Abstract
ABCG2 is an ATP-binding cassette (ABC) transporter preferentially expressed by immature human hematopoietic progenitors. Due to its role in drug resistance, its expression has been correlated with a protection role against protoporhyrin IX (PPIX) accumulation in stem cells under hypoxic conditions. We show here that zinc mesoporphyrin, a validated fluorescent heme analog, is transported by ABCG2. We also show that the ABCG2 large extracellular loop ECL3 constitutes a porphyrin-binding domain, which strongly interacts with heme, hemin, PPIX, ZnPPIX, CoPPIX, and much less efficiently with pheophorbide a, but not with vitamin B12. Kd values are in the range 0.5–3.5 μm, with heme displaying the highest affinity. Nonporphyrin substrates of ABCG2, such as mitoxantrone, doxo/daunorubicin, and riboflavin, do not bind to ECL3. Single-point mutations H583A and C603A inside ECL3 prevent the binding of hemin but hardly affect that of iron-free PPIX. The extracellular location of ECL3 downstream from the transport sites suggests that, after membrane translocation, hemin is transferred to ECL3, which is strategically positioned to release the bound porphyrin to extracellular partners. We show here that human serum albumin could be one of these possible partners as it removes hemin bound to ECL3 and interacts with ABCG2, with a Kd of about 3 μm.
Keywords: ABC Transporter, Cell Differentiation, Membrane Enzymes, Membrane Protein, Multidrug Transporters, ABCG2/BCRP, Hemin, Porphyrin
Introduction
ABCG2 is a membrane protein belonging to the human ABC2 transporter family (1). Also called breast cancer resistance protein (BCRP), it was discovered for conferring resistance to the anticancer drug mitoxantrone in breast cancer cell lines (2, 3). As other members of the family, it is involved in cell protection against xenobiotics, such as the anticancer drugs mitoxantrone, 7-ethyl-10-hydroxycamptothecin (SN-38), topotecan, and daunorubicin (4), as well as antiviral agents such as HIV-1 nucleoside reverse transcriptase inhibitors (5). As expressed in the brain-blood barrier, intestine, and placenta (6, 7), it constitutes a critical target for both reversing multidrug resistance (4, 8–12) and increasing drug bioavailability (13, 14).
In mammals, heme (ferrous PPIX; see structure in supplemental Fig. S1) and hemin (ferric PPIX) play a central role in cell metabolism. Heme is the prosthetic group of several hemoproteins involved in oxygen sensing, cell respiration and metabolism, and cell growth (for a review, see Ref. 15), whereas hemin is a potent erythroid cell differentiation inducer (16–18). Free heme is toxic, leading to hemolysis, proteolysis, lipid peroxidation, reactive oxygen species generation, and vascular inflammatory disorders (19). Poorly soluble, heme and hemin require extracellular and intracellular carriers, such as albumin and hemopexin, which bind hemin with a Kd of 10−8 and 10−12 m, respectively (20–22). Several reports have pointed out the role of ABCG2 in porphyrin detoxification, initiated by a study of Jonker and colleagues who described in Bcrp1−/− knock-out mice a marked sensitivity to the dietary chlorophyll-catabolite pheophorbide a and an accumulation of PPIX, the heme precursor in erythrocytes and plasma, leading to acute photosensitivity (23). ABCG2 is a hematopoietic stem cell marker (24), expressed at relatively high levels by immature human hematopoietic progenitors SP, 34+/38− or 34+/KDR+ populations (25). Levels of porphyrins are normally regulated by metabolic control mechanisms of their biosynthesis; however, ABCG2 was found to prevent PPIX accumulation in erythroid progenitor K562 cells submitted to hypoxic conditions of growth (26) and to regulate PPIX levels during erythroid differentiation (27). These data point toward a role of BCRP in porphyrin homeostasis and protection; however, the molecular mechanism by which porphyrins are transported and handled for controlling their toxicity remains unknown. In addition, although ABCG2 is recognized to export PPIX (26), its capacity to export either hemin or heme has not yet been demonstrated.
ABCG2 consists of a cytosolic nucleotide-binding domain fused to a transmembrane (TM) domain of six α-helices, giving an inverted domain organization by comparison to most other human ABC transporters (4). The minimal functional unit of ABCG2 is a homodimer (28–30). ABCG2 displays several topological and structural specificities, such as the presence of a large external loop, ECL3, located between TM5 and TM6. Approximately 70–80 residues long, ECL3 is large enough to constitute a full domain, although no specific role has been assigned to it. It contains three cysteine residues reported to be involved in both intermolecular (Cys603-Cys603) and intramolecular (Cys592-Cys608) disulfide bridges (31–34). ECL3 also contains a conformational epitope recognized by the 5D3 monoclonal antibody (24), the interaction of which is prevented upon reduction of the intramolecular disulfide bridge Cys603-Cys603 (35), making 5D3 a powerful conformation-sensing tool.
In the present report, we hypothesized that ABCG2 should transport hemin out of the cell, which could be particularly critical in the early stages of hematopoiesis when it is overexpressed, and hemin plays a role in differentiation. We found that ABCG2 transports zinc mesoporphyrin (ZnMP), a validated fluorescent heme analog (36, 37). We also hypothesized that hemin, once exported by ABCG2, should be transferred to an extracellular protein through interaction with an accessible domain of ABCG2 such as ECL3, which could transitorily interact with hemin after membrane translocation. We expressed the ECL3 domain in Escherichia coli and purified it in a folded state, as checked by biophysical methods. It was especially found to be able to interact with the 5D3 mAb only when it was oxidized. ECL3 displayed a high affinity for some, but not all, porphyrins, especially for heme and hemin, whereas it does not bind any of the known nonporphyrin substrates and inhibitors of ABCG2. We also found that hemin, when bound onto ECL3, can be rapidly transferred to human serum albumin (HSA).
EXPERIMENTAL PROCEDURES
Materials
The pET15b plasmid was from Novagen. The QuikChange site-directed mutagenesis kit was from Stratagene. Luria Bertani broth, isopropyl 1-thio-β-d-galactopyranoside, and ampicillin were from Euromedex. Foscholine 12 (FC12) was from Anatrace. Nickel-nitrilotriacetic acid (Ni-NTA) and gel filtration resins and materials for chromatography were from GE Healthcare. An Amicon ultracentrifuge filter device was from Millipore. 5D3-phycoerythrin was purchased from Clinisciences. Other products were from Sigma. Heme was prepared by incubating 0.2 mm hemin with 10 mm dithionite for 1 h at room temperature; the nearly immediate reduction was visible by eye.
Cell Culture
K562 cells, either expressing or not human ABCG2, were kindly provided by Drs. Sheng Zhou and Albert Sorentino and used as described in Ref. 27. K562 cells were pelleted by centrifugation for 5 min at 200 × g and washed with fresh DMEM. After centrifugation, cells were suspended in 5 ml of DMEM and seeded at 200 μl/well in a 96-well plate. The medium was removed by centrifugation, and 100 μl of ZnMP (10 μm final) was added with or without 100 μl of Ko143 (1 μm final). Accumulation of ZnMP was allowed for 30 min at 37 °C. Cells were then washed with PBS and centrifuged and then incubated for 1 h with or without Ko143. They were maintained on ice until analysis by flow cytometry. ZnMP accumulation was quantified with a FACScan flow cytometer (BD Biosciences), excitation at 488 nm and emission at 575 nm.
Hemin-Agarose Pulldown Assays
Experiments were performed as previously described by Krishnamurthy et al. (26), using K562 cells either expressing or not ABCG2 (27). Cells were lysed as described, and 50 μg of proteins were incubated at room temperature for 15 min in the presence of 0–2 ng/μl 5D3 antibody and 500 nm hemin-agarose. After incubation, the resin was pelleted by centrifugation, and unbound material was withdrawn. Pellets were washed with 1 ml of lysis buffer (150 mm NaCl, 10 mg/ml Triton X-100, and 50 mm Tris-Cl, pH 8.0), suspended in 20 μl of SDS sample loading buffer (38), and centrifuged at 10,000 × g for 2 min at 4 °C. Bound and unbound materials were analyzed on a 10% SDS-PAGE and transferred to a PVDF membrane, and ABCG2 was then revealed with the BXP21 monoclonal antibody (1/500, Millipore).
Expression Constructs
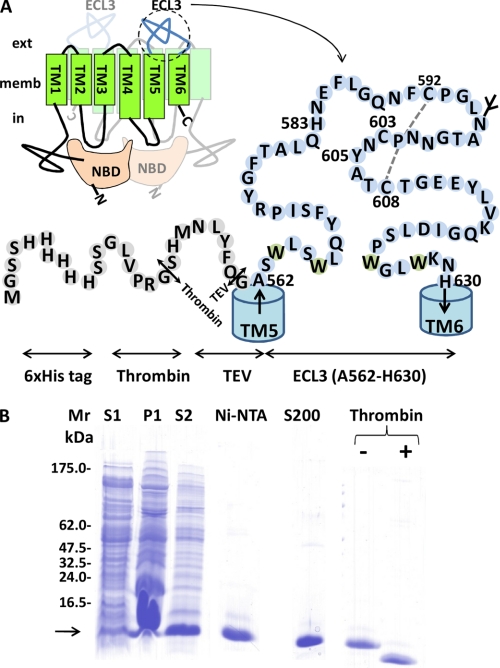
The bcrp1 gene encoding ABCG2, referenced as Q9UNQ0 at the UniProtKB/Swiss-Prot database, was used for constructs. The E. coli protein expression plasmid for producing ECL3, as displayed in Fig. 2, was constructed from alanine 562 to histidine 630 of the ABCG2 sequence by PCR using the plasmid pFastBac Dual-NinaA-BCRP (39), with the couple of forward and reverse primers Pfor and Prev displayed in Table 1. Pfor included a NdeI restriction site and encoded a tobacco etch virus (TEV) site (40); Prev included a BamHI restriction site. The PCR product was digested by NdeI and BamHI and inserted into the corresponding sites of the pET15b plasmid. The resulting plasmid pET15b-H6ThTe-ECL3 expressed the ECL3 (A562-H630) domain fused at its N terminus to a His6 tag through a thrombin site and a TEV site: MGSSHHHHHHSSGLVPRG/SHMNLYFQ/GA562-ABCG2-H630, where both cleavage sites are underlined, the cleavage position is indicated by “/,” and the ECL3 sequence is indicated in bold and italic characters. The single-point mutations H598A, C603A, and Y608A were introduced by site-directed mutagenesis using the corresponding primers displayed in Table 1.
FIGURE 2.
ECL3 design, expression, and purification. A, ECL3 localized between TM spans 5 and 6 extends from Ala562 to His630. It was fused to a His6 N-terminal tag followed by thrombin (Th) and TEV (Te) proteolytic cleavage sites, resulting in a fusion protein of 10,886 Da. The glycosylation site (in mammals) and disulfide bridge between Cys592 and Cys608 are indicated. B, Tricine SDS-PAGE is used to monitor expression and purification of ECL3. S1 and P1 correspond to the supernatant and pellet fractions obtained at low speed centrifugation of bacteria overexpressing the fusion protein. S2 corresponds to the soluble fraction after FC12 solubilization of P1. Ni-NTA and S200 correspond to ECL3 eluted from each resin. The protein was finally digested by thrombin (+ Thrombin) to remove the tag.
TABLE 1.
Primer sequences for generating the H6ThTeECL3 fusion protein and to introduce single mutations as detailed in the text
| Primer | 5′–3′ Sequence | Res. sites |
|---|---|---|
| Pfor | AAAAAAAAAACATATGAATCTTTATTTTCAGGGGGCATCTTGGCTGTCATGGC | NdeI |
| Prev | GGGGCTTGTGGAAGAATCACTAAGGATCCAATA | BamHI |
| H583Afor | GGATTTACGGCTTTGCAGGCCAATGAATTTTTGGGACAAAACTTCTGC | |
| H583Arev | GCAGAAGTTTTGTCCCAAAAATTCATTGGCCTGCAAAGCCGTAAATCC | |
| C603Afor | GCAACAGGAAACAATCCTGCCAACTATGCAACATGTACTGGCGAA | |
| C603Arev | TTCGCCAGTACATGTTGCATAGTTGGCAGGATTGTTTCCTGTTGC | |
| Y605Afor | GCAACAGGAAACAATCCTTGTAACGCCGCAACATGTACTGGC | |
| Y605Arev | GCCAGTACATGTTGCGGCGTTACAAGGATTGTTTCCTGTTGC |
The plasmid used for expressing full-length ABCG2 in insect cells was described previously (39); here, we extended from 6 to 10 the length of the N-terminal histidine tag and added a thrombin cleavage site. The resulting sequence H10Th-ABCG2 was MAHHHHHHHHHHLPRGSGGMDSPPPSGLVPRGS-S4-ABCG2-S655.
Expression and Purification of ECL3
The BL21(DE3) E. coli bacteria strain was transformed with the expression plasmid pET15b-H6ThTe-ECL3 including wild-type or mutated sequences. A single colony of freshly transformed cells was grown overnight at 37 °C in 5 ml of Luria Bertani broth containing 100 mg/liter ampicillin and then diluted into 2 liters of the same medium. The culture was incubated at 37 °C and shaken at 220 rpm until reaching an A600 nm of 0.6. Protein expression was then induced by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and continued for 3 h under the same conditions. TEV was expressed with the pRK793 plasmid, kindly given by Dr. David S. Vaugh (NCI, National Institutes of Health, Frederick, MD).
After expression of either wild-type or mutated ECL3, bacteria were harvested by centrifugation at 10,000 × g for 15 min. The pellet was suspended in 20 ml of 20 mm NaH2PO4, pH 7.0, 100 mm NaCl. Cells were disrupted by using a French press, and 2 m NaCl was added to the medium, which was shaken for 20 min at room temperature and then centrifuged at 10,000 × g for 20 min, at 4 °C, resulting in supernatant S1 and pellet P1 (see Fig. 2B). The pellet was suspended in 5 ml of 20 mm NaH2PO4, pH 7.0, 100 mm NaCl, 0.1 mm DTT, 29 mm FC12 and incubated for 1 h at room temperature under mild shaking. The solution was then centrifuged at 100,000 × g for 1 h at 4 °C, generating supernatant S2 (see Fig. 2B) which was loaded onto a 1-ml nickel Hi-trap chelating HP column previously equilibrated with 10 ml of 20 mm NaH2PO4, pH 7.0, 100 mm NaCl, 0.1 mm DTT, and 14.2 mm FC12. The resin was washed in two steps with 10 ml and 4 ml of the same buffer, lowering the FC12 concentration to 2.8 mm during the second step. The protein was eluted in 5 ml of the latter buffer with a linear gradient of 0–0.5 m imidazole. The pool of protein was concentrated with an Amicon ultracentrifuge filter device, cutoff 3,000 Da, and loaded onto a Superdex-200 gel filtration 10/300 column, equilibrated with 50 mm NaH2PO4, 2.8 mm FC12. Fractions of about 5–20 μg of protein were checked for homogeneity on a 10% Tricine SDS-PAGE, after half-dilution in a 2× SDS sample buffer (38) and incubation for 20 min at room temperature before electrophoresis. Fractions of interest were pooled and incubated overnight at 4 °C to allow thiol oxidation to occur. Protein concentration was determined by measuring A280 nm, using a molar extinction coefficient of 29,520 liters·mol−1·cm−1.
Bovine thrombin was added at a ratio of 10 units/mg of protein for removing the His6 tag and incubated for 5 h at room temperature. The efficacy of cleavage was checked by Tricine SDS-PAGE (see Fig. 2B). The His6 tag peptide was removed by a Ni-NTA-agarose column equilibrated with 20 mm NaH2PO4, pH 7.0, 150 mm NaCl, 2.8 mm FC12. Thrombin was removed on a 1-ml p-aminobenzamidine-agarose resin equilibrated with the same buffer.
Expression and Purification of ABCG2
ABCG2 was expressed in Hi5 cells as described in Ref. 39. All subsequent steps were performed at 4 °C. Cells were harvested by centrifugation at 5,000 × g for 10 min, and the pellet was suspended (7 ml/liter of culture) in PBS and centrifuged again. Cells were then suspended (25 ml/liter of culture) in 20 mm Tris-Cl, pH 8.0, 10 mm NaCl, 200 μm DTT, 1 mm MgCl2, protease inhibitors, and incubated for 1 h. Cells were disrupted by using a tight-fitting Dounce homogenizer and then centrifuged at 15,000 × g for 1 h. The resulting pellet was suspended (500 μl/liter of culture), in 10 mm HEPES, pH 8.0, 50 mm NaCl, 20% glycerol, 100 μm DTT, 1 mm EDTA, protease inhibitors, and stored in liquid nitrogen. The protein concentration was about 40 mg/ml, estimated by the bicinchoninic acid (BCA) method (41).
Membrane suspension was thawed, diluted to 10 mg/ml in 50 mm HEPES, pH 8.0, 2 m NaCl, 15% glycerol, 2 mm DTT, protease inhibitors, and incubated for 1 h before centrifugation at 15,000 × g for 1 h. The resulting pellet was suspended in 50 mm HEPES, pH 8.0, 250 mm NaCl, 15% glycerol, 40 mm dodecyl β-d-maltoside, 0.2 mm DTT, 20 mm imidazole, protease inhibitors, then incubated for 2 h and centrifuged at 100,000 × g for 1 h. The supernatant was applied to HiTrap chelating HP 1-ml column loaded with Ni2+ and equilibrated in 50 mm HEPES, pH 8.0, 250 mm NaCl, 15% glycerol, 10 mm dodecyl β-d-maltoside, 20 mm imidazole, protease inhibitors. The resin was washed with 16 bed volumes with the same buffer, and ABCG2 was eluted in the same buffer with a 0.02–1 m imidazole gradient. The protein was concentrated using an Amicon Ultra-15 concentrator device 50-kDa cutoff. The His10 tag was removed by thrombin cleavage as described above.
Absorbance Spectroscopy
A saturated solution of hemin was prepared by solubilizing about 10 mg in 100 μl of fresh 0.1 m KOH, followed by a dilution in 50 mm KH2PO4, pH 7.2, 100 mm NaCl. The solution was clarified by centrifugation at 11,000 × g for 3 min, and hemin concentration was determined at 385 nm using an extinction coefficient of 58,400 liters·mol−1·cm−1 (42). For assays, samples of 10 μm protein were subjected to UV-visible absorbance spectroscopy at room temperature on a DU 640 (Beckman) spectrophotometer, using 50 mm NaH2PO4, 2.8 mm FC12 buffer as blank. Spectra were recorded between 240 and 700 nm using a 1-cm path length cuvette, in a hemin concentration range of 0–50 μm. Difference spectra were obtained by subtracting the buffer spectrum from that of the protein·hemin complex. Spectra were normalized at 280 nm.
Ni-NTA-Agarose Pulldown Assays
Fifty microliters of Ni-NTA magnetic beads were equilibrated with 3 × 1 ml of 50 mm NaH2PO4, pH 7.0, 150 mm NaCl, 0.1% FC12 and then incubated with 200 μg of H6ECL3/100 μl (20 nmol) for 30 min 4 °C and mild agitation. The beads were washed three times with 600 μl of the same buffer, and the amount of unbound protein was quantified by the BCA method. H6ECL3 beads were loaded on a BioSprint 15 robot (Qiagen) to carry out the binding of hemin by incubating beads in 500 μl of 50 μm hemin in the same buffer for 20 min at room temperature. The incubation was followed by three washing steps with 500 μl of the same buffer containing 300 mm NaCl. The protein was eluted in 50 μl of the same buffer containing 300 mm NaCl and 300 mm imidazole. The amount of eluted protein and hemin were quantified by the BCA method and by absorbance, respectively. Bound and unbound materials were analyzed on a 16% SDS-PAGE and revealed by Coomassie Blue staining.
Circular Dichroism (CD)
Far-UV CD of purified ECL3 was performed on a Chirascan (Applied Photophysics). Spectra were recorded at room temperature using a 1-mm path length cuvette from 260 to 185 nm, with a step size of 1 nm. Spectra were recorded in the 190–250-nm wavelength range with a 0.2-nm increment, a bandwidth of 0.5 nm, and a 1-s integration time. Spectra were processed, corrected for baseline, and smoothed by Chirascan software on the basis of the Savitzky-Golay algorithm. The percent secondary structure was calculated by deconvulating data via the DichroWeb site (43) using methods developed by Provencher and Glockner (44), K2d (45), CDSSTR and a SELCON (46).
Fluorescence Spectroscopy
Experiments were performed at room temperature using a Photon Technology International Quanta Master I spectrofluorometer. All measurements were recorded in 500-μl quartz cuvettes (Hellma) of 0.5-cm path length. Data were corrected for buffer and the contribution to fluorescence and inner-filter effects of tested ligands by collecting the same spectra with N-acetyltryptophan (NATA). ECL3 (3 μm) in 50 mm NaH2PO4, pH 7.0, 2.8 mm FC12, 20% glycerol, or ABCG2 (0.1 μm) in 50 mm HEPES, pH 8.0, 250 mm NaCl, 15% glycerol, 10 mm dodecyl β-d-maltoside, 1.6 mm CHAPS, was excited at 295 nm, and the fluorescence emission was scanned from 310 to 370 nm. The concentration-dependent quenching studies for the different ligands (0–15 μm) were performed at least three times. Peak areas of corrected fluorescence spectra were fitted with the SigmaPlot software (version 11; Systat Software). Percentages of fluorescence quenching were calculated as a ratio of protein to NATA. Data were fitted assuming one binding site with different possible equations, as indicated,
 |
(47). Equation 2 takes into account the residual enzyme concentration and gives a better estimation of the Kd when substrate and enzyme concentration are close, but leads to error values in the same range.
which introduces a correction factor for unspecific binding.
Primary Structure Alignment
Primary structure alignments were performed with Clustal (48), carried out via the NPS@ bioinformatic server.
RESULTS
ZnMP Transport by ABCG2 in K562 Cells and Modulation by the 5D3 mAb of in Vitro ABCG2 Binding to Hemin-Agarose
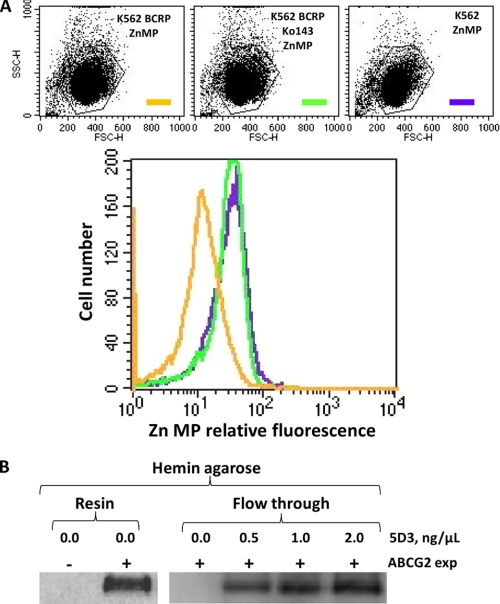
Because ABCG2 was shown to transport PPIX out of stem cells submitted to hypoxia (26), we hypothesized that the transporter could also be able to transport heme or hemin, which correspond to ferrous and ferric PPIX (see the corresponding chemical structure in supplemental Fig. S1). The experiment was carried out with erythroid K562 cells either transfected or not with the cDNA encoding ABCG2 (27), following the transport of the fluorescent ZnMP. Note that although ZnMP is a validated hemin analog (36, 37), its central zinc atom cannot coordinate with protein residues such as histidine in the same way as the iron atom in heme (49).
As shown in Fig. 1A, ZnMP accumulated in K562 control cells (green), whereas they remained at a lower level in cells expressing ABCG2 (orange). However, the latter showed the same level of accumulation as observed in control cells when incubated with 1 μm ABCG2 inhibitor Ko143 (50). Hemin and heme being rather cytotoxic, especially heme as due to the ferrous state of bound iron (15), we hypothesized that their efflux could require an intermediate transfer to a carrier via an extracellular domain of ABCG2, fully accessible to this carrier. ABCG2 indeed displays such a domain, which has been shown to interact with the 5D3 mAb (35). As a first experiment to test whether or not ECL3 could play such a role in heme/hemin transfer, we took advantage of the known capacity of the protein to bind in vitro onto hemin-agarose (26) to check whether 5D3 binding could alter domain interaction with the affinity column. Results are displayed in Fig. 1B. As shown on the left, ABCG2 expressed in K562 cells (Resin/0.0/+) could fully bind to the hemin-agarose resin, compared with control cells (Resin/0.0/−), no protein remaining in the flow-through fraction as shown in the right (Flow through/0.0/+). Addition of increasing amounts of 5D3 before incubation with the resin indeed prevented the binding of ABCG2 onto hemin-agarose; the protein accumulated in the flow-through fraction proportionally to the amount of added mAb. By revealing an antagonist effect between 5D3 binding onto ECL3 and ABCG2 binding onto hemin-agarose, this experiment suggested a heme-binding capacity of ECL3.
FIGURE 1.
Efflux of the heme analog Zn mesoporphyrin (ZnMP) by ABCG2-expressing cells, and decrease of ABCG2 binding to hemin-agarose by the 5D3 mAb raised against ECL3. A, ABCG2-expressing K562 cell transport of ZnMP (orange) compared with K562 control cells (green). Addition of the specific ABCG2 inhibitor Ko143 (1 μm) blocks the efflux of ZnMP (violet). B, modulation by the 5D3 mAb of in vitro ABCG2 binding to hemin-agarose. Whole cell lysates were subjected to a pulldown assay with hemin-agarose in the absence (Resin) or presence of increasing concentrations of 5D3 mAb increasing the amount of unbound ABCG2 (Flow through).
ECL3 Constitutes an Autonomously Folded Domain
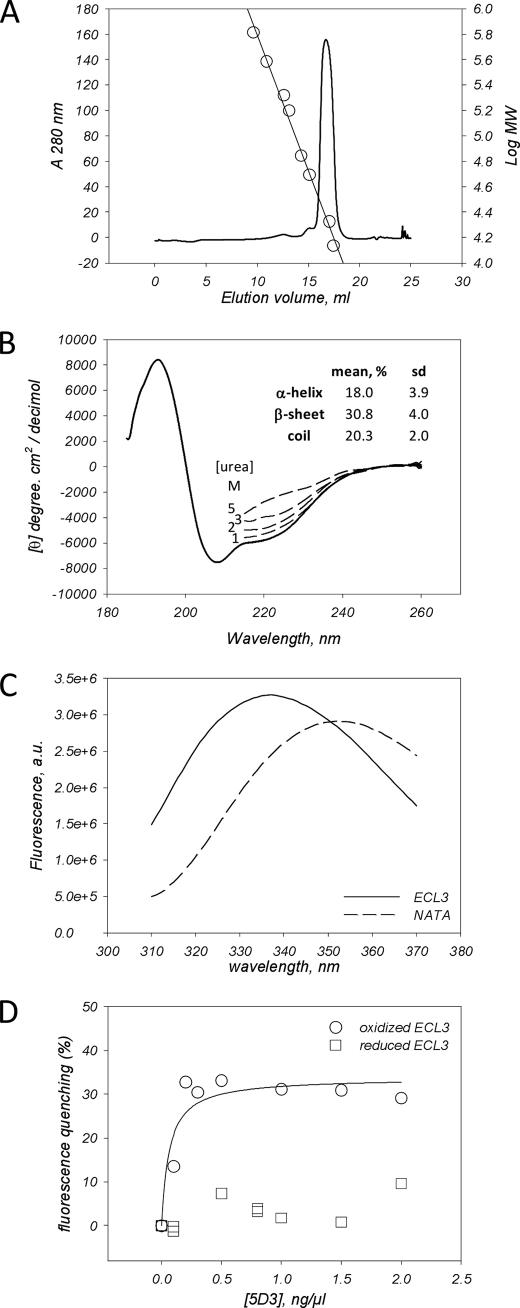
To further check the capacity of ECL3 itself to bind heme/hemin separately from the transport sites, we studied ECL3 as an isolated and purified domain, hypothesizing that it should be long enough to fold accurately when expressed alone. Using primary structure alignments, we selected the region extending from alanine 562 to histidine 630 as possible N and C termini of such a domain. These limits are close to those recently reported from various predictions, Trp567-Trp627 (51), Leu565-Asn629 (52), or Ser563-Gly618 (35). We noted the amphipathic character of both pairs of tryptophan residues 564–567, 624–627, located at each extremity of ECL3 and likely favoring the anchoring of the loop onto the membrane, rather than belonging to the TM helices 5 and 6. We generated a chimera including a His6 tag followed by thrombin and TEV (40) endoprotease cleavage sites at the N terminus of ECL3, which were inserted for later removing the nickel affinity tag after purification because histidine residues might be potential hemin ligands. The resulting fusion protein (Fig. 2A) was produced in E. coli and then purified as displayed in Fig. 2B. ECL3 was abundant in the insoluble fraction P1 (arrow in B) and was solubilized with FC12 (lane S2), showing that the domain, as designed, still displayed a significant hydrophobic character. The domain was then purified by nickel affinity and gel filtration chromatographies (lanes Ni-NTA, S200), reducing the detergent concentration to 3 mm, a value roughly equivalent to twice the critical micellar concentration. Reducing this concentration below the critical micellar concentration led to protein aggregation, indicating that detergent is required for maintaining the domain in solution. DTT was added during the solubilization step and then removed during the chromatographic steps for favoring the formation of the Cys592-Cys608 disulfide bridge, which occurred during overnight incubation at 4 °C. Later, thiol titration with Ellman's reagent (53) showed that one thiol remained free after this step. Thrombin fully digested the protein (lane Thrombin +) generating a protein that migrated at a position compatible with the predicted molecular mass of 8,945 Da. TEV could never cut, which might be due to the presence of detergent as reported before (54). ECL3 was finally purified by repeating both chromatographic steps and adding one for removing thrombin.
We next examined the folding of ECL3. As shown, the protein was eluted from a S200 10/300 gel filtration at 16.7 ml (Fig. 3A), corresponding to an apparent molecular mass of 23 kDa. Taking into account the presence of detergent micelles, this value is compatible with monomeric ECL3 (∼9 kDa) embedded inside a FC12 micelle (350 g/mol × ∼40 molecules = 14 kDa). The CD spectrum of the domain (Fig. 3B) was typical of a α + β polypeptide which, once deconvoluted, gave the average values of α-helix, β-sheet, and coil which are indicated in the inset. Increasing concentrations of urea induced a progressive loss of secondary structure. Intrinsic fluorescence of the domain (Fig. 3C), which contains four tryptophan residues (Fig. 2A), was also typical of a folded protein in which these tryptophan residues are buried, giving a maximal emission wavelength of 336 nm, much lower than the 354-nm value obtained with NATA used as a control. A further convincing result came from the capacity of the recombinant domain to interact with the 5D3 mAb (24), which has been raised against ECL3 and is sensitive to the oxidized state of the thiol bridge Cys592-Cys608 (35). As shown in Fig. 3D, the addition of increasing amounts of the 5D3 mAb quenched the tryptophan fluorescence and led to a saturation curve (circles), allowing us to estimate a Kd value of 0.06 ± 0.03 ng/μl, i.e. 4 ± 2 nm (all Kd values are summarized in supplemental Table S1), assuming an average IgG molecular mass of 150,000 Da. Because the binding of 5D3 was reported to be prevented upon reduction of the Cys592-Cys608 disulfide bridge (35), we reproduced a similar experiment with the reduced form of ECL3 (squares). As shown, the intrinsic fluorescence of reduced ECL3 was not modified by the mAb addition, suggesting no interaction. These experiments show that the ECL3 domain is self-folded and adopts a three-dimensional structure relevant to the same region within the native full-length ABCG2 transporter.
FIGURE 3.
ECL3 recombinant domain is folded and recognized by the conformational epitope recognizing 5D3 mAb. A, gel filtration on a Superdex-200 10/300 column of ECL3. The column was calibrated with molecular mass markers (right y log axis, circles) thyroglobulin (650 kDa), ferritin (380 kDa), catalase (210 kDa), aldolase (160 kDa), albumin (70 kDa), ovalbumin (50 kDa), chymotrypsinogen A (21 kDa), and ribonuclease (14 kDa), fitting the elution volume data with SigmaPlot. B, CD spectra of ECL3. The spectrum was fitted for estimating the percentage of each two-dimensional structure (see “Experimental Procedures”). The same experiment was done by adding 1–5 m urea (dashed lines). C, fluorescence spectra of ECL3 (straight line) and NATA (dashed line). D, intrinsic fluorescence quenching of ECL3 (1.5 μm), either oxidized (circles) or reduced (squares), by the 5D3 mAb, as measured at 336 nm and fitted with SigmaPlot using Equation 1.
ECL3 Specifically Binds Porphyrins
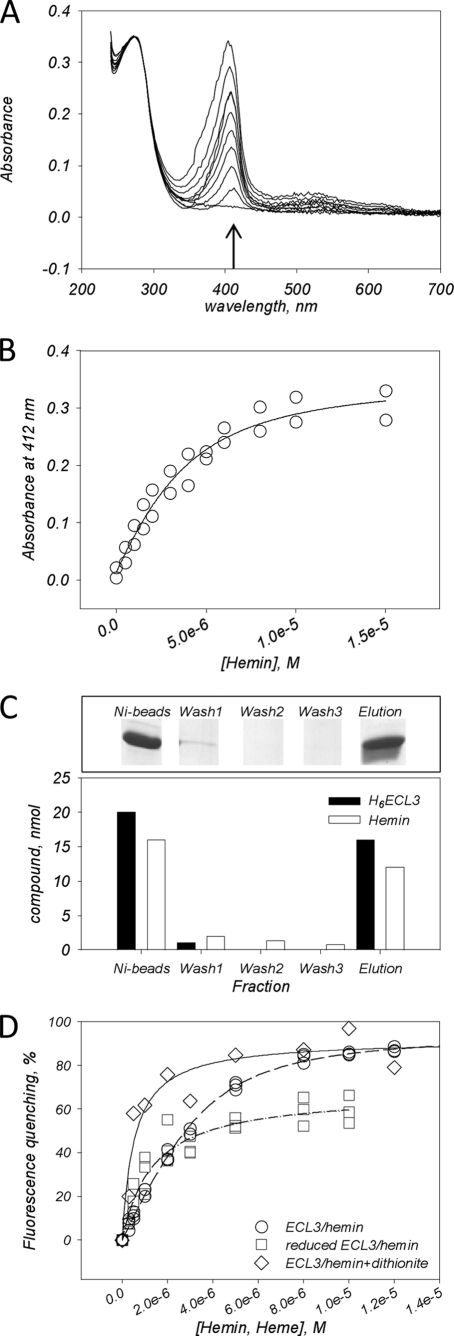
We first monitored the binding of hemin (see the structure of each tested compound in supplemental Fig. S1) by absorbance, following the evolution of the Soret peak at 412 nm which appears in the presence of ECL3 (Fig. 4A). The interaction led to an increase in absorbance at this wavelength when increasing the hemin concentration. The absorbance values, plotted in Fig. 4B, gave a saturation curve from which a Kd of 1.33 ± 0.72 μm was estimated (Kd values are summarized in supplemental Table S1), assuming one hemin bound per domain. This binding ratio was confirmed experimentally by pulldown experiments as displayed in Fig. 4C using Ni-NTA magnetic beads (see “Experimental Procedures”). Results show that 20 nmol of ECL3 bound to the beads binds 16 nmol of hemin, whereas after washing, elution leads to the removal of 16 nmol of ECL3 and 12 nmol of hemin. Both ratios suggest that up to one hemin binds to the domain. The ECL3-hemin interaction was also monitored by intrinsic fluorescence, leading to similar results (Fig. 4D, circles) with a Kd of 0.88 ± 0.18 μm. As shown, the maximal quenching of intrinsic fluorescence was approximately 90%. When hemin iron, present as a ferric state Fe3+, was reduced by dithionite to ferrous state, Fe2+, as in heme, a higher maximal quenching of intrinsic fluorescence was observed (Fig. 4D, diamonds), and the Kd of 0.47 ± 0.40 μm indicated a 2-fold higher affinity than for the oxidized form. We also monitored hemin binding to the reduced form of ECL3 (Fig. 4D, squares) which led to a similar saturation curve, although despite a lower maximal quenching (60%), the Kd of 1.03 ± 0.69 μm was close to that obtained with oxidized species. This suggested that reduction of the Cys592-Cys608 disulfide bridge does not dramatically alter hemin binding.
FIGURE 4.
Hemin and heme binding to ECL3 as monitored by UV absorbance, pulldown assay, and intrinsic fluorescence. A, UV-visible, corrected and normalized, spectra of oxidized hemin (0–15 μm) in the presence of ECL3 (3 μm). B, plot of the absorbance values at 412 nm (arrow in A) of hemin as a function of hemin concentration. C, pulldown assays of the hemin·H6ECL3 complex. The experiment was carried out as described under “Experimental Procedures,” leading to different fractions: Ni-beads, hemin·H6ECL3 complex bound to Ni-NTA magnetic beads; Wash 1–3, magnetic bead washing steps; Elution, imidazole elution step of hemin·H6ECL3 complex. Fractions were loaded on a 16% SDS-PAGE (upper panel), and protein (black bars) and hemin (white bars) amounts were quantified (lower panel). D, intrinsic fluorescence quenching of ECL3 (1–3 μm) with hemin (0–14 μm, circles) and heme as generated by dithionite reduction (0–14 μm, diamonds). Squares correspond to the interaction of hemin with reduced ECL3. Data were fitted using Equation 2 (see “Experimental Procedures”).
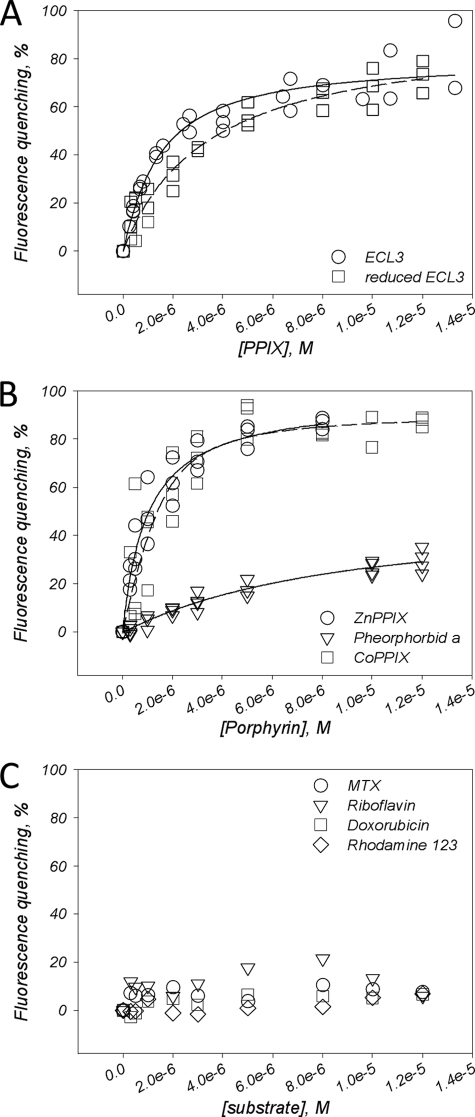
We then examined the interaction of other porphyrins, such as PPIX, ZnPPIX, CoPPIX, pheophorbide a, and vitamin B12, with ECL3. As shown in Fig. 5A, binding of PPIX when monitored by quenching of intrinsic fluorescence led to a saturation curve (circles) reaching about 80% and giving a Kd of 1.21 ± 0.71 μm. The same experiment carried out after reduction of ECL3 also led to a saturation curve of fluorescence quenching (squares), giving a Kd of 3.40 ± 0.01 μm. ZnPPIX, which usually reaches a high level in ferropenic anemia, as well as CoPPIX, were found to bind efficiently to ECL3, as shown in Fig. 5B (circles and squares, respectively), with respective Kd values of 0.80 ± 0.01 μm and 0.71 ± 0.38 μm. By contrast to these porphyrins, we observed a much reduced interaction of pheophorbide a (Fig. 5B, triangles), producing only 30% of maximal quenching, with a high Kd value of 3.42 ± 0.50 μm. No change at all of ECL3 intrinsic fluorescence was observed upon addition of either another porphyrin-type molecule, vitamin B12 (supplemental Fig S2A), which is not an ABCG2 substrate (55), or bilirubin, a product of heme catabolism (supplemental Fig S2B).
FIGURE 5.
Binding of porphyrin and nonporphyrin substrates to ECL3, as monitored by quenching of intrinsic fluorescence. A, binding of PPIX to ECL3, either oxidized (circles) or reduced (squares). B, binding of ZnPPIX (circles), CoPPIX (squares), and pheophorbide a (triangles) to oxidized ECL3. C, Interaction of mitoxantrone (circles), doxorubicin (squares), rhodamine 123 (diamonds), and riboflavin (triangles) with ECL3. Experiments were carried out and analyzed as described in the legend of Fig. 4D.
Finally, we measured the binding of nonporphyrin substrates of ABCG2, such as mitoxantrone (3) and riboflavin (56), and other drugs that bind to ABCG2 but are not transported, such as doxorubicin and rhodamine 123 (12). Results are displayed in Fig. 5C; as shown, all these compounds did not modulate the intrinsic fluorescence of ECL3, suggesting no interaction with the domain. We observed that the dual inhibitor of ABCG2 and ABCB1, GF120918, was able to bind but with very low affinity (Kd of 8 ± 6 μm) and unspecific contribution (supplemental Fig. S2C).
Single-point Mutations H583A and C603A of ECL3 Alter Hemin Binding
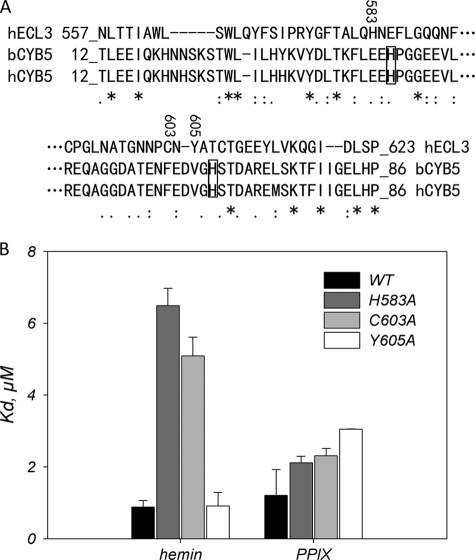
We noticed that ECL3 displays a significant sequence similarity with the heme-binding domain of cytochrome b5, with 14.3% identity and 15.6% similarity, as shown in Fig. 6A. The alignment showed, in addition, that residues His583, Cys603, and Tyr605 of ECL3 are close to the histidine residues of cytochrome b5 (boxed in Fig. 6A) which are involved in the strong chelation of hemin iron. We mutagenized these residues into alanine and monitored by intrinsic fluorescence the binding capacity of the mutant proteins toward hemin and PPIX. The quenching curves are displayed in supplemental Fig. S3 and summarized in Fig. 6B. As shown, the most striking differences were observed for hemin binding because the H583A and C603A mutations lowered the binding affinity 5–6-fold. In contrast, the binding of PPIX was hardly affected by the same mutations. These differences suggest that iron may be involved in hemin binding.
FIGURE 6.
Single-point mutations H583A and C603A, but not Y605A, in ECL3 dramatically alter hemin binding but hardly affect PPIX binding. A, primary structure alignment of ECL3 with the heme-binding domain of cytochrome b5. The score of homology is as follows: asterisk, identity; colon, strong similarity; period, weak similarity; no symbol for no similarity. Histidine residues of cytochrome b5 involved in iron chelation are boxed. The residues His583, Cys603, and Tyr605, which are replaced by an alanine by site-directed mutagenesis, are numbered. B, dissociation constants for hemin and PPIX binding to H583A, C603A, and Y605A mutant ECL3. Values are taken from the fit of data displayed in supplemental Fig. S3 and summarized in supplemental Table S1.
ABCG2 First Saturated with Mitoxantrone or Doxorubicin Binds Hemin but Not Pheophorbide a
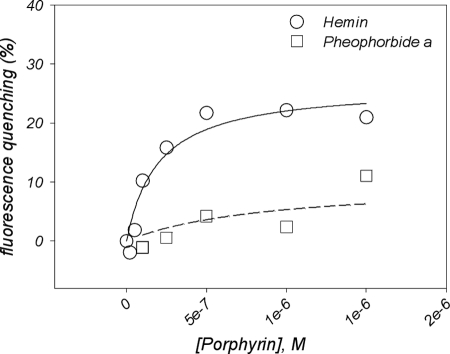
We then checked whether porphyrins could bind to ECL3 within the whole ABCG2 transporter. A preliminary binding experiment of hemin alone on ABCG2 probed by intrinsic fluorescence was carried out (supplemental Fig. S4), leading to an apparent Kd of 0.3 ± 0.2 μm. Because the protein contains more than one transport site in the membrane region to which porphyrins such as pheophorbide a (23) or PPIX (26) bind, we saturated these transport sites with nonporphyrin substrates, such as mitoxantrone and doxorubicin, which additionally do not bind to ECL3, as shown in Fig. 5C. The whole ABCG2 transporter was expressed in insect cells, solubilized in β-d-dodecyl maltoside and purified as previously described (12, 39) and detailed here under “Experimental Procedures.” ABCG2 (0.1 μm) was first incubated with 5 μm mitoxantrone or doxorubicin, a concentration large enough to saturate the transports sites, based on the respective Kd values of 0.9 and 1.6 μm previously reported (12). Then, increasing concentrations of hemin (Fig. 7, circles) or pheophorbide a (Fig. 7, squares) were added and their effects monitored by intrinsic florescence quenching. As shown, hemin induced a fluorescence quenching of about 25%, leading to a saturation curve from which a Kd value of 0.19 ± 0.07 μm could be estimated, two times better than for isolated ECL3. On the contrary, a much poorer interaction was detected with pheophorbide a (Kd >8 μm), suggesting no additional binding site for the compound.
FIGURE 7.
Effect of previous saturation of ABCG2 drug-binding sites by drug substrates on the ability to bind other substrates further. The full-length transporter was expressed and purified, and fluorescence quenching experiments were carried out as detailed under “Experimental Procedures.” ABCG2 (0.1 μm) was preincubated with 5 μm mitoxantrone and doxorubicin before addition of hemin (circles) or pheophorbide a (squares). Fluorescence experiments were carried out and analyzed as detailed in the legend of Fig. 3C.
HSA Removes Hemin Bound to the ECL3 Domain or ABCG2
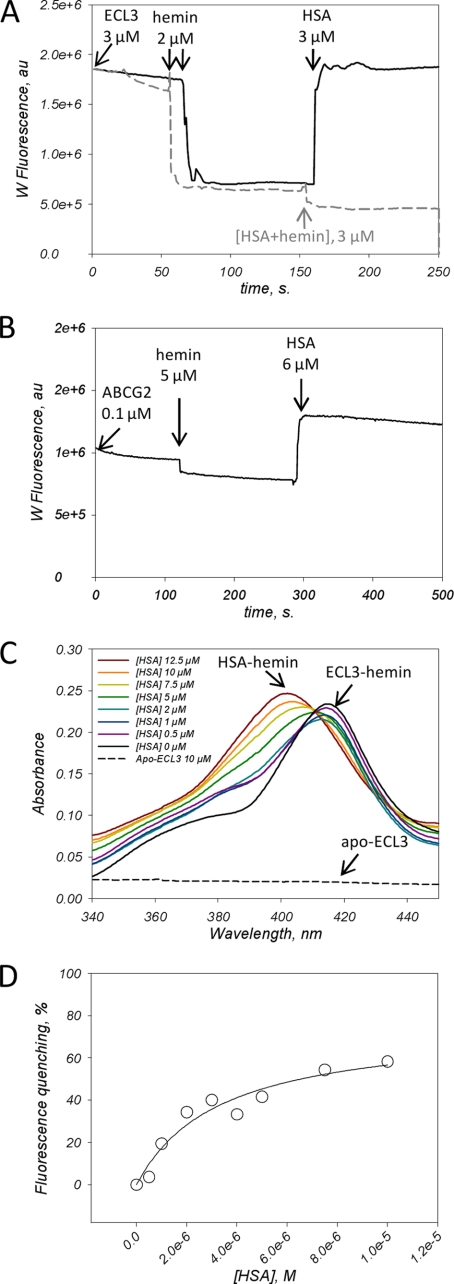
The fact that ECL3 is extracellularly located and binds hemin and related porphyrins makes it potentially able to interact with serum hemin-binding proteins such as HSA and hemopexin (20, 21). Both proteins bind hemin with a Kd of 10−8 and 10−12 m, respectively (20–22) (see also supplemental Fig. S5) and have been reported to be involved in heme homeostasis (22). In the present study we have tested such a possible interaction with HSA and the transfer of hemin from ECL3/ABCG2 to that serum protein. Results are displayed in Fig. 8. In a first experiment, we monitored the binding and release of hemin by recording the intrinsic fluorescence of ECL3 (Fig. 8A). As shown, the addition of 2 μm hemin to 3 μm ECL3 induced a fast fluorescence quenching (dark trace) of about 50–60% as expected from Fig. 4D. In addition, the fluorescence signal remained stable with time after the addition of hemin (see the dark trace e.g. between 100 and 150 s), indicating that the hemin is stably bound to the domain. Further addition of 3 μm apoHSA fully reverted ECL3 fluorescence to its initial level, as quickly as for hemin binding, indicating that hemin was released from the domain. Note that the intrinsic fluorescence signal of ECL3 was corrected from the contribution of HSA as detailed in supplemental Fig. S6. The addition of a preformed HSA·hemin complex at the same concentration did not modify the fluorescence of the ECL3·hemin complex (dashed gray trace in Fig. 8A), indicating no change in hemin interaction with ECL3. The same type of experiment was carried out using the whole ABCG2 preloaded with mitoxantrone and doxorubicin, as described above. As displayed in Fig. 8B, comparable results were obtained, with, however, a larger increase of ABCG2 fluorescence signal upon addition of HSA. These experiments suggested that hemin is released from ECL3 upon addition of HSA. We then monitored the absorbance of hemin bound to ECL3 in the presence of increasing concentrations of HSA. As shown in Fig. 8C, addition of 5 μm hemin to 10 μm apo-ECL3 (dashed black trace) generated the expected Soret peak at 412 nm of the ECL3·hemin complex (dark trace), with the addition of two shoulders at 360 and 385 nm corresponding to 1 μm unbound hemin. Despite this, upon addition of HAS, the peak at 412 nm decreased, even at low (0.5–1 μm) concentrations of HSA, with a new Soret peak at 402 nm of the HSA·hemin complex (57). This result is in agreement with a transfer of hemin from ECL3 to HSA which also supposed an interaction between HSA and ABCG2. We probed such an interaction by monitoring the intrinsic fluorescence signal of ABCG2 in the presence of increasing HSA concentrations. This led to a typical saturation curve displayed in Fig. 8D, reaching to approximately 60% quenching and a rather high Kd of 3.3 ± 1.1 μm, suggesting that such an interaction occurs between both proteins.
FIGURE 8.
Hemin transfer from ECL3 to HSA. A, intrinsic fluorescence of ECL3 (3 μm) was recorded with time before and after addition at the indicated times of 2 μm hemin, and then 3 μm HSA, either alone (black trace) or complexed to hemin (gray dashed trace). ECL3 intrinsic fluorescence was recorded at 336 nm. The contribution of HSA to the fluorescence signal was corrected as detailed in supplemental Fig. S6. B, intrinsic fluorescence of ABCG2 (0.1 μm) preincubated with 5 μm mitoxantrone and doxorubicin was recorded with time before and after addition at the indicated times of 5 μm hemin and then 6 μm HSA. The ABCG2 intrinsic fluorescence was recorded at 328 nm. The contribution of HSA to the fluorescence signal was corrected as detailed in supplemental Fig. S7. C, ECL3 to HSA hemin transfer monitored by UV absorbance. The experiment was carried out as described in Fig. 4 with 10 μm apoECL3 (dashed trace), 5 μm hemin and 0–12.5 μm HSA (black to brown traces). The Soret bands at 402 and 412 nm resulting from the interaction of hemin with HSA and ECL3, respectively, are indicated. D, interaction of HSA with ABCG2 monitored by quenching of ABCG2 intrinsic fluorescence. Fluorescence experiments were carried out and analyzed as detailed in the legend of Fig. 3C, correcting the intrinsic fluorescence of ABCG2 with that of NATA in the presence of the same concentrations of HSA.
DISCUSSION
We show that ABCG2 exports heme, formally its fluorescent analog ZnMP (36, 37), and binds porphyrins, preferably the metal-chelated forms, to its large extracellular domain ECL3 and can release them to HSA. Bound compounds are restricted to porphyrins because nonporphyrin molecules, such as mitoxantrone, riboflavin, or doxorubicin (the latter binds to ABCG2 (12) but is only transported by the mutated R482T/G transporter (4)) do not bind to the domain. Heme and hemin appear to be the preferred ligands, because (i) they display the highest binding affinities for both the isolated ECL3 domain (0.5–1 μm) and the full-length ABCG2 transporter (0.2 μm); (ii) the H583A and C603A single-point mutations alter more markedly the binding of hemin than that of metal-free corresponding PPIX; and (iii) pheophorbide a, which is transported by ABCG2 (26), displays a low binding affinity for ECL3 (3 μm), which is even lower for the full protein (>8 μm) when saturated with MTX or doxorubicin, suggesting that pheophorbide a will not bind to ECL3 under physiological conditions.
ECL3 is topologically located downstream from the membrane transport-binding sites and close to them (4, 51). It is consequently better positioned to bind heme or hemin coming from the transport sites of ABCG2 than those already present in the extracellular space. The relatively low affinity of ECL3, either isolated or within ABCG2, for hemin and heme (micromolar to submicromolar range) reinforces that hypothesis, especially when taking into account the affinity of albumin for hemin in the same compartment, which is 100-fold higher (10−8 m). The extracellular location of ECL3 likely favors a role for delivering hemin, e.g. to a serum protein such as albumin (or hemopexin, not yet tested) (22), a hypothesis strengthened by our experiments showing that apoHSA releases hemin bound to ECL3 by direct interaction with ABCG2.
To date, ABCG2 has been identified to export pheophorbide a (23) and PPIX (26, 27) for preventing intracellular accumulation of these compounds. Our data show that it also transports hemin and heme, in agreement with protein binding to a hemin-agarose resin (26). Such an efflux is also likely related to protecting from damages that such molecules may produce (19), being more toxic than metal-free porphyrins (15). In that context, ECL3 is strategically located to handle these compounds after membrane translocation and before their extracellular release. FLVCR, a membrane protein of the MFS family, has been shown to export hemin out of the cell (37, 58), making the ABCG2-mediated efflux of hemin apparently redundant. One possible explanation could lie in the expression pattern of these proteins during hematopoiesis: ABCG2, which is a hematopoietic stem cell marker (24), is particularly expressed at high levels in the early stages of hematopoiesis (25), whereas the level of FLVCR is particularly high during erythropoiesis. This suggests complementary roles of the transporters.
Supplementary Material
Acknowledgments
We thank Aurélie Badillo, Roland Monseret, and François Penin (Institute of Protein Binding and Chemistry (IBCP)) for help in carrying out CD experiments and spectra analyses. We thank Annie Chaboud and Isabelle Grosjean from the “Production et Analyze des Protéines” platform of the IFR 128 for their help in magnetic bead pulldown assays and ABCG2 expression in insect cells. CD analyses were performed on the same platform. We thank Dr. David S. Vaugh (NCI, National Institutes of Health) for the TEV protease-producing bacteria and Dr. Sheng Zhou and Dr. Albert Sorentino for the K562 cells. We thank Drs Richard. Haser (IBCP), Léa Payen (Lyon I, Rockfeller), and Alain Desbois (CEA Saclay, France) for discussions.
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and University Lyon 1 (UMR5086). This work was also supported by Agence Nationale de la Recherche (ANR) Contracts ANR-06-BLANC-0420, ANR-06-PCVI-0019-01, and ANR-09-PIRI-0002-01, the Association pour la Recherche sur le Cancer (ARC), which funded the thesis of E. D.-M., and the Ligue Nationale Contre le Cancer Labelisation 2009.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7, Table S1, and additional references.
- ABC
- ATP-binding cassette
- BCRP
- breast cancer resistance protein
- FC12
- foscholine 12
- Ni-NTA
- nickel-nitrilotriacetic acid
- HSA
- human serum albumin
- NATA
- N-acetyltryptophanamide
- PPIX
- protoporphyrin IX
- TEV
- tobacco etch virus
- TM
- transmembrane
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ZnMP
- zinc mesoporphyrin.
REFERENCES
- 1.Dean M., Hamon Y., Chimini G. (2001) J. Lipid Res. 42, 1007–1017 [PubMed] [Google Scholar]
- 2.Ross D. D., Yang W., Abruzzo L. V., Dalton W. S., Schneider E., Lage H., Dietel M., Greenberger L., Cole S. P., Doyle L. A. (1999) J. Natl. Cancer Inst. 91, 429–433 [DOI] [PubMed] [Google Scholar]
- 3.Litman T., Brangi M., Hudson E., Fetsch P., Abati A., Ross D. D., Miyake K., Resau J. H., Bates S. E. (2000) J. Cell Sci. 113, 2011–2021 [DOI] [PubMed] [Google Scholar]
- 4.Sarkadi B., Ozvegy-Laczka C., Német K., Váradi A. (2004) FEBS Lett. 567, 116–120 [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Baba M. (2005) Antivir. Chem. Chemother. 16, 213–216 [DOI] [PubMed] [Google Scholar]
- 6.Maliepaard M., Scheffer G. L., Faneyte I. F., van Gastelen M. A., Pijnenborg A. C., Schinkel A. H., van De Vijver M. J., Scheper R. J., Schellens J. H. (2001) Cancer Res. 61, 3458–3464 [PubMed] [Google Scholar]
- 7.van Herwaarden A. E., Schinkel A. H. (2006) Trends Pharmacol. Sci. 27, 10–16 [DOI] [PubMed] [Google Scholar]
- 8.Ahmed-Belkacem A., Pozza A., Muñoz-Martínez F., Bates S. E., Castanys S., Gamarro F., Di Pietro A., Pérez-Victoria J. M. (2005) Cancer Res. 65, 4852–4860 [DOI] [PubMed] [Google Scholar]
- 9.Ahmed-Belkacem A., Macalou S., Borrelli F., Capasso R., Fattorusso E., Taglialatela-Scafati O., Pietro A. D. (2007) J. Med. Chem. 50, 1933–1938 [DOI] [PubMed] [Google Scholar]
- 10.Boumendjel A., Macalou S., Ahmed-Belkacem A., Blanc M., Di Pietro A. (2007) Bioorg. Med. Chem. 15, 2892–2897 [DOI] [PubMed] [Google Scholar]
- 11.Nicolle E., Boumendjel A., Macalou S., Genoux E., Ahmed-Belkacem A., Carrupt P. A., Di Pietro A. (2009) Adv. Drug Deliv. Rev. 61, 34–46 [DOI] [PubMed] [Google Scholar]
- 12.Pozza A., Perez-Victoria J. M., Sardo A., Ahmed-Belkacem A., Di Pietro A. (2006) Cell Mol. life Sci. 63, 1912–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seamon J. A., Rugg C. A., Emanuel S., Calcagno A. M., Ambudkar S. V., Middleton S. A., Butler J., Borowski V., Greenberger L. M. (2006) Mol. Cancer Ther. 5, 2459–2467 [DOI] [PubMed] [Google Scholar]
- 14.Bihorel S., Camenisch G., Lemaire M., Scherrmann J. M. (2007) J. Neurochem. 102, 1749–1757 [DOI] [PubMed] [Google Scholar]
- 15.Tsiftsoglou A. S., Tsamadou A. I., Papadopoulou L. C. (2006) Pharmacol. Ther. 111, 327–345 [DOI] [PubMed] [Google Scholar]
- 16.Sassa S. (1988) Semin. Hematol. 25, 312–320 [PubMed] [Google Scholar]
- 17.Baliga B. S., Mankad M., Shah A. K., Mankad V. N. (1993) Cell Prolif. 26, 519–529 [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y., Hon T., Zhang L. (1999) Biochem. Biophys. Res. Commun. 258, 87–93 [DOI] [PubMed] [Google Scholar]
- 19.Kumar S., Bandyopadhyay U. (2005) Toxicol. Lett. 157, 175–188 [DOI] [PubMed] [Google Scholar]
- 20.Muller-Eberhard U., Fraig M. (1993) Am. J. Hematol. 42, 59–62 [DOI] [PubMed] [Google Scholar]
- 21.Wardell M., Wang Z., Ho J. X., Robert J., Ruker F., Ruble J., Carter D. C. (2002) Biochem. Biophys. Res. Commun. 291, 813–819 [DOI] [PubMed] [Google Scholar]
- 22.Taketani S. (2005) Tohoku J. Exp. Med. 205, 297–318 [DOI] [PubMed] [Google Scholar]
- 23.Jonker J. W., Buitelaar M., Wagenaar E., Van Der Valk M. A., Scheffer G. L., Scheper R. J., Plosch T., Kuipers F., Elferink R. P., Rosing H., Beijnen J. H., Schinkel A. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15649–15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou S., Schuetz J. D., Bunting K. D., Colapietro A. M., Sampath J., Morris J. J., Lagutina I., Grosveld G. C., Osawa M., Nakauchi H., Sorrentino B. P. (2001) Nat. Med. 7, 1028–1034 [DOI] [PubMed] [Google Scholar]
- 25.Scharenberg C. W., Harkey M. A., Torok-Storb B. (2002) Blood 99, 507–512 [DOI] [PubMed] [Google Scholar]
- 26.Krishnamurthy P., Ross D. D., Nakanishi T., Bailey-Dell K., Zhou S., Mercer K. E., Sarkadi B., Sorrentino B. P., Schuetz J. D. (2004) J. Biol. Chem. 279, 24218–24225 [DOI] [PubMed] [Google Scholar]
- 27.Zhou S., Zong Y., Ney P. A., Nair G., Stewart C. F., Sorrentino B. P. (2005) Blood 105, 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia A., Schäfer H. J., Hrycyna C. A. (2005) Biochemistry 44, 10893–10904 [DOI] [PubMed] [Google Scholar]
- 29.Polgar O., Ozvegy-Laczka C., Robey R. W., Morisaki K., Okada M., Tamaki A., Koblos G., Elkind N. B., Ward Y., Dean M., Sarkadi B., Bates S. E. (2006) Biochemistry 45, 5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polgar O., Robey R. W., Morisaki K., Dean M., Michejda C., Sauna Z. E., Ambudkar S. V., Tarasova N., Bates S. E. (2004) Biochemistry 43, 9448–9456 [DOI] [PubMed] [Google Scholar]
- 31.Özvegy C., Litman T., Szakács G., Nagy Z., Bates S., Váradi A., Sarkadi B. (2001) Biochem. Biophys. Res. Commun. 285, 111–117 [DOI] [PubMed] [Google Scholar]
- 32.Kage K., Tsukahara S., Sugiyama T., Asada S., Ishikawa E., Tsuruo T., Sugimoto Y. (2002) Int. J. Cancer 97, 626–630 [DOI] [PubMed] [Google Scholar]
- 33.Kage K., Fujita T., Sugimoto Y. (2005) Cancer Sci. 96, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henriksen U., Fog J. U., Litman T., Gether U. (2005) J. Biol. Chem. 280, 36926–36934 [DOI] [PubMed] [Google Scholar]
- 35.Özvegy-Laczka C., Laczkó R., Hegedus C., Litman T., Várady G., Goda K., Hegedus T., Dokholyan N. V., Sorrentino B. P., Váradi A., Sarkadi B. (2008) J. Biol. Chem. 283, 26059–26070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Worthington M. T., Cohn S. M., Miller S. K., Luo R. Q., Berg C. L. (2001) Am. J. Physiol. Gastrointest. Liver Physiol. 280, G1172–G1177 [DOI] [PubMed] [Google Scholar]
- 37.Quigley J. G., Yang Z., Worthington M. T., Phillips J. D., Sabo K. M., Sabath D. E., Berg C. L., Sassa S., Wood B. L., Abkowitz J. L. (2004) Cell 118, 757–766 [DOI] [PubMed] [Google Scholar]
- 38.Montigny C., Penin F., Lethias C., Falson P. (2004) Biochim. Biophys. Acta 1660, 53–65 [DOI] [PubMed] [Google Scholar]
- 39.Trometer C., Falson P. (2010) in Heterologous Expression of Membrane Proteins: Methods and Protocols (Mus-Veteau I. ed) pp. 105–117, Humana Press, Totowa, NJ [Google Scholar]
- 40.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 41.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 42.Li S. D., Su Y. D., Li M., Zou C. G. (2006) Acta Biochim. Biophys. Sin. 38, 63–69 [DOI] [PubMed] [Google Scholar]
- 43.Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 45.Andrade M. A., Chacón P., Merelo J. J., Morán F. (1993) Protein Eng. 6, 383–390 [DOI] [PubMed] [Google Scholar]
- 46.Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 47.Falson P., Penin F., Divita G., Lavergne J. P., Di Pietro A., Goody R. S., Gautheron D. C. (1993) Biochemistry 32, 10387–10397 [DOI] [PubMed] [Google Scholar]
- 48.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheidt W., Lee Y. (1987) in Metal Complexes with Tetrapyrrole Ligands I, pp. 1–70, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 50.Allen J. D., van Loevezijn A., Lakhai J. M., van der Valk M., van Tellingen O., Reid G., Schellens J. H., Koomen G. J., Schinkel A. H. (2002) Mol. Cancer Ther. 1, 417–425 [PubMed] [Google Scholar]
- 51.Li Y. F., Polgar O., Okada M., Esser L., Bates S. E., Xia D. (2007) J. Mol. Graph. Model 25, 837–851 [DOI] [PubMed] [Google Scholar]
- 52.Hazai E., Bikádi Z. (2008) J. Struct. Biol. 162, 63–74 [DOI] [PubMed] [Google Scholar]
- 53.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 54.Mohanty A. K., Simmons C. R., Wiener M. C. (2003) Protein Expr. Purif. 27, 109–114 [DOI] [PubMed] [Google Scholar]
- 55.Jonker J. W., Merino G., Musters S., van Herwaarden A. E., Bolscher E., Wagenaar E., Mesman E., Dale T. C., Schinkel A. H. (2005) Nat. Med. 11, 127–129 [DOI] [PubMed] [Google Scholar]
- 56.van Herwaarden A. E., Wagenaar E., Merino G., Jonker J. W., Rosing H., Beijnen J. H., Schinkel A. H. (2007) Mol. Cell. Biol. 27, 1247–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monzani E., Bonafè B., Fallarini A., Redaelli C., Casella L., Minchiotti L., Galliano M. (2001) Biochim. Biophys. Acta 1547, 302–312 [DOI] [PubMed] [Google Scholar]
- 58.Keel S. B., Doty R. T., Yang Z., Quigley J. G., Chen J., Knoblaugh S., Kingsley P. D., De Domenico I., Vaughn M. B., Kaplan J., Palis J., Abkowitz J. L. (2008) Science 319, 825–828 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.