphatase (16, 17); and two S-adenosyl-l-methionine (AdoMet)-dependent methyltransferases carried by nsp14 (N7-MTase) and nsp16 (2′O-MTase) (18–20). In addition, some of these nsps have thus far no known function nor enzymatic activity, and/or their functions remain more elusive (nsp1, nsp2, nsp4, nsp6, nsp9, nsp10, and nsp11), even if some of them may regulate host cellular functions (nsp1, nsp4, and nsp6) or enzymatic activities from the replication-transcription complex (10).
RNAs from mouse hepatitis virus (MHV), a member of the coronavirus genus, and from toroviruses carry a 5′ cap structure (21–23), and all coronaviruses encode three enzymes involved in the capping pathway in their genome: nsp13 helicase/RNA triphosphatase, nsp14 N7-MTase, and the nsp16 2′O-MTase. Although no CoV guanylyltransferase (16–23) activity has been identified, CoVs are likely to follow the canonical capping pathway involving (i) an RNA triphosphatase that removes the 5′ γ-phosphate group of the mRNA; (ii) a guanylyltransferase that catalyzes the transfer of GMP to the remaining 5′-diphosphate terminus; and (iii) an N7-MTase that methylates the cap guanine at the N7-position, producing the 7MeGpppN “cap 0 structure.” Cap 1 structure formation requires an additional 2′O-MTase, that methylates the ribose 2′O-position of the first nucleotide of the mRNA. The involvement of nsp14 N7-MTase and of nsp16 2′O-MTase in the capping pathway was recently demonstrated biochemically (1, 18, 20). Moreover, both nsp14 and -16 play crucial roles for efficient RNA synthesis within the SARS-CoV replicon and for transcription/replication of MHV-CoV (13, 24).
Several genome-wide analyses have been reported in which viral protein interactions by mammalian or yeast two-hybrid (Y2H) systems were studied (25–27). The identified protein-protein interaction networks involve non-structural and structural proteins as well as accessory proteins, suggesting a key role for protein interactions in replication-transcription complex assembly (25–27). Furthermore, these data emphasized the complex protein-protein interaction network used by the SARS-CoV for both replication-transcription complex assembly and expression of multiple activities involved in the transcription/replication of its genome.
Among protein-protein interactions recently identified within the SARS proteome, interactions between nsp10 and nsp16 were found bidirectionally in both yeast and mammalian two-hybrid systems (25, 26). The crystal structure of nsp10 shows that it belongs to the zinc finger protein family (22, 28, 29). nsp10 has no known enzymatic activity but may have a role in the regulation of enzymatic activities at different steps of the viral transcription/replication or by playing an architectural role. This hypothesis is supported by the micromolar affinity binding of both MHV and SARS-CoV nsp10 to single-stranded RNA (28, 30). In MHV, nsp10 plays a critical role in RNA synthesis, and a temperature-sensitive nsp10 Q65E mutation causes a defect in minus-strand RNA synthesis, whereas plus-strand synthesis is unaffected (31). Furthermore, the role of nsp10 in MHV replication was confirmed by alanine-scanning mutagenesis of residues conserved among CoVs (22). Viable mutants synthesized lower amounts of viral RNA, and lethal mutants delineated a core structure of nsp10 surrounding the zinc fingers (22). Also nsp10 acts as an essential co-factor triggering nsp16 2′O-MTase activity, suggesting its involvement in the regulation of viral RNA capping (1). Altogether, these studies suggest that the nsp10 and nsp16 protein-protein complex might define a new target for antiviral molecules against pathogenic CoVs, such as the SARS-CoV.
In the absence of a nsp10-nsp16 protein complex crystal structure, we have merged several approaches to define the nsp10-nsp16 interaction at the molecular level, coupling reverse yeast two-hybrid (RY2H) technology with bioluminescence resonance energy transfer (BRET), molecular modeling, pull-down experiments, and NMR. Using mutagenesis and functional assays, we have mapped key nsp10 residues involved in the interaction with nsp16 and in regulating the SARS-CoV nsp16 RNA cap 2′O-MTase activity. In particular, we have identified a continuous specific surface of ∼830 A2 on nsp10 involved in its interaction with nsp16.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Transfection
HEK 293T cells were grown in accordance with ATCC recommendations in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. Cells were transfected using Fugene® 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocol in a 10-cm dishes or 6-well plates. Cells were plated at 5 × 106 or 300,000 cells/10-cm dish and 6-well plate, respectively, 8 h prior to transfection. Cells were transfected with a total amount of 10 and 1.5 μg of DNA per 10-cm dish and 6-well plate, respectively, by adding pUC19 vector. For the 10-cm dish, 4 μg of pNRLuc-nsp16 and 2 μg of pNEYFP-nsp10 were used. Transfections in 6-well plates were made with 300 ng of pNRLuc-nsp16 vectors and various amounts of pNEYFP-nsp10 vectors (50, 100, 300, 600, and 900 ng). Cells were incubated at 37 °C, 5% CO2 for 48 h, and a BRET assay was then performed.
Plasmids
All of the cloning experiments were performed using Gateway® technology (Invitrogen). Name, sequence, and position of all of the primers used in this study are indicated in Table 1. For RY2H experiments, the ORFs of nsp16 and nsp10 with a STOP codon were cloned into pDBa and pAD, respectively. For BRET experiments, pNEYFP-GW vector was obtained by ligating Gateway Cassette B into the SmaI site of the pEYFP-C1 plasmid (Clontech). The pNRLuc-GW vector was obtained by ligating Gateway Cassette C.1 into the EcoRV site of the hpRLuc-C2 plasmid (BioSignal Packard). Each plasmid fuses EYFP or Renilla luciferase (RLuc) proteins to the N terminus of nsp10 or nsp16. Mutated nsp10 ORF isolated from RY2H were transferred into BRET vectors using the Gateway® technology (Invitrogen). nsp10 mutants were cloned into Escherichia coli expression plasmids (pDEST14) by PCR using mutated pNEYFP-nsp10 plasmids as template and reintroduced into pDest14 expression vector.
TABLE 1.
Sequences of primers used in this study
| Primer name | Primer sequence | Primer location |
|---|---|---|
| bp | ||
| AD | CGCGTTTGGAATCACTACAGGG | |
| DB | GGCTTCAGTGGAGACTGATATGCGTCGCG | |
| TERM | GGAGACTTGACCAAACCTCTG | |
| GWF (attB1) | GGGGACAAGTTTGTACAAAAAAGCAGGCTTC | |
| GWR (attB2) | GGGGACCACTTTGTACAAGAAAGCTGGGTC | |
| nsp10-GWF | GWFGCTGGAAATGCTACAGAAGTACCT | 1–24 |
| nsp10-GWR | GWRTTACTGCATCAAGGGTTCGCGGAGTT | 394–417 |
| nsp10GWFkozATG | GWFGCCACCATGGCTGGAAATGCTACAGAAGTACCT | 1–24 |
| nsp10GWRNoSTOP | GWRCCTGCATCAAGGGTTCGCGGAGTT | 394–417 |
| nsp16-GWF | GWFGCAAGTCAAGCGTGGCAACCA | 1–21 |
| nsp16-GWR | GWRTTAGTTGTTAACAAGAATATCACTTGAAACC | 866–897 |
| nsp16GWFkozATG | GWFGCCACCATGGCAAGTCAAGCGTGGCAACCA | 1–21 |
| nsp16GWRNoSTOP | GWRCGTTGTTAACAAGAATATCACTTGAAACC | 866–897 |
| nsp10-V42A | CCAATCACCAACTGTGCGAAGATGTTGTGTACAC | 109–142 |
| nsp10-M44A | CCAACTGTGTGAAGGCGTTGTGTACACACACTGG | 116–149 |
| nsp10-L45A | CCAACTGTGTGAAGATGGCGTGTACACACACTGG | 116–149 |
| nsp10-G69A | GGACCAAGAGTCCTTTGCTGGTGCTTCATGTTGTCTG | 189–225 |
| nsp10-G70A | GTCCTTTGGTGCTGCTTCATGTTGTCTGTATTG | 193–230 |
| nsp10-S72A | GTCCTTTGGTGGTGCTGCATGTTGTCTGTATTGTAG | 198–233 |
| nsp10-R78A | ATGTTGTCTGTATTGTGCATGCCACATTGACCATCC | 216–251 |
| nsp10-K93A | GGATTCTGTGACTTGGCAGGTAAGTACGTCCAAATACC | 262–299 |
| nsp10-G94A | GATTCTGTGACTTGAAAGCTAAGTACGTCCAAATACC | 263–299 |
| nsp10-K95A | CTGTGACTTGAAAGGTGCGTACGTCCAAATACCTAC | 267–302 |
| nsp10-Y96A | CTGTGACTTGAAAGGTAAGGCCGTCCAAATACCTACCACTTGTGCTAATGACCC | 267–329 |
| nsp10-Y96F | CTGTGACTTGAAAGGTAAGTTCGTCCAAATACCTACCACTTGTGCTAATGACCC | 267–329 |
| nsp10-Y96V | CTGTGACTTGAAAGGTAAGGTCGTCCAAATACCTACCACTTGTGCTAATGACCC | 267–329 |
| nsp10-Y96I | CTGTGACTTGAAAGGTAAGATCGTCCAAATACCTACCACTTGTGCTAATGACCC | 267–329 |
| nsp10-Q65E | GCTAACATGGACGAAGAGTCCTTTGGTGGTGC | 176–212 |
Antibodies
Anti-GFP antibody (mix of clones 7.1 and 13.1) was purchased from Roche Applied Science. Anti-Renilla luciferase antibodies (MAB4400 and MAB4410) were purchased from Chemicon. Secondary antibodies coupled to horseradish peroxidase were purchased from Dako.
BRET Assay
BRET assays were performed on living cells, as described by Issad and Jockers (32). In each experiment, transfections of pNRLuc-nsp16 alone or plus pEYFP were performed as controls. Coelenterazine H (Tebu-Bio) was added at a 5 μm final concentration and incubated at room temperature. BRET measurements were performed at 25 °C by sequentially integrating luminescence signals at 480 and 530 nm for 1 s. The BRET ratio is defined as follows, (emission at 530 nm − emission at 485 nm × Cf)/emission at 485 nm, where Cf corresponds to emission at 530 nm/emission at 485 nm for the Rluc fusion protein expressed alone in the same experimental conditions. All experiments were performed more than three times.
Generation of the Full-length Enriched Mutated Allele Library of nsp10 and nsp16
The full-length enriched mutated allele libraries of nsp10 and nsp16 were generated using the SureFrameTM allele library construction kit (Invitrogen). This technology consists of a modified Gateway® donor vector that allows cloning and expression of PCR products as N-terminal fusions to the kanamycin resistance gene. When plating the library onto Luria broth (LB) containing kanamycin, only alleles coding for full-length proteins will confer kanamycin resistance and produce colonies. They will constitute the enriched mutated allele library. First, ORFs encoding nsp10 and nsp16 without STOP codon were created by PCR using the Platinium Taq HiFi (Invitrogen) and primers Nsp10GWR NoSTOP + nsp10-GWF and Nsp16GWR NoSTOP + nsp16-GWF (15 cycles). PCR products were then cloned into pDONR201 using a BP reaction, sequenced, and subsequently transferred into pAD (LR reaction). Libraries were created by amplifying nsp10 NoSTOP and nsp16 NoSTOP by PCR using the Platinium Taq HiFi (Invitrogen) with primers AD and TERM (35 cycles). The mutagenic PCR was performed in 25 tubes within a volume of 20 μl to maximize the number of independent mutations (total volume 500 μl). The PCR products were subsequently cloned into the pDONR-Express using the BP reaction and then transformed into One Shot® TOP10 Electrocomp bacteria (Invitrogen) and plated onto an LB agar plate containing spectinomycin (100 μg/ml), kanamycin (40 μg/ml), and isopropyl β-d-1-thiogalactopyranoside (1 mm). The nsp10 and nsp16 full-length enriched mutated allele libraries contained 34,000 and 43,000 independent clones, respectively. These libraries were transferred into pAD to perform RY2H screens (LR reaction).
Reverse Yeast Two-hybrid Screens
RY2H screens were performed as described by Walhout and Vidal (33–35). Libraries were covered more than 10 times by each screen (400,000 and 460,000 clones were screened for nsp10 and nsp16, respectively). Following transformation and plating, yeasts were incubated at 30 °C for 5 days. Positive clones were then isolated, and their phenotypes were assessed on medium lacking URA or HIS as well as their β-galactosidase activity, using a semiautomatic procedure as described previously (33). Mutated alleles from clones growing on 5-fluorootic acid plates but not on uracil plates were amplified by PCR and sequenced (36). All yeast media were prepared as described (33–35). Mutated nsp10 alleles were then transferred into the pNEYFP-GW vector using GatewayTM technology for BRET assays.
Reagents
AdoMet was purchased from New England BioLabs, and the [3H]AdoMet was purchased from PerkinElmer Life Sciences.
Cloning of the SARS-CoV nsp10 and nsp16 Genes
The SARS-CoV nsp10 and nsp16-coding sequences were amplified by RT-PCR from the genome of SARS-CoV Frankfurt-1 (accession number AY291315) as described previously (37). The nsp10 and nsp16 genes (encoding residues 4231–4369 and 6776–7073 of replicase pp1ab) were cloned using Gateway® technology into expression vector pDest14 (pDest14/6His-nsp10 and pDest14/6His-nsp16) to produce recombinant proteins carrying an N-terminal His6 tag.
Expression and Purification of SARS-CoV nsp10 and nsp16 Proteins
E. coli C41 (DE3) cells (Avidis SA, France), containing the pLysS plasmid (Novagen), were transformed with the various expression vectors and grown in 2YT medium containing ampicillin and chloramphenicol. Protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 500 μm (nsp10) or 50 μm (nsp16), when the A600 value of the culture reached 0.5. nsp16 expression was performed during 16 h at 17 °C, whereas nsp10 expression was incubated at 37 °C during 4 h. Bacterial cell pellets were frozen and resuspended in lysis buffer (50 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgSO4, 5 mm β-mercaptoethanol (only for nsp10) supplemented with 1 mm PMSF, 20 mm imidazole, 10 μg/ml DNase I, and 0.5% Triton X-100. After sonication and clarification, proteins were purified by IMAC (HisPurTM cobalt resin; Thermo Scientific) and eluted with lysis buffer supplemented with 250 mm imidazole. The nsp10 protein was next loaded on a HiLoad 16/60 Superdex 200 gel filtration column (GE Healthcare) and eluted with 10 mm HEPES, pH 7.5, 150 mm NaCl. The protein fractions were concentrated to around 2 mg/ml and stored at −20 °C in the presence of 50% glycerol. For NMR experiments, 15N-labeled nsp10 proteins were grown and induced on M9 minimum medium supplemented with 15NH4Cl and further purified as described above. For pull-down assays, SARS-CoV nsp10-nsp16 complex was produced in E. coli in a dual promotor expression plasmid kindly provided by Bruno Coutard (Architecture et Fonction des Macromolécules Biologiques, France). In this backbone, SARS CoV nsp10 can be expressed under a tet promoter and encodes a protein in fusion with an N-terminal Strep tag, whereas nsp16 is expressed under a T7 promoter and encodes a protein in fusion with an N-terminal His6 tag. The single point mutants in the nsp10 gene were generated by PCR using the QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. E. coli C3016 cells (Biolabs) were transformed with the various expression vectors and grown in 2YT medium containing ampicillin and chloramphenicol. Protein expression was induced by adding 50 μm isopropyl 1-thio-β-d-galactopyranoside and 200 μg/liter anhydrotetracycline; then cells were incubated for 16 h at 24 °C. Bacterial cell pellets were frozen and resuspended in lysis buffer (50 mm HEPES, pH 7.5, 500 mm NaCl, 5 mm MgSO4), supplemented with 1 mm PMSF, 10 μg/ml DNase I, and 0.5% Triton X-100. After sonication and clarification, proteins were purified by chromatography with Strep-Tactin-Sepharose (IBA GmbH, Göttingen, Germany). After three washes in high salt buffer (1 m NaCl) and three washes in low salt buffer (500 mm NaCl), bound proteins were eluted with 2.5 mm d-desthiobiotin in binding buffer. After analyzing the purified protein complex by SDS-PAGE, the intensities of Coomassie-stained bands were quantified using ImageJ (National Institutes of Health) software.
NMR Experiments
1H-15N heteronuclear HSQC NMR experiments were carried out on a 600-MHz Bruker spectrometer (with cryoprobe) at 288 K. The samples were prepared in a volume of 550 μl, at 20 μm concentration, in 50 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgSO4, and 5 mm β-mercaptoethanol, supplemented by 50 μl of D2O. The NMR parameters used to run the experiments were 256 scans, TD1 2048, and TD2 128.
RNA Synthesis and Purification
Short capped RNAs (7MeGpppAC4) were synthesized in vitro using bacteriophage T7 DNA primase and were purified by HPLC as described previously (38).
Radioactive Methyltransferase and Filter Binding Assay
MTase activity assays were performed in 40 mm Tris-HCl, pH 8.0, 5 mm DTT, 1 mm MgCl2, 1 μm 7MeGpppAC4, 10 μm AdoMet, and 0.03 μCi/μl [3H]AdoMet (GE Healthcare). In the standard assay, nsp10 and nsp16 were added at final concentrations of 1.2 μm and 200 nm, respectively. Reaction mixtures were incubated at 30 °C and stopped after the indicated times by a 10-fold dilution of the reaction mixture in 100 μm ice-cold S-adenosyl-l-homocysteine. Samples were kept on ice and then transferred to glass fiber filtermats (DEAE filtermat; Wallac) by a filtermat harvester (Packard Instruments). Filtermats were washed twice with 0.01 m ammonium formate, pH 8.0, twice with water, and once with ethanol, dried, and transferred into sample bags. Betaplate Scint (Wallac) scintillation fluid was added, and the methylation of RNA substrates was measured in counts/min by using a Wallac 1450 MicroBeta TriLux liquid scintillation counter.
RESULTS
Delineation of the nsp10 Surface Involved in Its Interaction with nsp16
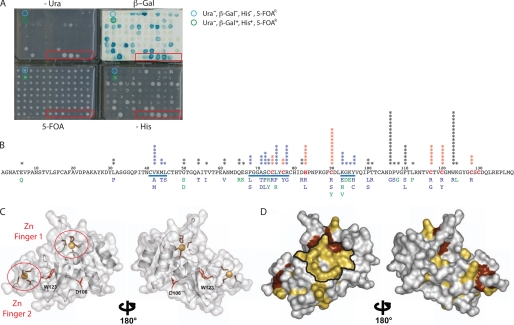
We used RY2H with nsp10 and nsp16 to isolate interaction-defective alleles (IDAs) and thereby delineate their surface of interaction (25, 26). IDAs are alleles that contain mutations affecting their ability to interact with their wild type binding partners, leading to the identification of specific amino acid residues involved in the interaction between nsp10 and nsp16 (39). nsp16 was used as a bait to screen a library of potential nsp10 IDAs generated by PCR-mutagenesis and selected to express full-length proteins (see “Experimental Procedures”). From this screen, 133 independent full-length nsp10 IDAs were determined (Fig. 1A) (see below). As expected, all clones with a mutated allele were resistant to 5FAO and were therefore unable to grow on medium lacking uracil (Fig. 1A). The 5FOAR phenotypes could be due to loss of nsp10-nsp16 interaction or to reduced expression of nsp10 through instability or misfolding. Thus, we excluded residue changes that were involved in chelating zinc (Cys74, Cys77, His83, and Cys90 and Cys117, Cys120, Cys128, and Cys30 in the first and second zinc fingers, respectively; Fig. 1, B and C) and changes at internal residues (D106G, W123R) (Fig. 1, C and D). The remaining residue changes defined three clusters forming a small region on one side of nsp10 (Fig. 1, C and D).
FIGURE 1.
Identification of nsp10 IDAs unable to interact with nsp16 in RY2H. A, phenotypic assays were performed with clones isolated from the RY2H screen in a 96-well plate format using a semiautomated protocol (-Leu-Trp-His + 25 mm 3-AT, SC-Leu-Trp + 0.2% 5-FOA and SC-Leu-Trp-Ura and β-galactosidase activity) (45). Seven controls of known phenotypes were included (red box). B, mutations within positive clones from RY2H screen were identified by sequencing and reported on the nsp10 sequence. The stars indicate the number of times the mutated alleles were isolated. Stars in blue represent residues that are within the delineated potential surface of interaction. Mutants in blue are Ura−, β-Gal−, His−, and 5-fluorootic acid-resistant, and mutants in green are Ura−, β-Gal−/+, His−/+, and 5FOAR. Cysteine and histidine residues involved in chelating the zinc are in red. The underlined sequences represent groups of mutated amino acids exposed on the protein surface. C, ribbon representation of the monomeric nsp10 backbone structure (Protein Data Bank code 2FYG). Residues involved in zinc chelation are circled in red. Residues Asp106 and Trp123 are not exposed on the protein surface, as shown in a red stick representation on the nsp10 backbone. D, all mutations corresponding to IDAs identified by the RY2H screen are highlighted in yellow on the surface of nsp10 (Protein Data Bank code 2FYG). The zinc finger residues are shown in dark red, and the potential surface of interaction between nsp10 and nsp16 is delineated in black.
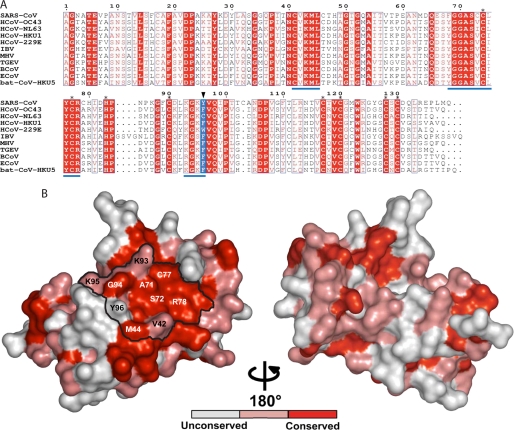
At this stage, it is interesting to note that absolutely conserved residues among CoVs tend to be on the same side of nsp10 as the potential interacting surface found by RY2H (Fig. 2, A and B). The three clusters of residues identified by RY2H are present within or close to these very well conserved boxes, suggesting their involvement in crucial function(s) among CoVs (Figs. 1D and 2A). Furthermore, when mapped on the nsp10 crystal structure, these clusters occupy mainly the conserved side and form a continuous surface consisting of an area of 830 Å2, extremely well conserved among CoVs (Figs. 1D and 2, A and B).
FIGURE 2.
nsp10 protein conservation among coronavirus groups 1, 2, and 3. A, sequence alignment of nsp10 proteins derived from genome sequences of the following: SARS-CoV, SARS coronavirus (group 2b, NC_004718), HCoV-OC43, human coronavirus OC43 (group 2a, NC_005147), HCoV-NL63, human coronavirus NL63 (group 1, NC_005831), HCoV-HKU1, human coronavirus HKU1 (group 2a, NC_006577), HCoV-229E, human coronavirus 229E (group 1, NC_002645), IBV, infectious bronchitis virus (group 3a, NC_001451), MHV, mouse hepatitis virus (group 2a, NC_006852), TGEV, transmissible gastroenteritis virus (group 1, NC_002306), BCoV, bovine coronavirus (group 2a, NC_003045), ECoV, equine coronavirus (group 2a, NC_010327), and bat coronavirus, bat-CoV-HKU5–1 (group 2c, NC_009020). The black triangle points to residue 96, colored in blue. The sequences were aligned using the ESPript program (46). Stars indicate residues involved in coordinating zinc atoms. Underlined sequences in blue correspond to clusters of mutations obtained by RY2H. B, nsp10 from SARS-CoV (Protein Data Bank code 2FYG) is depicted in a surface representation. From the alignment above, absolutely conserved residues are shown in red, whereas conserved residues and non-conserved residues are shown in pink and white, respectively. The different amino acids identified as being part of nsp10-nsp16 interaction are named, and the potential surface of interaction is delineated in black.
Thus, using the RY2H system, we have identified specific residues conserved among CoVs onto a specific and limited surface area of nsp10. These residues delineate an ∼830-Å2 surface involved in the interaction with nsp16.
Impact of nsp10 IDAs on the Interaction with nsp16 in Mammalian Cells
Interaction of nsp10 with nsp16 has already been detected in mammalian cells using the two-hybrid system and confirmed using pull-down assays (26). However, mammalian two-hybrid systems detect interaction within the cell nucleus, whereas these viral proteins are localized in the cytoplasm during infection. Therefore, we made use of a BRET assay to detect interaction of nsp10 with nsp16 in their native mammalian intracellular environment. ORFs encoding nsp10 proteins were fused to the C terminus of EYFP or RLuc, and BRET was measured in HEK 293T cells following transfection of the corresponding plasmids. Under these experimental conditions, we were able to detect a BRET signal only when nsp10 was fused to EYFP and nsp16 was fused to RLuc (data not shown). The specificity of this interaction was assessed by BRET donor saturation assays (supplemental Fig. 1). A specific BRET signal, characterized by a hyperbolic curve, was detected only when nsp16 was fused to RLuc, with a BRETmax of 130 milli-arbitrary units (supplemental Fig. 1). No BRET signal was obtained with EYFP alone, even at the highest concentrations.
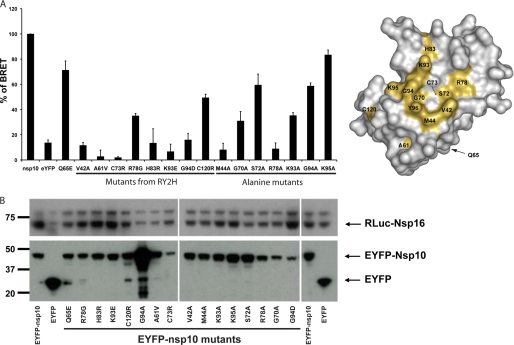
Then we investigated the ability of nsp10 IDAs to interact with nsp16 using this assay (Fig. 3A, left). As anticipated, IDAs were expressed at different levels, and thus the amount of plasmid encoding each IDA was adjusted to normalize protein expression levels (data not shown; see “Experimental Procedures”). Most of the nsp10 IDAs identified by RY2H failed to interact with nsp16, except R78G and C120R, which still retained limited binding (BRET values of 35 and 50% of wild type nsp10, respectively) (Fig. 3, A and B). These data show that the nsp10 surface residue IDAs we identified fail to interact with nsp16 in mammalian cells when expressed at levels equivalent to wild type nsp10.
FIGURE 3.
Characterization of the nsp10 IDAs unable to interact with nsp16 by BRET in mammalian cells. A, left, BRET assays were performed with IDAs identified by RY2H and with alanine mutants. The Q65E mutant was used because it inhibits MHV replication. The experiments were performed three times, and the effect of each mutation was compared with the interaction of wild type nsp10 (100% interaction). Right, the different mutations from the left panel are depicted in gold on the nsp10 surface. B, Western blot analysis indicates protein expression levels. Levels of RLuc-nsp16 and EYFP-nsp10 were determined with anti-luciferase and anti-GFP antibodies, respectively.
Effect of nsp10 Alanine Mutations on the Interaction with nsp16 in Mammalian Cells
To further define the surface of interaction, we mutated to alanine amino acids identified by RY2H as well as other amino acids covering the anticipated surface of interaction (Fig. 2B) and tested them using the BRET assay. Because an alanine residue substitution eliminates the side chain beyond the β-carbon without altering the main-chain conformation or imposing a straining electrostatic or steric effect and because all of these residues are surface residues, the mutated protein structures should not be drastically different from wild type nsp10 (40) (Fig. 3A, right). In addition, we made a G70A change in a surface residue within the putative nsp16 interaction region. Gly70 is of interest because it forms a hydrogen bound with a structural water molecule, suggesting also a potential role for this residue in the architecture of the complex (Protein Data Bank code 2FYG) (Fig. 3A, right). We also included a previously identified Q65E change, which affects MHV RNA synthesis (Fig. 3A, right) (31). Most of the alanine substitutions and the Q65E change caused a loss of nsp16 interaction as defined by our BRET assay. S72A, G94A, and K95A changes resulted in only modest losses of nsp16 interaction (BRET signal ranging from 60 to 80%; Fig. 3A). Thus, these data support the direct involvement of Val42, Met44, Ala61, Gly70, Cys73, Arg78, Lys93, Gly94, and Tyr96 in forming a binding surface of interaction with nsp16 (Fig. 3A, right). Because mutations may impact nsp10 structure, we performed 1H-15N HSQC NMR studies with two mutant proteins that no longer interact with nsp16: V42A, identified in RY2H, and M44A, an “alanine mutant.” By this analysis, these two mutant proteins have no significant structural changes, substantiating our conclusion of the direct involvement of these two residues in the binding of nsp16. (supplemental Fig. 2, A and B).
The Hydroxyl Group of Tyr96 Is a Critical Binding Determinant
Analysis of amino acid composition of protein-protein interfaces shows that some residues, such as tyrosine, arginine, and tryptophan, are found more frequently than others (41). Among the residues found to be involved in the nsp10-nsp16 interaction, amino acids Arg78 and Tyr96 are located on the edge of the interacting surface (Fig. 2, A and B). Arg78 is conserved among all CoV nsp10 proteins. However, Tyr96 is unique to SARS-CoV, whereas Phe96 is found in most other CoV sequences. Of note, residue 96 is a cysteine in HCoV-NL63 nsp10 and a tryptophan in HCoV-229E nsp10 (Cys96 and Trp96, respectively) (Fig. 2A). The apparent plasticity of Tyr96, compared with Arg78, led us to further characterize the requirements for Tyr96 in the interaction with nsp16 at a molecular level. This residue stands on the edge of the very well conserved nsp10 surface area described above and could thus be specific to the different nsp10 partners.
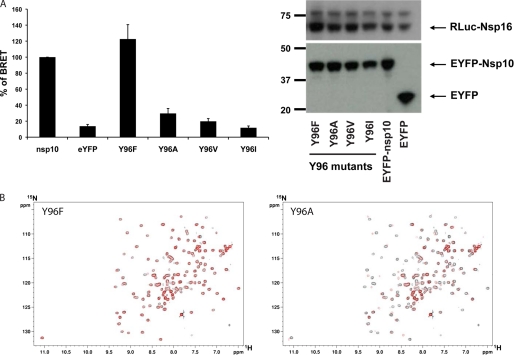
To assess the role of the Tyr96 hydroxyl group, we first mutated the tyrosine into phenylalanine because most of the nsp10 proteins harbor this residue (Fig. 2A). Surprisingly, this mutant enhances the BRET signal (BRET signal 120% of the wild type), suggesting an increase in the binding affinity between these two proteins (Fig. 4A, left). Thus, the hydroxyl group has a negative effect on the interaction with nsp16.
FIGURE 4.
Characterization of the nsp10 Tyr96 mutations by BRET. A, BRET assays were performed with Tyr96 mutants (left). The experiments were performed three times, and the effect of each mutation was compared with the interaction of wild type nsp10 (100% interaction). Western blot analysis indicates homogeneous levels of protein expression. Levels of RLuc-nsp16 and EYFP-nsp10 were determined with anti-luciferase and anti-GFP antibodies, respectively (right). B, 1H-15N heteronuclear HSQC NMR experiments were carried out on a 600-MHz Bruker spectrometer (with cryoprobe) and compared with the wild type spectrum. Spectra of 15N-labeled nsp10 wild type protein and main mutants (Y96F and Y96A) are shown in black and red, respectively. Overall spectra are superimposable, demonstrating that the global architecture of the protein is conserved.
To assess the role of the aromatic moiety and a potential effect of a hydrophobic side chain, Tyr96 was then mutated into alanine, valine, and isoleucine. The Y96A and Y96V mutants were able to interact weakly with nsp16 as judged by BRET values close to 30 and 20%, respectively (Fig. 4A, left). In contrast, Y96I completely failed to interact with nsp16. Thus, Tyr96 mutations into non-aromatic hydrophobic residues decreased the interaction. This inhibition seems proportional to the length of the side chain, reflecting a possible steric hindrance (Fig. 4A, left). These results point out the importance of the aromatic property of the phenyl group of Tyr96 at the surface of nsp10 for interaction with nsp16, which is confirmed by the conservation of this aromatic residue at position 96 in most coronaviruses. In these experiments, all mutants were expressed at a similar level in HEK 293T cells as detected by Western blot (Fig. 4A, right). This result also demonstrates that the Y96F mutation does not have a structural impact on the nsp10 conformation because nsp10 Y96F is interacting more tightly with nsp16 than wild type. The integrity of two nsp10 representative mutated proteins, Y96F and Y96A, was indeed confirmed using heteronuclear 1H-15N HSQC NMR studies, which ruled out any effect of these mutations on the structure of nsp10 (Fig. 4B and supplemental Fig. 2).
The 2′O-MTase Activity of nsp16 Correlates with Its Interaction with nsp10 Mutants
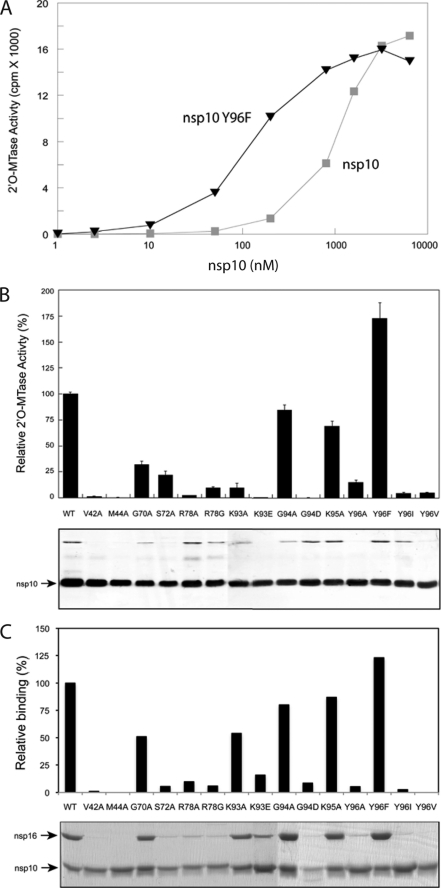
SARS-CoV nsp10 was recently found to be a nsp16 helper protein; nsp10 turns on the otherwise inactive 2′O-MTase activity of nsp16 (1). We therefore analyzed the functional consequences of nsp10 mutations on nsp16 2′O-MTase activity. The nsp16 2′O-MTase activity was first determined by incubating 200 nm purified nsp16 with an increasing concentration of wild type nsp10 (1 nm to 3.2 μm), in the presence of a short capped RNA substrate (7MeGpppAC4; Fig. 5A, square). Fifty percent of the maximal 2′O-MTase activity was obtained when 200 nm purified nsp16 was incubated with 1200 nm nsp10 (Fig. 5A). These conditions were chosen to assess the stimulating or inactivating effect of nsp10 mutant proteins on nsp16 2′O-MTase activity. Under these conditions, any slight alteration of nsp10 binding strength to nsp16 should result in a significant increase or decrease of nsp16 2′O-MTase activity (Fig. 5A). For this purpose, 14 His6 tag nsp10 mutants were produced in E. coli and purified. The bottom panel of Fig. 5B shows that the mutant proteins migrated in SDS-PAGE at a molecular mass similar to that of wild type nsp10 protein (15 kDa) upon SDS-PAGE with a minor contaminant detected around 50 kDa. Fig. 5B shows that one mutant protein overstimulates nsp16 2′O-MTase activity (Y96F), whereas almost all of the others were not active in potentiating the nsp16 2′O-MTase activity. Overall, there was excellent correlation between the BRET analysis and the ability of nsp10 mutant proteins to stimulate nsp16 enzyme activity; all nsp10 mutant proteins with reduced nsp16 affinity as judged by BRET assay (BRET value ranging from 10 to 50%) also lose their stimulating effect on nsp16 2′O-MTase activity (V42A, M44A, G70A, R78A/G, K93A/E, G94D, and Y96I/V/A) (Figs. 3A and 4A). Furthermore, two mutant proteins, G94A and K95A, that bind nsp16 with a slightly reduced affinity in our BRET assay (60 and 80%, respectively) are still able to stimulate nsp16 2′O-MTase activity (compare Figs. 3A and 5B). These data confirm that the binding of nsp10 to nsp16 is absolutely required to stimulate the nsp16-mediated 2′O-MTase.
FIGURE 5.
Effect of nsp10 mutations on the nsp16 2′O-MTase activity. A, the effect of increasing nsp10 concentration (■, wild type; ▲, Y96F) was determined on nsp16 (200 nm) 2′O-MTase activities measured as in B. B, the upper panel shows the 2′O-MTase activity of each mutant protein. A 1200 nm concentration of each nsp10 mutant protein was incubated with N7MeGpppAC4 and in the presence of [3H]AdoMet methyl donor for 30 min, as described under “Experimental Procedures.” The 2′O-MTase activity obtained in the presence of wild type nsp10 control protein was arbitrarily set to 100%. The bar graph presents the results of three independent experiments. Corresponding purified His6-tagged nsp10 mutant proteins analyzed by SDS-PAGE and visualized by Coomassie Blue staining are shown in the bottom of the panel. C, pull-down assay was performed with nsp10 protein co-expressed with nsp16 in E. coli. Following cell lysis, nsp10 was purified with a Strep-Tactin-Sepharose column, and the amount of nsp16 interacting with nsp10 was visualized by Coomassie Blue staining. Intensities of Coomassie-stained bands were quantified using ImageJ (National Institutes of Health) software. The binding activities were then compared with wild type nsp10 interaction with nsp16, which was arbitrarily set to 100%. Error bars, S.D.
We note that the S72A mutant protein binds nsp16 (BRET value of 60%) but weakly activates nsp16 2′O-MTase activity (compare Figs. 3A and 5B). This result may either reflect an impact of this mutation on the protein folding in mammalian cells or indicate a crucial role of this residue in the nsp16 stimulation process. To further clarify this, we performed pull-down assays to assess the binding efficiency of each nsp10 mutant expressed in E. coli (Fig. 5C). In these conditions, no binding of S72A mutant protein to nsp16 was detected, consistent with its absence of nsp16 2′O-MTase-stimulating activity (Fig. 5B). In contrast, the G70A and K93A mutant proteins bind nsp16 as efficiently as they do when assayed by BRET (50% here versus 40% when detected by BRET), although their nsp16 2′O-MTase activity is reduced to a greater extent (Fig. 5B). This apparent discrepancy could reflect the requirement of an affinity threshold to induce an active complex with a full enzymatic activity.
To confirm the extent of the interaction surface delineated by RY2H (Fig. 1D), we generated new single alanine mutants in the nsp10 gene by targeting residues outside the identified surface of interaction (supplemental Fig. 3, A and B). These new mutant proteins were tested by pull-down experiments for their binding to nsp16. As expected, all of them bind nsp16 with a relative binding activity above 65% (supplemental Fig. 3A). It must be noted that none of these residues were identified by RY2H (Fig. 1B), confirming the surface of an area of ∼830 Å2 (described in Figs. 1D and 2, A and B).
Interestingly, we observed a strong stimulating effect of the Y96F mutation on nsp16 2′O-MTase activity. This mutation was also shown to enhance the interaction with nsp16 as described in the BRET assay (120% of wild type) and validated by the pull-down experiment (Fig. 5C). To further compare wild type nsp10 and the Y96F mutant, we incubated increasing concentrations of nsp10 with nsp16 and measured 2′O-MTase activity. Fig. 5A indicates that wild type and Y96F nsp10 stimulate the nsp16 2′O-MTase activity in a dose-dependent manner. A shift of 50% activity toward a 10-fold lower concentration was observed for the mutant (∼100 nm) relative to wild type protein (∼1 μm), although both plateau at similar 2′O-MTase activity values. Altogether, these results demonstrate that the stimulation of nsp16 2′O-MTase activity by nsp10 mutant proteins tends to correlate with their affinity as determined by BRET and pull-down assay. Moreover, our results indicate that the interaction between nsp10 and nsp16 is essential to trigger nsp16 2′O-MTase activity.
DISCUSSION
The SARS-CoV nsp16 protein was recently shown to be inactive when expressed alone, but its 2′O-MTase activity, playing a role in RNA viral synthesis, is turned on by the addition of its nsp10 partner (1). In this paper, we have identified residues on the nsp10 surface involved in its interaction with nsp16. The role of these surface residues was further studied in mammalian cells by alanine-scanning mutagenesis coupled to BRET methodology and pull-down assay. Key amino acids, or hot spots, regulating the formation of the nsp10-nsp16 complex were confirmed. The effect of nsp10 mutant proteins was further analyzed in terms of nsp16 2′O-MTase activation. Interestingly, all nsp10 mutant proteins that fail to activate nsp16-mediated 2′O-MTase activity have a nsp16-binding activity below 50%. Our results reveal a strong correlation between nsp10 binding to nsp16 and activation of nsp16 cap MTase activity.
Because our initial screen by RY2H could not discriminate IDAs that were misfolded or underexpressed, we also mutated surface residues outside the identified interaction surface of nsp10 (supplemental Fig. 3, A and B) and tested their binding to nsp10 by pull-down assay. As expected, all of these mutants were still able to interact with nsp16 (supplemental Fig. 3, A and B). This result confirms the extent of the interaction surface identified by RY2H (Fig. 1B).
We have also performed an RY2H experiment with nsp10 as a bait to isolate nsp16 IDAs and identified 53 bona fide IDAs (supplemental Fig. 4A). Because the nsp16 tridimensional structure has not yet been solved, we could not propose a potential surface of interaction. Nevertheless, when mapped on the nsp16 primary sequence, these IDAs cover most of the protein sequence but fall into one main cluster (supplemental Fig. 4A). This distribution is comparable with the previous results obtained with nsp10 IDAs, and this cluster might delineate a surface on nsp16 (supplemental Fig. 4A). Furthermore, as observed with nsp10, this single cluster encompasses stretches of highly conserved residues among different coronaviruses (supplemental Fig. 4B). In the absence of tridimensional data on nsp16, no potential surface of interaction could be identified.
On nsp10, we found that residues involved in the activation of the complex activity cluster preferentially on one side, forming a ∼830 Å2 surface area, which is well conserved among all CoV groups (Fig. 2, A and B). Because a 2′O-MTase activity is present in feline coronavirus nsp16 alone, albeit at lower level than that of the SARS-CoV nsp10-nsp16 complex, it would be of interest to test if both the interaction and stimulation are also conserved (18). Furthermore, assessing the effect of nsp10 on the stimulation of nsp16 2′O-MTase activity in other CoVs might open a path to the development of small molecule protein-protein interaction inhibitors that could specifically inhibit the 2′O-MTase activity of nsp16 and limit the replication capacity of CoVs, including SARS-CoV.
Most of the residues identified here are localized within a central core of nsp10 defined by Donaldson et al. (22) on MHV nsp10. This central core is resistant to mutations because any modification within this region induces a lethal phenotype (22). It is therefore tempting to hypothesize that the phenotype observed by Donaldson et al. (22) reflects the importance of the nsp10-nsp16 interaction during CoV replication. Moreover, most of the residues involved in the interaction between nsp10 and nsp16 are also involved in its stimulating effect on the nsp16 2′O-MTase activity. This result strongly suggests that a non-transient physical association between nsp10 and nsp16 is essential to express full nsp16 2′O-MTase activity (Figs. 3A, 4B, and 5B) and to regulate an efficient virus replication. This hypothesis is strengthened by the G70A and K93A mutant proteins that retain 50% of binding to nsp16 in pull-down assay but are unable to efficiently stimulate nsp16 2′O-MTase activity. We note a residual 2′O-MTase activity of G70A mutant protein compared with K93A. Because our pull-down assay was performed in the absence of RNA substrate, we wondered if this difference could reflect a potential role of the RNA substrate in the interaction. To address this issue, nsp10 and nsp16 were incubated alone or in combination with mGpppRNA. In these experimental conditions, we could not detect any mGpppRNA binding activity when proteins were incubated alone (data not shown). In contrast, when both proteins are present, the complex is able to bind mGpppRNA, and cross-linking experiments identify nsp16 as the unique RNA-binding protein, suggesting that nsp10 binding induces a conformational change in nsp16, resulting in mGpppRNA binding activity (data not shown). These results also indicate that nsp10 does not bind mGpppRNA by itself, alone or in complex.
Our study also highlights the aromatic essence of the Tyr96 residue, which plays a crucial role in the nsp16-nsp10 interaction and induction of nsp16 2′O-MTase activity. This residue is specific to SARS-CoV nsp16 and is a phenylalanine in most other Coronaviridae homologues except in HCoV-NL3 and HCov-229E, where a cysteine and a tryptophan are found, respectively (Fig. 2A). To assess at the molecular level the role of the hydroxyl and phenyl groups in the interaction between nsp10 and nsp16, the Tyr96 was mutated successively into phenylalanine, alanine, valine, and isoleucine (Fig. 4A). Interestingly, removing the hydroxyl group resulted in an enhanced binding to nsp16 correlated with an increased 2′O-MTase activity (Figs. 4A and 5B). In contrast, the absence of the phenyl group completely abrogates nsp16 enzymatic activity and partially decreases the binding to nsp16 (Figs. 4B and 5B). Heteronuclear 1H-15N HSQC NMR studies of different key mutant proteins ruled out any drastic effect of mutations on the tridimensional structure (Fig. 4B and supplemental Fig. 2). These results demonstrate that the decreased interactions observed in the RY2H, BRET, and pull-down experiments are not related to a loss of protein architecture but rather to a loss of specific protein-protein interactions.
It is puzzling that residue 96 is a tyrosine only in SARS-CoV and a phenylalanine in most of other CoVs. Based on the results presented here, one could hypothesize that the tyrosine residue is required to dampen the efficiency of the nsp16 2′O-MTase activity or to fine tune the efficiency of association of nsp10 to nsp16. Indeed, a phenylalanine induces both a strong increase of MTase activity and tighter association, which could be detrimental for viral replication for unknown reasons. More experiments using nsp10 and nsp16 from different CoVs may cast light on a possible conserved role of this interaction as well as on the protein network involved in the replication of the different CoVs.
These results allow us to propose that specific “hot spots” of the surface of nsp10 can be targeted to disrupt the complex and lead to inhibition of the 2′O-MTase activity of nsp16. Indeed, cap methylation genes of many (+)-RNA viruses, such as alphaviruses and flaviviruses, were shown to play a crucial role for efficient virus replication (20, 24, 42–44). In the case of CoVs, a functional and genetic analysis performed on MHV temperature-sensitive mutants mapped to the N7-MTase domain of CoV nsp14 and in the 2′O-MTase nsp16 indicated that both are involved in positive strand RNA synthesis by previously formed replicase-transcriptase complexes (13). The importance of cap MTase for viral RNA synthesis is also supported by data obtained by mutagenesis of MTase catalytic residues in SARS-CoV RNA replicon systems (20, 24). Therefore, cap MTases constitute a new attractive antiviral target. Accordingly, the development of small molecules specifically inhibiting protein-protein interaction and the 2′O-MTase activity of nsp16 could be envisaged. Furthermore, because this surface of interaction is conserved among CoVs, molecules or peptides inhibiting SARS-CoV nsp10-nsp16 interaction might be developed and extended to the inhibition of the nsp10-nsp16 interaction in other viruses when this interface of interaction is conserved (30).
Supplementary Material
Acknowledgments
We thank Bruno Coutard, Karen Dalle, Violaine Lantez, and Séverine Blanc for excellent technical assistance and fruitful discussion and Gerard Zurawsky for excellent scientific advice.
This work was supported, at its initial phase, by the VIZIER integrated project (LSHG-CT-2004-511960) of the European Union 6th Framework and the Euro-Asian SARS-DTV Network (SP22-CT-2004-511064) from the European Commission specific research and technological development program “Integrating and Strengthening the European Research Area” and then by the French National Research agency, under reference “ANR-08-MIEN-032,” the Fondation pour la Recherche Médicale (Programme Équipe FRM) (to B. C.), and Direction Générale de l'Armement Contract 07co404.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- CoV
- coronavirus
- SARS
- severe acute respiratory syndrome
- AdoMet
- S-adenosyl-l-methionine
- EYFP
- enhanced yellow fluorescent protein
- 2′O-MTase
- 2′-O-methyltransferase
- MHV
- mouse hepatitis virus
- RY2H
- reverse yeast two-hybrid
- BRET
- bioluminescence resonance energy transfer
- IDA
- interaction-defective allele
- RLuc
- Renilla luciferase
- N7-MTase
- (guanine-N7)-methyltransferase.
REFERENCES
- 1.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E. J., Canard B., Decroly E. (2010) PLoS Pathog. 6, e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorbalenya A. E., Enjuanes L., Ziebuhr J., Snijder E. J. (2006) Virus Res. 117, 17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiris J. S., Guan Y., Yuen K. Y. (2004) Nat. Med. 10, S88–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H. D., Rappuoli R. (2003) Nat. Rev. Microbiol. 1, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masters P. S. (2006) Adv. Virus Res. 66, 193–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasternak A. O., Spaan W. J., Snijder E. J. (2006) J. Gen. Virol. 87, 1403–1421 [DOI] [PubMed] [Google Scholar]
- 7.Sawicki S. G., Sawicki D. L., Siddell S. G. (2007) J. Virol. 81, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hemert M. J., van den Worm S. H., Knoops K., Mommaas A. M., Gorbalenya A. E., Snijder E. J. (2008) PLoS Pathog. 4, e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J. (2008) PLoS Biol. 6, e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlman S., Netland J. (2009) Nat. Rev. Microbiol. 7, 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y., Lu X., Denison M. R. (1995) J. Virol. 69, 3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker S. C., Yokomori K., Dong S., Carlisle R., Gorbalenya A. E., Koonin E. V., Lai M. M. (1993) J. Virol. 67, 6056–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawicki S. G., Sawicki D. L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S. G. (2005) PLoS Pathog. 1, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.te Velthuis A. J., Arnold J. J., Cameron C. E., van den Worm S. H., Snijder E. J. (2010) Nucleic Acids Res. 38, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imbert I., Guillemot J. C., Bourhis J. M., Bussetta C., Coutard B., Egloff M. P., Ferron F., Gorbalenya A. E., Canard B. (2006) EMBO J. 25, 4933–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov K. A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A. E., Ziebuhr J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minskaia E., Hertzig T., Gorbalenya A. E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5108–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A. E., Snijder E. J., Canard B. (2008) J. Virol. 82, 8071–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Grotthuss M., Wyrwicz L. S., Rychlewski L. (2003) Cell 113, 701–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai M. M., Stohlman S. A. (1981) Adv. Exp. Med. Biol. 142, 69–82 [DOI] [PubMed] [Google Scholar]
- 22.Donaldson E. F., Sims A. C., Graham R. L., Denison M. R., Baric R. S. (2007) J. Virol. 81, 6356–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Vliet A. L., Smits S. L., Rottier P. J., de Groot R. J. (2002) EMBO J. 21, 6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almazán F., Dediego M. L., Galán C., Escors D., Alvarez E., Ortego J., Sola I., Zuñiga S., Alonso S., Moreno J. L., Nogales A., Capiscol C., Enjuanes L. (2006) J. Virol. 80, 10900–10906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbert I., Snijder E. J., Dimitrova M., Guillemot J. C., Lécine P., Canard B. (2008) Virus Res. 133, 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M. L., Enjuanes L., Guo D. (2008) PLoS ONE 3, e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Brunn A., Teepe C., Simpson J. C., Pepperkok R., Friedel C. C., Zimmer R., Roberts R., Baric R., Haas J. (2007) PLoS ONE 2, e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseph J. S., Saikatendu K. S., Subramanian V., Neuman B. W., Brooun A., Griffith M., Moy K., Yadav M. K., Velasquez J., Buchmeier M. J., Stevens R. C., Kuhn P. (2006) J. Virol. 80, 7894–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X. C., Bartlam M., Rao Z. (2006) J. Virol. 80, 7902–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthes N., Mesters J. R., Coutard B., Canard B., Snijder E. J., Moll R., Hilgenfeld R. (2006) FEBS Lett. 580, 4143–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donaldson E. F., Graham R. L., Sims A. C., Denison M. R., Baric R. S. (2007) J. Virol. 81, 7086–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Issad T., Jockers R. (2006) Methods Mol. Biol. 332, 195–209 [DOI] [PubMed] [Google Scholar]
- 33.Thalappilly S., Suliman M., Gayet O., Soubeyran P., Hermant A., Lecine P., Iovanna J. L., Dusetti N. J. (2008) Proteomics 8, 3071–3081 [DOI] [PubMed] [Google Scholar]
- 34.Walhout A. J., Temple G. F., Brasch M. A., Hartley J. L., Lorson M. A., van den Heuvel S., Vidal M. (2000) Methods Enzymol. 328, 575–592 [DOI] [PubMed] [Google Scholar]
- 35.Walhout A. J., Vidal M. (2001) Methods 24, 297–306 [DOI] [PubMed] [Google Scholar]
- 36.Armstrong C. M., Li S., Vidal M. (eds) (2005) Modular Scale Yeast Two-hybrid Screening, Vol. 4, Academic Press, Inc., San Diego, CA [Google Scholar]
- 37.Campanacci V., Egloff M. P., Longhi S., Ferron F., Rancurel C., Salomoni A., Durousseau C., Tocque F., Brémond N., Dobbe J. C., Snijder E. J., Canard B., Cambillau C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1628–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peyrane F., Selisko B., Decroly E., Vasseur J. J., Benarroch D., Canard B., Alvarez K. (2007) Nucleic Acids Res. 35, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal M. (1997) in The Yeast Two-hybrid System (Bartel P. L., Fields S. eds) pp. 109–147, Oxford University Press, New York [Google Scholar]
- 40.Cunningham B. C., Wells J. A. (1989) Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 41.Bogan A. A., Thorn K. S. (1998) J. Mol. Biol. 280, 1–9 [DOI] [PubMed] [Google Scholar]
- 42.Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T. S., Zhou Y., Li H., Shi P. Y. (2006) J. Virol. 80, 8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H. L., O'Rear J., Stollar V. (1996) Virology 217, 527–531 [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K. A., Shi P. Y., Li H. (2007) J. Virol. 81, 3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lembo F., Lecine P. (2009) TECAN J. 1, 14–15 [Google Scholar]
- 46.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.