Abstract
Cysteine synthesis in bacteria and plants is catalyzed by serine acetyltransferase (SAT) and O-acetylserine (thiol)-lyase (OAS-TL), which form the hetero-oligomeric cysteine synthase complex (CSC). In plants, but not in bacteria, the CSC is assumed to control cellular sulfur homeostasis by reversible association of the subunits. Application of size exclusion chromatography, analytical ultracentrifugation, and isothermal titration calorimetry revealed a hexameric structure of mitochondrial SAT from Arabidopsis thaliana (AtSATm) and a 2:1 ratio of the OAS-TL dimer to the SAT hexamer in the CSC. Comparable results were obtained for the composition of the cytosolic SAT from A. thaliana (AtSATc) and the cytosolic SAT from Glycine max (Glyma16g03080, GmSATc) and their corresponding CSCs. The hexameric SAT structure is also supported by the calculated binding energies between SAT trimers. The interaction sites of dimers of AtSATm trimers are identified using peptide arrays. A negative Gibbs free energy (ΔG = −33 kcal mol−1) explains the spontaneous formation of the AtCSCs, whereas the measured SAT:OAS-TL affinity (KD = 30 nm) is 10 times weaker than that of bacterial CSCs. Free SAT from bacteria is >100-fold more sensitive to feedback inhibition by cysteine than AtSATm/c. The sensitivity of plant SATs to cysteine is further decreased by CSC formation, whereas the feedback inhibition of bacterial SAT by cysteine is not affected by CSC formation. The data demonstrate highly similar quaternary structures of the CSCs from bacteria and plants but emphasize differences with respect to the affinity of CSC formation (KD) and the regulation of cysteine sensitivity of SAT within the CSC.
Keywords: Amino Acid, Plant, Protein Chemistry, Protein Structure, Protein-Protein Interactions, Cysteine Biosynthesis, Metabolic Regulation, O-Acetylserine Thiol Lyase, Cysteine Synthase Complex, Serine Acetyltransferase
Introduction
Cysteine biosynthesis in plants and bacteria is catalyzed by a two-step process. Serine acetyltransferase (SAT2; EC 2.3.1.30) activates serine by transfer of the acetyl moiety from acetyl coenzyme A to form O-acetylserine (OAS). Then OAS accepts sulfide by catalysis of OAS (thiol)-lyase (OAS-TL; EC 2.5.1.47). This fixation of free sulfide from assimilatory sulfate reduction or external sulfide sources is the exclusive entry of reduced sulfur into cellular metabolism. SAT and OAS-TL form the hetero-oligomeric cysteine synthase complex (CSC). In enterobacteria and plants, the interaction of SAT and OAS-TL is stabilized by the presence of sulfide, although the addition of OAS dissociates the two enzymes (1, 2). Plant and bacterial OAS-TLs are dimers that are catalytically inactive in the CSC but become fully active upon dissociation of the complex by OAS (1, 3). However, these properties do not seem to relate to metabolic regulation of cysteine synthesis in enterobacteria. In Escherichia coli, regulation of cysteine synthesis is mainly achieved by control of the cysteine regulon that includes OAS-TL and the genes encoding for proteins catalyzing sulfate uptake and reduction but not bacterial SAT. Bacterial SAT is constitutively expressed but strongly inhibited by cysteine (KI = 1.1 μm cysteine). In the presence of cysteine, SAT of E. coli (EcSAT) is almost inactive. At low cysteine concentrations, EcSAT is released from cysteine inhibition and produces OAS. OAS spontaneously converts to N-acetylserine, which binds to the CysB repressor and acts as an inducer. The N-acetylserine-CysB complex dissociates from the DNA, and the genes of the Cys-operon are expressed (4). In contrast, plant SAT and OAS-TL are part of a metabolic flux control system in cytosol, plastids, and mitochondria that contributes to the genetic regulation of cellular sulfate uptake and reduction (2, 5). Interestingly, at normal sulfur supply, all SAT is found in the CSC. Due to this interaction, OAS-TL is inactivated allowing OAS, the product of the SAT reaction and limiting factor for cysteine biosynthesis, to dissociate from the complex. A high excess of free OAS-TL dimers in plants converts OAS and sulfide to cysteine. During sulfur starvation, the sulfide level declines as a consequence of reduced sulfate assimilation, which results in accumulation of OAS and the loss of stabilization of the CSC by sulfide (2, 3).
The three-dimensional structures of plant SAT and OAS-TL proteins obtained by x-ray crystallography and of plant SAT proteins obtained by modeling appear to be very similar to their enterobacterial counterparts. This observation also applies to their kinetic properties (2, 6–8). The SAT C terminus was identified as one interaction site between bacterial SAT and OAS-TL (9) and the SAT and OAS-TL from the cytosol of Arabidopsis thaliana and Glycine max (10–12). Deletion of the C-terminal 20 amino acids of EcSAT strongly reduced binding to EcOAS-TL and inhibition by cysteine (9, 13). Feedback sensitivity of plant SAT is also conferred to a large extent by a methionine residue near the C terminus (14). The affinity of full-length EcSAT to EcOAS-TL in the complex was determined as KD <2 nm (15, 16) with SAT forming a central dimer of trimers associated with two OAS-TL homodimers. The affinities of SAT and OAS-TL in plant CSCs were reported to be considerably lower at 25 and 41 nm (3, 17). Recently, the quaternary structure of the cytosolic CSC from G. max was found to differ from that in bacteria and to consist of one SAT trimer associated with three OAS-TL dimers that bind with affinities from KD = 0.2 to 78 nm (12).
Despite strong similarities between the CSCs from bacteria and plants, the regulatory function of the CSC seems to be completely different, because plant SAT is regulated by complex formation (3), whereas EcSAT is not affected (1). Here, we use analytical ultracentrifugation (AUC), size exclusion chromatography (SEC), and isothermal titration calorimetry (ITC) to explore quaternary composition of plant CSCs from Arabidopsis and G. max and define fundamental regulatory differences between plant and bacterial CSCs.
EXPERIMENTAL PROCEDURES
General Cloning
PCR and cloning of DNA fragments were performed according to Ref. 18. Deletion of AtSATm (At3g13110) proteins was introduced by selective amplification using PCR with primer p_210 in combination with p_ΔC10 or p_ΔC15 for SATΔC10 and SATΔC15 (supplemental data 1). The resulting PCR fragments were cloned in the vector pET-32a (Novagen, Germany). The same strategy was applied for cloning of full-length AtSATc (At5g56760), GmSATc (Glyma16g03080), and histidine-tagged AtOAS-TLc (At4g14880) using the primer combinations p_1366, p_1367, p_1364, p_1365, p_202, and p_203 (supplemental data 1). PCR fragments encoding for AtSATc, GmSATc, and AtOAS-TLc were cloned in the vectors pET-28a and pET-30a, respectively.
Expression and Purification of Proteins
Mature histidine-tagged AtSATm and native AtOAS-TLm (At3g59760) were co-expressed in HMS 174(DE3) bacteria and purified via immobilized metal affinity chromatography as described in Ref. 19 as CSC. Free histidine-tagged AtSATm was eluted after on-column dissociation of the immobilized CSC with 10 mm OAS. In addition, free AtOAS-TLc and AtOAS-TLm were purified via their affinity to SAT as described in Ref. 8 using the same pET vector constructs. AtSATc and GmSATc were purified via their affinity to OAS-TL, after immobilization of histidine-tagged AtOAS-TLc on a HiTrapTM Chelating HP column (GE Healthcare) as bait. Elution of mature AtSATc and GmSATc was achieved by dissociation of CSC with 10 mm OAS in 50 mm Tris, pH 7, 0.25 m sodium chloride for size exclusion chromatography or 0.1 m HEPES, pH 7, 0.25 m sodium chloride for analytical ultracentrifugation.
Enzyme Assays for SAT and OAS-TL
For determination of SAT activity, purified recombinant SAT protein (∼1 ng) was assayed according to Ref. 19. OAS-TL activity was determined as described previously (8).
Size Exclusion Chromatography
Molecular weights were determined by size exclusion chromatography (SEC) using a Pharmacia FPLCTM system (GE Healthcare) that was connected to a Superdex200 HiLoadTM 16/60 prep grade column (GE Healthcare) and equilibrated in 50 mm Tris, pH 7, 0.25 m sodium chloride. Calibration of the Superdex 200 column was performed with the high and low molecular weight gel filtration calibration kits (GE Healthcare) according to the manufacturer's instructions. The protein amounts were varied between 0.5 nmol for SAT hexamers and up to 17 nmol of mixtures of SAT hexamers and OAS-TL dimers.
Analytical Ultracentrifugation
Centrifugation was performed in 50 mm Tris, pH 8.5, 0.25 m sodium chloride for AtSATm and AtOAS-TLm and in 0.1 m HEPES, pH 7, 0.25 m sodium chloride for AtSATc, GmSATc, and AtOAS-TLc. Protein concentration varied between 0.5 and 5 mg ml−1. Formation of heterogeneous CSC composed of the GmSATc hexamer and the AtOAS-TLc dimer was achieved by addition of 1 mm sulfide and 12-fold excess of the AtOAS-TLc dimer. Sedimentation velocity profiles were monitored at 280 nm for SAT and at 412 nm for OAS-TL and CSC. The samples were centrifuged at 45,000 rpm and 4 °C in a Beckman Optima XL-A centrifuge equipped with the AN-60 Ti rotor. Sedimentation coefficient and molecular weight distributions were analyzed by the C(s) method implemented in the Sedfit software package (20). AUC data were presented as molecular weight distributions (C(M)). Buffer density and viscosity corrections were made according to the data published previously (21).
Isothermal Titration Calorimetry
Experiments were performed using a VP-ITC Microcal calorimeter connected to a cell with a 1.44-ml volume (Microcal, Northhampton, MA). All proteins were dialyzed extensively against ITC buffer (50 mm Tris, pH 8.5, 0.25 m NaCl) prior to all titrations. A typical titration at 20 °C consisted of 24 injections of 6–12-μl aliquots of 20 μm of the AtOAS-TLm dimer or 55 μm of the AtOAS-TLc dimer into 0.7 μm of the AtSATm hexamer or 1.7 μm the AtSATc hexamers at time intervals of 5 min to ensure that the titration peak returned to the base line. The ITC data were analyzed using the program Origin version 5.0 provided by the manufacturer.
Comparative Modeling, Computation of Binding Energies and Molecular Dynamics Simulation
A description of the computational procedures is provided in supplemental data 2.
Screening for Interaction Domains of SAT with Custom-made Peptide Arrays
Custom-made peptide arrays of SAT were produced by solid phase peptide synthesis based on the Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry as described previously (22). The sequence of mature AtSATm was presented as oligopeptides consisting of 15 residues with one amino acid lap from peptide to peptide immobilized on a membrane (22). [35S]Met-labeled AtSATm was synthesized by feeding of [35S]Met (16.5 MBq) to HMS 174 (DE3) E. coli cells expressing histidine-tagged SAT in M9 medium without methionine (18) and purified as described above. The peptide arrays were probed for 2 h with [35S]Met-labeled AtSATm (0.3 ng ml−1) in binding buffer (50 mm Tris, pH 8, 0.25 m NaCl) supplied with 2.5% BSA. Binding specificity was ensured by washing three times with binding buffer (50 mm Tris, pH 8, 0.25 m NaCl). Signal spots were detected by autoradiography.
Determination of Molecular Weight of the Native Cytosolic CSC from Arabidopsis
Leaves of 7-week-old soil-grown A. thaliana (ecotype Col-0) plants were harvested and ground in liquid nitrogen to a fine powder. Proteins were extracted with 50 mm Tris, pH 7, 0.15 m sodium chloride, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 10 mm DTT, 0.5 mm PMSF for 15 min and separated from cell debris by centrifugation at 20 °C and 25,000 × g for 20 min. Soluble proteins (3 mg) were separated in the presence or absence of 10 mm OAS by SEC as described (fraction size 1 ml). AtSATc was immunologically detected in each fraction using an AtSATc-specific antiserum after separation of fractionated proteins by SDS-PAGE and transfer of proteins to nitrocellulose (0.22 μm, Applichem, Darmstadt, Germany) as described previously (18). The antiserum was demonstrated to be specific by comparison of signals determined in crude extracts of wild type and a T-DNA knock-out line of AtSATc (supplemental data 3).
RESULTS
Quaternary Structure of the Mitochondrial CSC and Its Subunits in Arabidopsis
The mitochondrial SAT provides the bulk of OAS in Arabidopsis (23) and was selected for analysis. Experiments were carried out with affinity-purified recombinant AtSATm and AtOAS-TLm. The transit peptides were not included in the construct. Apparent molecular sizes were determined by molecular SEC as 72 ± 5 kDa for AtOAS-TLm (n = 7), 208 ± 12 kDa for AtSATm (n = 6), and 343 ± 45 kDa for CSC (n = 7) (Fig. 1A). Based on the calculated molecular weights, these sizes correspond to a dimer of AtOAS-TLm (calculated molecular mass for AtOAS-TLC of 33.8 kDa), a hexamer of AtSATm (calculated molecular mass for AtSATm of 35.1 kDa), and a 2:1 ratio of the AtOAS-TLm dimer to the AtSATm hexamer in the CSC. These findings were verified using AUC and revealed molecular masses of 72 ± 2 kDa for AtOAS-TLm (n = 4) and 199 ± 3 kDa for AtSATm (n = 3, Fig. 1B). In two of the six AtSATm purifications analyzed by SEC, a protein complex was detected with a molecular mass of 350 kDa, which perfectly fits to the calculated size of the recombinant CSC of 345.8 kDa. Most likely, a small contamination of the AtSATm preparation with AtOAS-TLm that was co-expressed to increase yield resulted in CSC formation. Furthermore, we applied ITC to analyze the association of AtSATm and AtOAS-TLm (Fig. 1C). The best fit of the experimental data (supplemental data 4) was achieved by applying a “one set of identical binding site” model as follows: A + nB → Abn, where A is the AtSATm hexamer, and B is the AtOAS-TLm dimer (supplemental data 5). Titrations of AtSATm with AtOAS-TLm yielded a stoichiometry of n = 2.20 ± 0.04 corresponding to a ratio of two AtOAS-TLm dimers to one AtSATm hexamer. The dissociation constant was determined as KD = 30 ± 6 nm. The reaction enthalpy and Gibbs free energy were strongly negative (Table 1), indicating an exergonic process that confirms the observed spontaneous formation of the CSC. The reaction enthalpy contributed by far more to ΔG than entropy, emphasizing the importance of the transition from free AtOAS-TLm dimers and AtSATm hexamers to CSC-bound subunits.
FIGURE 1.
Quaternary structure of the AtCSCm and its subunits. A, AtOAS-TLm, AtSATm, and AtCSCm were purified independently and resolved by SEC as described under “Experimental Procedures.” Gray vertical lines represent elution volume of proteins used for calibration (1, thyroglobulin, 669 kDa; 2, ferritin, 440 kDa; 3, catalase, 232 kDa; 4, β-amylase, 200 kDa; 5, aldolase, 158 kDa; 6, alcohol-dehydrogenase, 150 kDa; 7, BSA 66 kDa). B, molecular weights of CSC subunits were independently confirmed by AUC. C, association kinetic of the AtCSCm was analyzed by ITC. The release of heat by subsequent injection of AtOAS-TLm dimer to a AtSATm hexamer is plotted against the molar ratio of AtOAS-TLm dimer to AtSATm hexamer. The solid line represents a fit to data using the one set of equal binding sites model.
TABLE 1.
Kinetic properties of CSCs from plants and enterobacteria
Stoichiometry and kinetic properties of CSC from S. typhimurium, H. influenzae and E. coli are shown in Refs. 1, 15, and 16, respectively. Specific SAT activity and inhibition by cysteine of bacterial SAT were determined with SAT of E. coli in the presence (CSC) or absence of EcOAS-TL (SAT alone).
|
A. thaliana in |
Enterobacteria in cytoplasm | ||
|---|---|---|---|
| Cytosol | Mitochondria | ||
| N | 2.07 ± 0.01 | 2.20 ± 0.04 | 2a,b |
| KD | 28 ± 2 nm | 30 ± 6 nm | 2.6 ± 0.8c and <2 nmb |
| DH | −28.4 ± 0.1 kcal mol−1 | −35.5 ± 0.9 kcal mol−1 | |
| T·ΔS | −18.1 kcal mol−1 | −25.2 kcal mol−1 | |
| ΔG | −10.3 kcal mol−1 | −10.3 kcal mol−1 | |
| CSC specific SAT activity | 10.14 ± 1.23 units mg−1 | 6.99 ± 0.94 units mg−1 | 10.18 ± 1.85 units mg−1 |
| IC50 cysteine | 50 μm | 250 μm | 0.25 μm |
| SAT alone specific SAT activity | 11.72 ± 1.59 units mg−1 | 7.09 ± 0.64 units mg−1 | 7.68 ± 1.86 units mg−1 |
| IC50 cysteine | 30 μm | 65 μm | 0.25 μm |
Identification of SAT-SAT-binding Sites
The association of SAT trimers into a hexamer implies specific binding sites. The SAT-SAT interaction was therefore mapped using AtSATm peptide arrays in combination with [35S]Met-labeled AtSATm. 15-Mer oligopeptides moving in one amino acid steps along the entire AtSATm sequence (without leader peptide) were synthesized and immobilized on a membrane. Each peptide spot thus had a 14-amino acid overlap with the previous one. The binding assay was repeated twice with freshly prepared [35S]Met-AtSATm protein and independently synthesized SAT peptide arrays (Fig. 2, A and B). The peptide stretch identified corresponded to part of the helix α4 in the N-terminal AtSATm domain (supplemental data 6). Homology modeling of AtSATm to EcSAT (24) suggests that this α-helix is in the same position as the corresponding helix of EcSAT (Fig. 2, C and D). These data are consistent with crystallographic studies (25) demonstrating the involvement of the helix α4 in dimerization of two EcSAT trimers.
FIGURE 2.
Identification of SAT-SAT interaction domains using peptide arrays. A and B, custom-made SAT peptide arrays were probed independently with [35S]Met-labeled AtSATm. In both cases, signal stretches corresponding to part of helix α4 of AtSATm were evident (MNY-92LFDLFSGVLQGN103). C, homology modeling of AtSATm to HiSAT revealed that helix α4 shown in space-filling representation is in the same position as the corresponding helix of HiSAT. Helix α4 of HiSAT is known to be involved in the dimerization of bacterial SAT trimers by crystallographic studies. D, side view on α and δ monomer of SAT hexamer that interact via helix α4. The β, χ, ϵ, and γ monomers of the HiSAT hexamer are hidden for clarity.
Quaternary Structure of Cytosolic SAT and CSC in Arabidopsis
Because SAT of soybean was reported to be trimeric (12), we investigated AtSATc to explore potential compartment-specific differences in quaternary structure of CSCs from Arabidopsis. The recombinant purified protein was subjected to AUC and revealed a molecular size of 183 ± 18 kDa for AtSATc, providing evidence for the hexameric structure of AtSATc (calculated molecular mass for AtSATc of 32.8 kDa, n = 2, Fig. 3A).
FIGURE 3.
Quaternary structure of the AtCSCc and its subunits. A, recombinant AtSATc hexamer (0.5 nmol) was purified and resolved by AUC. B, association kinetics of the AtCSCc was analyzed by ITC. The release of heat by subsequent injection of AtOAS-TLc dimer to an AtSATc hexamer is plotted against the molar ratio of AtOAS-TLc dimer to AtSATc hexamer. The solid line represents a fit to the data using the one set of equal binding sites model. C, soluble leaf proteins of 6-week-old Arabidopsis plants were resolved by SEC in the absence or presence of 10 mm OAS to determine the molecular weight of cytosolic CSC and free AtSATc from Arabidopsis, respectively. Equal amounts of fractions (1 ml) corresponding to retention volume 55 to 73 ml were subjected to immunological detection of AtSATc using a specific antiserum.
Similarly to the analysis of mitochondrial CSC, ITC was also applied to analyze the interaction of AtSATc with AtOAS-TLc (Fig. 3B and supplemental data 4). A stoichiometry of n = 2.07 ± 0.01 was determined upon titration of AtSATc with AtOAS-TLc corresponding to a molar ratio of two AtOAS-TLm dimers to one AtSATm hexamer. The dissociation constant, reaction enthalpy, entropy, and Gibbs free energy were clearly negative and comparable with the respective values for the mitochondrial CSC formation (Table 1).
To ask whether endogenous CSC has the same stoichiometry as the recombinant enzymes, the molecular size of the native CSCc was analyzed. Soluble proteins from Arabidopsis leaves were separated by SEC in the presence and absence of OAS. AtSATc was detected by Western blotting (see “Experimental Procedures”). An apparent molecular mass of 342 kDa ± 8 (n = 3) in the absence of OAS corresponded to AtSATc in the CSC, whereas free AtSATc was found in the presence of 10 mm OAS with an apparent molecular mass of 185 kDa (n = 1). Furthermore, the abundance of SAT and OAS-TL in purified recombinant mitochondrial and cytosolic CSC was tested by silver staining after separation by SDS-PAGE. In both cases, higher abundance of SAT with respect to OAS-TL was detected, compatible with a lower OAS-TL to SAT ratio as would be expected from a 4:6 ratio of monomers (supplemental data 7).
Quaternary Structure of Cytosolic SAT in Soybean
Species-specific differences could be a cause for different quaternary structures of Arabidopsis and soybean SATs and hence the composition of the respective CSCs (12). This hypothesis was addressed by investigation of cytosolic SAT from G. max under the same conditions as Arabidopsis AtSATc. RNA from G. max L. Merr. cv. Williams82 as used in Ref. 12 was isolated to generate a full-length cDNA. Comparison of the resulting cDNA sequence with cDNA corresponding to GenBankTM accession number AF452452.1 (cytosolic SAT of soybean published in Ref. 12) revealed considerable differences but a perfectly matched genomic DNA from the soybean sequencing project that also used G. max L. Merr. cv. Williams82 (Glyma16g0308 (supplemental data 6) (26)). Repeated cloning of cDNA from G. max L. Merr. cv. Williams82 and additionally from G. max L. Merr. cv. Effi confirmed the precise match with the respective entry in the soybean genomic database (Glyma16g03080 (26)). The cytosolic SAT from G. max used here is therefore named GmSATc in contrast to AF452452.1 (12). The differences in the amino acid sequence of AF452452.1 are at residues 149 and 175–192 and correspond to an amino acid composition that is significantly different from known SATs (supplemental data 6).
SEC of the GmSATc obtained from cv. Williams82 in this work revealed a molecular size of 232 kDa (n = 2; Fig. 4A). Using AUC, the molecular mass of GmSATc was determined as 180 kDa (n = 2; Fig. 4B). The heterologous CSC composed of GmSATc and AtOAS-TLc was used for AUC because plant SATs and OAS-TLs from different species are known to interact (3, 27). The molecular weights of the three detectable protein species correspond well to the AtOAS-TLc dimer, the GmSATc hexamer, and a heterologous CSC composed of one hexamer of GmSATc and two dimers of AtOAS-TLc (n = 1, Fig. 4C). GmSATc therefore shows a hexameric composition, and the corresponding CSC probably consists of a decameric structure with most likely two OAS-TL dimers.
FIGURE 4.
Quaternary structure of the GmSATc. A, recombinant GmSATc hexamer (0.5 nmol) was resolved by SEC. B, molecular weight of the GmSATC hexamer (1.6 nmol) was confirmed by AUC. C, GmSATc (1.3 nmol) and cytosolic OAS-TL from Arabidopsis (15.5 nmol AtOAS-TLc) were mixed and subjected to AUC. All experiments were performed as described under “Experimental Procedures.”
Modeling the Quaternary Structure of SAT
In view of the apparent discrepancy between the quaternary structures of AtSATm and AF452452.1 (12), the binding energies of two SAT trimers in a SAT hexamer were calculated for several SATs (Table 2 and supplemental data 2). The binding energies of the SAT trimers from plants were consistently about 30% higher than those for bacterial SATs, mostly due to differences in electrostatic binding energy. Nevertheless, the binding energies demonstrate that the existence of SAT in a hexameric state is favorable in all cases. In particular, the results suggest that, as in E. coli, Haemophilus influenzae, and A. thaliana, the oligomeric state of the SATs in G. max is also likely to be a homohexamer.
TABLE 2.
Calculated binding energies for dimerization of SAT trimers in enterobacteria and plants
HiSAT and EcSAT represent the sole SAT isoforms of H. influenzae and E. coli, respectively. GmSATc relates to the cytosolic SAT from G. max, which is encoded by gene Glyma1603080. AF452452.1 is the cytosolic SAT of G. max studied in Ref. 12. GmSATo corresponds to a putatively organelle-localized isoform in G. max and is encoded by gene Glyma18g08910.
| Isoform | Localization | Electrostatic energy | Apolar energy | Binding energy |
|---|---|---|---|---|
| kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| HiSAT | Cytoplasm | −2 | −128 | −130 |
| EcSAT | Cytoplasm | 4 | −123 | −118 |
| AtSATc | Cytosol | 32 | −114 | −82 |
| AtSATm | Mitochondria | 26 | −108 | −82 |
| GmSATc | Cytosol | 37 | −118 | −81 |
| AF452452.1 | Cytosol | 36 | −123 | −87 |
| GmSATo | Organelle | 27 | −113 | −86 |
The flexibility of the SAT trimers of the two isoforms of cytosolic SAT from G. max was assessed in light of their sequence differences. Molecular dynamics simulation (supplemental data 2) revealed higher flexibility in AF452452.1, accompanied by local structural distortion, mainly in the β-sheet domain and less apparent in the α-helical domain (supplemental data 8). These results suggest that, due to reduced stability, the hexamerization of AF452452.1 might be less favorable than that of GmSATc. Moreover, one can speculate that the higher flexibility may lead to a higher proportion of OAS-TL in the CSC for AF452452.1, as found experimentally (12).
SAT C Terminus Represses SAT Activity and Modulates Feedback Inhibition
The C terminus of SAT proteins was shown to be a major determinant for the interaction with OAS-TL (11, 13, 19). Because the domain involved in feedback inhibition to cysteine is in the vicinity of the C terminus (9, 11, 14, 28), deletions of the 10 or 15 C-terminal amino acids were introduced into mitochondrial AtSATm (Fig. 5A) designated as SATΔC10 and SATΔC15, respectively.
FIGURE 5.
C-terminal deletion of AtSATm affects enzymatic activity and inhibition by cysteine. A, full-length AtSATm and the C-terminally truncated proteins SATΔC10 and SATΔC15 were purified twice and analyzed for their maximal enzymatic activity (n = 8). B, feedback inhibition of SAT to cysteine was tested. Asterisks indicate significant differences between AtSATm and the truncated SAT proteins (p < 0.05). Statistical analysis was performed with the Student's t test implemented in SigmaStat 3.0 (SPSS).
SATΔC15 showed 2-fold increased activity (p < 0.001, n = 8), and SATΔC10 activity was also significantly (p = 0.01, n = 8) increased. Kinetic analysis of SATΔC15 showed that only Vmax but not Km was affected by the deletion (supplemental data 9). This deletion analysis of free AtSATm suggests that the C terminus, although predicted to be flexible (25) and not attached to the β-sheet domain, represses SAT enzymatic activity. Interestingly, SATΔC15 eliminated the methionine residue Met376 involved in cysteine feedback inhibition, and SATΔC10 reached close to this residue. Accordingly, the deletion of the C-terminal 10 amino acids resulted in decreased sensitivity to feedback inhibition by cysteine (Fig. 5B).
Differential Feedback Inhibition by Cysteine in Free and CSC-bound SAT Proteins
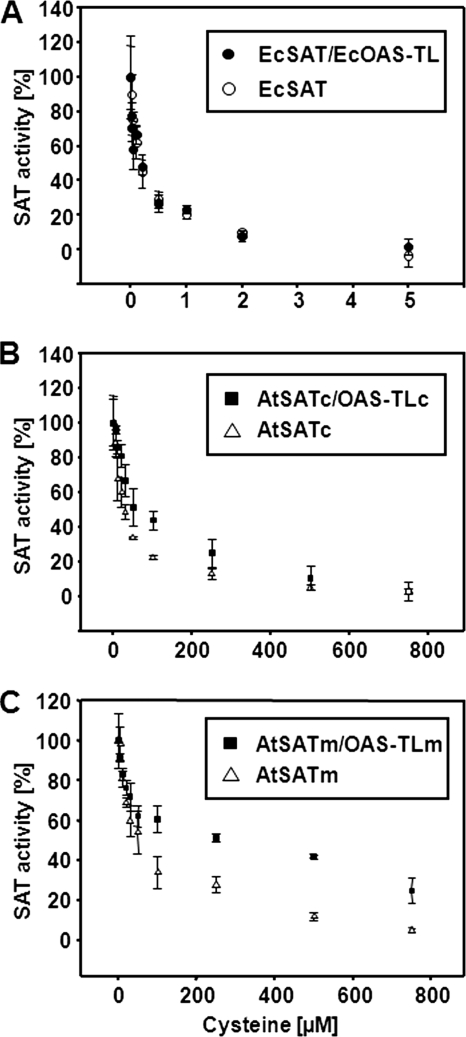
The possible dual role of the SAT C terminus in OAS-TL binding and control of SAT activity prompted the investigation of feedback sensitivity of free and complex bound SAT proteins to cysteine. Analysis of purified recombinant EcSAT showed inhibition with an IC50 = 0.25 μm by cysteine and almost complete inhibition at higher cysteine concentrations (Fig. 6A). SAT in the bacterial CSC was inhibited in the same way by increasing cysteine concentrations (Table 1). In contrast, the inhibition constant of free AtSATc was IC50 = 30 μm (Fig. 6B) and thus statistically significantly higher compared with AtSATc bound in AtCSCc (IC50 = 50 μm). The difference in feedback inhibition was even larger for free and CSC-bound AtSATm (IC50 = 65 and 250 μm, respectively). Complete inhibition could not be achieved for SAT activity in the complex even at high cysteine concentrations (Fig. 6C). The three free SAT proteins showed specific activities from 7 to 11 nmol mg−1 min−1 (Table 1). Taken together, AtSATm/c are less sensitive to feedback inhibition by cysteine when bound in the CSC compared with the free forms. In contrast, feedback inhibition of EcSAT is apparently independent of its oligomeric state.
FIGURE 6.
Cysteine inhibition of plant and bacterial SATs within the CSC. A–C, recombinant EcSAT (A), AtSATc (B), and AtSATm (C) were purified and tested for inhibition of SAT activity by cysteine in free or CSC associated state (n = 4). CSC formation was achieved by co-expression of SAT with its native interaction partner in E. coli followed by histidine-affinity purification of the spontaneously formed CSC. Cysteine concentration was varied between 0 and 0.005 mm for inhibition of bacterial SAT, whereas the concentration of cysteine was increased up to 0.8 mm to efficiently inhibit plant SATs. For comparison, activity of plant and bacterial SAT is shown as percentage of noninhibited free or complex associated SAT.
DISCUSSION
In this study, three independent approaches demonstrate quaternary structures of a dimer for AtOAS-TLm, a homohexamer for AtSATm in the absence of AtOAS-TLm, and a 2:1 ratio of OAS-TL dimer to SAT hexamer in the assembled mitochondrial and cytosolic CSCs from Arabidopsis. This composition supports findings of a recent modeling and docking study that identified a 2:1 ratio of AtOAS-TLm dimer to AtSAT hexamer as the energetically and geometrically most likely composition (24). In this model, one AtSATm trimer binds one AtOAS-TLm dimer. The hexameric core of such a 2:1 complex is also in agreement with the functional mapping of the helix α4 of AtSATm as a determinant of dimerization of SAT trimers, in analogy to the equivalent helix in the HiSAT (SAT of Haemophilus influenzae) crystal structure (6).
A hexameric structure was also found for cytosolic AtSATc from Arabidopsis. Notably, the determination of the molecular size of native AtSATc from Arabidopsis leaf extracts was used for the first time to verify the analyses of recombinant SATs from bacteria and plants. These results rule out compartment-specific differences between mitochondrial and cytosolic SATs and also CSC composition. Still, species-specific differences might account for the discrepancies found in comparison with cytosolic AF452452.1 (12). Isolation of the corresponding cDNA from soybean failed. It repeatedly yielded a sequence that differed in 19 amino acids and was in accordance with the soybean genome database (26). Analysis of the encoded protein termed GmSATc (Glyma16g03080) of this sequence fully confirmed the hexameric structure of SAT. For this reason, the difference in β-sheet strands 4–6 of AF452452.1 in comparison with GmSATc and other known SATs (supplemental data 6) seems to be responsible for the unique trimeric organization of AF452452.1 and the difference in stoichiometry of the AF452452.1-GmOAS-TLc complex to bacterial and plant CSCs (Figs. 1, 3, and 4) (1, 6,12, 24, 25, 29). The altered β-sheet domain of AF452452.1 is not expected to be directly involved in the dimerization of SAT trimers or interaction of SAT with OAS-TL (6, 11, 29). Nevertheless, several mutations in the β-sheet domain of AtSATm have been experimentally proven to affect the SAT-SAT interaction and also CSC formation (19). Molecular dynamics simulation of the GmSATc and of the modeled AF452452.1 structure (supplemental data 8) indicate destabilization of the β-sheet domain in AF452452.1, which may lead to the observed trimeric organization and the 3:1 molar ratio in the CSC formed by AF452452.1. In agreement with this hypothesis, a point mutation in the β-sheet domain of bacterial SAT, named BB1, causes a total disruption of CSC formation and impairs SAT-SAT interaction (19, 30). When the BB1 mutation was introduced into AtSATm, it had the same effect, although it is far away from the C-terminal tail and the α-helical N terminus of SAT (supplemental data 6) (6, 8–11, 24). These examples demonstrate the significance of the conserved SAT β-sheet domain for self-oligomerization and interaction with OAS-TL.
Further support for the generally hexameric structure of SATs is provided by calculation of the binding energy for SAT trimer-trimer association (Table 2). Bacterial and plant SATs, including GmSATc and GmSATo, each fall into distinct groups with overall properties that favor dimerization of trimers as opposed to the existence of single trimers. However, it should be cautioned that this approach did not distinguish AF 452452.1 from the other modeled plant SATs.
The molecular weight of a heterologous CSC made up of GmSATc and AtOAS-TLc was in agreement with a hexameric GmSATc as the core that binds 2 OAS-TL dimers (2:1 ratio). A point of caution must be added to this interpretation, because a GmSATc trimer that binds three OAS-TL dimers (3:1 ratio) as postulated in Ref. 12 would have an indistinguishable molecular weight.
ITC experiments further corroborate a CSC stoichiometry of one AtSATm or AtSATc hexamer and two corresponding OAS-TL dimers (Table 1). They demonstrate that CSC formation is a spontaneous and mainly enthalpy-driven process. The large negative values of enthalpy indicate a very specific recognition of the interaction partners SAT and OAS-TL. The remarkable decrease of entropy points to a significant degree of conformational change, which occurs upon complex formation. Because ΔS is negative, the order of the entire system increases during complex formation. Potentially the increase in order is a result of fixation of the flexible C terminus of AtSATm and AtSATc in the active sites of AtOAS-TLm and AtOAS-TLc, respectively. This increase in order counteracts the enthalpy-driven spontaneous formation of the entire complex and might be a possible explanation for the finding that the C-terminal decapeptide of Arabidopsis cytosolic AtSATc appears to have a higher affinity than the full-length AtSATc protein (31).
Furthermore, the dissociation constants of bacterial CSCs were reported to be 10 times lower than those of plant CSCs (16, 17). The comparatively high KD value of the plant CSCs had previously been determined by surface plasmon resonance (17) and enzymatic measurements (3). Using ITC, this high KD value could be independently confirmed for the Arabidopsis cytosolic and mitochondrial CSCs. The discrepancy to the much lower KD value of the bacterial CSC suggests differences in the affinity of bacterial and plant SAT/OAS-TL interaction, which is mediated by the C terminus of SATs. The exception so far is the CSC of G. max formed by AF452452.1 (12) with its 3:1 ratio, which displayed three KD values ranging from 0.3 to 78 nm (12). Remarkably, the C-terminal 10 amino acids that are responsible for the interaction with OAS-TL are the only part of SAT that is not highly conserved between bacterial and plant SATs (supplemental data 6).
The feedback sensitivity of free SAT to cysteine was described to be 1–2 orders lower in magnitude in bacteria than plants (4, 14). Direct comparison of free and complex bound SATs confirmed this difference and revealed indistinguishable feedback sensitivity for bacterial SAT but enhanced sensitivity of free SAT over bound SAT for the Arabidopsis proteins. This difference was reported to be even larger in the CSC formed by AF452452.1 (12) and is thus likely to contribute to the regulation of the rate of cysteine synthesis. A regulation of different feedback sensitivity to cysteine of phosphorylated versus nonphosphorylated SAT has been reported for GmSerat2.1 (32). This enzyme is associated with the cytosol and plastids and phosphorylated by a calcium-dependent protein phosphorylase in response to oxidative stress (32). Interestingly, the phosphorylation site is located in the C terminus of GmSerat2.1.
In conclusion, evolution seems to have favored conservation of the quaternary structures of the CSC in bacteria and higher plants with apparently different functions. In plants, the CSC serves as a regulatory platform for its subunits, but the bacterial CSC lacks regulatory function for SAT. The functional differences of the CSC in these two types of organisms can be explained by the low conservation of the C-terminal residues of SAT, which largely determine cysteine inhibition of SAT and the association kinetics of the CSC.
Supplementary Material
This work was supported by the Landesgraduiertenförderung Baden-Württemberg (Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology, BioQuant Graduate School Molecular Machines), the Schmeil-Stiftung, Heidelberg, and the Deutsche Forschungsgemeinschaft Grants He1848/5-2 and He1848/13-1 (to H. B. and C. H.) and by the Klaus Tschira Foundation (to A. F.-S. and R. C. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data 1–9.
- SAT
- serine acetyltransferase
- AUC
- analytical ultracentrifugation
- CSC
- cysteine synthase complex
- ITC
- isothermal titration calorimetry
- OAS
- O-acetylserine
- OAS-TL
- O-acetylserine(thiol)lyase
- EcSAT
- E. coli SAT
- SEC
- size exclusion chromatography
- AtSATm
- mitochondrial SAT from A. thaliana
- AtSATc
- cytosolic AtSAT
- GmSAT
- G. max SAT.
REFERENCES
- 1.Kredich N. M., Becker M. A., Tomkins G. M. (1969) J. Biol. Chem. 244, 2428–2439 [PubMed] [Google Scholar]
- 2.Wirtz M., Hell R. (2006) J. Plant Physiol. 163, 273–286 [DOI] [PubMed] [Google Scholar]
- 3.Droux M., Ruffet M. L., Douce R., Job D. (1998) Eur. J. Biochem. 255, 235–245 [DOI] [PubMed] [Google Scholar]
- 4.Kredich N. M. (1996) in Escherichia coli and Salmonella Typhimurium. Cellular and Molecular Biology (Neidhardt F. C., Curtiss R., Ingraham J. L., Lin E. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umberger E. eds) pp. 514–527, American Society for Microbiology, Washington, D. C [Google Scholar]
- 5.Sirko A., Błaszczyk A., Liszewska F. (2004) J. Exp. Bot. 55, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 6.Olsen L. R., Huang B., Vetting M. W., Roderick S. L. (2004) Biochemistry 43, 6013–6019 [DOI] [PubMed] [Google Scholar]
- 7.Tai C. H., Nalabolu S. R., Jacobson T. M., Minter D. E., Cook P. F. (1993) Biochemistry 32, 6433–6442 [DOI] [PubMed] [Google Scholar]
- 8.Wirtz M., Droux M., Hell R. (2004) J. Exp. Bot. 55, 1785–1798 [DOI] [PubMed] [Google Scholar]
- 9.Mino K., Yamanoue T., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (1999) Biosci. Biotechnol. Biochem. 63, 168–179 [DOI] [PubMed] [Google Scholar]
- 10.Bogdanova N., Hell R. (1997) Plant J. 11, 251–262 [DOI] [PubMed] [Google Scholar]
- 11.Francois J. A., Kumaran S., Jez J. M. (2006) Plant Cell 18, 3647–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumaran S., Yi H., Krishnan H. B., Jez J. M. (2009) J. Biol. Chem. 284, 10268–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mino K., Hiraoka K., Imamura K., Sakiyama T., Eisaki N., Matsuyama A., Nakanishi K. (2000) Biosci. Biotechnol. Biochem. 64, 1874–1880 [DOI] [PubMed] [Google Scholar]
- 14.Noji M., Inoue K., Kimura N., Gouda A., Saito K. (1998) J. Biol. Chem. 273, 32739–32745 [DOI] [PubMed] [Google Scholar]
- 15.Campanini B., Speroni F., Salsi E., Cook P. F., Roderick S. L., Huang B., Bettati S., Mozzarelli A. (2005) Protein Sci. 14, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao C., Moriga Y., Feng B., Kumada Y., Imanaka H., Imamura K., Nakanishi K. (2006) Biochem. Biophys. Res. Commun. 341, 911–916 [DOI] [PubMed] [Google Scholar]
- 17.Berkowitz O., Wirtz M., Wolf A., Kuhlmann J., Hell R. (2002) J. Biol. Chem. 277, 30629–30634 [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Book 1, pp. 5.50–51, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19.Wirtz M., Berkowitz O., Droux M., Hell R. (2001) Eur. J. Biochem. 268, 686–693 [DOI] [PubMed] [Google Scholar]
- 20.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C. eds) The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 22.Petersen G., Hahn C., Gehring U. (2001) J. Biol. Chem. 276, 10178–10184 [DOI] [PubMed] [Google Scholar]
- 23.Haas F. H., Heeg C., Queiroz R., Bauer A., Wirtz M., Hell R. (2008) Plant Physiol. 148, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman-Salit A., Wirtz M., Hell R., Wade R. C. (2009) J. Mol. Biol. 386, 37–59 [DOI] [PubMed] [Google Scholar]
- 25.Pye V. E., Tingey A. P., Robson R. L., Moody P. C. (2004) J. Biol. Chem. 279, 40729–40736 [DOI] [PubMed] [Google Scholar]
- 26.Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D. L., Song Q., Thelen J. J., Cheng J., Xu D., Hellsten U., May G. D., Yu Y., Sakurai T., Umezawa T., Bhattacharyya M. K., Sandhu D., Valliyodan B., Lindquist E., Peto M., Grant D., Shu S., Goodstein D., Barry K., Futrell-Griggs M., Abernathy B., Du J., Tian Z., Zhu L., Gill N., Joshi T., Libault M., Sethuraman A., Zhang X. C., Shinozaki K., Nguyen H. T., Wing R. A., Cregan P., Specht J., Grimwood J., Rokhsar D., Stacey G., Shoemaker R. C., Jackson S. A. (2010) Nature 463, 178–183 [DOI] [PubMed] [Google Scholar]
- 27.Wirtz M., Hell R. (2007) Plant Cell 19, 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denk D., Böck A. (1987) J. Gen. Microbiol. 133, 515–525 [DOI] [PubMed] [Google Scholar]
- 29.Hindson V. J., Moody P. C., Rowe A. J., Shaw W. V. (2000) J. Biol. Chem. 275, 461–466 [DOI] [PubMed] [Google Scholar]
- 30.Becker M. A., Tomkins G. M. (1969) J. Biol. Chem. 244, 6023–6030 [PubMed] [Google Scholar]
- 31.Kumaran S., Jez J. M. (2007) Biochemistry 46, 5586–5594 [DOI] [PubMed] [Google Scholar]
- 32.Liu F., Yoo B. C., Lee J. Y., Pan W., Harmon A. C. (2006) J. Biol. Chem. 281, 27405–27415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.