Abstract
Abnormalities in the process of endocytosis are classically linked to malignant transformation through the deficient down-regulation of signaling receptors. The present study describes a non-classical mechanism that does not require internalization by which endocytic proteins affect cell migration and basement membrane invasion. Specifically, we found that the endocytic adaptor epsin binds and regulates the biological properties of the signaling molecule RalBP1 (Ral-binding protein 1). Epsin interacted with the N terminus of RalBP1 via its characteristic epsin N-terminal homology (ENTH) domain. A combination of siRNA-mediated knock-down and transfection of siRNA-resistant constructs in fibrosarcoma cells demonstrated that impairment of the epsin-RalBP1 interaction led to cell migration and basement membrane invasion defects. We found the ENTH domain was necessary and sufficient to sustain normal cell migration and invasion. Because all the epsin endocytic motifs reside in the C-terminal part of the molecule, these results suggest that this novel regulatory circuit does not require endocytosis. In addition, cells depleted of epsin-RalBP1 complex displayed deficient activation of Rac1 and Arf6 suggesting a signaling function for this novel interaction. Further, overexpression of either epsin or RalBP1 enhanced migration and invasion of fibrosarcoma cells. Collectively, our results indicate that epsin regulates RalBP1 function in Rac1- and Arf6-dependent pathways to ultimately affect cell migration and invasion. We propose that the observed up-regulation of both epsin and RalBP1 in certain cancers contributes to their invasive characteristics.
Keywords: Adaptor Proteins, Cell Migration, Endocytosis, Signal Transduction, Trafficking, Cell Invasion, Epsin, RalBP1
Introduction
It is widely accepted that the processes of endocytosis and signaling are intimately linked (1–3). Further, it is now recognized that internalization can lead to both the termination and initiation of signaling events (3). Consequently, abnormalities in endocytosis are expected to affect signaling pathways and therefore, to contribute to the onset of diseases, particularly cancer (1, 4).
Signaling termination by endocytosis is a straightforward concept based on the removal and destruction of cell surface signaling complexes (3). As predicted by this mechanism, deficiencies in the down-regulation of receptor tyrosine kinases (RTKs) are known to contribute to signaling deregulation that leads to malignant transformation (4). In addition to termination, endocytosis can also lead to signaling activation. This notion recognizes the existence of signaling activity originated in the endosomal compartment (signaling endosome hypothesis, see Ref. 2 for a review).
One of the well-known signaling pathways requiring endocytosis for its activation is the cell fate determination Notch pathway (5). It is well established that the endocytic protein epsin is essential for triggering this pathway, i.e. Notch-ligand internalization (6, 7). Further, abnormalities in the regulation of the Notch juxtacrine signaling system are associated with tumor formation (8).
Importantly, the epsin family of endocytic adaptors has also been found to lead to signaling activation via another mechanism that does not require receptor internalization. Specifically, we discovered that in Saccharomyces cerevisiae, cell polarity-dependent processes such as cell division rely on epsin function integrity (9, 10). We found this signaling role of epsins to be dependent on the interaction of the ENTH3 (epsin N-terminal homology) domain with GTPase-activating proteins (GAPs) for the RhoGTPase and cell polarity-organizer protein Cdc42 (9, 10). However, whether this epsin signaling function was conserved in mammals remained to be established.
By combining siRNA-mediated knock-down and transfection of siRNA-resistant constructs, we show here that epsins are required for the cell polarity-dependent process of cell migration and invasion. Complementary experiments indicated that overexpression of epsins enhanced fibrosarcoma migration and invasion through the basement membrane. Our results also demonstrated that this novel role of epsins in cell migration and invasion depends on the interaction of their ENTH domain with the N terminus of the Cdc42/Rac1 GAP and signaling molecule RalBP1 (ral-binding protein 1). Further, our data suggest that this novel epsin-mediated migration/invasion pathway does not require receptor internalization but involves Arf6 and Rac1 activation. We propose that the observed up-regulation of either epsins or RalBP1 in certain cancers contributes to their invasive characteristics.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture Conditions, and Reagents
HT1080 and NIH3T3 cell lines were acquired from ATCC. Cells were cultured in DMEM, streptomycin/penicillin, 2 mm l-glutamine, and 10% fetal bovine serum (HT1080) or 10% calf serum (NIH3T3). For fibronectin stimulation, surfaces were coated with bovine plasma fibronectin (Biomedical Technologies) for 2 h at 37 °C. Transfections of siRNA and plasmid DNA were carried out as previously described (11) with RNAiMax (Invitrogen) and TransIT-LT1 (Mirus) reagents, respectively. Details about siRNAs, plasmids, and antibodies used in this study can be found in supplemental Tables S1–S3.
Immunofluorescence
For colocalization analysis, cells were extracted at 4 °C for 45 s in 30 mm Hepes pH 7.4, 100 mm KCl, 5 mm MgCl2, 5 mm EGTA, 0.03% saponin, and then fixed in ice-cold 3% formaldehyde in PBS. Cells were immunolabeled and imaged as previously described (11).
Protein Binding Assays
Immunoprecipitation: cells were lifted and re-seeded on dishes coated with 10 μg/ml fibronectin at low density for 3 h to promote adhesion and cell motility. Plates were rinsed with 4 °C PBS and cells lysed with 400 μl/plate ice-cold lysis buffer: 50 mm Tris-HCl, pH 7.5, 1% IGEPAL, 100 mm KCl, 100 mm NaCl, 5 mm MgCl2, 2 mm EGTA, 1 mm DTT, 1 mm Na3VO4, 1 mm NaF, and Complete Protease Inhibitor (Roche). Lysates were pre-clarified and immunoprecipitations were carried-out for 1 h at 4 °C using BSA-blocked beads. Beads were then washed three times with lysis buffer and boiled in protein sample buffer. Epitope-tagged proteins were immunoprecipitated with anti-HA, anti-Flag, or anti-Myc EZ-view Red affinity beads (Sigma). GFP-tagged proteins were immunoprecipitated with the indicated antibodies pre-bound to protein A- Sepharose (Zymed Laboratories Inc.) and endogenous proteins were immunoprecipitated with indicated antibodies pre-bound to protein G-Sepharose (Zymed Laboratories Inc.).
In Vitro Binding Assays
Bacterially produced recombinant proteins were purified according to standard protocols in PBS, 0.1% Tween, 5% glycerol, 1% BSA. 7 μg of GST, or GST-RalBP1 fusion protein was immobilized on glutathione beads and incubated with 1–5 μm His6-ENTH (WT or YTQV) in the presence or absence of PtdIns (4,5)bisphosphate diC4 (Echelon) for 1 h at room temperature. After washing, beads were boiled in protein sample buffer. SDS/PAGE and immunoblotting were carried out as described before (11).
Surface Plasmon Resonance (SPR) Experiments
Binding experiments using SPR technology were conducted according to standard procedures (12). Briefly, purified GST-RalBP1 was immobilized to a Biacore 3000 chip and 5.5–0.028 μm His6-ENTH1 was flowed on the RalBP1 chips and binding in resonance units (Ru) was measured. Dissociation was measured upon flow of buffer onto preformed [ENTH1·RalBP1] complex on the chip. Association (kon) and dissociation (koff) kinetic constants were estimated by non-linear regression and the interaction affinity was calculated as Kd = koff/kon.
Cell Migration and Invasion Assays
Migration assays: 104 cells were trypsinized and recovered in complete media for 1 h. Cells were applied on 8-μm pore transwell inserts (Corning Inc) coated with 100 μg/ml BSA and 10 μg/ml fibronectin on the upper and bottom side of the insert membrane, respectively. Inserts were placed in wells containing complete media and allowed to migrate for 4 h (NIH3T3) or 2.5 h (HT1080) before fixation in 3% formaldehyde. Cell migration was quantified as described previously (11). Alternatively, membranes were stained with 0.1% Crystal Violet in 200 mm Hepes pH 6.5 for 5 min, rinsed, and dried. Stain was then extracted for 5 min in 10% acetic acid and the absorbance measured at 560 nm.
Invasion Assays
Cells were resuspended at 3 × 105 cells/ml in DMEM, penicillin/streptomycin, and 0.5% BSA. Invasion through basement membrane-coated filters (Cell BioLabs) was allowed for 20 h and quantified as described above.
GTPase Activation Assays
HA-Arf6-transfected cells were resuspended as described above and incubated an additional 1 h in medium containing 0.2% BSA before seeding on 5 μg/ml fibronectin for 5–10 min after attachment. Cell were lysed, and Arf6 was pulled-down by the VHS+GAT domain as previously described (13).
To assay Rac1 activation, cells were seeded on fibronectin-coated plates as with Arf6 activation experiments. Cells were lysed, and activated Rac1 was pulled-down with the GST-PAK CRIB domain according to Ref. 14.
RESULTS
Epsin ENTH Domain Directly Interacts with the N Terminus of RalBP1
We asked whether the epsin novel regulatory function on RhoGTPase signaling (9, 10) would be conserved in mammals and what cellular processes would be under its control. First, we sought to test whether mammalian ENTH domain interacts with RhoGAPs. We considered the Cdc42/Rac1 GAP RalBP1 as a likely epsin binding candidate because it is involved in endocytosis of the same cargoes as the epsins (15, 16), interacts with epsin binding elements of the endocytic machinery (AP2 and Reps2/POB1; Refs. 17, 18) and has been found in complexes with epsin1 (17, 19). RalBP1 is a ubiquitously expressed, plasma membrane-associated protein (20, 21), involved in drug resistance (22, 23) and NIH3T3 fibroblast migration (14).
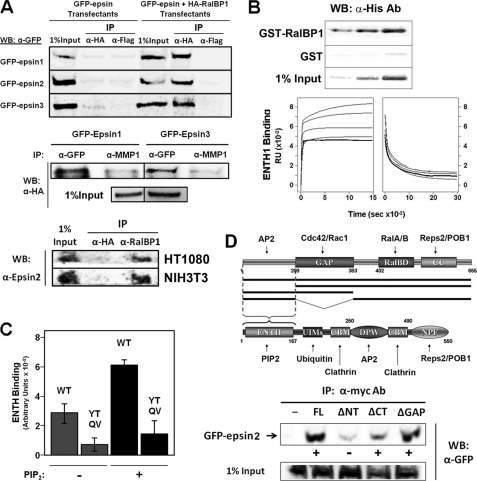
In agreement with previous reports, and supporting the idea of epsin-RalBP1 interplay, we observed that RalBP1 could be co-immunoprecipitated with all epsin paralogs from human HT1080 cell lysates (Fig. 1A) and mouse NIH3T3 (data not shown). Further supporting the existence of this interaction in vivo, we successfully immunoprecipitated endogenous epsin2 with endogenous RalBP1 from HT1080 and NIH3T3 lysates (Fig. 1A). Given that the ENTH domain of epsin binds RhoGAPs in Saccharomyces cerevisiae (9, 10), we tested whether recombinant and purified ENTH domain from rat epsin1 (epsin11–168) could directly interact with purified GST-RalBP1 fusion protein. As shown in Fig. 1B GST-RalBP1, but not GST, was able to pull-down the ENTH domain in a dose-dependent manner. Further, the direct interaction between purified proteins was confirmed using SPR (Fig. 1B, lower panels) and analytical ultracentrifugation (data not shown). The SPR experiments yielded an affinity (Kd) value of 0.2 μm for the ENTH domain-RalBP1 interaction (Fig. 1B, lower panels). This affinity lies within a range of binding strength described for proteins involved in endocytic and signaling regulatory networks (24, 25). In agreement with known RhoGAP-ENTH domain interactions (9, 10), our results showed that binding of ENTH to RalBP1 is impaired by mutations in the Tyr-101, Thr-105-loop7 ENTH domain regions (Y101R, T105D, Q110A, and V112A) (Fig. 1C). It should be noted that the ENTH domain is the region with highest homology among epsin paralogs (>74% identity, including strict conservation of the Tyr-101-Thr-105 and loop 7 regions).
FIGURE 1.
The epsins bind the mammalian Cdc42/Rac1 GAP RalBP1. A, upper panel: lysates from HT1080 cells expressing HA-RalBP1 and GFP-epsin were immunoprecipitated with anti-HA or anti-Flag (negative control) antibodies. Bound epsins were detected by immunoblot with anti-GFP antibody. Medium panel, lysates from A were immunoprecipitated with an anti-GFP or anti- MT-MMP1 (negative control) rabbit polyclonal antibodies. Bound RalBP1 was detected by immunoblotting with an anti-HA antibody. Lower panel, lysates from HT1080 and NIH3T3 cells were immunoprecipitated with an anti-RalBP1 or an anti-HA (negative control) antibody. Bound epsin2 was detected by Western blot using a specific antibody. B, upper panel: different concentrations of purified His6-ENTH1 were incubated with GST or GST-RalBP1 immobilized on beads. Bound-ENTH1 was detected by immunoblotting with an anti-His6 antibody. Lower panels, formation and dissociation of [ENTH1·RalBP1] complex detected by surface plasmon resonance. Rate constant estimates after non-linear regression: kon = 0.1 μm−1 s−1 and koff = 0.02 s−1 (i.e. Kd = 0.2 μm). C, binding of 5 μm His6-ENTH domain WT or YTQV (Y101R, T105D, Q110A, and V112A) mutant to immobilized GST-RalBP1was done as in B, upper panel, in the presence (+) or absence (−) of 50 μm PtdIns(4,5)P2 (PIP2). Band intensity was quantified by densitometry and the resulting values represented as the mean ± S.D. of triplicates. D, mapping of the epsin binding regions on RalBP1. Upper panel, schemes represent the domain composition of RalBP1 and epsin. Names of known interaction partners are indicated next to their mapped binding sites. Black bars represent RalBP1 truncations tested for binding to epsins. FL: full-length; ΔNT: RalBP1209–655; ΔCT: RalBP11–383; ΔGAP: RalBP1Δ209–383. Lower panel, ability of the different Myc-tagged truncations to bind or not to GFP-epsin2 was tested by co-immunoprecipitation and immunoblotting and is represented by + or −, respectively. GAP: GTPase-activating protein domain; RalBD: Ral binding domain; CC: coiled-coil; UIM: ubiquitin-interacting motif; CBM: clathrin binding motif; DPW and NPF: Asp-Pro-Trp and Asn-Pro-Phe motifs for binding to AP2- and EH-containing proteins, respectively.
Using co-immunoprecipitation and deletion constructs, we mapped the epsin-binding region to the N-terminal end of RalBP1 (Fig. 1D). Interestingly, the N terminus of RalBP1 is also involved in binding to another element of the endocytosis machinery (and epsin interaction partner): the adaptor complex AP2 (17). These results suggest that the RalBP1 N-terminal region is important for its association with the endocytic machinery.
Epsin and RalBP1 Colocalize in Peripheral Membrane Structures
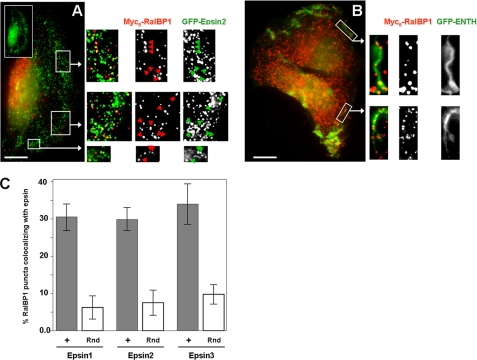
The epsin ENTH domain associates with the plasma membrane via interaction with phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)P2) (26). Moreover, it has been demonstrated that PtdIns(4,5)P2 binding leads to a substantial conformational change in the ENTH domain (27). Therefore, we asked whether the novel interaction between epsins and the membrane-located RalBP1 is affected by PtdIns(4,5)P2. Our experiments indicated that the presence of PtdIns(4,5)P2 improved RalBP1 binding by the ENTH domain (Fig. 1C). This result suggests that the RalBP1-epsin interaction would preferentially occur at the plasma membrane where both proteins and PtdIns(4,5)P2 are significantly enriched (20, 28). In fact, RalBP1 partially co-localized with GFP-ENTH in peripheral membrane structures (Fig. 2B) suggesting that in vivo, this novel interaction occurs at the plasma membrane. Further, full-length epsin also co-localized with RalBP1 in peripheral and puncta structures (e.g. GFP-epsin2, see Fig. 2A). In all cases, we noted that co-localization accounted for ∼30% (i.e.; limited to a subset) of RalBP1-positive structures (Fig. 2C). We observed that colocalization appeared to be a dynamic process and was enhanced in migratory cells or following cell stimulation with serum or by adhesion to fibronectin substrates (data not shown).
FIGURE 2.
GFP-epsin/ENTH domain partially co-localizes with RalBP1 at the plasma membrane in ruffles and puncta. HT1080 cells co-transfected with Myc6-RalBP1 and GFP-epsin2 (A) or GFP-ENTH (B) were extracted with saponin, fixed, and immunolabeled. Arrows highlight some areas of co-localization. The inset shows polarization of GFP-epsin2 toward the leading edge. Scale bar: 10 microns. C, percentage of RalBP1 puncta colocalizing with each of the epsins was quantified. Approximately 200 puncta per cell were counted in 6 different cells. To estimate the probability of random colocalization, the image corresponding to the RalBP1 channel was rotated 180° and random signal overlap was assessed (Rnd, open bars).
Epsins and RalBP1 Are Required for Cell Migration of Fibroblasts and Fibrosarcoma Cells
Because RalBP1 plays a crucial role in NIH3T3 fibroblast motility (14) and is up-regulated in invasive tumors (22, 29), we asked whether the novel RalBP1-epsin interaction was involved in cell migration. To address this question, we used a knock-down/rescue strategy. Specifically, we depleted either RalBP1 or epsins and transfected WT or mutated forms of each protein in both NIH3T3 fibroblasts and HT1080 fibrosarcoma cells. Epsin3 has been reported to be expressed only in migrating keratinocytes and basal carcinomas (30). Because, we confirmed that neither NIH3T3 nor HT1080 cells expressed epsin3 (supplemental Fig. S1A), this epsin paralog was not targeted for knock-down. The efficiency of the knock-downs and protein levels of transfectants were verified by immunoblotting (supplemental Fig. S1, B–D).
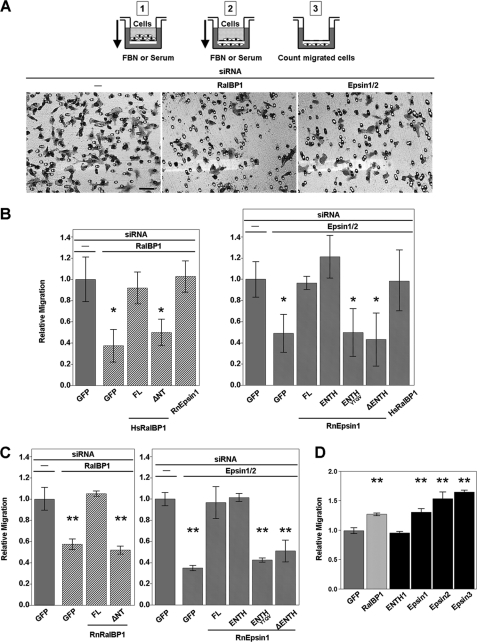
In agreement with previous reports (14), RalBP1 knock-down in NIH3T3 cells led to a significant impairment in their ability to migrate through fibronectin-coated transwell filters (Fig. 3, A and B). Similar results were obtained in human HT1080 fibrosarcoma cells (Fig. 3C). Importantly, this phenotype was specifically due to RalBP1 deficiency as siRNA-resistant full-length RalBP1 was able to re-establish cell migration (ref (14). and Fig. 3, B and C). We also found that RalBP1 lacking a region that includes the ENTH binding site (ΔNT) was not capable of sustaining normal cell migration in RalBP1-depleted cells (Fig. 3, B and C). Interestingly, we found that the epsin1- and epsin2-double knock-down cells also exhibited migration deficiencies (Fig. 3). Ruling-out off-target effects, siRNA-resistant full-length epsin1 was capable of rescuing the migration phenotype of epsin-depleted cells (Fig. 3, B and C).
FIGURE 3.
Epsins promote cell migration. A, cells seeded in the upper chamber (1) are allowed to migrate toward a gradient of fibronectin (FBN) or serum (2). After swabbing non-migrated cells in the upper chamber, the migrated cells (filter lower face) are counted by microscopy (3) or by staining with crystal violet and measuring the A560 nm of extracted stain. Fields from a representative experiment of crystal violet-stained membranes are shown. Scale bar: 50 microns. B and C, RalBP1 or epsins were knocked-down (KD) in NIH3T3 (B) or HT1080 (C) cells and assayed for migration. Expression of siRNA-resistant proteins is indicated by a label under the corresponding bar (FL: full-length). D, HT1080 cells overexpressing the indicated proteins were assayed for cell migration. Experiments were performed in triplicate. Statistical significance was determined by the t test with Bonferroni correction (panel B: *, p < 0.08/4 and p < 0.08/6; panel C: **, p < 0.05/3 and p < 0.05/5; panel D: **, p < 0.05/5).
Although RalBP1/epsin knock-down affected cell migration stimulated by adhesion onto fibronectin-coated substrates (haptotaxis) and by soluble ligands (chemotaxis), we determined that effects on haptotaxis were more severe than on chemotaxis (supplemental Fig. S2). This observation is in agreement with the described role of RalBP1 in Rac1 activation and cell migration following cell adhesion (14). Therefore, we adopted a haptotaxis scheme for our cell migration experiments.
We also found that single epsin knock-down had a more moderate effect on cell migration than the double epsin1 and 2 knock-down (data not shown), supporting the idea that epsins are redundant in their role in cell migration. This observation is not completely unexpected considering the shared ability of the different epsin paralogs to bind RalBP1 (Fig. 1) through the highly conserved, RalBP1 binding, ENTH domain (30, 31). Consistently, the ENTH domain (i.e. the RalBP1 binding unit) was necessary and sufficient for rescuing these phenotypes (Fig. 3, B and C). It should be noted that the epsin determinants involved in endocytosis are located in the C-terminal region of the molecule (see Fig. 1D), i.e. absent in the ENTH domain construct (epsin11–168).
We also found that simultaneous knock-down of epsins and RalBP1 showed no additive effects over the independent knock-down of the targets (supplemental Fig. S2B). This result indicates that epsins and RalBP1 are not in parallel pathways, further supporting the idea that their interaction is directly involved in sustaining cell migration.
Therefore, our results suggest that the ENTH-specific function in cell migration does not require endocytosis. In fact, we found that the internalization of β1-integrins, which is most likely to play a role in haptotaxis, was not significantly affected in either epsin or RalBP1 knock-down (supplemental Fig. S3A). Because epsin has been implicated in EGF receptor internalization, we studied the uptake of fluorescently tagged EGF. Our results indicate that internalization of TMR-EGF is not affected in cells expressing GFP-ENTH compared with GFP transfectants (supplemental Fig. S3, C and D).
Further supporting the role of the ENTH-RalBP1 interaction in cell migration, mutations in the RalBP1 binding region (ENTHYTQV) render the ENTH domain unable to rescue the phenotype (Fig. 3, B and C). Furthermore, an epsin truncation lacking the ENTH domain (ΔENTH: epsin1166–550) cannot alleviate the cell migration defects (Fig. 3, B and C).
We also reasoned that, because epsin depletion by siRNA-mediated knock-down typically leaves ∼10–20% residual dose of epsins (see supplemental Fig. S1), increasing the intracellular concentration of RalBP1 will raise the levels of [epsin·RalBP1] complex by mass action (i.e. by a “high-copy suppression” mechanism). We predicted that if the epsin-RalBP1 interaction was involved in cell migration, then the elevated levels of the [epsin·RalBP1] complex should lead to the suppression of the phenotype. Indeed, epsin knock-down cells transfected with RalBP1 displayed less severe migratory defects (Fig. 3B). The same rationale was applied to RalBP1-depleted cells expressing epsin with similar results (Fig. 3B). Interestingly, epsin or RalBP1 overexpression on their own led to enhanced cell migration by fibrosarcoma cells (Fig. 3D).
Epsin Is Required for Proper Activation of Rac1 and Arf6
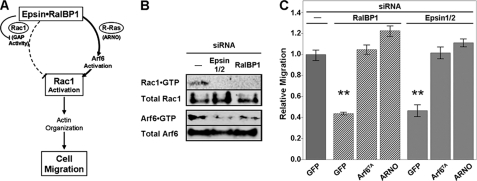
The RhoGTPase Rac1 controls the actin rearrangements required for cell migration and invasion (32). The RalBP1 effect on Rac1 activation status depends on the balance between its Rac1GAP inhibitory function (21, 33) and the Arf6-mediated activation of Rac1 initiated by RalBP1 recruitment of the Arf6-GEF ARNO (14). We hypothesized that epsin binding would tilt the balance between these opposing functions of RalBP1. Specifically, because it is known that epsin binding inhibits GAP activity (9), we speculated that epsin binding would favor RalBP1/ARNO-mediated activation of Arf6 and consequently Rac1 activation as well ((14) and Fig. 4A). Therefore, we tested the prediction that epsin-interference would affect the levels of GTP-bound (i.e. activated) Arf6 and Rac1 (Fig. 4A).
FIGURE 4.
Rac1 and Arf6 activation depend on epsin function. A, working model for the role of [epsin·RalBP1] complex in Rac1-mediated cell migration. RalBP1 can promote both deactivation (via GAP activity) and activation (via an Arf6-dependent pathway) of Rac1. Upon binding, epsin inhibits RalBP1 GAP activity and promotes Rac1 activation which is required for cell migration. B, lysates from mock or siRNA-treated HT1080 were used for CRIB or GGA3 (VHS+GAT) pull-down experiments to determine the levels of activated Rac1 and Arf6, respectively. C, RalBP1 and epsin KD cells were transfected with GFP, HA-Arf6T157A (TA), or Myc-ARNO. Transwell migration was determined as in Fig. 3A and compared with controls. Statistical significance was determined by the t test with Bonferroni correction (**, p < 0.05/6).
In agreement with this hypothesis, epsin knock-down led to deficiencies in [HA-Arf6·GTP] levels as detected by pull-down with the Arf binding region of GGA3 (Fig. 4B). As expected (14), similar results were obtained for RalBP1 knock-down (Fig. 4B). Further, the cell migration deficiencies displayed by either epsin or RalBP1 knock-down cells were rescued by overexpressing an activated, fast cycling, mutant of Arf6 (Arf6T157A: Arf6TA) or its GEF ARNO (Fig. 4C). Epsin-depleted cells also exhibited decreased levels of activated Rac1 as measured by pull-down with the CRIB (Cdc42 and Rac-interactive binding) domain of PAK1B and by in cellula determination with Rac1 biosensors (Fig. 4 and supplemental Fig. S4, respectively).
The Epsin-RalBP1 Interaction Is Required for HT1080 Invasion through Basement Membrane
Because cell migration is fundamental to cancer cell invasion and metastasis, we investigated whether interference with the novel epsin-RalBP1 interaction would affect the ability of the fibrosarcoma cell line HT1080 to invade through basement membrane. We performed invasion assays toward a gradient of serum using transwell filters coated with basement membrane (Fig. 5A). Results mirrored those obtained for cell migration, as knock-down of either RalBP1 or epsins impaired the ability of HT1080 cells to invade through basement membrane (Fig. 5, A and B). Expression of siRNA-resistant epsin, ENTH domain or RalBP1 rescued the cell invasion deficiency, ruling out off-target effects (Fig. 5B). In contrast, HT1080 knock-down cells expressing epsin or RalBP1 variants impaired for the ability to form the [epsin·RalBP1] complex showed a significant decrease in their ability to invade through basement membrane (Fig. 5B). Collectively, these results indicate that the epsin-RalBP1 interaction is required for fibrosarcoma invasion through basement membrane (Fig. 5, A and B). Because fluorescent gelatin degradation assays showed no significant difference between epsin- or RalBP1-depleted and control cells (data not shown), we conclude that the invasion defects described above are mostly due to cell migration abnormalities. Importantly, we also found that over-expression of RalBP1 or epsins, particularly epsin 2 and 3, enhanced the invasiveness of this human fibrosarcoma (Fig. 5C).
FIGURE 5.
The epsin-RalBP1 interaction is required for fibrosarcoma cell invasion. A, HT1080 cells were seeded on top of basement membrane (BM) and allowed to migrate toward serum. After 20 h, cells in the upper chamber were swabbed, and the cells that invaded through BM were fixed and stained with rhodamine-phalloidin and DAPI. Panels show random fields taken from cells depleted of RalBP1, epsins or mock-treated, that invaded through the BM. Scale bar: 50 microns. B and C, quantitation of results from three independent experiments. Mean ± S.E. are shown (panel B: **, p < 0.05/3 and p < 0.05/4; panel C: **, p < 0.05/5 using the t test with Bonferroni correction).
DISCUSSION
This study is the first to report a direct and essential role of the epsin family of endocytic adaptors in cell migration and invasion through basement membrane. Moreover, our results suggest that this novel epsin function does not require membrane internalization, but is dependent on binding to the Cdc42/Rac1 GAP RalBP1 and affects the activation of Arf6 and Rac1.
Knock-down experiments revealed that depletion of epsin impaired cell migration and invasion (Figs. 3 and 5). Importantly, rescue experiments with siRNA-resistant constructs confirmed the specificity of the knock-down results and ruled out off-target effects (Figs. 3 and 5). These results indicate that analogous to RalBP1 (14), epsins are involved in cancer cell migration. In fact, epsin1 and epsin2 are enriched in the leading edge of migratory cells (34) and our results extend those observations to migrating HT1080 fibrosarcoma cells (Fig. 2A, inset). Furthermore, previous studies showed that epsin3 expression is up-regulated in migrating keratinocytes (30, 35). Interestingly, we found that overexpression of epsins enhanced both migration and invasion of fibrosarcoma cells (Figs. 3D and 5C). These observations are particularly relevant considering that epsins have been recently found to be up-regulated in some non-small cell carcinomas and breast cancers (35, 36). In contrast, endocytic proteins such as Numb and Reps2/POB1, are down-regulated in malignant cells suggesting a suppressor function (37, 38). However, the endocytic and epsin-interacting protein Intersectin, can induce malignant transformation, and similar to epsin, it affects RhoGTPase and Ras signaling pathways (39).
Although it is known that endocytosis is required for cell migration (1, 40, 41), our results are the first to implicate the epsins in this process. Importantly, the epsin ENTH domain (i.e. lacking the main endocytic determinants) was necessary and sufficient to fulfill this novel epsin function (Figs. 3–5). However, whereas full-length epsin overexpression showed significant ability to enhance both migration and invasion in HT1080 cells, the isolated ENTH domain showed weak or nonexistent effect (Figs. 3D–5C). We speculate that the lack of endocytic determinants makes PtdIns(4,5)P2 binding the main localization determinant for the ENTH domain. As expected, the ENTH domain showed preferential localization to the leading edge and membrane ruffles where PtdIns(4,5)P2 is found broadly enriched (Fig. 2B). Therefore, we hypothesize that even when the ENTH domain was “diluted” over a more extensive membrane area overexpression was enough to render a critical concentration at specific sites to act upon RalBP1 and to sustain cell migration. Our results also indicate that the ENTH membrane “dilution” rendered its effective concentration at specific RalBP1 sites insufficient to enhance cell migration above normal levels. Because we have observed that the C-terminal end of epsin significantly contributes to protein localization (Fig. 2), we speculate that epsin full-length potency relies on its superior protein targeting capabilities. However, we cannot rule out a potential role of C-terminal determinants in a yet-to-be-determined pro-migration/invasion activity.
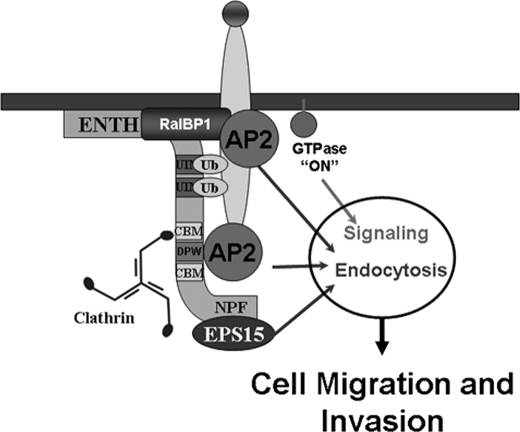
We envision that although the endocytic properties of epsin are not critical for its role in migration/invasion, its localization at endocytic sites might be important. Further, the dual role of epsin in signaling and endocytosis also provides a novel mechanism for the synchronization of these processes in time and space (Fig. 6).
FIGURE 6.
Working model for the coordination of the epsin functions on endocytosis and signaling. Ligand binding to its specific receptor triggers recruitment of epsin as well as other endocytic proteins. Epsin interaction with RalBP1 leads to GTPase activation and signaling at sites of endocytosis (see text and Fig. 4A for details). The scheme does not depict all known interactions between the proteins represented in the scheme, and not all interactions depicted occur at the same time.
Our data indicate that the critical role of the ENTH domain in cell migration and invasion is mediated by its interaction with RalBP1 (Figs. 3 and 5). Specifically, epsin constructs competent for RalBP1-binding (i.e. epsin full-length and ENTH domain) were capable of alleviating the migration/invasion abnormalities due to epsin knock-down whereas an N-terminal truncation (ΔENTH) was not (Figs. 3 and 5). Along the same lines, full-length RalBP1, but not the epsin binding-impaired RalBP1ΔNT, rescued the cell migration deficiencies triggered by RalBP1 depletion (Fig. 3B). Further, a “high-copy suppression” strategy designed to enhance the formation of the [epsin·RalBP1] complex in knock-down background by mass action, also alleviated the cell migration phenotype because of protein depletion (Fig. 3B). In agreement with a role in cell migration, co-localization between the interaction partners could be detected in peripheral ruffle-like structures (Fig. 2). Further, colocalization and immunoprecipitation of epsin with RalBP1 were feasible only following stimulation for cell migration. Although the ENTH-RalBP1 interaction displays a reasonable affinity, similar to other interactions involved in trafficking and signaling, it is likely to be subjected to constant regulation to ensure its transient characteristics to be suited to dynamic processes such as cell migration. In fact, we believe that the transient character of the interaction and the low abundance of epsin-RalBP1 complex were responsible for the weak signals detected by immmunoprecipitation.
The novel epsin-interacting protein RalBP1 has been found to be overexpressed in invasive cancer (29) and thoroughly demonstrated to play an important role in processes such as cell detoxification (23), cell survival (42), and embryo development (43, 44). RalBP1 has been previously linked to endocytosis of the same RTKs as epsin (15, 16) and has been found in complexes containing endocytic proteins such as AP2, Reps2/POB1 (17, 18), and epsin1 (19). Further, RalBP1 has been involved in promoting epsin1 regulation by Cdk1 phosphorylation (19). Because RalBP1 is also a known GAP for Cdc42 and Rac1 (21, 33, 45), our results support the idea that in mammals, like in yeast, epsins can affect cell polarity-dependent processes by binding RhoGTPase GAPs (9, 10). Moreover, we have been able to detect the [epsin·RalBP1] complex and verify its role in cell migration using other cell lines such as HeLa and Huvec,4 suggesting that this novel interaction is ubiquitous.
RalBP1 is known to have dual effects on Rac1 activation. (i) As a GAP, it leads to GTPase inactivation. (ii) As an R-Ras effector it activates Rac1 via an Arf6-dependent pathway, through the recruitment of the Arf-GEF ARNO (Fig. 4A). Because the consequence of the epsin-GAP interaction is the inhibition of GAP activity (9), we hypothesized that upon epsin binding, the Arf6-mediated activation of Rac1 should be predominant (Fig. 4A). Therefore, we predicted that interference of the epsin-RalBP1 interaction will lead to decreased levels of activated Rac1 (due to unrepressed RalBP1 GAP activity) and Arf6 (because of diminished R-Ras-dependent ARNO binding by RalBP1). Our results indeed confirmed these predictions (Fig. 4, A and B and supplemental Fig. S4). Further supporting our hypothesis, bypassing of the Arf6 deficiency (by overexpressing Arf6T157A or the Arf6-GEF ARNO) re-established normal cell migration in both epsin and RalBP1 knock-down cells (Fig. 4C). These results suggested that, similar to RalBP1 knock-down ((14) and Fig. 4), epsin depletion leads to Arf6-dependent cell migration defects. Altogether, our results indicate that the absence of [epsin·RalBP1] complex leads to Arf6 activation deficiency, lack of Rac1 activation, and cell migration and invasion abnormalities.
Consistent with the idea of a link between the endocytic machinery and GTPase regulators, other endocytic adaptor proteins have been described to recognize GAPs, e.g. the ARFGAP SMAP2 by AP180 (46), the RasGAP Dab2IP by Dab1/2 (47, 48) and CamGAP by CIN85 (49). These findings along with our own, suggest that GTPase regulation by endocytic proteins might be a generalized mechanism in cellular signaling.
This study reports a novel functional link between the endocytic machinery and the processes of cell migration and invasion. The relevance of these findings for cancer research is 2-fold: on the one hand, our data provide a new mechanism likely to contribute to the invasiveness of cancers that display up-regulated levels of epsins and/or RalBP1. On the other hand, this work points to these proteins as suitable targets for the development of therapeutics, particularly for control of metastatic cancers. In fact, RalBP1 regulation is already considered a feasible target for new anticancer therapies (22, 50). Our studies showed that epsins, similar to RalBP1, are also involved in motility of and invasion by malignant cells suggesting a role for epsins in tumor growth and metastasis.
Supplementary Material
Acknowledgments
We thank Elizabeth Taparowsky, Chris Staiger, Henry Chang, and Debarati Mukherjee (Purdue University) for stimulating discussions and critical reading of the manuscript. We thank Claudia B. Hanna for excellent technical assistance.
This work was supported by an American Cancer Society-Purdue Center for Cancer Research institutional grant and by start-up funds from the Dept. of Biological Sciences, Purdue University (to R. C. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S3.
B. G. Coon and R. C. Aguilar, unpublished observations.
- ENTH
- epsin N-terminal homology
- GAP
- GTPase-activating protein
- SPR
- surface plasmon resonance.
REFERENCES
- 1.Polo S., Di Fiore P. P. (2006) Cell 124, 897–900 [DOI] [PubMed] [Google Scholar]
- 2.Sorkin A., von Zastrow M. (2009) Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorkin A., von Zastrow M. (2002) Nat. Rev. Mol. Cell Biol. 3, 600–614 [DOI] [PubMed] [Google Scholar]
- 4.Lanzetti L., Di Fiore P. P. (2008) Traffic 9, 2011–2021 [DOI] [PubMed] [Google Scholar]
- 5.Nichols J. T., Miyamoto A., Weinmaster G. (2007) Traffic 8, 959–969 [DOI] [PubMed] [Google Scholar]
- 6.Chen H., Ko G., Zatti A., Di Giacomo G., Liu L., Raiteri E., Perucco E., Collesi C., Min W., Zeiss C., De Camilli P., Cremona O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13838–13843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Struhl G. (2004) Development 131, 5367–5380 [DOI] [PubMed] [Google Scholar]
- 8.D'Souza B., Miyamoto A., Weinmaster G. (2008) Oncogene 27, 5148–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilar R. C., Longhi S. A., Shaw J. D., Yeh L. Y., Kim S., Schön A., Freire E., Hsu A., McCormick W. K., Watson H. A., Wendland B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4116–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee D., Coon B. G., Edwards D. F., 3rd, Hanna C. B., Longhi S. A., McCaffery J. M., Wendland B., Retegui L. A., Bi E., Aguilar R. C. (2009) J. Cell Sci. 122, 2453–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coon B. G., Mukherjee D., Hanna C. B., Riese D. J., 2nd, Lowe M., Aguilar R. C. (2009) Hum. Mol. Genet. 18, 4478–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Northup J. (2004) Methods Mol. Biol. 261, 93–112 [DOI] [PubMed] [Google Scholar]
- 13.Santy L. C., Casanova J. E. (2001) J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldfinger L. E., Ptak C., Jeffery E. D., Shabanowitz J., Hunt D. F., Ginsberg M. H. (2006) J. Cell Biol. 174, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakashima S., Morinaka K., Koyama S., Ikeda M., Kishida M., Okawa K., Iwamatsu A., Kishida S., Kikuchi A. (1999) EMBO J. 18, 3629–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazic M., Bertelsen V., Pedersen K. W., Vuong T. T., Grandal M. V., Rødland M. S., Traub L. M., Stang E., Madshus I. H. (2009) Traffic 10, 235–245 [DOI] [PubMed] [Google Scholar]
- 17.Jullien-Flores V., Mahé Y., Mirey G., Leprince C., Meunier-Bisceuil B., Sorkin A., Camonis J. H. (2000) J. Cell Sci. 113, 2837–2844 [DOI] [PubMed] [Google Scholar]
- 18.Ikeda M., Ishida O., Hinoi T., Kishida S., Kikuchi A. (1998) J. Biol. Chem. 273, 814–821 [DOI] [PubMed] [Google Scholar]
- 19.Rossé C., L'Hoste B., Offner N., Picard A., Camonis J. (2003) J. Biol. Chem. 278, 30597–30604 [DOI] [PubMed] [Google Scholar]
- 20.Yadav S., Singhal S. S., Singhal J., Wickramarachchi D., Knutson E., Albrecht T. B., Awasthi Y. C., Awasthi S. (2004) Biochemistry 43, 16243–16253 [DOI] [PubMed] [Google Scholar]
- 21.Cantor S. B., Urano T., Feig L. A. (1995) Mol. Cell. Biol. 15, 4578–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Awasthi S., Singhal S. S., Awasthi Y. C., Martin B., Woo J. H., Cunningham C. C., Frankel A. E. (2008) Clin. Cancer Res. 14, 4372–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singhal S. S., Yadav S., Singhal J., Sahu M., Sehrawat A., Awasthi S. (2008) FEBS Lett. 582, 3408–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brett T. J., Traub L. M., Fremont D. H. (2002) Structure 10, 797–809 [DOI] [PubMed] [Google Scholar]
- 25.Praefcke G. J., Ford M. G., Schmid E. M., Olesen L. E., Gallop J. L., Peak-Chew S. Y., Vallis Y., Babu M. M., Mills I. G., McMahon H. T. (2004) EMBO J. 23, 4371–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. (2001) Science 291, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 27.Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. (2002) Nature 419, 361–366 [DOI] [PubMed] [Google Scholar]
- 28.Chen H., Fre S., Slepnev V. I., Capua M. R., Takei K., Butler M. H., Di Fiore P. P., De Camilli P. (1998) Nature 394, 793–797 [DOI] [PubMed] [Google Scholar]
- 29.Smith S. C., Oxford G., Baras A. S., Owens C., Havaleshko D., Brautigan D. L., Safo M. K., Theodorescu D. (2007) Clin. Cancer Res. 13, 3803–3813 [DOI] [PubMed] [Google Scholar]
- 30.Spradling K. D., McDaniel A. E., Lohi J., Pilcher B. K. (2001) J. Biol. Chem. 276, 29257–29267 [DOI] [PubMed] [Google Scholar]
- 31.Rosenthal J. A., Chen H., Slepnev V. I., Pellegrini L., Salcini A. E., Di Fiore P. P., De Camilli P. (1999) J. Biol. Chem. 274, 33959–33965 [DOI] [PubMed] [Google Scholar]
- 32.Kurisu S., Suetsugu S., Yamazaki D., Yamaguchi H., Takenawa T. (2005) Oncogene 24, 1309–1319 [DOI] [PubMed] [Google Scholar]
- 33.Jullien-Flores V., Dorseuil O., Romero F., Letourneur F., Saragosti S., Berger R., Tavitian A., Gacon G., Camonis J. H. (1995) J. Biol. Chem. 270, 22473–22477 [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Ding S. J., Wang W., Jacobs J. M., Qian W. J., Moore R. J., Yang F., Camp D. G., 2nd, Smith R. D., Klemke R. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Dai Z., Sadee W., Hancock W. S. (2006) Mol. Pharm. 3, 566–578 [DOI] [PubMed] [Google Scholar]
- 36.Pawlowski K. M., Krol M., Majewska A., Badowska-Kozakiewicz A., Mol J. A., Malicka E., Motyl T. (2009) J. Physiol. Pharmacol. 60, 85–94 [PubMed] [Google Scholar]
- 37.Colaluca I. N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., Di Fiore P. P. (2008) Nature 451, 76–80 [DOI] [PubMed] [Google Scholar]
- 38.Oosterhoff J. K., Penninkhof F., Brinkmann A. O., Grootegoed J. A., Blok L. J. (2003) Oncogene 22, 2920–2925 [DOI] [PubMed] [Google Scholar]
- 39.Wang J. B., Wu W. J., Cerione R. A. (2005) J. Biol. Chem. 280, 22883–22891 [DOI] [PubMed] [Google Scholar]
- 40.Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P. P. (2008) Cell 134, 135–147 [DOI] [PubMed] [Google Scholar]
- 41.Jones M. C., Caswell P. T., Norman J. C. (2006) Curr. Opin. Cell Biol. 18, 549–557 [DOI] [PubMed] [Google Scholar]
- 42.Singhal S. S., Roth C., Leake K., Singhal J., Yadav S., Awasthi S. (2009) Biochem. Pharmacol. 77, 1074–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lebreton S., Boissel L., Iouzalen N., Moreau J. (2004) Mech. Dev. 121, 1481–1494 [DOI] [PubMed] [Google Scholar]
- 44.Boissel L., Houssin N., Chikh A., Rynditch A., Van Hove L., Moreau J. (2007) Dev. Biol. 312, 331–343 [DOI] [PubMed] [Google Scholar]
- 45.Park S. H., Weinberg R. A. (1995) Oncogene 11, 2349–2355 [PubMed] [Google Scholar]
- 46.Natsume W., Tanabe K., Kon S., Yoshida N., Watanabe T., Torii T., Satake M. (2006) Mol. Biol. Cell 17, 2592–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z., Tseng C. P., Pong R. C., Chen H., McConnell J. D., Navone N., Hsieh J. T. (2002) J. Biol. Chem. 277, 12622–12631 [DOI] [PubMed] [Google Scholar]
- 48.Homayouni R., Magdaleno S., Keshvara L., Rice D. S., Curran T. (2003) Brain Res. Mol. Brain Res. 115, 121–129 [DOI] [PubMed] [Google Scholar]
- 49.Sakakibara T., Nemoto Y., Nukiwa T., Takeshima H. (2004) FEBS Lett. 566, 294–300 [DOI] [PubMed] [Google Scholar]
- 50.Singhal S. S., Singhal J., Yadav S., Dwivedi S., Boor P. J., Awasthi Y. C., Awasthi S. (2007) Cancer Res. 67, 4382–4389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.