Abstract
LptC is a conserved bitopic inner membrane protein from Escherichia coli involved in the export of lipopolysaccharide from its site of synthesis in the cytoplasmic membrane to the outer membrane. LptC forms a complex with the ATP-binding cassette transporter, LptBFG, which is thought to facilitate the extraction of lipopolysaccharide from the inner membrane and release it into a translocation pathway that includes the putative periplasmic chaperone LptA. Cysteine modification experiments established that the catalytic domain of LptC is oriented toward the periplasm. The structure of the periplasmic domain is described at a resolution of 2.2-Å from x-ray crystallographic data. The periplasmic domain of LptC consists of a twisted boat structure with two β-sheets in apposition to each other. The β-sheets contain seven and eight antiparallel β-strands, respectively. This structure bears a high degree of resemblance to the crystal structure of LptA. Like LptA, LptC binds lipopolysaccharide in vitro. In vitro, LptA can displace lipopolysaccharide from LptC (but not vice versa), consistent with their locations and their proposed placement in a unidirectional export pathway.
Keywords: Bacteria, Cell Surface, Crystal Structure, Endotoxin, Lipopolysaccharide (LPS), Membrane Biogenesis, Membrane Proteins, Membrane Trafficking, LPS Export, Lpt Proteins
Introduction
The outer membrane (OM)3 of Gram-negative bacteria, such as Escherichia coli, is comprised of an asymmetric lipid bilayer with phospholipids in the inner leaflet and glycolipids, predominately lipopolysaccharide (LPS), in the outer leaflet (1). LPS is an essential component of the OM in most Gram-negative pathogens, and its unique structural features contribute to the effective permeability properties of the OM (2–4). In E. coli and many other Gram-negative bacteria, LPS is composed of the lipid A moiety, typically a highly conserved diglucosamine-based phospholipid, that is linked to the long chain polysaccharide known as O-antigen via a core oligosaccharide (5).
The structure and many elements of the biosynthesis of LPS have been established (reviewed in Refs. 4–6). However, the precise mechanism(s) of transport and assembly at the cell surface still remain(s) obscure. The lipid A-core region is synthesized in consecutive steps at the cytoplasmic face of the inner membrane (IM) (5) and is exported across the IM by the ATP-binding cassette (ABC) transporter MsbA (7, 8). O-antigen is synthesized independently by one of the three different pathways and is ligated to the lipid A-core moiety by the WaaL ligase at the periplasmic face of the IM (5). The complete LPS molecule then interacts with the LPS transport (Lpt) machinery (6). It is believed to be extracted from the IM by an ABC protein complex comprised of LptB (a predicted ABC protein) (9, 10), LptF (11), LptG (11), and LptC (12, 13). Each of these proteins is essential for LPS transport and viability in E. coli. Recent data have established that LptBFGC forms a stable complex in a 2:1:1:1 stoichiometry (13). Once LPS is released from the IM, its transit across the periplasm is proposed to be facilitated by LptA, a soluble periplasmic protein (9). However, the exact mechanism by which LPS is moved across the periplasm and through the peptidoglycan layer is still unknown. The final steps in the pathway are attributed to two OM proteins: LptD, a protein with predicted β-barrel structure, and LptE, a lipoprotein (14–17). Both proteins are essential in E. coli and required for LPS assembly on the cell surface. LptD and LptE form a stable interaction in a 1:1 complex (16). LptE also binds LPS in vitro (16).
Two models have been proposed for LPS transport across the periplasm (9, 18). In the first, oligomers of LptA form a proteinaceous bridge that physically connects the IM and OM, allowing for direct transfer of LPS to the OM. Evidence supporting this model was provided by the formation of LptA filaments when crystallized in the presence of LPS (18). In the filament, LptA monomers interact in a head-to-tail fashion, forming a linear filament with four monomers constituting one turn of a left-handed helix (18). The proposal is also consistent with continued LPS export detected in spheroplasts of E. coli (19). In addition, the entire Lpt machinery was recently identified in E. coli membrane fractions that contained markers of both inner and outer membranes, providing additional evidence that the Lpt proteins form a trans-envelope complex (20). The second model proposes that the LPS export pathway is analogous to the Lol-mediated lipoprotein transport, and the similarity between the various components in the two systems is striking. In the lipoprotein trafficking pathway, LolCDE represents an ABC protein complex required for the release of nascent lipoproteins from the IM to a periplasmic chaperone, LolA (21, 22). LolA transfers its cargo to LolB in the OM (23). LptBFG may be the functional equivalent of LolCDE, but the lipoprotein export pathway has no obvious component analogous to LptC.
To gain further understanding of the LPS export pathway, the properties of LptC were investigated. The data reported here describe the crystal structure of the soluble periplasmic portion of LptC at 2.2-Å resolution and the structural relationships between LptA and LptC. Like LptA, LptC binds LPS in vitro, and the results suggest that LptC transfers LPS to LptA in a unidirectional manner.
MATERIALS AND METHODS
Bacterial Strains and Recombinant DNA Techniques
All bacterial strains and plasmids used in this study are listed in Table 1. Recombinant DNA and molecular biology techniques were performed as described previously (24).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| W3110 | Wild type, F−, λ− | E. coli Genetic Stock Center (Yale) |
| XL-1 Blue | recA1 endA1 gyrA96thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIqZΔM15::Tn10 (TetR)) | Stratagene |
| Rosetta(DE3)pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pLysSRARE (CamR) | Novagen |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL StrrendA1 nupG | Invitrogen |
| CWG904 | TOP10 carrying the plasmid, pWQ480; Apr | This work |
| CWG905 | Rosetta(DE3)pLysS carrying the plasmid, pWQ481; Kmr | This work |
| CWG906 | TOP10 carrying the plasmid, pWQ482; Apr | This work |
| CWG907 | TOP10 carrying the plasmid, pWQ483; Apr | This work |
| CWG908 | TOP10 carrying the plasmid, pWQ484; Apr | This work |
| CWG909 | TOP10 carrying the plasmid, pWQ485; Apr | This work |
| Plasmids | ||
| pBAD24 | l-Arabinose-inducible plasmid; Apr | (25) |
| pET28a | Cloning/expression vector; Kmr | Novagen |
| pWQ480 | pBAD24 derivative encoding LptC-His6; Apr | This work |
| pWQ481 | pET28a derivative encoding His6-LptC24–191; Kmr | This work |
| pWQ482 | pBAD24 derivative encoding LptC24–191-TEV-His6; Apr | This work |
| pWQ483 | pBAD24 derivative encoding LptC-His6 with insertion of cysteine (C2); Apr | This work |
| pWQ484 | pBAD24 derivative encoding LptC-His6 with insertion of cysteine (C186); Apr | This work |
| pWQ485 | pBAD24 derivative encoding LptA-TEV-His6; Apr | This work |
Construction of Plasmids
Primers used for cloning are listed in Table 2. Plasmid pWQ480 was constructed by amplifying lptC from E. coli W3110 with primers MT5 and MT6 and cloned into the EcoRI and XbaI sites of pBAD24 (25). The reverse primer incorporates the sequence for a C-terminal hexahistidine (His6) tag. Using plasmid pWQ480 as the template, single-cysteine derivatives of LptC were made by the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Primers MT37 and MT38 were used to introduce the codon for a single Cys residue between codons 1 and 2 of LptC to generate pWQ483. Primers MT39 and MT40 were used to introduce the codon for a single cysteine residue between codons 185 and 186 of LptC, generating pWQ484. A 507-bp DNA fragment, encoding residues 24–191 of LptC (191 residues in total for LptC), was amplified from W3110 with primers MT30 and MT27 and cloned into the NdeI and BamHI sites of pET28a (Novagen), producing the plasmid pWQ481. The absence of the transmembrane helix in this construct yielded a soluble cytoplasmic protein. The His6-LptC(24–191) construct includes an N-terminal thrombin/His6 affinity tag. To facilitate removal of the His6 tag, a version of LptC(24–191) containing a tobacco etch virus (TEV) protease recognition site was constructed by amplifying lptC from W3110 with primers MT28 and MT56. The PCR fragment was then cloned into the EcoRI and XbaI sites of pBAD24 to generate the plasmid pWQ482. A removable C-terminal His6-tagged version of LptA, also containing a TEV protease recognition site, was constructed by amplifying lptA from W3110 with primers MT1 and MT55 and cloned into the EcoRI and XbaI sites of pBAD24 to generate the plasmid pWQ485. All plasmids and codon changes were confirmed by DNA sequencing.
TABLE 2.
Oligonucleotides used for cloning
Primers are 5′ → 3′.
| Name | Sequence | Description |
|---|---|---|
| MT5 | GCGCGCGAATTCACCATGAGTAAAGCCAGACGT | FPa: LptC-His6 |
| MT6 | GCGCGCTCTAGATCAGTGGTGGTGGTGGTGGTGAGGCTGAGTTTGTTTGTT | RP: LptC-His6 |
| MT30 | GAGCTACATATGGCCGAAAAAGACGATACC | FP: His6-LptC24–191 |
| MT27 | GATGCTGGATCCTTAAGGCTGAGTTTGTTT | RP: His6-LptC24–191 |
| MT28 | CAGGAGGAATTCACCATGAATGATCCCACCTATAAAAGC | FP: LptC24–191-TEV-His6 |
| MT56 | GCGCGCTCTAGACTAATGATGATGATGATGATGACCCTGAAAATACAGGTTTTCACCGGTACCAGGCTGAGTTTGTTTGTTTTG | RP: LptC24–191-TEV-His6 |
| MT37 | CAGGAGGAATTCACCATGTGTAGTAAAGCCAGACGTTGGG | FP: LptC-His6 (C2) |
| MT38 | CCCAACGTCTGGCTTTACTACACATGGTGAATTCCTCCTG | RP: LptC-His6 (C2) |
| MT39 | GAACATCCTATGAAATTCAATGTAACAAACAAACTCAGCC | FP: LptC-His6 (C186) |
| MT40 | GGCTGAGTTTGTTTGTTACATTGAATTTCATAGGATGTTC | RP: LptC-His6 (C186) |
| MT1 | GCGCGCGAATTCACCATGAAATTCAAAACAAAC | FP: LptA-TEV-His6 |
| MT55 | GCGCGCTCTAGACTAATGATGATGATGATGATGACCCTGAAAATACAGGTTTTCACCGGTACCATTACCCTTCTTCTGTGCCGG | RP: LptA-TEV-His6 |
a FP, forward primer; RP, reverse primer.
Membrane Topology of LptC
The topology of LptC was determined by site-directed fluorescence labeling of precisely located Cys residues, following the strategy of Fu and Maloney (26) with some minor modifications. E. coli strains CWG904, CWG907, and CWG908 were grown at 37 °C in LB medium containing 100 μg/ml ampicillin for 18 h. Each culture was diluted 1:100 in 50 ml of fresh medium and grown until mid-logarithmic phase (A600 ∼0.6). Expression of LptC-His6 from the pBAD promoter (25) was then induced for 3 h by adding l-arabinose to a final concentration of 0.2%. Each culture was divided into three equal aliquots. After harvesting by centrifugation, cells from aliquot one were resuspended in 5 ml of buffer A (50 mm KH2PO4, pH 8, containing 100 mm K2SO4) containing freshly prepared Oregon green 488 maleimide carboxylic acid (OGM) (40 μm) (Molecular Probes Inc.). The cell suspension was incubated for 20 min at 23 °C before the labeling reaction was quenched with β-mercaptoethanol (6 mm). Cells from aliquot two were prepared as described above, except the cells were labeled with freshly prepared methanethiosulfonate ethyltrimethylammonium (MTSET) (2 mm) (Toronto Research Chemicals) for 15 min at 23 °C instead of OGM. Both cell aliquots were then subjected to three cycles of centrifugation (10,000 × g, 10 min) and resuspension/washing in buffer A. The third aliquot of cell suspension remained untreated. All three aliquots were resuspended in a lysis solution (10 mm Tris HCl, pH 7.5, containing 500 μg/ml lysozyme, 40 μg/ml DNase, and 5 mm EDTA) and incubated at 37 °C for 30 min to initiate cell rupture. Membrane ghosts were obtained by a 10-fold dilution into distilled water, and cytoplasmic proteins were removed by three cycles of centrifugation (10,000 × g, 10 min) and membrane resuspension/washing in buffer A. Membranes obtained from aliquots two and three were resuspended with buffer A containing OGM (40 μm). After a 20-min incubation at 23 °C, the labeling reactions were quenched by the addition of β-mercaptoethanol (6 mm) and then washed three times with buffer A. Membranes from all three aliquots were immediately analyzed by 12% polyacrylamide gels by SDS-PAGE (27). After electrophoresis, the fluorescence profile from the SDS-PAGE gel was visualized using the ChemiDoc XRS system (Bio-Rad), and the protein contents of each lane were evaluated by staining the same gel with SimplyBlue SafeStain (Invitrogen).
Purification of His6-LptC(24–191), LptC(24–191)-TEV-His6, and LptA-TEV-His6
CWG905 was grown at 37 °C in LB containing kanamycin (50 μg/ml) for 18 h. This culture was diluted 1:100 in fresh medium and grown until mid-logarithmic phase (A600 ∼0.6). Expression of His6-LptC(24–191) was induced for 3 h by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mm. CWG906 was grown in LB containing ampicillin (100 μg/ml), and expression of LptC(24–191)-TEV-His6 was induced with 0.2% l-arabinose for 3 h. CWG909 was grown at 37 °C in M9 minimal medium containing ampicillin (100 μg/ml) as described previously (24), and expression of LptA-TEV-His6 was induced with 0.2% l-arabinose for 1 h. Cells were harvested by centrifugation (5,000 × g, 10 min), resuspended in buffer B (20 mm NaH2PO4, pH 7.5, containing 300 mm NaCl), and disrupted by passage through a French press. Unbroken cells and cell debris were removed by centrifugation (12,000 × g, 20 min). The membrane fraction was removed from the cleared lysate by ultracentrifugation (100,000 × g, 1 h). Soluble proteins were purified from the supernatant by using HIS-Select nickel affinity gel (Sigma) as described previously (24). Elution fractions were monitored by 12% polyacrylamide gels by SDS-PAGE. The pooled fractions containing purified protein were desalted by using a PD-10 column (GE Healthcare), eluted into buffer C (20 mm Tris, pH 7.5, 300 mm NaCl), and finally concentrated by using a Vivaspin 15R column (10,000 molecular weight cut-off: Vivascience). Protein concentrations were determined by the bicinchoninic acid method (28) using bovine serum albumin as the standard. Removal of the His6 tag using recombinant TEV protease (Invitrogen) was performed according to the manufacturer's directions.
Analytical Ultracentrifugation
Purified His6-LptC(24–191) was diluted to 0.75, 0.5, and 0.25 mg/ml in 10 mm Tris buffer, pH 7.5, containing 150 mm NaCl. The samples were analyzed using an Optima XL-1 analytical ultracentrifuge (Beckman Coulter). Experiments were carried out at 45,000 rpm for 16 h at 16 °C. Measurements of A280 nm were carried out at 5-min intervals during the ultracentrifugation.
Crystallization Conditions
Purified His6-LptC(24–191) was further refined by gel filtration using a HiPrep 16/60 Sephacryl S-100 HR column in buffer C, and the peak fractions were pooled and concentrated to 20 mg/ml. Screening of protein crystallization conditions was performed using instrumentation from Cartesian Dispensing Systems (Genomic Solution). The diamond-shaped crystals took 2 weeks to grow in 0.04 m K2SO4, 16% PEG 8000, and 20% glycerol at 20 °C, using the sitting-drop vapor diffusion method with 2 μl of protein and 2 μl of the crystallization solution. The selenomethionine derivative of His6-LptC(24–191) was obtained using the methionine biosynthesis inhibition method (29). Selenomethionine-labeled His6-LptC(24–191) was purified as described above in the presence of 2 mm β-mercaptoethanol. The selenomethionine-derivative His6-LptC(24–191) crystals were obtained in 0.04 m K2SO4, 20% PEG 8000, and 20% glycerol after 2 weeks of incubation at 20 °C, using the same sitting-drop vapor diffusion method employed for the native protein.
Data Collection and Structure Determination
Selenomethionine-labeled His6-LptC(24–191) crystals were protected by a cryoprotectant containing 20% PEG 8000, 0.04 m K2SO4, and 25% glycerol, and the data were collected at the Diamond IO3 Light Source at the peak wavelength of 0.9764 Å at −100 K. The dataset was indexed, integrated, and scaled using MOSFLM and Scala (30, 31). The His6-LptC(24–191) crystal belongs to the space group P43212 with the cell dimensions of a = b = 93.91 Å, c = 58.60 Å, α = β = γ = 90° with one monomer of the LptC in an asymmetry unit cell. The phases for His6-LptC(24–191) were initially calculated at the Diamond IO3 Light Source using SHELXD (32) and reevaluated using SOLVE (33) with the single-wavelength anomalous dispersion (SAD) data. The positions for two selenium atoms were identified in the asymmetric unit. The phases were improved, and the initial model was built using RESOLVE (34). The phases from 2.8 Å were extended to the high resolution (2.2 Å) using DM (58) and the model was partially built using ARP/wARP (35). The complete model was built using Coot (36), and structural refinement was carried out using REFMAC 5 (37). The structure was evaluated using MolProbity (38).
In Vitro LPS Binding Assay
The in vitro LPS binding assays were carried out as described previously (24). The binding substrate, smooth LPS from E. coli serotype O9a, was isolated using the hot water-phenol method (39). LPS was examined by SDS-PAGE and visualized by silver staining (40) as described previously by Hitchcock and Brown (41). An in vitro LPS transfer assay was developed based on the assay described above, and a strategy was used to examine the Lol system (24, 42). Briefly, assays were carried out in 500-μl reactions in buffer C containing 500 μg of purified LptA-TEV-His6 or LptC-TEV-His6 and 150 μg of purified O9a LPS. The reactions were incubated at 23 °C for 1 h on a rotary shaker to allow formation of LPS·protein complexes. HIS-Select nickel affinity gel (Ni2+-NTA resin) (200 μl, washed in 1 ml of buffer C; Sigma) was added to the reaction mixture and incubated for another hour to facilitate binding of the LPS·protein complexes via the His6 tag. Next, the reaction mixtures were centrifuged at 13,000 × g for 1 min, and the supernatant was collected (designated as FT in Figs. 2, 5, and 6). The resin was then washed four times (W1–W4) with buffer C to ensure complete removal of any unbound LPS, protein, and LPS·protein complexes. Subsequently, 500 μg of TEV-digested (i.e. non-His6-tagged) LptA or LptC(24–191) was added to the reaction mixture, and incubation was continued for 1 h at 23 °C. The reaction mixture was centrifuged again, as described above, and the supernatant was collected (FT2). The resin was then washed four times (W5–W8) with 1 ml of buffer C. The remaining immobilized protein (LptA-TEV-His6 or LptC-TEV-His6) was eluted with 500 μl of buffer C containing 300 mm imidazole. A final wash with 500 μl of buffer C containing 500 mm imidazole was employed to ensure that all protein was eluted.
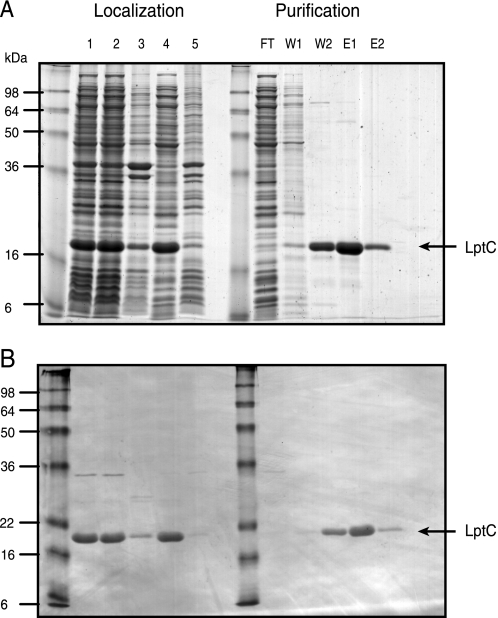
FIGURE 2.
Cellular localization, purification, and characterization of the periplasmic domain of LptC. A and B, His6-LptC(24–191) was expressed in E. coli CWG905. Fractions were separated by SDS-PAGE, and proteins were visualized using either SimplyBlue SafeStain (A) or Western immunoblotting with anti-His5 monoclonal antibody (B). The His6-LptC(24–191) construct migrates as a 21-kDa band (indicated by the arrow on the right), consistent with the predicted value of 21,300 Da. Lane 1 contains whole cell lysate from induced E. coli CWG905 cells. Lanes 2 and 4 represent the soluble fractions after centrifugation at 15,000 and 100,000 × g, respectively. Lanes 3 and 5 represent the membrane fractions after centrifugation at 12,000 and 100,000 × g, respectively. The purification of His6-LptC(24–191) by Ni2+-NTA affinity chromatography is shown on the right side of A and B. FT, flow-through with unbound proteins; W, wash; E, elution with imidazole.
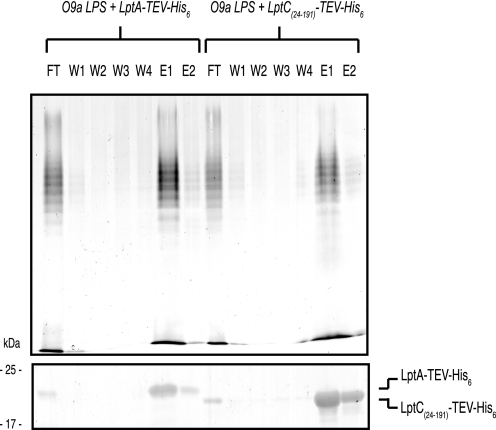
FIGURE 5.
In vitro LPS binding by LptC. The ability of LptA-TEV-His6 or LptC(24–191)-TEV-His6 to bind to purified O9a LPS was assessed by its co-elution from Ni2+-NTA affinity chromatography resin. LPS was examined by SDS-PAGE and silver staining (upper) after proteinase K digestion of the elution fractions. LptA-TEV-His6 and LptC(24–191)-TEV-HIS6 protein in each elution fraction were also examined by SDS-PAGE and stained with SimplyBlue SafeStain (lower). FT, flow-through; W, wash in buffer C; E, elution using buffer C containing 300 mm imidazole.
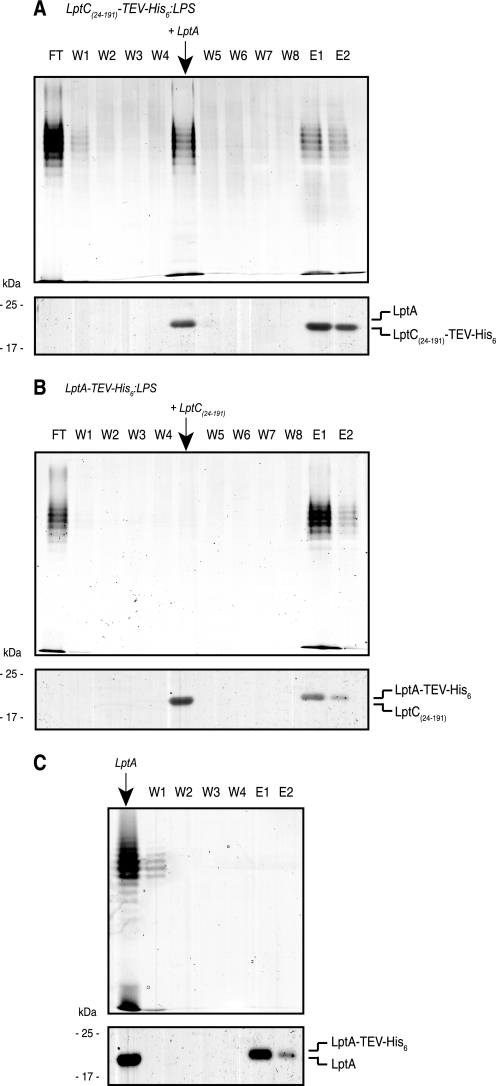
FIGURE 6.
In vitro transfer of LPS from LptC to LptA. A, the dissociation of O9a LPS from LptC(24–191)-TEV-His6·LPS complexes by LptA is shown. The LptC(24–191)-TEV-His6·LPS complex was formed and immobilized on Ni2+-NTA resin. After washing in buffer C to remove unbound material (W1–W4), LptA was added, and the eluted material was collected. The remaining resin was washed in buffer C (W5–W8) followed by buffer C containing 300 mm imidazole (E1–E2). A sample from each fraction was taken for SDS-PAGE. The upper part of each panel shows LPS in proteinase K-digested fractions, stained by silver nitrate. The lower part shows proteins stained by SimplyBlue SafeStain. B, in the converse experiment, LptC(24–191) was unable to dissociate LPS from LptA-TEV-His6·LPS. C, the fraction eluted by LptA from LptC(24–191)-TEV-His6·LPS complexes was immediately mixed with resin containing immobilized LptA-TEV-His6 to capture any free LPS. The resin was washed in buffer (W1–W3) and buffer containing imidazole (E1–E2). No LptA-TEV-His6·LPS complexes were identified in the capture strategy, suggesting that LPS remains associated with LptA after its dissociation from LptC(24–191)-TEV-His6·LPS complexes. LPS was examined by SDS-PAGE and silver staining (upper) after proteinase K digestion of the elution fractions. Proteins in each elution fraction were also examined by SDS-PAGE and stained with SimplyBlue SafeStain (lower).
To investigate whether the addition of TEV-digested (non-His6-tagged) LptA resulted in the transfer of LPS from LptC to LptA or dissociation of LPS from LptC(24–191), the FT2 fraction was collected in a tube containing an equal amount of LptA-TEV-His6 (500 μg) immobilized on Ni2+-NTA resin in buffer C. The reaction mixture was incubated for 30 min at 23 °C and washed four times with buffer C, and the LptA-TEV-His6 protein was eluted from the Ni2+-NTA resin with buffer C containing 300 mm imidazole, as described above.
RESULTS
Periplasmic Orientation of LptC
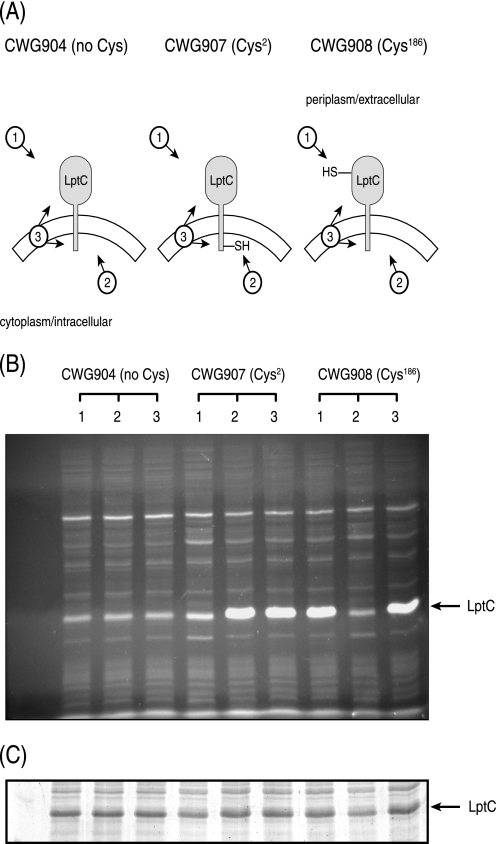
Analysis of the primary sequence of E. coli LptC using the ExPASy tools (43) predicted a single transmembrane helix (Trp7-Asp29) and a large soluble domain, consistent with LptC being an IM protein as proposed previously (12). To probe the topology and orientation of LptC, an established site-directed fluorescence cysteine-labeling approach was applied as described previously (26). There are no native Cys residues in LptC, so single Cys residues were introduced near the N terminus (Cys2) and C terminus (Cys186) by site-directed mutagenesis.
Three experimental conditions were used during the site-directed fluorescent labeling (Fig. 1A). In the first condition, intact cells were labeled with the membrane-impermeable fluorescent probe, OGM, which will label any exposed Cys-containing proteins exposed outside the IM. After quenching by β-mercaptoethanol, the cells were lysed, and membrane ghosts were prepared. In the second condition, intact cells were preincubated with the membrane-impermeable blocking agent, MTSET, which prevents any subsequent labeling of exposed Cys residues by OGM. MTSET reagent was removed from the cell suspension by repeated washes with buffer A prior to the preparation of membrane ghosts, and then OGM was used to label any newly exposed cytoplasmic Cys residues. In the final condition, membrane ghosts were directly treated with OGM, allowing both periplasmic and cytoplasmic exposed Cys residues to be labeled. A low level of background fluorescence was detected in a band co-migrating with LptC in each sample, including the wild-type protein (in CWG904), which has no Cys residues and provides the negative control. However, specific labeling yielded a substantially stronger signal. Cys186 was labeled in conditions 1 and 3, but not in condition 2, indicating a periplasmic location (Fig. 1B). In contrast, Cys2 was labeled in conditions 2 and 3, but not in condition 1, indicating a cytoplasmic location. From these results, it is apparent that the N terminus of LptC (and Cys2) is oriented toward the cytoplasm and the large soluble domain of LptC (and Cys186) is oriented toward the periplasm.
FIGURE 1.
Determination of the topology of LptC. A, the graphic indicates the predicted OGM accessibility of cysteine derivatives of LptC under three experimental conditions. B, the fluorescent proteins generated under the three conditions were separated by SDS-PAGE and are shown. C, to verify equal expression and loading for each of the tested LptC constructs, the same gel was stained with SimplyBlue SafeStain. The arrow to the right of B and C indicates the migration of LptC. In condition 1, exposed Cys residues in intact cells were labeled by OGM before a quenching by β-mercaptoethanol and preparation of membrane ghosts. In condition 2, exposed Cys residues were blocked by preincubation with MTSET, which was removed prior to preparation of membrane ghosts and OGM labeling of newly exposed Cys residues. In condition 3, membrane ghosts were labeled directly with OGM.
Purification of the Periplasmic Domain of LptC (His6-LptC(24-191))
The predicted transmembrane domain (Trp7-Asp29) of LptC is located near the N terminus. Due to the difficulties in maintaining LptC in a soluble form and potential complications associated with crystallizing proteins with transmembrane regions, we opted to solve the structure of the periplasmic region of LptC as an alternative to the full-length LptC protein. Deletion of the first 23 amino acids in His6-LptC(24–191) yielded a stable soluble form (Fig. 2A, left side, lanes 2 and 4). Only a minor trace of His6-LptC(24–191) was detected in the corresponding membrane fractions (Fig. 2B, left side, lanes 3 and 5).
His6-LptC(24–191) was purified to near homogeneity by Ni2+-NTA affinity chromatography, and its identity was verified by Western immunoblotting with anti-pentahistidine (His5) monoclonal antibody (Fig. 2, A and B). The migration of the purified protein on SDS-PAGE was consistent with the predicted molecular mass of 21,300 Da. In gel filtration chromatography, His6-LptC(24–191) elutes at the size expected for a monomer (data not shown). The oligomeric state of purified His6-LptC(24–191) in solution was confirmed using analytical ultracentrifugation. In Tris buffer (pH 7.5), the predominant His6-LptC(24–191) species (95.1% of the loading concentration) sedimented with an s value of 2.6 s and an estimated molar mass of 25.4 kDa (supplemental Fig. S1). This is compatible with a monomer and consistent with the gel filtration data.
Determination of the Structure of the Periplasmic Domain of LptC (His6-LptC(24–191))
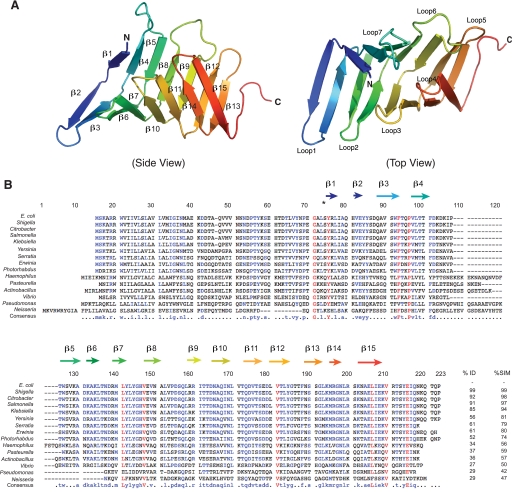
The crystal structure of the periplasmic domain of LptC was solved at 2.2-Å resolution using the SAD method. Data and structural refinement statistics are listed in Table 3. The asymmetric unit contained a single monomer of LptC. Residues 59–184 were observed in the electron density map, but two regions (residues 24–58 and 185–191) were disordered. The structure of the periplasmic domain of LptC consists of a twisted boat structure with two β-sheets coming into apposition (Figs. 3A and 7); one β-sheet contains seven antiparallel β-strands, and the other consists of eight antiparallel β-strands. The smallest angle formed between the two β-sheets (∼60°; measured in PyMOL) occurred at the center of the structure. The largest angles were observed at the termini (∼85°), indicating that the structure of LptC is slightly open at the both ends, similar to LptA (18) (Figs. 3A and 4). An amino acid sequence alignment of E. coli LptA and LptC using ClustalW (44) revealed a sequence identity score of 4 (less than 10%) between the two proteins (supplemental Fig. S2A). However, secondary structural predictions of E. coli LptA and LptC(24–191) (43, 44) indicate that the two proteins have high structural similarities (score = 84) (supplemental Fig. S2B). This is consistent with a structural comparison between LptA and LptC(24–191) using the Dali server (45), which yielded a Z-score of 15.1 with r.m.s.d. of 2.1 over 117 residues.
TABLE 3.
Data collection and structure validation
| Data collection | |
| Wavelength (Å) | 0.9764 |
| Resolution (highest shell, Å) | 50.90-2.20 (2.26-2.20) |
| Space group | P43212 |
| Cell constants (Å) | a = 94.21, b = 94.21, c = 60.44, α = β = γ = 90° |
| Unique reflections | 14,286 (1043) |
| Average redundancy | 6.7 (6.8) |
| I/σ | 15.1 (2.3) |
| Completeness (%) | 99.8 (99.9) |
| Anomalous complete (%)a | 99.8 (100) |
| Rmergeb | 0.084 (0.604) |
| SAD phasing statistics | |
| Selenium ions per assymetric unit | 2 |
| Figure of merit (SOLVE 2.8 Å) | 0.35 |
| Figure of merit (RESOLVE 2.8 Å) | 0.66 |
| Refinement | |
| Rfactor | 20.45 |
| Rfree | 22.22 |
| r.m.s.d. bonds (Å) / angles (°) | 0.011/1.283 |
| B-factor deviationc | |
| Bonds/angles (Å2): | |
| Main chain | 0.729/1.118 |
| Side chains | 1.832/2.845 |
| Residues in Ramachandran cored (%) | 98.39 (1.61% in allowed regions) |
| Protein atoms | 1029 |
| Water atoms | 71 |
| Average B-factor (Å2) | 40.12 |
| Protein Data Bank accession code | 3MY2 |
| Molprobity score | 95th percentile |
a Anomalous completeness corresponds to the fraction of possible eccentric reflections generated from the anomalous diffraction in the dataset for which an anomalous difference has been measured. The anomalous completeness is the percent of the reflections in the dataset.
b Rmerge = Σhkl Σ|Ii − <I>|/Σhkl ΣiIi, where Ii is an intensity for the ith measurement of a reflection with indices hkl and <I> is the weighted mean of the reflection intensity.
c Isotropic thermal factor restraints.
d There are two residues, Lys162 and Gly113, that are not at the core of the Ramachandran plot, but they are in allowed regions.
FIGURE 3.
Structure of the periplasmic domain of LptC. A, a ribbon diagram of a single His6-LptC(24–191) molecule at 2.2-Å resolution. The structure of the periplasmic domain of LptC is composed of a series of 15 antiparallel β-strands that wind back along the path of the preceding peptide stretch throughout the length of the protein, resembling the structure of LptA. B, an alignment of the sequences of selected LptC homologues; predicted secondary structure features are identified. Residue numbering corresponds to LptC from Neisseria meningitidis (without gaps). Alignment was performed with MultAlin (59). Residues with high sequence identity or similarity are shown as colors, and the overall identity (%ID) and similarity (%SIM) are reported. Non-conserved residues are shown as black letters.
FIGURE 7.
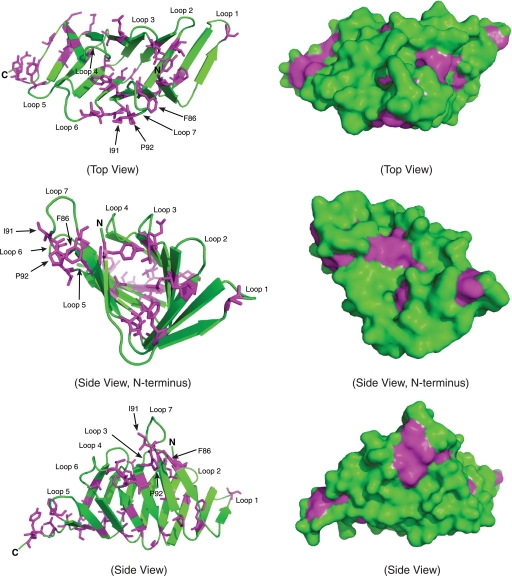
Ribbon diagrams and surface representations of the conserved hydrophobic residues of LptC. The magenta and green colors represent the highly conserved hydrophobic residues and non-conserved residues of LptC, respectively. Hydrophobic residues in the core, as well as loop 7, could potentially adopt different conformations to accommodate LPS binding.
FIGURE 4.
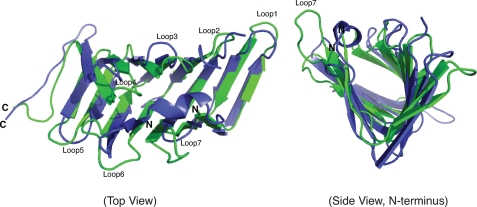
Structural comparison of LptA and His6-LptC(24–191). Ribbon diagrams of LptA (blue; 2.15 Å) (Protein Data Bank accession code: 2R19) and His6-LptC(24–191) (green; 2.2 Å) are superimposed.
LptC Binds to LPS in Vitro
We have previously shown that LptA binds structurally diverse LPS substrates in vitro, and available data suggest that it interacts specifically with the lipid A domain of LPS (24). Because LptC has been implicated in LPS transport and is structurally similar to LptA, the ability of LptC to bind LPS was investigated using the strategy developed for LptA. Purified LPS from E. coli serotype O9a was incubated with purified LptC(24–191)-TEV-His6 or, as a positive control, purified LptA-TEV-His6. The His6-tagged protein was then repurified from the reaction mixture by Ni2+-NTA affinity chromatography resin and examined for the presence of bound LPS. Previous work has already established that purified LPS does not bind to Ni2+-NTA affinity chromatography resin under these conditions, and LPS binding does not occur with irrelevant His6-tagged proteins (16, 24, 46). As shown in Fig. 5, incubating LptC(24–191)-TEV-His6 with LPS resulted in a substantial amount of LPS binding, with the complex eluting with imidazole (Fig. 5, fractions E1 and E2). The extent of binding was qualitatively similar to the LptA-TEV-His6 control (Fig. 5) and to what we observed previously with a different construct, LptA-His6 (24). Like LptA, LptC also binds smooth LPS from other serotypes, as well as rough LPS from E. coli W3110 (data not shown). This result indicates that O-antigenic polysaccharide is not required for substrate recognition and is consistent with LptC binding to the hydrophobic domain of LPS, as has been shown for LptA (24).
Evidence for Directional Transfer of LPS From LptC to LptA in Vitro
Sorting of lipoproteins to the OM requires the ABC transporter LolCDE, which mediates the detachment of OM-specific lipoproteins from the IM and delivers them to the periplasmic carrier protein LolA (21, 22). By analogy, a potential role of LptC is to facilitate the transfer of LPS from the IM to a periplasmic chaperone, LptA. To investigate whether LptC can transfer LPS to LptA, LPS was incubated with LptC-TEV-His6, and the complex was immobilized in Ni2+-NTA affinity chromatography resin. Unbound or excess LPS molecules were removed by repeated washes with buffer C (Fig. 6A, W1–W4). Purified TEV-digested (non-His6-tagged) LptA was then added to the reaction mixture, resulting in the release of substantial quantities of unbound LPS and LptA in solution (fraction LptA). After washing the resin with buffer C (W5–W8), the remaining LptC-TEV-His6·LPS was eluted with 300 mm imidazole (E1–E2). In the converse experiment (Fig. 6B), purified untagged LptC(24–191) was added to immobilized LptA-TEV-His6·LPS complexes. In this case, no LPS was displaced; it remained immobilized on the resin and was only eluted (with LptA-TEV-His6) after the application of 300 mm imidazole (E1–E2).
These results indicate that LptA can displace LPS from LptC(24–191)·LPS complexes (but not vice versa), consistent with a directionality in the export pathway. To examine whether the observed effect reflects only displacement, or transfer of LPS between LptC and LptA, a capture-based approach was used to distinguish between free and bound LPS (Fig. 6C). The His6-LptC(24–191)·LPS complex was formed and immobilized on Ni2+-NTA resin. Non-His6-tagged LptA was added to reaction mixture, resulting in a release of LPS and LptA. This fraction was immediately mixed with LptA-TEV-His6 that was immobilized on Ni2+-NTA resin, with the expectation that this would capture any LPS molecules that were free in solution. The fractions obtained from washes in buffer C (W1–W3) and buffer C containing 300 mm imidazole were examined by SDS-PAGE. The final elution with imidazole released LptA-TEV-His6 but no LPS, suggesting that the starting material contained no free LPS. The results are consistent with a transfer of LPS from LptC to LptA, rather than a simple displacement effect.
DISCUSSION
LPS is an essential component of the OM in most Gram-negative bacteria, and general aspects of its structure and biosynthesis are relatively well understood. In contrast, the components involved in LPS transport from the site of biosynthesis in the IM to the bacterial cell surface have only recently been identified (9, 11, 12, 15–17, 47), and the precise functions of the various transport proteins have yet to be resolved. One of the central questions has been whether the process involves a periplasmic scaffold formed by LptA and linking the IM components (LptBFGC) with those in the OM (LptDE). The alternative model invokes LptA as a soluble periplasmic chaperone in a system showing extensive similarity to the transport of OM lipoproteins (21, 23, 48, 49). Recent reports support the former model (20).
LptC is a bitopic membrane protein that has been implicated in LPS transport, and, recently, it was shown to be part of a complex that includes an ABC protein (LptB) as well as two integral IM proteins (LptFG) (13). The complex is proposed to be required for LPS extraction of the IM (12). It has been speculated that LptC serves as the docking site for LptA at the IM (50). The data reported here do not preclude this scenario. Although the Lol system lacks an equivalent of LptC, the transfer of LPS cargo from an IM-bound ABC protein complex to a periplasmic protein resembles the sequence of events in the Lol system for lipoprotein transport. OM lipoproteins are released from the IM by an ABC protein complex (LolCDE) in the presence of a periplasmic chaperone protein, LolA (22). Lipoproteins destined for the OM initially bind to LolE, which is transferred to LolC. LolC then releases the lipoprotein in an ATP-dependent manner to LolA, which then delivers its cargo to LolB in the OM (22). The lipoprotein transfer from LolA to LolB has been shown to occur in a “mouth-to-mouth” manner (23), and it has been suggested that a similar mode may exist for lipoprotein transfer from LolCDE to LolA (22). By comparison, LPS may be initially released from the IM by LptBFG in an ATP-dependent process and then transferred via LptC to LptA in an affinity-driven manner. If this is correct, LptA would deliver the LPS molecule to the LptDE complex in the OM; LptE has been demonstrated to bind LPS (16).
The x-ray structure of the periplasmic domain of LptC (at resolution of 2.2 Å) reveals an overall structure sharing striking similarities with LptA. Both contain consecutive antiparallel β-strands (15 in LptC and 16 in LptA) that form a twisted boat structure (Fig. 4). Although LptC and LptA share limited primary sequence similarities (supplemental Fig. S2A), structural comparison of the two proteins using the Dali server (45) revealed a Z-score of 15.1 with an r.m.s.d. of 2.1 for over 117 residues. However, there are some slight differences in the structures. For example, the opening between the two β-sheets is slightly larger in LptC, and the conformations of the C termini of the two proteins are different (Fig. 4). The N terminus of LptA contains a short α-helix that is sandwiched between the two β-sheets (Fig. 4). In contrast, the corresponding N terminus of LptC is disordered. Another notable difference between the two proteins is that loop 2 in LptC is much shorter than its counterpart from LptA; however, loop 7 from LptC is much longer than the comparable loop in LptA (Fig. 4). In the context of similarities between the Lpt and Lol pathways, it is interesting to note that LolA and LolB, which provide consecutive lipoprotein binding steps in the pathway, also share a very similar fold despite limited similarity in their primary sequences (Z-score of 10.6 with an r.m.s.d. of 3.2 over 134 residues) (48).
Mammalian LPS-binding proteins such as MD-2 and CD14 have a similar binding cavity size (∼15 × 8 × 10 Å) (51), and co-crystal structures are available for MD-2 with bound lipid IVA, or the mimic compound, eritoran (52, 53). Well defined substrate-binding pockets are also evident in LolA and LolB (23, 51, 52). In contrast, the structures of LptA and LptC do not reveal an obvious cavity on their surfaces that is deep enough to accommodate the fatty acyl chains of lipid A. LptC does possess many conserved hydrophobic residues that form a hydrophobic core along the protein that could potentially serve as an LPS-binding site (Figs. 3B and 7). Ribbon and surface representations of LptC both demonstrate that most of the conserved hydrophobic residues are oriented toward the interior cavity of the protein (Fig. 7). The protein would presumably need to undergo a conformational change to form an internal binding pocket for lipid A, and that pocket would also have to accommodate the side chains of the hydrophobic amino acid residues (Fig. 7). Local rearrangements that result in possible LPS-binding sites are also feasible (Fig. 7). For example, all of the residues in loop 7 (residues 86–92) (Fig. 7 and supplemental Fig. S2D) have B-factors ranging from 50 to 60, whereas neighboring residues have B-factors that range only from 30 to 40. Interestingly, this loop region shares limited homology (ClustalW (44) score = 21) to the peptide LPS antagonist LBP-14 (RVQGRWKVRASFFK), a synthetic fragment derived from the LPS-binding protein that has been shown to interact with LPS (54) (supplemental Fig. S2D). The functional significance of this similarity remains unclear. Recent isolation of a temperature-sensitive mutant of E. coli, designated MB2, revealed two amino acid substitutions (V60D and V132A) in LptA, as well as the addition of two amino acid substitutions (S44R and M108R) in LptC (55). However, only the expression of LptA was able to rescue this mutant at 44 °C (55). V132A is located in loop 5 of LptA; however, V60D is located near a region that also shares homology to LBP-14 (ClustalW (44) score = 21), as well as to the loop 7 region of LptC (Fig. 4 and supplemental Fig. S2C). Several hydrophobic residues located in loop 7 are exposed in LptC, Phe86, Ile91, and Pro92, indicating that loop 7 may play an important role in LPS binding (Fig. 7 and supplemental Fig. S2D). However, additional mutagenesis and structural studies of LptC are needed to determine the important residues required for LPS binding. Ultimately, this question can only be resolved by a crystal structure of LptC (or LptA) with bound LPS. Unfortunately, attempts to crystallize the complexes have, so far, been unsuccessful.
The model for LPS transport that invokes an LptA-containing periplasmic bridge is heavily influenced by the filaments of LptA oligomers obtained from crystallization conditions that included LPS (Protein Data Bank accession code: 2R1A) (18). In our hands, LptA behaves as a monomer in solution, but the localization of LptA and (potentially) its oligomeric status may be influenced by the level of expression (20). We were able to obtain one crystal form of LptA with four molecules in the asymmetric unit (data not shown). The four LptA molecules in the asymmetric unit were organized as two sets of dimers arranged in a head-to-tail fashion as reported previously (18), and its structure is superimposable with the reported structure of LptA (Protein Data Bank accession code: 2R19) (Z-score of 23.9 with r.m.s.d. of 1.2 for over 133 residues). Interestingly, for reconstruction of the crystal lattice of both LptA structures (our data and the previously reported structure (2R19)), which were grown in the absence of LPS, we observed LptA molecules arranged in a head-to-tail fashion, resembling the eight-molecule (filamentous) form of LptA (2R1A) previously reported to only be induced in the presence of LPS (18). Thus, it is possible that the chain-like arrangements of LptA monomers previously observed for 2R1A are not exclusively dependent on LPS but are the result of crystal packing. However, we have been unable to detect the eight-molecule form of LptA (or any higher order complexes) in solution using size exclusion chromatography, regardless of the presence or absence of added LPS. There is support from protein localization (16) and spheroplast (19) experiments for the “scaffold” assembly model. However, under the experimental conditions used here, we were unable to detect any stable interactions between the LptC and LptA derivatives in pulldown experiments (data not shown). Collectively, the current protein structure data cannot unequivocally distinguish between either of the proposed assembly models.
The essential requirement for LPS in many bacteria, including prominent Gram-negative pathogens (3–5), as well as the periplasmic location of much of the transport machinery, suggests that the Lpt pathway may be an interesting potential target for new therapeutic interventions. Interestingly, bioinformatic surveys and examination of the COGs (Clusters of Orthologous Groups of proteins) database (56, 57) indicate that not all components of the pathway are conserved in other bacteria. Several bacterial species lack a structural gene for lptC; examples include Helicobacter pylori, Campylobacter jejuni, Mesorhizobium loti, Caulobacter crescentus, Aquifex aeolicus, Thermotoga maritima, Deinococcus radiodurans, Rickettsia prowazekii, Chlamydia trachomatis, Treponema pallidum, and Borrelia burgdorferi. Interestingly, these same organisms do not have homologues of lptE. Presumably in these organisms, LptBFG delivers LPS directly to LptA, which then delivers directly to the OM β-barrel protein, LptD (16, 50). Regardless, although many components of the LPS machinery are conserved, there may be subtle variations in the process across Gram-negative bacteria.
Supplementary Material
Acknowledgments
We thank the staff at the IO3 Diamond Light Source for assistance in the data collection.
This work was supported by a grant from the Canadian Institutes of Health Research (to C. W.) and a Wellcome Trust Career Development Fellowship (to C. D.).
The atomic coordinates and structure factors (code 3MY2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- OM
- outer membrane
- IM
- inner membrane
- ABC
- ATP-binding cassette
- TEV
- tobacco etch virus
- OGM
- Oregon green 488 maleimide carboxylic acid
- MTSET
- methanethiosulfonate ethyltrimethylammonium
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- SAD
- single-wavelength anomalous dispersion
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1.Mühlradt P. F., Golecki J. R. (1975) Eur. J. Biochem. 51, 343–352 [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H. (2003) Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetz C. R., Guan Z., Ingram B. O., Six D. A., Song F., Wang X., Zhao J. (2009) J. Lipid Res. 50, (suppl.) S103–S108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz N., Kahne D., Silhavy T. J. (2009) Nat. Rev. Microbiol. 7, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doerrler W. T., Gibbons H. S., Raetz C. R. (2004) J. Biol. Chem. 279, 45102–45109 [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z., White K. A., Polissi A., Georgopoulos C., Raetz C. R. (1998) J. Biol. Chem. 273, 12466–12475 [DOI] [PubMed] [Google Scholar]
- 9.Sperandeo P., Cescutti R., Villa R., Di Benedetto C., Candia D., Dehò G., Polissi A. (2007) J. Bacteriol. 189, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperandeo P., Pozzi C., Dehò G., Polissi A. (2006) Res. Microbiol. 157, 547–558 [DOI] [PubMed] [Google Scholar]
- 11.Ruiz N., Gronenberg L. S., Kahne D., Silhavy T. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperandeo P., Lau F. K., Carpentieri A., De Castro C., Molinaro A., Dehò G., Silhavy T. J., Polissi A. (2008) J. Bacteriol. 190, 4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narita S., Tokuda H. (2009) FEBS Lett. 583, 2160–2164 [DOI] [PubMed] [Google Scholar]
- 14.Bos M. P., Tefsen B., Geurtsen J., Tommassen J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9417–9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun M., Silhavy T. J. (2002) Mol. Microbiol. 45, 1289–1302 [DOI] [PubMed] [Google Scholar]
- 16.Chng S. S., Ruiz N., Chimalakonda G., Silhavy T. J., Kahne D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5363–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T., McCandlish A. C., Gronenberg L. S., Chng S. S., Silhavy T. J., Kahne D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suits M. D., Sperandeo P., Dehò G., Polissi A., Jia Z. (2008) J. Mol. Biol. 380, 476–488 [DOI] [PubMed] [Google Scholar]
- 19.Tefsen B., Geurtsen J., Beckers F., Tommassen J., de Cock H. (2005) J. Biol. Chem. 280, 4504–4509 [DOI] [PubMed] [Google Scholar]
- 20.Chng S. S., Gronenberg L. S., Kahne D. (2010) Biochemistry 49, 4565–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narita S., Matsuyama S., Tokuda H. (2004) Arch. Microbiol. 182, 1–6 [DOI] [PubMed] [Google Scholar]
- 22.Narita S., Tokuda H. (2006) FEBS Lett. 580, 1164–1170 [DOI] [PubMed] [Google Scholar]
- 23.Okuda S., Tokuda H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5877–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran A. X., Trent M. S., Whitfield C. (2008) J. Biol. Chem. 283, 20342–20349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D., Maloney P. C. (1998) J. Biol. Chem. 273, 17962–17967 [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 29.Doublié S. (1997) Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 30.Evens P. R. (1997) Joint CCP4 and ESF-EACBM Newsletter 33, 22–24 [Google Scholar]
- 31.Leslie A. G. (1998) J. Appl. Crystallogr. 30, 1036–1040 [Google Scholar]
- 32.Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terwilliger T. C. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamzin V. S., Wilson K. S. (1997) Methods Enzymol. 277, 269–305 [DOI] [PubMed] [Google Scholar]
- 36.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37.Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 38.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic. Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westphal O., Jann K. (1965) Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 40.Tsai C. M., Frasch C. E. (1982) Anal. Biochem. 119, 115–119 [DOI] [PubMed] [Google Scholar]
- 41.Hitchcock P. J., Brown T. M. (1983) J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taniguchi N., Tokuda H. (2008) J. Biol. Chem. 283, 8538–8544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003) Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuthbertson L., Kimber M. S., Whitfield C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohtsu I., Kakuda N., Tsukagoshi N., Dokyu N., Takagi H., Wachi M., Aono R. (2004) Biosci. Biotechnol. Biochem. 68, 458–461 [DOI] [PubMed] [Google Scholar]
- 48.Takeda K., Miyatake H., Yokota N., Matsuyama S., Tokuda H., Miki K. (2003) EMBO J. 22, 3199–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi N., Matsuyama S., Tokuda H. (2005) J. Biol. Chem. 280, 34481–34488 [DOI] [PubMed] [Google Scholar]
- 50.Sperandeo P., Dehò G., Polissi A. (2009) Biochim. Biophys. Acta 1791, 594–602 [DOI] [PubMed] [Google Scholar]
- 51.Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 52.Kim H. M., Park B. S., Kim J. I., Kim S. E., Lee J., Oh S. C., Enkhbayar P., Matsushima N., Lee H., Yoo O. J., Lee J. O. (2007) Cell 130, 906–917 [DOI] [PubMed] [Google Scholar]
- 53.Ohto U., Fukase K., Miyake K., Satow Y. (2007) Science 316, 1632–1634 [DOI] [PubMed] [Google Scholar]
- 54.Pristovsek P., Simcic S., Wraber B., Urleb U. (2005) J. Med. Chem. 48, 7911–7914 [DOI] [PubMed] [Google Scholar]
- 55.Ma B., Reynolds C. M., Raetz C. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13823–13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. (2000) Nucleic Acids Res. 28, 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatusov R. L., Natale D. A., Garkavtsev I. V., Tatusova T. A., Shankavaram U. T., Rao B. S., Kiryutin B., Galperin M. Y., Fedorova N. D., Koonin E. V. (2001) Nucleic Acids Res. 29, 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter 31, 34–38 [Google Scholar]
- 59.Corpet F. (1998) Nucleic Acids Res. 22, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.