Abstract
We show that the length of a loop in the β-knee, between the first and second cysteines (C1-C2) in integrin EGF-like (I-EGF) domain 2, modulates integrin activation. Three independent sets of mutants, including swaps among different integrin β-subunits, show that C1-C2 loop lengths of 12 and longer favor the low affinity state and masking of ligand-induced binding site (LIBS) epitopes. Shortening length from 12 to 4 residues progressively increases ligand binding and LIBS epitope exposure. Compared with length, the loop sequence had a smaller effect, which was ascribable to stabilizing loop conformation, and not interactions with the α-subunit. The data together with structural calculations support the concept that the C1-C2 loop is an entropic spring and an emerging theme that disordered regions can regulate allostery. Diversity in the length of this loop may have evolved among integrin β-subunits to adjust the equilibrium between the bent and extended conformations at different set points.
Keywords: Biophysics, Immunochemistry, Immunology, Integrin, Protein Structure
Introduction
Integrins are heterodimeric adhesion receptors that regulate adhesion to and migration across and along the extracellular matrix and neighboring cells (1, 2). Furthermore, integrins transmit bidirectional signals across the plasma membrane (3). In “inside-out” signaling, interactions of integrin cytoplasmic domains with the actin cytoskeleton mediated by proteins including talins and kindlins activate integrins for ligand binding (4–6). In “outside-in” signaling, ligand binding to the extracellular domain and separation of the α- and β-subunit transmembrane and cytoplasmic domains, together with integrin clustering induced by binding to multivalent ligands, set in motion intracellular signaling cascades that regulate growth and differentiation and inhibit apoptosis (3, 7, 8).
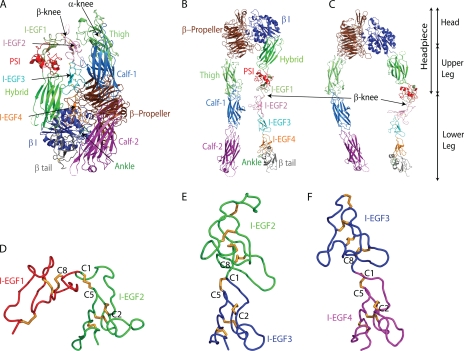
The nature of the integrin conformational changes involved in bidirectional signal transmission has been described by recent structural, biochemical, and biophysical studies (3, 9). In the resting state, integrins are in a bent conformation (Fig. 1A). Integrin activation is associated with extension, which involves a striking unbending at the β-knee between the I-EGF1 and 2 domains and at the α-knee between the thigh and calf-1 domains (Fig. 1, B and C). Extension brings the ligand binding head about 200 Å further above the cell surface and into an orientation more favorable for binding to ligand on other cells and in the extracellular matrix. Although the lower integrin legs and particularly the β-leg are flexible, there are two overall extended conformations, with the headpiece either closed, as in the bent conformation (Fig. 1B), or open (Fig. 1C). Integrin activation is therefore associated not only with extension but also with conversion of the headpiece to the open conformation, which has high affinity for ligands.
FIGURE 1.
Overall integrin structure and the I-EGF domains in the β-leg. A–C, cartoons of ectodomain conformations, based on the bent αIIbβ3 structure (4). A, bent ectodomain. B, extended conformation with the closed headpiece. C, extended conformation with the open headpiece. D–F, chain traces of tandem pairs of I-EGF domains from αVβ3 (25) after structural superposition using SSM (49) on the second module of each tandem. Thus I-EGF2 in D, I-EGF3 in E, and I-EGF4 in F are shown in the same orientation, after separation in the horizontal direction on the page.
Integrin extension can be induced (or stabilized) by transmembrane domains separation in the plane of the membrane, followed by separation of the lower legs, destabilizing their large hydrophilic interface with the headpiece (4, 10). Extension can alternatively be induced (or stabilized) by ligand binding, which induces swing of the hybrid domain out of the interface between the headpiece and lower legs and also requires a change in orientation between the hybrid, I-EGF1, and I-EGF2 domains in the β-knee, further destabilizing the headpiece-lower leg interface (4, 11, 12).
The β-knee and lower β-leg have an essential role in integrin activation for several reasons. They have a central location in the bent conformation. The lower β-leg is buried in a crevice between the upper β-leg on one side and the α-subunit on the other (Fig. 1A) (4). Many mutations that induce integrin activation map to the lower β-leg showing the importance of its burial in the crevice for maintenance of the resting state (13–17). Many activating and activation reporter or ligand-induced binding site (LIBS)2 antibodies also map to the lower β-leg, demonstrating the strong association between exit of the β-leg from its cleft and integrin activation (17–21). Furthermore, integrins bind to talin and the actin cytoskeleton through the β-subunit cytoplasmic domain (22). A recent model of integrin activation, based on the known lateral movement across the cell surface of integrins associated with the actin cytoskeleton, suggests that lateral pulling on the lower β-leg helps to strip it out of the crevice. Moreover, once ligand is bound and resists pulling, steered molecular dynamics suggest that the lateral pulling force stabilizes swing-out of the hybrid domain and the high affinity integrin state (4). Thus, the β-leg has a critical role not only in maintaining the resting state, but also in transmitting the force (i.e. the signal) between the actin cytoskeleton and the ligand binding site that induces (or stabilizes) the extended, open integrin conformation with high affinity for ligand.
The recent determination of the integrin αIIbβ3 structure, which included for the first time the structure of I-EGF domains 1 and 2 in the context of the complete ectodomain, resulted in new concepts about integrin EGF domains, and hypotheses about how the dramatic change in orientation at the integrin knee is achieved (4). EGF domains are small, 35–50 residue domains containing at least two β-strands with an intervening β-hairpin turn that form a small β-sheet, and six conserved cysteines that form intra-domain disulfides and help to provide stability by compensating for the very small hydrophobic core of the domain (23). Integrin EGF domains differ from classical EGF domains in containing one extra disulfide which brings the number of cysteines to eight (denoted C1–C8 in each domain). The extra disulfide links the C1 at the N terminus of each domain to the C5 in the β-hairpin turn between the two β-strands (24) (Fig. 1, D–F). At each domain junction, only one residue intervenes between C8 of the previous domain and C1 of the next domain (a C8-X-C1 linkage, Fig. 1, D–F)). It might be expected that this short linkage would limit interdomain flexibility to only one type of rotation between C8 and X (at the phi angle) and one type of rotation between X and C1 (at the psi angle). However, comparison among different I-EGF domains demonstrated that this is not the case (4). Indeed, in integrin extension at the I-EGF1/I-EGF2 interface, much greater rearrangement was found in the tip of I-EGF2, than at the domain junction per se at the C8-X-C1 junction. This was accomplished by backbone movement in a long loop connecting C1 to C2 in I-EGF2 and by change in the chi angles within the C1-C5 disulfide, i.e. in the rotameric configuration of the linked cysteine side chains. Because movement could occur at two positions, the linkage between I-EGF domains was termed gimbal-like (4).
The current study concerns a central question in integrin structure and function: what structural features modulate or regulate the equilibrium between the bent resting state and the active, extended state? It is commonly thought that the equilibrium between these states is set differently among different integrin family members with differences in the proportion of the bent and extended state under basal conditions. Such differences among integrins may be regulated either by the differences among the α-subunits, e.g. in the β1 or β2 integrins, or by differences among the β-subunits, e.g. in the αV integrins, which associate with five different β-subunits. In the past, it has been assumed that such an equilibrium would be regulated by complementarity between interfaces. Indeed, a recent finding that deletion of part of the C1-C2 loop in I-EGF2 of β3 was activating was interpreted in terms of removal of an important stabilizing interface between this loop and the αV-subunit thigh domain (25). However, we show below that no such stable interface exists.
Here we make a surprising observation based on an extensive set of C1-C2 loop mutations. We demonstrate that the length of the C1-C2 loop in I-EGF2, which varies among different β-integrins, dramatically modulates the equilibrium between the bent and the extended integrin conformations; shortening the loop can super-activate integrins to higher levels than previously achieved with Mn2+ or activating antibodies. We know of no other example where an allosteric equilibrium is regulated by the length of a loop; however, disordered regions are increasingly recognized as important in regulating allostery (26). The results demonstrate that the I-EGF C1-C2 loop acts as an entropic spring, and suggest that in the evolution of integrin diversity, the length of this loop may be a key factor in determining the equilibrium between the bent and the extended integrin conformations.
MATERIALS AND METHODS
Computational Remodeling of I-EGF2 C1C2 Loop
The program Rosetta was used to design the most stable loops equal in length (12 residues) or shorter (10, 9, 8, or 7 residues) than the native loop between Cys-473 and Cys-486 using fragments from the protein data bank (39). Native Ser-474, Asp-484, and Glu-485 were retained for loops of 12, 10, 9, and 8 residues in length (Fig. 2D). The 20,000 lowest energy structures built for each loop length at centroid level (i.e. representing side chains a single centroid atom) were selected with weights of 1.0 for vdw, env, pair, cb, co, sheet, sspair, rsigma, rg, and rama scores (39). In the subsequent all-atom stage, only the identities of the non-native amino acids in the remodeled loop were allowed to change. The models were ranked by their scores at the full atom level (40), and the lowest energy sequence for each length was chosen for experimental testing.
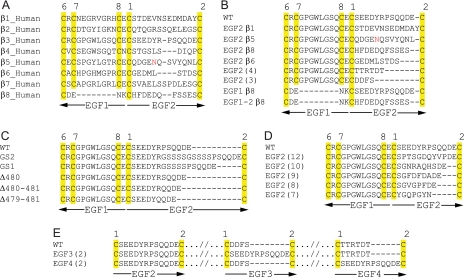
FIGURE 2.
Wild-type sequence at the β-knee and the mutants. Alignments show sequences at the junction between I-EGF1 and I-EGF2 (A–D) and at the C1-C2 loops of I-EGF modules 2–4 (E). A, wild-type human sequences. B, swaps from different integrin subunits and I-EGF modules. C, insertions and deletions. D, computationally designed constructs. E, swaps into I-EGF3 and I-EGF4.
Plasmid Construction and Transient Transfection
cDNAs coding for full-length αIIb and αV were cloned into pEF1/V5-HisA; cDNA coding for full-length β3 was cloned into pCDNA3.1/Myc-His(+) (10). All mutants were generated by QuickChange site-directed mutagenesis (Stratagene) or by overlap PCR (41). All constructs were confirmed by sequencing. 293T cells were transiently transfected using 25 kDa linear polyethyleneimine (Polysciences, Warrington, PA) (42). Cell surface expression and ligand binding were assessed 48 h later.
Antibodies
The activating anti-αIIb mAb PT25–2 (43) was a generous gift from M. Handa (Keio University Hospital, Tokyo, Japan), LIBS β3 mAb AP5 (28) was from the Fifth International Leukocyte Workshop, non-blocking β3 mAb AP3 was from the American Type Culture Collection and ligand-mimetic PAC-1 IgM was from Becton Dickinson (San Jose, CA).
Soluble Ligand Binding
PAC-1 IgM (BD Biosciences) and human fibrinogen (Fg) (Enzyme Research Laboratories, South Bend, IN) binding to αIIbβ3 and αVβ3 transfectants was determined by a method adapted from Pampori et al. (44). AP3 and Fg were fluorescently labeled according to the manufacturer's instructions with Alexa 488 (Molecular Probes, Eugene, OR) and R-phycoerythrin (Dojindo Molecular Technologies, Rockville, MD), respectively. Transiently transfected cells were resuspended in 20 mm HEPES-buffered saline (pH 7.4) containing 5.5 mm glucose and 1% bovine serum albumin (Sigma) and incubated at room temperature for 30 min with 10 μg/ml Fg-PE or 10 μg/ml PAC-1 in the presence of either 1) 5 mm EDTA, 2) 1 mm Ca2+/1 mm Mg2+ or 3) 0.1 mm Ca2+/1 mm Mn2+ plus 10 μg/ml activating mAb PT25–2 for αIIbβ3 or after preactivation with 5 mm DTT (Ref. 45) for αVβ3. For αVβ3, there was an additional half-hour pre-incubation with 5 mm DTT added as stated above in group 3; all groups were then centrifuged and resuspended in buffer containing Fg-PE and lacking DTT. Cells were washed once and stained on ice for 30 min with Alexa 488-labeled AP3 and, for PAC-1 binding, with PE-conjugated anti-mouse IgM (Santa Cruz Biotechnology, Santa Cruz, CA). Cells were then washed once and subjected to fluorescent flow cytometry. Binding was measured as the mean fluorescence intensity (MFI) of PE-conjugated anti-mouse IgM or Fg after subtraction of the MFI in presence of EDTA. The MFI of Alexa 488-AP3 staining was corrected by subtraction of MFI from binding in presence of EDTA. Positive controls were αIIb G991FFKR/GAAKR and αV G989FFKR/GAAKR mutants cotransfected with wild-type β3 (46).
LIBS Epitope Exposure
LIBS epitope exposure was determined as described (47). Briefly, cells were resuspended in 20 mm HEPES-buffered saline (pH 7.4) containing 5.5 mm glucose and 1% bovine serum albumin (Sigma) and incubated at room temperature for 30 min in the presence of 1 mm Ca2+/1 mm Mg2+ or 0.1 mm Ca2+/1 mm Mn2+ plus 100 μm GRGDSP peptide (Telios Pharmaceuticals, San Diego, CA). Then AP5, or control AP3 antibody was added to a final concentration of 10 μg/ml. After 30 min on ice, cells were washed and stained with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG and subjected to fluorescent flow cytometry. Epitope exposure was measured as specific MFI, after subtraction of the MFI of mock transfectants.
Free Cysteine Labeling and Western Blot
Transiently transfected cells were labeled with 1-biotinamido-4-(4′-[maleimidoethyl-cyclohexane]-carboxamido)butane (biotin-BMCC) and lysed with detergent as described (48). Lysates were pre-cleared with protein G beads for 1 h at 4 °C. Supernatants were then immunoprecipitated with AP3 mAb (∼16 h at 4 °C) followed by protein G beads for 1 h at 4 °C. Samples were subjected to non-reducing SDS 7.5% PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were probed with horseradish peroxidase (HRP)-conjugated streptavidin (Zymed Laboratories Inc. laboratories Inc., San Fransisco, CA) or with anti-HisTag antibody for 1 h at room temperature, followed by washing and horseradish peroxidase-conjugated rabbit anti-IgG (GE Healthcare) for 1 h at room temperature. Detection was with the enzymatic chemiluminescence Western blotting kit (Pierce) and a luminescent image analyzer (LAS 4000, Fujifilm). The intensity of each band was determined using ImageJ 1.42n (NIH). The ratio of the intensity of biotin-BMCC and anti-His bands (after subtraction of the intensity of bands of same area from mock transfectant) was taken as proportional to the number of free cysteines/mol assuming one free cysteine/mol for the β3V332C mutant.
Loop Length and Integrin Conformation
The contribution of loop length to the entropic stabilization of the bent and extended conformations between two tandem I-EGF domains was estimated by counting the number of instances in the protein data base of a polypeptide chain of the same length that could link C1 and C2 in I-EGF2. Segments that could link C1 and C2 in I-EGF3 or in I-EGF4 were also examined as examples of an extended conformation. Every segment of i to i + n residues where n was the fragment length of 4 to 22 residues, was taken from a data base of 5621 structures that were filtered for high resolution and low identity in sequence. Holding all coordinates of the two I-EGF domains fixed and deleting residues between C1 and C2, a fragment was inserted as a new polyalanine segment after the position of the first cysteine. This yielded a loop conformation that was continuous with the protein chain on the N-terminal side after C1, but broken on the C-terminal side before the second cysteine (C2). The goodness of fit of the loop conformation was evaluated by the repulsive portion of the Rosetta full-atom Lennard Jones potential (40) to assess atomic clashes created by the loop, and a chain-break score to evaluate the C-terminal loop closure. The chain-break score is a ten atom RMSD computed between the N, CA, C, O, and CB atoms of two constructed ghost residues and their corresponding real residues. The first ghost residue is built off of the C-terminal end of the inserted loop. The second ghost residue is built off of the N-terminal end of C2 in the native protein. Each ghost residue is built using the coordinates internal to the native structure, i.e. using the orientation between the last residue in the loop and C2 in the native structure. Evaluating this “handshake-like” RMSD yields zero for a fully closed loop conformation and a positive number otherwise. All fragments of each length in our structure data base were evaluated and the number of fragments is reported that yielded both a clash score of <1000 and a chain-break RMSD of <4.0 Å.
RESULTS
I-EGF2 C1C2 Swaps
We have systematically tested the effect of varying the sequence and length of the loop between C1 and C2 in I-EGF2 at the gimbal-like connection between I-EGF1 and I-EGF2 in the β-knee. In one group of mutants, the C1-C2 loop from each β-subunit that can associate with the αV-subunit, i.e. β1, β5, β6, and β8, was substituted for the same loop in the β3-subunit (Fig. 2B). This series of β-subunits covers the full range of diversity in C1-C2 loop length found among the eight integrin β-subunits, from 13 residues in β1 to 9 residues in β6 (Fig. 2A). The I-EGF2 chimeric β-subunits were designated EGF2β1 to EGF2β8 (Fig. 2B); all were expressed in association with the αIIb- and αV-subunits at levels comparable to wild-type (Fig. 3A).
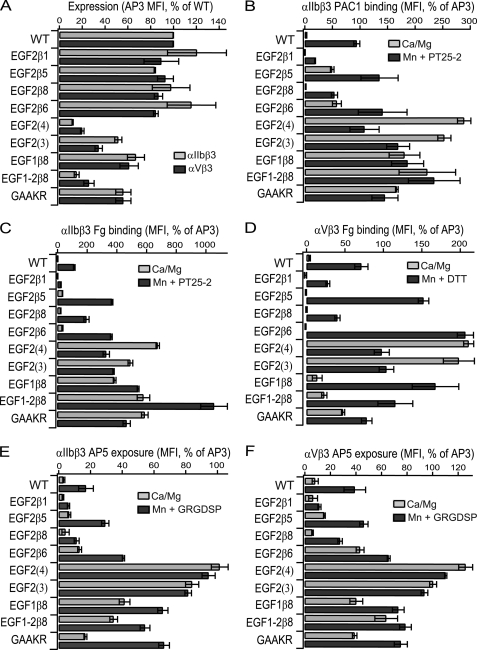
FIGURE 3.
Effects of loop swap mutants on cell surface expression, ligand binding and LIBS exposure. A, antibody and ligand binding to 293T cells co-transfected with the indicated β3 mutants and wild-type or GFFKR mutant αIIb- or αV-subunits was measured by fluorescence flow cytometry. All results are expressed as MFI, as % of MFI of binding to the same transfectants of AP3 antibody to the β3-subunit, except AP3 binding is expressed as % wild type in panel A. Data are mean and S.D. of three independent experiments.
The effect on soluble ligand binding activity was assayed by fluorescent flow cytometry using the ligand-mimetic PAC-1 antibody for αIIbβ3 (Fig. 3B) and fibrinogen for αIIbβ3 and αVβ3 (Fig. 3, C and D). Ligand binding by wild-type αIIbβ3 and αVβ3 was low, and was markedly stimulated in presence of Mn2+ and activating αIIb-specific PT25–2 mAb for αIIbβ3 and Mn2+ and DTT for αVβ3 (Fig. 3, B–D). Mutation of the conserved juxtamembrane GFFKR sequence to GAAKR in αIIb or αV also greatly stimulated ligand binding. Swapping the 13 residue β1 C1-C2 loop for the 12 residue β3 C1-C2 loop markedly reduced the level of ligand binding stimulated by Mn2+/PT25–2 and Mn2+/DTT (termed Mn2+-stimulated below) (Fig. 3, B–D). The level of ligand binding by the 11-residue β8 swap was comparable to wild-type. By contrast, swaps of the 12-residue β5 loop, which contains an NQS N-glycosylation sequon (Fig. 2A), and the 9-residue β6-loop resulted in PAC-1 binding in absence of stimulation (Fig. 3B), and augmented Mn2+-stimulated binding of fibrinogen (Fig. 3, C and D).
The C1-C2 loops of the I-EGF3 and I-EGF4 domains are markedly shorter than those of I-EGF2, as previously pointed out (4). Therefore, the 4-residue loop of β3 I-EGF3 and 6-residue loop of β3 I-EGF4 were substituted in the EGF2(3) and EGF2(4) mutants, respectively (Fig. 2B). These mutations decreased expression (Fig. 3A). In general, activating integrin mutations tend to decrease surface expression (27); e.g. the GAAKR mutation (Fig. 3A). The EGF2(4) and EGF2(3) swaps were highly activating (Fig. 3, B–D). In basal conditions, in absence of Mn2+, each swap showed 3–6-fold more PAC-1 or fibrinogen binding than wild-type in the presence of Mn2+. Addition of Mn2+ did not further enhance ligand binding.
I-EGF1 C7-C8 Loop Swaps
The β8 integrin subunit is unusual for specifically lacking the C7 and C8 cysteines and hence the C7-C8 disulfide bond in I-EGF1 (Fig. 2A). Therefore, we substituted the region from β8 corresponding to C7, C8, and the residues in between into β3 (EGF1β8) and both this region and the C1-C2 loop from β8 into β3 (EGF1–2β8).
Expression of EGF1β8 was at ∼60% of wild type and EGF1–2β8 was at ∼30% of wild type in both αIIbβ3 and αVβ3 (Fig. 3A). Basal ligand binding activity by αIIbβ3 was strongly stimulated by both swaps (Fig. 3, B and C) while stimulation was less marked for αVβ3 (Fig. 3D).
LIBS Epitope Exposure in C1-C2 and C7-C8 Swaps
AP5 antibody recognizes a LIBS epitope in the β3 PSI domain (28). LIBS epitope exposure was examined as a surrogate for integrin extension (Fig. 3, E and F). The results mirrored those seen for ligand binding. Binding comparable to wild type was observed for EGF2β1, β5, and β8 while binding to EGF2β6 was enhanced. In Mn2+, binding to EGF2β1 was reduced about 3-fold compared with wild type (Fig. 3, E and F), similarly to the lower Mn2+-stimulated ligand binding seen with EGF2β1 (Fig. 3, B–D). In resting conditions, AP5 epitope exposure was markedly increased for EGF1β8 and EGF1–2β8 mutants. Basal epitope exposure was greatest in the EGF2(3) and EGF2(4) mutants, which expressed amounts of the AP5 LIBS epitope comparable to the constitutively expressed AP3 epitope (Fig. 3, E and F). This correlated with the greatest ligand binding by the same mutants (Fig. 3, B–D).
I-EGF2 C1-C2 Loop Insertion and Deletion Mutants
Overall, the above studies suggest an inverse correlation between the length of the C1-C2 loop and integrin activation. This suggestion was tested with further types of C1-C2 loop mutations. One type of I-EGF2 C1-C2 loop mutant was designed by inserting one (GS1) or two (GS2) blocks of Gly-Ser-Ser-Ser-Ser residues or deleting 1, 2, or 3 residues (Δ480, Δ480–481, and Δ479–481) (Fig. 2C). Surface expression of αIIbβ3 was similar to wild type for all mutants, except Δ479–481 was expressed at ∼50% of wild type (Fig. 4A). Results were similar for αVβ3, except GS2 and Δ480–481 were expressed at ∼60% of wild type.
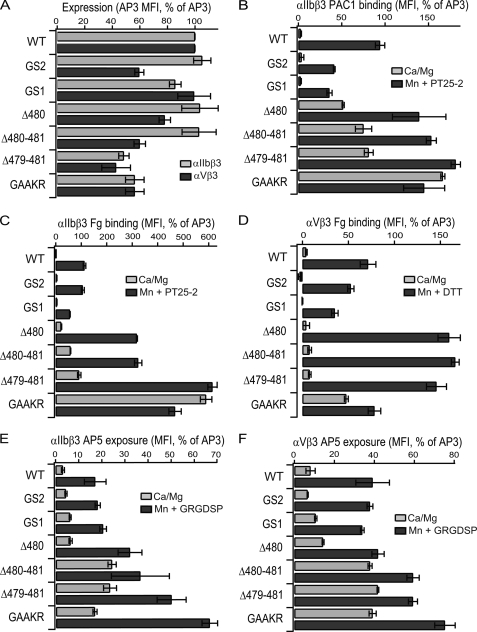
FIGURE 4.
Effects of insertion/deletion mutants on cell surface expression, ligand binding and LIBS exposure. A, measurements were as described in the Fig. 3 legend. Data are mean and S.D. of three independent experiments.
Increasing the length of the C1-C2 loop in the GS1 and GS2 mutants had no significant effect on ligand binding in resting conditions (Fig. 4, B–D). In Mn2+, some decrease in PAC-1 and Fg binding was seen with GS1 (Fig. 4, C and D).
Decreasing the length of the C1-C2 loop enhanced ligand binding (Fig. 4, B–D). In resting conditions, ligand binding to αIIbβ3 was progressively increased (5–20-fold) in the Δ480, Δ480–481, and Δ479–481 mutants (Fig. 4, B and C). Fibrinogen binding by the αVβ3 Δ480–481 and Δ479–481 mutants was significantly (p < 0.05) increased by 2-fold (Fig. 4D). Furthermore ligand binding in Mn2+ by the Δ480, Δ480–481, and Δ479–481 mutants was enhanced compared with wild type (Fig. 4, B–D).
Conformational changes were monitored by exposure of the AP5 epitope. The GS1 and GS2 insertion mutants showed little or no difference in AP5 exposure compared with wild type in either basal or Mn2+-stimulated conditions (Fig. 4, E and F). In contrast, markedly increased AP5 exposure was seen under both conditions for both the Δ480–481 and Δ479–481 mutants (Fig. 4, E and F).
Computational Redesign
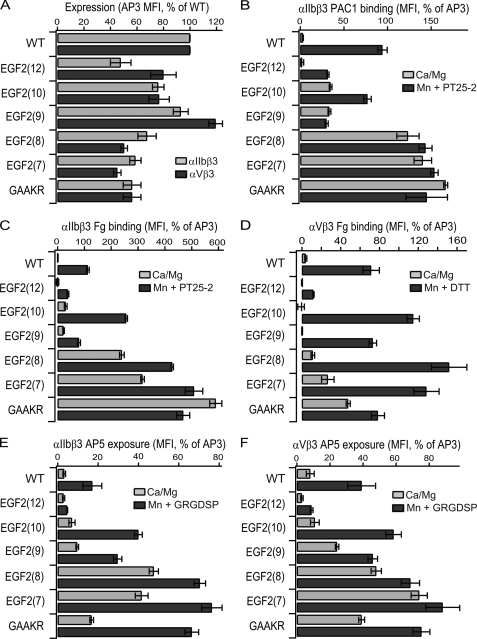
The above results strongly supported the inverse correlation between the length of the C1-C2 loop in I-EGF2 and a shift in the equilibrium from the bent toward the extended integrin conformation. However, loop sequence and hence structure might also modulate the equilibrium. To examine this possibility, we used Rosetta to predict loop sequences and structures that had the lowest energy for loops of 7–12 residues in length (Fig. 2D). All mutants were expressed at levels of 50–100% of wild type (Fig. 5A).
FIGURE 5.
Effects of computationally designed mutants in the I-EGF2 C1-C2 loop on cell surface expression, ligand binding, and LIBS exposure. Measurements were as described in the Fig. 3 legend. Data are mean and S.D. of three independent experiments.
Mutant EGF2(12), which has a length equal to wild-type, showed little or no basal ligand binding and Mn2+-stimulated binding that was markedly reduced, by 50–80% relative to wild type (Fig. 5, B–D). Thus stabilization was achieved relative to wild type.
In contrast, it appeared that it was difficult to stabilize the bent conformation with loop lengths of 7–10 residues. The designs with 9 and 10 residues were comparable to wild type with αVβ3 (Fig. 5D) and basally more active with αIIbβ3 (Fig. 5, B and C). The designs with 7 and 8 residues were basally active with both αIIbβ3 and αVβ3 (Fig. 5, B–D) and also more active than wild type in Mn2+ (Fig. 5, B–D).
Similar trends were seen with LIBS epitope exposure. The 12-residue design was resistant to epitope exposure by Mn2+ and the 10- and 9-residue designs showed exposure comparable to or greater than wild-type (Fig. 5, E and F). In contrast, the designs with 7 and 8 residues showed constitutive AP5 epitope exposure (Fig. 5, E and F).
Free Cysteine Labeling
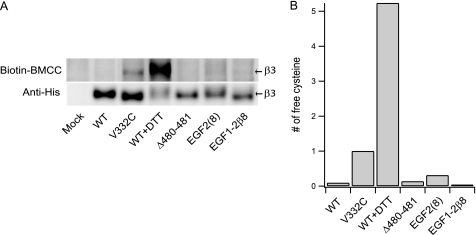
An alternative explanation for integrin activation by shortening of the C1-C2 loop would be that shortening had the unintended consequence of preventing disulfide formation by either the C1 or C2 cysteines; reduction of cysteines in the β-subunit or their mutation can cause activation (29). To test this possibility, selected transfectants were treated with biotin-BMCC to covalently label free cell surface cysteines, lysed, and subjected to immunoprecipitation with AP3 antibody, non-reducing SDS-PAGE, and Western blotting (Fig. 6). Representative Δ480–481, EGF2(8), and EGF1–2β8 mutants, all of which were active under basal conditions, showed labeling with biotin-BMCC comparable to wild type (Fig. 6A). By contrast, the β3V332C mutant that has one mutationally introduced cysteine, and wild-type cells treated with 10 mm DTT, showed specific labeling (Fig. 6A). Using blotting with anti-His6 tag as a measure of the amount of β3, and assuming a stoichiometry of 1 Cys/mol for β3V332C, the amount of free cys in each of the mutants was estimated (Fig. 6B). The amount of free Cys was much less than the 2 Cys/mol expected for failure to form one disulfide bond, showing that the activation seen in this study cannot be explained by disruption of disulfide bond formation.
FIGURE 6.
Quantification of free cysteines in αIIbβ3 mutants. A, cells co-transfected with αIIb and the indicated β3 mutants were labeled with biotin-BMCC, lysed, and subjected to immunoprecipitation with AP3 antibody, non-reducing SDS-PAGE, and Western blotting with HRP-conjugated streptavidin to detect biotin-BMCC-labeled free cysteines or anti-His Tag antibody and HRP-conjugated rabbit anti-IgG to detect the His6-tagged β3-subunit. The β3V332C mutant and wild-type transfectants pretreated with 10 mm DTT for 1 h at 37 °C served as positive controls. B, number of free cysteines was calculated from the intensity ratio of the biotin-BMCC and anti-His bands after subtraction of the intensity of a band of same area.
Swaps of the I-EGF3 and I-EGF4 C1-C2 Loops
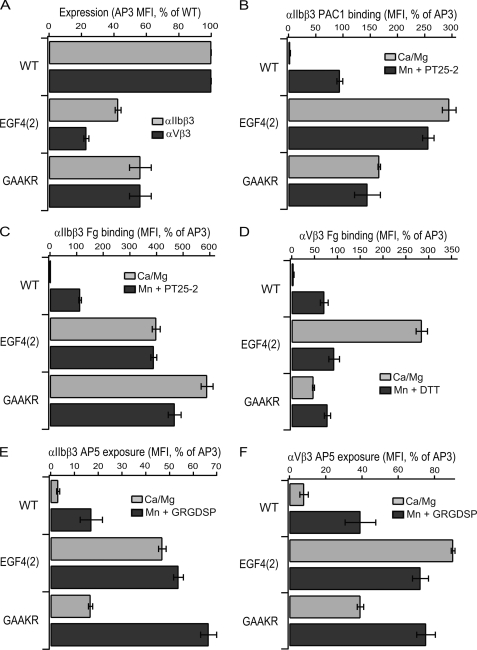
The orientations at the I-EGF2/I-EGF3 and I-EGF3/I-EGF4 interfaces in the bent integrin conformation are extended, and the C1-C2 loops at these interfaces are short (Fig. 1, E and F). We hypothesized that short C1-C2 loops were important for these extended orientations and for burial of the lower β-leg in its crevice in the bent conformation, and that therefore, introduction of long C1-C2 loops would be activating. The C1-C2 loop of I-EGF3 is buried in the crevice; replacement of the 4-residue C1-C2 loop of I-EGF3 with the 12-residue C1-C2 loop of I-EGF2 abolished cell surface expression. By contrast, the C1-C2 loop of I-EGF4 is partially exposed; replacement of this 6-residue loop with the 12-residue I-EGF2 C1-C2 loop in the EGF4(2) mutant was compatible with expression of ∼ 30% of normal levels of αIIbβ3 and αVβ3 on the cell surface (Fig. 7A). The EGF4(2) αIIbβ3 and αVβ3 mutants showed basal ligand binding that was strongly activated, to levels equivalent to or much higher than GAAKR mutants (Fig. 7, B–D). The AP5 LIBS epitope was also highly exposed under basal conditions (Fig. 7, E and F).
FIGURE 7.
Effects of swap of the I-EGF4 C1C2 loop on cell surface expression, ligand binding, and LIBS exposure. Measurements were as described in the Fig. 3 legend. Data are mean and S.D. of three independent experiments.
DISCUSSION
Previously, it was noted that the length of the C1-C2 loop in I-EGF2 was markedly longer than in I-EGF3 or I-EGF4, and it was suggested that this longer length enabled greater flexibility at the I-EGF1/2 interface than at the I-EGF2/3 or I-EGF3/4 interface (4); however, it was completely unanticipated that the length of the C1-C2 loop would have a major role in regulating the equilibrium between the bent and extended integrin conformations. Here, using three independent series of swap mutants of the I-EGF2 domain C1-C2 loop, we have surprisingly, yet unequivocally, demonstrated that the length of the C1-C2 loop inversely correlates with integrin activation. Short C1-C2 loops favor integrin activation, as measured by ligand binding, and integrin conformational change, as measured by LIBS epitope exposure; conversely, long C1-C2 loops favor low affinity for ligand and lack of conformational change as measured by LIBS epitope masking.
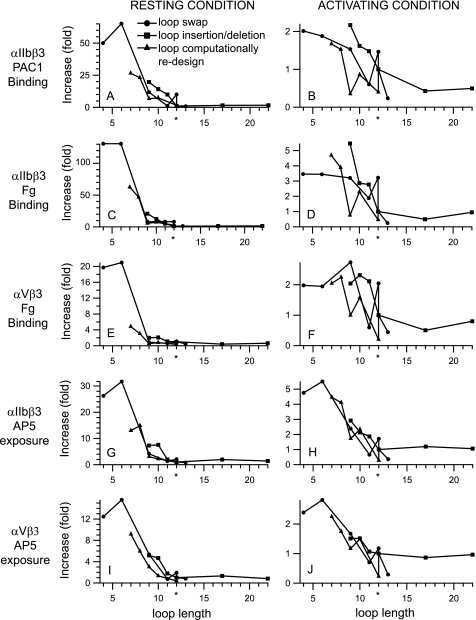
The overall correlation between I-EGF2 C1-C2 loop length and integrin activation was highly robust, because it was demonstrated for binding of the ligand PAC-1 and fibrinogen to αIIbβ3 (Fig. 8, A–D), binding of fibrinogen to αVβ3 (Fig. 8, E and F) and exposure of the AP5 LIBS epitope in both αIIbβ3 and αVβ3 (Fig. 8, G–J). Furthermore, swaps with sequences from other β-subunits and other I-EGF domains, deletions and insertions, and computationally designed sequences all revealed the inverse correlation between length and activation (Fig. 8). Enhancement of ligand binding and LIBS exposure was most marked when measured under basal conditions in Mg2+ and Ca2+ where it ranged from 12–130-fold at the shortest loop lengths tested (Fig. 8, A, C, E, G, and I). Ligand binding and LIBS exposure had previously been considered to be maximal for αIIbβ3 in Mn2+ and the activating antibody PT25–2, and for αVβ3 in Mn2+ and DTT; however, these measures were also increased by 2–5-fold by loop shortening (Fig. 8, B, D, F, H, and J).
FIGURE 8.
Effect of β-knee loop length on integrin activation. Data from Figs. 3–5 are plotted as a function of the length of the C1-C2 loop in I-EGF2. The length of the wild-type loop in β3 is shown with an asterisk. Ligand binding or LIBS exposure is calculated relative to wild type under resting conditions or under activating conditions (Mn2+ and PT25–2 antibody for αIIbβ3 or Mn2+ and DTT for αVβ3).
Although length was of paramount importance, loop sequence also made a definite, if lesser contribution to the conformational equilibrium. When loops of equal length were compared, the computationally-designed loops fulfilled their intended function of stabilizing the bent orientation between I-EGF1 and I-EGF2, as shown by lesser ligand binding and LIBS epitope exposure under basal conditions (Fig. 8, A, C, E, G, and I), and in most cases under activating conditions as well (Fig. 8, B, D, F, H, and J).
Not only could loop shortening activate, but loop lengthening and computational design could also suppress activation. It was difficult to assess decreases in ligand binding or epitope exposure in the resting state, because these measures were already low basally. However, decreases in Mn2+-stimulated measures were noted for several mutants. Replacement of the 12-residue β3 loop with the 13-residue β1 loop decreased Mn2+-stimulated ligand binding and LIBS epitope exposure by αIIbβ3 and αVβ3 by 2–4-fold (x̄ = 3.3-fold). The GS1 and GS2 mutants, with loop length of 17 and 22 residues, decreased ligand binding by αIIbβ3 and αVβ3 about 2-fold, (x̄ = 1.8-fold). However, no suppression of LIBS exposure epitope was seen, perhaps in accord with the relatively modest effect on ligand binding. No computationally designed sequences longer than the wild-type sequence of 12-residues were tested. However, it was notable that the computationally designed 12-residue sequence suppressed Mn2+-stimulated αIIbβ3 and αVβ3 ligand binding and LIBS epitope exposure by 2–5-fold (x̄ = 3.5-fold).
These findings show that the length and to a lesser extent the sequence of a loop at the integrin β-knee play a critical role in regulating the equilibrium between the low-affinity and high-affinity integrin states. In general, there was excellent agreement between activation of ligand binding and exposure of the AP5 LIBS epitope, in accord with many other lines of evidence showing that the bent integrin conformation in which LIBS epitopes are masked corresponds to the low-affinity state, and that the extended integrin conformation with the open headpiece in which LIBS epitopes are exposed corresponds to the high-affinity state (3, 9).
The AP5 mAb binds to the PSI domain, which is adjacent to the I-EGF1 domain in the upper β-leg. The PSI domain is partially masked in the bent conformation by the I-EGF2 and I-EGF3 domains in the lower β-leg (Fig. 1A). Thus exposure of the AP5 epitope is likely to be associated with integrin extension (Fig. 1, A and B), headpiece opening (Fig. 1, B and C), or both. Since headpiece opening requires disruption of the bent conformation, we assume that AP5 epitope exposure requires integrin extension. The extent of this extension may be lesser than shown in Fig. 1, A–C, and integrin extension would not require a fully extended orientation between I-EGF domains 1 and 2, such as is seen between I-EGF domains 2 and 3 and between I-EGF domains 3 and 4 (Fig. 1, D–F).
It is not difficult to understand why the length of the C1-C2 loop in I-EGF2 should be important in regulating the conformational equilibrium between the bent and extended conformations, because the course of the polypeptide chain has to bend around the tip of I-EGF2 in the bent conformation (Fig. 1D), whereas it can take a much more direct path in the extended conformation, as seen in I-EGF3 and I-EGF4 (Fig. 1, E and F). However, it is perhaps more difficult to understand the less important role of loop sequence. The commonly thought of structural elements that regulate allostery are structurally complementary interfaces that contribute both enthalpically and entropically to stability. For example, interfaces between the integrin headpiece and lower legs, and between the α- and β-subunit transmembrane and juxtamembrane domains, stabilize the bent conformation relative to the extended conformation (10, 14, 15, 30, 31).
The familiar concept of stabilizing interfaces was invoked previously, when it was found that a seven-residue deletion in the C1-C2 loop of the β3 I-EGF2 domain activated αVβ3 (25). It was concluded that activation was due to disruption of a stabilizing interface between the β3 I-EGF2 C1-C2 loop and the αV thigh domain (25). In the same manuscript, no extension of αVβ3 by this deletion or RGD-mimics known to extend and open integrins by EM (32) were reported in fluorescence lifetime imaging microscopy (25); however, no fluorescence lifetime decay curves were presented, making it difficult to assess key aspects of the fluorescence resonance energy transfer (FRET) data, including whether one or two lifetimes were present. Although fit to a single lifetime was used (25), a fit to bi-exponential decay of unquenched (no FRET) and quenched (FRET) populations is more appropriate for FRET experiments (33). Furthermore, autoquenching from labeling each Fab with 3–7 fluorophores, and lack of determination of the Förster radius for the donor and acceptor pair, complicated the results. Recently, several groups besides our own have reported that extension is associated with integrin activation (34, 35).
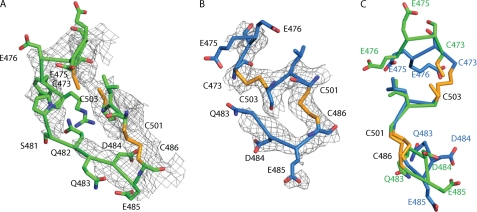
Inspection of the αVβ3 electron density shows that the C1-C2 loop in EGF-1 is highly flexible, and reveals no evidence for a stable interface with the thigh domain. The backbone cannot be traced with certainty, and there is no backbone density at residues Ser-481 and Gln-482 at the 1 σ level (Fig. 9A). Moreover, 8 of the 12 residues in the C1-C2 loop, and additionally Cys-473 and Cys-486, are Ramachandran outliers (3IJE PDB record). This unusually high proportion of outliers (0.2% is average for high resolution structures) is a symptom either of an incorrect backbone trace, or the lack of a single backbone conformation. In an oversight in the molecular model, the sulfur atoms of cysteine residues 473 and 503 are not covalently connected; continuous density between the cysteine side chains shows that they are disulfide bonded to form the C1-C5 disulfide (Fig. 9A). There is no density for the side chains of Glu-475, Glu-476, Asp-477, Tyr-478, Arg-479, Ser-481, Gln-482, Gln-483, or Glu-485 in the C1-C2 loop of I-EGF2 in the αVβ3 crystal structure (Fig. 9A). In contrast, density is very well defined for all six side chains of the C1-C2 loop of I-EGF4. The lack of order of the C1-C2 loop side chains in I-EGF2 demonstrates that they do not participate in a stable interface. Thus, they cannot provide hydrogen bonds or salt bridges to stabilize interaction with the thigh domain as previously suggested (25). Moreover, of the putative 11 stabilizing interactions, shown with dashed lines in Fig. 4D of (25), 5 actually represent repulsive interactions, including two each between carboxyl side chains of αV Asp-550 and β3 Glu-500, and between the carbonyl oxygen of αV Phe-548 and side chain of β3 Glu-476.
FIGURE 9.
Flexibility of the C1-C2 loop of I-EGF2. A and B, C1-C2 loop and the region containing the C1-C5 and C2-C4 disulfides of αVβ3 (A) and αIIbβ3 (B), with 2Fo-Fc electron density contoured at 1 σ. C, residues in common between A and B, after superposition on all residues present in the I-EGF2 domains. Carbons of αVβ3 and αIIbβ3 are green and blue respectively, with the entire side chain of Cys in yellow.
The mobility of the C1-C2 loop of I-EGF2 is further illustrated by its partial disorder in integrin αIIbβ3 (Fig. 9B). There is no discernable electron density for residues 476–482, which were therefore omitted from the molecular model. Furthermore, after superposition of the I-EGF2 domains from the αVβ3 and αIIbβ3 crystal structures, the markedly different conformations of the ordered portions of their C1-C2 loops common to the two structures, at 473–476 and 483–485, are evident (Fig. 9C). This difference in backbone conformation further illustrates the flexibility of the C1-C2 loop of I-EGF2. Different examples of integrin αXβ2 in crystals also differ markedly in I-EGF2 C1-C2 backbone conformation (17).
Further evidence argues against a specific interface between the β3 I-EGF2 C1-C2 loop and the thigh domain in integrins in general, and specifically in αIIbβ3 or αVβ3. We found that loops with inserted Gly-Ser-Ser-Ser-Ser repeats, a 12-residue designed loop, and a β1 loop were all more stabilizing than the native β3 loop. Insertion of the Gly-Ser-Ser-Ser-Ser sequence must have disrupted the native backbone conformation of the loop. Moreover, the computational designs, including for the stabilizing 12-residue loop, were based on the structure of αIIbβ3 only. The 12-residue designed loop stabilized αVβ3 as well, despite major differences in the structure of the αIIb and αV thigh domains adjacent to the C1-C2 loop. The β1-subunit associates with αV but not αIIb, and therefore the stabilizing β1 loop, which stabilizes αIIbβ3 as well as αVβ3, is under no evolutionary selection for complementarity with αIIb. We therefore conclude that although specific I-EGF2 C1-C2 loop sequences can stabilize the bent, low-affinity integrin conformation, the stabilization is unrelated to specific interaction with the αV or αIIb thigh domains.
Entropic differences between states are increasingly recognized as contributing to difference in free energy between alternative conformational states. Furthermore, intrinsically disordered regions can regulate allostery by being linked by equilibria to conformational change, without any requirement that shape shifting be transmitted through such disordered regions (26). In the absence of enthalpic differences, states with greater disorder and hence greater entropy have lower free energy and are thus favored relative to more ordered states. For example, integrin extension will be accompanied by an increase in entropy, because of the increased number of interdomain orientations that are accessible in this more flexible state, helping to lower the free energy of the extended state and offset the loss of the stabilizing interfaces described above.
The notion that the number of conformational states accessible to the I-EGF2 C1-C2 loop differed in a length-dependent manner for the bent and extended conformations was tested using the protein structure data base. The number of polypeptide segments of a given length in the data base capable of connecting C1 and C2, without introducing major clashes and bad stereochemistry, was used as a surrogate for the number of entropic conformational states accessible to a given loop length. As models for the bent conformation, we used two independent examples of the C1-C2 loop from each of the β3- and β2-subunits (Fig. 10A). As models for the extended conformation, we used examples of the C1-C2 loop from I-EGF3 and IEGF4 from the β3- and β2-subunits (Fig. 10B).
FIGURE 10.
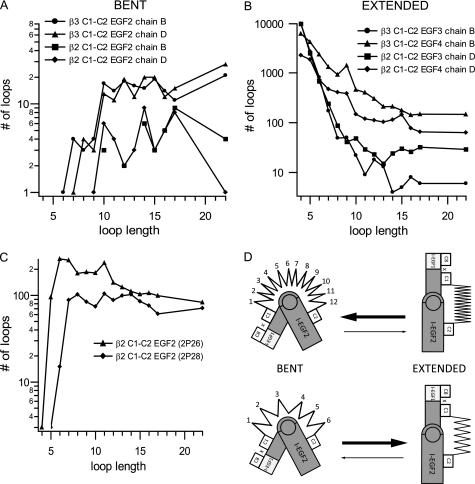
Regulation of integrin conformation by an entropic spring in the β-knee. A–C, as a surrogate for loop entropy, the number of different loops in the Protein Data Bank that could span C1 and C2 in different integrin conformational states was estimated as a function of loop length. A, bent conformation, sampled with the C1-C2 loop of I-EGF2 of two independent αIIbβ3 molecules (chains B and D of 3FCS) and αXβ2 molecules (chains B and D of 3K6S). B, extended conformation, sampled with the C1-C2 loops of I-EGF3 and I-EGF4 of αIIbβ3 and αXβ2. C, relaxed conformation, sampled with the C1-C2 loop of I-EGF2 of β2-leg fragment structures 2P26 and 2P28. D, schematic of an entropic spring in the β-knee. A large number of residues (spring coils) pushes the conformation toward the bent state. A smaller number of residues (spring coils) pulls the conformation toward the extended state.
The effect of loop length on number of accessible states (Fig. 10B) is in excellent agreement with the effect of loop length on activation (Fig. 8). At the shortest loop lengths tested here of 4 and 6 residues, the extended conformation was highly favored, with a large number of accessible states (Fig. 10B); furthermore, there were no or few states accessible for the bent conformation (Fig. 10A). This agreed with the strong activation by 4 and 6-residue loops (Fig. 8). As loop length increased to 11 or 12 residues, the number of extended states gradually decreased until it plateaued at 12–22 residues (Fig. 10A). Conversely, the number of accessible bent states increased from 6 to 10 residues, and then plateaued at 10 to-22 residues (Fig. 10B). This is in excellent agreement with the decrease in integrin activation from a loop length of 6 residues to a length of 10–12 residues, and the lack of any marked further effect of lengthening to 17 and 22 residues (Fig. 8). We caution these calculations are only approximations, and that only the overall trends, and not the specific number of states, are meaningful. Furthermore, enthalpic contributions will favor specific backbone conformations and sequences, consistent with some effect of sequence on the conformational equilibrium.
These results further suggest that when integrins extend, the orientation between I-EGF1 and 2 is not likely to approach as fully an extended conformation as seen between I-EGF2 and 3 or I-EGF3 and 4, because of the high entropic cost. Fragments of the β2 leg crystallized in the absence of any constraints from the bent conformation adopt conformations intermediate between the bent I-EGF1/2 interface and extended I-EGF2/3 and I-EGF3/4 interfaces, and thus the fragments provide two examples of the range of I-EGF1 and 2 orientations that could be sampled in the extended conformation (Fig. 10C). Interestingly, these “relaxed” orientations permitted a substantially larger ensemble of loop conformations than the bent conformation, and exhibited less loop length dependence than either the bent or extended conformations (Fig. 10, A and B).
Thus, we have discovered a novel mechanism for modulating integrin activation. The I-EGF2 C1-C2 loop acts as an entropic spring that connects to and regulates the orientation between the headpiece and the lower legs (Fig. 10D). The equilibrium length of the spring in most integrin β-subunits is optimal for the bent conformation. Shortening the spring shifts the equilibrium toward extension. We are unaware of other examples where the length of an entropic spring regulates a conformational or allosteric equilibrium.
Our results on mutations at the β-knee underline the importance of knee extension in integrin activation. The shortest I-EGF2 C1-C2 loops tested, in the EGF2(4) and EGF2(3) mutants, activated ligand binding under basal Ca2+/Mg2+ conditions to levels 2–5-fold higher than seen with wild-type αIIbβ3 or αVβ3 in Mn2+/PT25–2 Ab or Mn2+/DTT, respectively. The level of activation, and LIBS epitope exposure, was greater than or similar to that seen with GFFKR/GAAKR mutants, and LIBS epitope exposure approached 100% compared with a constitutively expressed epitope, suggesting maximal extension. Together with evidence for a lack of a specific interface between the C1-C2 loop and α-subunit thigh domains, these results add yet further support for the importance of integrin extension in activation of ligand binding.
The lesser but still significant contribution of sequence to stabilizing the low affinity state and hindering LIBS exposure may be to select for loop conformations that are better suited to span C1 and C2 in the bent than in the extended conformation. The most stable loops we found were the β1 loop and a computationally designed 12-residue loop.
One loop sequence tested here from β5 has an N-linked glycosylation sequon. The ability of N-linked glycans to stabilize proteins has been ascribed to lessening of the number of conformational states accessible to the unfolded polypeptide chain, because of the large volume excluded by the glycan (36). The lesser number of states accessible to the N-glycosylated β5 loop might explain why at 12-residues it was less stabilizing than the β3 loop of the same length (Fig. 3).
We have shown that the length and sequence of an entropic spring in the β-knee is important for tuning the threshold for integrin activation. This β-knee spring is similar in importance to domain-domain and α-β interfaces in integrins that regulate the conformational equilibrium between the active and resting states. We speculate that the length and sequence of the entropic spring in the β-knee is evolutionarily adapted to different set points in this equilibrium for different β-subunits. The β1-subunit has to associate with 13 different α-subunits, 9 more than the next most frequently used β2-subunit. It will be difficult to co-evolve stable interfaces in the bent conformation with so many different α-subunits, consistent with the constitutive activity of many of the β1 integrins. We speculate that the length and sequence of the I-EGF2 C1-C2 loop in the β1-subunit have evolved to compensate for this difficulty, and help make β1 integrins more stable in the bent conformation than they would otherwise be.
The substantial variation in C1-C2 loop length among β-subunits that associate with αV may in part contribute to differences in basal activation status among αV integrins. Why αV needs so many β-subunits is currently not known. αVβ6 is the most important αV integrin for activation of TGF-β in airways (37), and it is interesting that the β6 I-EGF2 C1-C2 loop was the most activating native integrin loop tested. It will be interesting to compare αV integrins for their basal activation status. Although the β6 C1-C2 loop is activating when transplanted into the β3-subunit, it remains possible that other interfaces in αVβ6 might compensate by being more stabilizing than in αVβ3. Such differences might result in different activation kinetics or differences in the pathways between the bent and extended open conformations among αV integrins.
It will be interesting to learn whether the absence of specific disulfide-bonded loops in I-EGF1 and the lower β-leg is an evolutionary mechanism for activating integrins that do not associate with the actin cytoskeleton. Uniquely among β-subunits, β8 and β4 lack the NPXY motif required for association with talin and the actin cytoskeleton. Like αVβ6, αVβ8 is important in activating TGF-β (38). The β8-subunit is unique in lacking the C7-C8 disulfide and loop in I-EGF1 (Fig. 2A). Making β3 β8-like in this region in either the EGF1β8 or EGF1–2β8 mutants was highly activating, similar to the GFFKR/GAAKR mutation. The β8-subunit also lacks a loop and a disulfide in the β-tail domain. The β4-subunit lacks the C1-C5 disulfide in I-EGF3 and the disulfide in the β-ankle. Otherwise, all disulfides in I-EGF1 and the lower β-leg are invariant among integrin β-subunits. It will be interesting to explore whether the integrin β8- and β4-subunits are basally more activated than other β-subunits that associate with the αV and α6-subunits, respectively, and whether absence of conserved disulfides and disulfide-bonded loops play an important role in basal activation status of integrins.
Acknowledgments
We are grateful to Drs. Thomas Schurpf, Jieqing Zhu, and Sladjana Prisic for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-48675.
- LIBS
- ligand-induced binding site
- MFI
- mean fluorescence intensity
- FRET
- fluorescence resonance energy transfer.
REFERENCES
- 1.Hynes R. O. (1992) Cell 69, 11–25 [DOI] [PubMed] [Google Scholar]
- 2.Schwartz M. A., Schaller M. D., Ginsberg M. H. (1995) Annu. Rev. Cell Dev. Biol. 11, 549–599 [DOI] [PubMed] [Google Scholar]
- 3.Luo B.-H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., Springer T. A. (2008) Mol. Cell 32, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moser M., Legate K. R., Zent R., Fassler R. (2009) Science 234, 895–899 [DOI] [PubMed] [Google Scholar]
- 6.Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., Calderwood D. A. (2003) Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 7.Miyamoto S., Akiyama S. K., Yamada K. M. (1995) Science 267, 883–885 [DOI] [PubMed] [Google Scholar]
- 8.Coller B. S., Shattil S. J. (2008) Blood 112, 3011–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Askari J. A., Buckley P. A., Mould A. P., Humphries M. J. (2009) J. Cell Sci. 122, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takagi J., Petre B. M., Walz T., Springer T. A. (2002) Cell 110, 599–611 [DOI] [PubMed] [Google Scholar]
- 11.Takagi J., Strokovich K., Springer T. A., Walz T. (2003) EMBO J. 22, 4607–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao T., Takagi J., Coller B. S., Wang J. H., Springer T. A. (2004) Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanhoorelbeke K., De Meyer S. F., Pareyn I., Melchior C., Plancon S., Margue C., Pradier O., Fondu P., Kieffer N., Springer T. A., Deckmyn H. (2009) J. Biol. Chem. 284, 14917–14918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vomund A. N., Stuhlsatz-Krouper S., Dimitry J., Song Y., Frazier W. A. (2008) Biochemistry 47, 11616–11624 [DOI] [PubMed] [Google Scholar]
- 15.Kamata T., Handa M., Sato Y., Ikeda Y., Aiso S. (2005) J. Biol. Chem. 280, 24775–24783 [DOI] [PubMed] [Google Scholar]
- 16.Zang Q., Springer T. A. (2001) J. Biol. Chem. 276, 6922–6929 [DOI] [PubMed] [Google Scholar]
- 17.Xie C., Zhu J., Chen X., Mi L., Nishida N., Springer T. A. (2010) EMBO J. 29, 666–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C., Ferzly M., Takagi J., Springer T. A. (2001) J. Immunol. 166, 5629–5637 [DOI] [PubMed] [Google Scholar]
- 19.Luo B. H., Strokovich K., Walz T., Springer T. A., Takagi J. (2004) J. Biol. Chem. 279, 27466–27471 [DOI] [PubMed] [Google Scholar]
- 20.Mould A. P., Travis M. A., Barton S. J., Hamilton J. A., Askari J. A., Craig S. E., Macdonald P. R., Kammerer R. A., Buckley P. A., Humphries M. J. (2005) J. Biol. Chem. 280, 4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang R. H., Tng E., Law S. K., Tan S. M. (2005) J. Biol. Chem. 280, 29208–29216 [DOI] [PubMed] [Google Scholar]
- 22.Legate K. R., Fässler R. (2009) J. Cell Sci. 122, 187–198 [DOI] [PubMed] [Google Scholar]
- 23.Wouters M. A., Rigoutsos I., Chu C. K., Feng L. L., Sparrow D. B., Dunwoodie S. L. (2005) Protein Sci. 14, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takagi J., Beglova N., Yalamanchili P., Blacklow S. C., Springer T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11175–11180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong J. P., Mahalingham B., Alonso J. L., Borrelli L. A., Rui X., Anand S., Hyman B. T., Rysiok T., Müller-Pompalla D., Goodman S. L., Arnaout M. A. (2009) J. Cell Biol. 186, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilser V. J., Thompson E. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8311–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo B. H., Karanicolas J., Harmacek L. D., Baker D., Springer T. A. (2009) J. Biol. Chem. 284, 3917–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda S., Tomiyama Y., Pelletier A. J., Annis D., Honda Y., Orchekowski R., Ruggeri Z., Kunicki T. J. (1995) J. Biol. Chem. 270, 11947–11954 [DOI] [PubMed] [Google Scholar]
- 29.Kamata T., Ambo H., Puzon-McLaughlin W., Tieu K. K., Handa M., Ikeda Y., Takada Y. (2004) Biochem. J. 378, 1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau T. L., Kim C., Ginsberg M. H., Ulmer T. S. (2009) EMBO J. 9, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J., Luo B. H., Barth P., Schonbrun J., Baker D., Springer T. A. (2009) Mol. Cell 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagi J., Springer T. A. (2002) Immunol. Rev. 186, 141–163 [DOI] [PubMed] [Google Scholar]
- 33.Festy F., Ameer-Beg S. M., Ng T., Suhling K. (2007) Molecular bioSystems. 3, 381–391 [DOI] [PubMed] [Google Scholar]
- 34.Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A., Ginsberg M. H. (2010) J. Cell Biol. 188, 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Askari J. A., Tynan C. J., Webb S. E., Martin-Fernandez M. L., Ballestrem C., Humphries M. J. (2010) J. Cell Biol. 188, 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shental-Bechor D., Levy Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8256–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 38.Travis M. A., Reizis B., Melton A. C., Masteller E., Tang Q., Proctor J. M., Wang Y., Bernstein X., Huang X., Reichardt L. F., Bluestone J. A., Sheppard D. (2007) Nature 449, 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohl C. A., Strauss C. E., Misura K. M., Baker D. (2004) Methods Enzymol. 383, 66–93 [DOI] [PubMed] [Google Scholar]
- 40.Kuhlman B., Dantas G., Ireton G. C., Varani G., Stoddard B. L., Baker D. (2003) Science 302, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 41.Higuchi R., Krummel B., Saiki R. K. (1988) Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., Behr J. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokuhira M., Handa M., Kamata T., Oda A., Katayama M., Tomiyama Y., Murata M., Kawai Y., Watanabe K., Ikeda Y. (1996) Thromb. Haemost. 76, 1038–1046 [PubMed] [Google Scholar]
- 44.Pampori N., Hato T., Stupack D. G., Aidoudi S., Cheresh D. A., Nemerow G. R., Shattil S. J. (1999) J. Biol. Chem. 274, 21609–21616 [DOI] [PubMed] [Google Scholar]
- 45.Ni H., Li A., Simonsen N., Wilkins J. A. (1998) J. Biol. Chem. 273, 7981–7987 [DOI] [PubMed] [Google Scholar]
- 46.Zhu J., Boylan B., Luo B.-H., Newman P. J., Springer T. A. (2007) J. Biol. Chem. 16, 11914–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo B. H., Springer T. A., Takagi J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo B. H., Takagi J., Springer T. A. (2004) J. Biol. Chem. 279, 10215–10221 [DOI] [PubMed] [Google Scholar]
- 49.Krissinel E., Henrick K. (2004) Acta. Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]