Abstract
The paired box homeodomain Pax6 is crucial for endocrine cell development and function and plays an essential role in glucose homeostasis. Indeed, mutations of Pax6 are associated with diabetic phenotype. Importantly, homozygous mutant mice for Pax6 are characterized by markedly decreased β and δ cells and absent α cells. To better understand the critical role that Pax6 exerts in glucagon-producing cells, we developed a model of primary rat α cells. To study the transcriptional network of Pax6 in adult and differentiated α cells, we generated Pax6-deficient primary rat α cells and glucagon-producing cells, using either specific siRNA or cells expressing constitutively a dominant-negative form of Pax6. In primary rat α cells, we confirm that Pax6 controls the transcription of the Proglucagon and processing enzyme PC2 genes and identify three new target genes coding for MafB, cMaf, and NeuroD1/Beta2, which are all critical for Glucagon gene transcription and α cell differentiation. Furthermore, we demonstrate that Pax6 directly binds and activates the promoter region of the three genes through specific binding sites and that constitutive expression of a dominant-negative form of Pax6 in glucagon-producing cells (InR1G9) inhibits the activities of the promoters. Finally our results suggest that the critical role of Pax6 action on α cell differentiation is independent of those of Arx and Foxa2, two transcription factors that are necessary for α cell development. We conclude that Pax6 is critical for α cell function and differentiation through the transcriptional control of key genes involved in glucagon gene transcription, proglucagon processing, and α cell differentiation.
Keywords: Chromatin Immunoprecipitation (ChiP), Gene Regulation, Pancreatic Islet, siRNA, Transcription Factors, Glucagon Biosynthesis, MafB/cMaf, NeuroD1/Beta2, Pax6, Primary α-cells
Introduction
Type 2 diabetes is a common disease characterized by insulin resistance and insulin deficiency, hyperglucagonemia, and dysregulation of glucagon secretion, reflecting dysfunction of both α (glucagon) and β (insulin) cells of the endocrine pancreas. The molecular determinants of the basic abnormalities leading to islet dysfunction are poorly understood but the relationships between the α and β cells and their secretory products are starting to emerge as critical for glucose homeostasis.
Indeed, complementary to β cell dysfunction, it has been well established that α cell dysregulation contributes to the maintenance of fasting and postprandial hyperglycemia in type 2 diabetes (1). Thus, better understanding of α cell differentiation and function especially the control of glucagon biosynthesis and secretion could bring new therapeutic advances. Although the determinants of pancreatic α cell differentiation and functions are poorly characterized, key roles for transcription factors Pax6, Arx, and Foxa2 have been demonstrated through the analyses of homozygous mutant mice for their coding genes showing no or very few α cells (2–5).
Pax6 is a transcription factor with two DNA-binding domains (a paired box and a homeobox) and a proline-serine-threonine (PST)-rich transactivation domain at the C terminus (6). It has been implicated in the development of the eye, central nervous system (7), and the pancreas (8). Pax6 is present very early in the developing mouse pancreas at embryonic day 9.0 in both the dorsal and ventral pancreatic buds and is restricted to endocrine cells during development and adulthood (3).
Mice with a targeted disruption of Pax6, as well as Pax6 mutant mice die soon after birth (2, 3, 9). The pancreas of these mice is characterized by the absence of α cells and a markedly decreased number of β, δ, and γ cells (PP) (2, 3). More recently, in a pancreas-specific Pax6 knock-out mouse model, a drastic diminution of glucagon- and insulin-positive cells was also observed, but surprisingly PP- and somatostatin-producing cells were not affected (9). Furthermore, these mice failed to form islets suggesting that normal islet morphogenesis requires a critical number of cells, or alternatively that Pax6 might potentially have a direct role in islet morphogenesis through the control of genes that mediate cell-cell adhesion. Pax6 thus has a crucial role in endocrine cell development and particularly for α cells, although the molecular mechanisms remain unknown (2).
As with Pdx1, NeuroD1/Beta2, HNF-1α and -β, and HNF-4α, Pax6 mutations are associated with diabetes mellitus. Indeed, heterozygous mutations of Pax6 are associated with glucose intolerance or early-onset diabetes mellitus in humans (10, 11). Furthermore, conditional inactivation of Pax6 in the mouse pancreas is characterized by a diabetic phenotype with hyperglycemia and hypoinsulinemia reflecting an essential role of Pax6 in the regulation of glucose homeostasis and endocrine cell function (9).
We and others have shown by transfection studies that Pax6 activates glucagon gene expression through the transactivation of the G1 and G3 elements of the rat gene promoter (12–14). More recently, we demonstrated that Pax6 interacts synergistically with cMaf on the α cell-specific G1 element to activate glucagon gene expression (15).
In addition to Pax6, development and differentiation of glucagon-producing cells depend on a variety of transcription factors, among them Arx, Nkx2.2, Isl1, NeuroD1/Beta2, Foxa1, and Foxa2 as well as MafB and cMaf. Although Pax6, Arx, and Foxa2 are the most critical inasmuch as mutant mice lacking any one of them have no α cells (3–5), mice deficient for Nkx2.2, NeuroD1/Beta2, Isl1, MafB, cMaf, Sox4, and Foxa1 also have a significant decrease of glucagon-positive cells (16–23). In addition, NeuroD1/Beta2, Isl1, MafB, cMaf, Foxa1, and Foxa2 have been proposed to regulate Glucagon gene expression in glucagon-producing cell lines (15, 24–27). However, the architecture of α cell organization in the islets, the restricted number of α cells, and the unavailability of an adequate primary α cell model have considerably hampered progress in the molecular determinants of glucagon gene transcription as well as differentiation and function.
The aim of the present study was to better define and characterize the role of Pax6 in α cell function and understand the lack of α cell observed in the absence of Pax6. Inasmuch as the presence of glucagon is the most obvious evidence for differentiation of α cells, we investigated whether Pax6 could regulate genes coding for transcription factors involved in glucagon gene transcription. We developed two Pax6-deficient models, a partial knockdown with specific Pax6 siRNA in primary rat α cells and cultured glucagon-producing cell lines and selected clones expressing a dominant-negative form of Pax6 in hamster InR1G9-glucagon-producing cells.
We now report that Pax6 controls the expression of the MafB, cMaf, and NeuroD1/Beta2 genes in primary rat α cells and in the glucagon-producing cell line αTC1.9. Furthermore, we confirm that Pax6 is crucial for glucagon biosynthesis in rat islets through the direct and indirect controls of the Proglucagon and PC2 genes, respectively. Pax6 is able to bind and activate the promoter regions of the MafB, cMaf, and NeuroD1/Beta2 genes through specific sites as assessed by electrophoretic mobility shift assays (EMSA), chromatin immunoprecipitation (ChIP), mutational analyses, and transfection experiments in BHK-21 cells. Furthermore, the transcriptional activities of the promoters of these three genes are significantly decreased in Pax6-deficient cells (InR1G9 stable clones expressing a dominant negative form of Pax6 constitutively). Taken together, we identify three new Pax6 target genes in the adult pancreatic α cell, which are involved in α cell differentiation.
We conclude that Pax6 regulates directly or indirectly the transcription of the Proglucagon, PC2, MafB, cMaf, and NeuroD1/Beta2 genes, which are critical for α cell development and differentiation as well as glucagon biosynthesis and thus provide a potential explanation for the dramatic absence of pancreatic α cells in Pax6 knock-out mice.
EXPERIMENTAL PROCEDURES
Islets Isolation
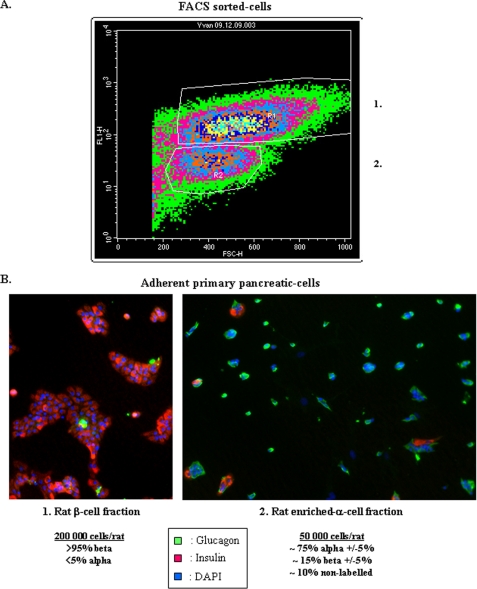
Briefly, islets of Langerhans were isolated by collagenase digestion of pancreases from male adult Sprague-Dawley rats followed by Ficoll purification using a modification of previously described procedures (28, 29). β Cells were separated from non-β cells by autofluorescence-activated sorting using a FACStar-Plus cell sorter (BD Bioscience), as previously described (29–31). We obtained two major populations characterized by immunohistochemistry, β cells and an enriched-α cell fraction (Fraction 1 and 2, see Fig. 1). Fraction 1 was composed almost exclusively by insulin-positive cells (>95%), whereas fraction 2 by a majority of glucagon-positive cells (∼75% ±5%). As noted, we obtained about 200,000 and 50,000 β and enriched-α cells, respectively, per rat. 16 h after isolation, enriched α cells were cultured on extracellular matrix-coated plates derived from 804G cells (Laminin-5-rich extracellular matrix, 804G-ECM) in DMEM containing 10% fetal calf serum (FCS), 11.2 mm d-Glucose, 2 mm l-Glutamine and antibiotics (32).
FIGURE 1.
Characterization of primary islet α cells. A, primary rat β cells are separated from non-β cells by autofluorescence-activated cell sorting using a FACStar Plus cell sorter (BD Bioscience). Two major populations were obtained, fractions 1 and 2, which correspond to β cells and non-β cells (enriched α cells). B, characterization of FACS-sorted cells by immunohistochemistry. Endocrine cells fixed with 4% paraformaldehyde were treated with 0.1% Triton X-100 and immunolabeled using polyclonal rabbit glucagon antibody (number AB932, Chemicon International) combined with monoclonal mouse insulin (number I2018, Sigma) antibodies. We also use DAPI to count total cells. Fraction 1 is composed almost exclusively of insulin positive cell (>95%) and fraction 2 is composed by a majority of glucagon-positive cells (∼75% ± 5%).
Cell Culture
The glucagon producing cell αTC1.9 cells were grown in DMEM (Invitrogen) supplemented with 3 g/liter glucose, 10% fetal calf serum, 2 mm glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. The hamster InR1G9 (33) as well as the non-islet Syrian baby hamster kidney (BHK-21)2 cell lines were grown in RPMI 1640 (R-6504, Sigma, Basel, CH) supplemented with 5% fetal bovine serum and 5% newborn calf serum, 2 mm glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Empty vector and Pax6-DN306-expressing InR1G9 stable clones were grown in the same medium supplemented with 400 μg/ml of G418/geneticin/disulfate (Brunschwig, Amsterdam, NL).
Materials and Plasmids
GluCAT vector constructs corresponding to the 5′-flanking region of the rat glucagon gene were previously described (34). The cDNA coding for a dominant-negative form of human Pax6 (Pax6-DN306) cloned into the pRC-CMV expression vector was obtained from Dr. S. Singh (35).
RNA Interference
Two different specific sequences of siRNA were designed by Invitrogen for Pax6 against rat and mouse mRNA sequences (Stealth RNAi, 25-mers). The siRNA named siPax6–614 and siPax6–1007 recognize specifically segments 614/638 and 1007/1031 of the Pax6 mRNA sequence. These siRNA were designed in two exons common for the Pax6 isoforms. Appropriate scramble siRNA were obtained at the same time (the percentage of GC content was identical to Pax6 siRNA).
For primary rat islets, primary adherent enriched α cells (about 40,000 per condition) were transfected with 100 nm of the mixed Pax6 siRNAs using 1 μl of Lipofectamine 2000 (Invitrogen) in 200 μl of serum-free medium. Transfections were performed twice sequentially (spaced by 24 h). After the second transfection, medium was replaced by growth medium (11.2 mm d-glucose + 10% FCS) for 48 h. αTC1.9 cells grown in 6-well plates were transfected in the same conditions with 100 nm of the siRNA mixture (siPax6–614 and siPax6–1007) or scramble siRNA. Total RNA and nuclear or cellular extracts were isolated 96 h after the first transfection.
Pax6-DN306 Stable InR1G9 Clones
InR1G9 cells were transfected with Lipofectamine 2000 reagent according to the manufacturer's specifications. Briefly, 1 day before transfection InR1G9 cells were plated in 10-cm dishes at 80–90% confluence in antibiotic-free RPMI medium. Next, cells were transfected with 10 μg of Pax6-DN306 cDNA or 10 μg of pRC-CMV in fresh medium without antibiotics (OptiMEM, Invitrogen). Medium was replaced by RPMI with antibiotics 16 h after transfection. Cells were divided after 48 h (1:3) in new culture dishes. Two days later, the antibiotic geneticin/G418 was added to the medium (600 μg/ml) to start clone selection. We obtained about 100 clones; to get individual colonies cloning rings were used. Trypsin was added to each cloning ring for 5 min. The reaction was stopped by the addition of complete medium. Finally, each colony was seeded to a well in a 48-well plate. Each clone was verified by the immunodetection of Pax6-DN306.
Western Blot Analyses
Nuclear extracts were isolated as described (36, 37) from native InR1G9 cells and stable clones (named A5/C5 and B6/C4) expressing either the empty vector or Pax6-DN306. Twenty μg of each protein extract were resolved on a 10% SDS-polyacrylamide gel and transferred electrophoretically to polyvinylidene difluoride membranes. Immunoblotting was performed with polyclonal antibodies to rabbit Pax6 diluted 1:1000 (gift from Dr S. Saule) and goat anti-rabbit IgG conjugated with horseradish peroxidase diluted 1:2000 (number sc2030, Santa Cruz Biotechnology, Santa Cruz, CA). The signal was detected with Super Signal West Pico Trial Kit (Pierce). Protein loading was normalized by immunodetection of rabbit TFIIE-α diluted 1:500 (number C17, Santa Cruz). At least three independent experiments were performed with nuclear cell extracts.
In-Cell Western Analyses
The In-Cell Western (Licor) is an immunocytochemical assay performed directly into culture plates. Target-specific primary antibodies and infrared-labeled secondary antibodies were used to detect target proteins in fixed cells, and the fluorescent signal from each well was quantified. In-Cell Western blots were used simultaneously to detect two targets at 700 and 800 nm using two spectrally distinct dyes. We thus quantified glucagon with specific antibodies (AB932, Chemicon International) in InR1G9 stable clones and normalized by TFIIE-α (antibody F-2, sc-133065, Santa Cruz Biotechnology). We also used secondary antibodies (anti-rabbit IR680 and anti-mouse IR780) to reveal the respective content of glucagon and TFIIE-α. At least three independent experiments were performed.
Measurements of Glucagon Content
Primary rat α cells (about 40,000 cells/condition) were removed and frozen in 1 ml of acid/ethanol mixture. An aliquot of the medium was used to measure glucagon using a radioimmunoassay kit following the manufacturer's recommendations (Glucagon RIA Kit, GL-32K, Millipore).
RNA Preparation and RT-PCR Analysis
Total RNA was isolated from primary rat α cells and αTC1.9 cells using TRIzol Reagent (Invitrogen) according to the manufacturer's specifications. Different targets were analyzed by real time RT-PCR using QuantiTect SYBR Green kit (Qiagen, Basel, CH) and Light-Cycler (Roche Diagnostics) using specific primers (supplemented Table S1). Results were corrected by β-actin and TBP (TATA-box binding protein) mRNA levels.
Promoter Analysis
For all transfections cell lines were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. Glucagon promoter transactivation was studied by transfection of the −292 to +50 bp sequence of the 5′-flanking region of the rat Glucagon promoter (−292GluCAT) into BHK-21 and InR1G9 stable clones. Promoter activities for the rat cMaf (−320/+700 bp) and mouse MafB (−660/+329 bp) (gift from Dr. M. Sakai), and mouse NeuroD1/Beta2 (−2200/+11 bp) (gift from Dr. T. Ming-Jer) were also analyzed in BHK-21 and αTC1.9 cells.
Promoter activities were analyzed by luciferase (LUC) or chloramphenicol acetyltransferase (CAT) activities. Cells were cotransfected with a plasmid containing the human placental alkaline phosphatase gene (pSV2A-placental alkaline phosphatase) to monitor transfection efficiency. Overexpression of mouse Pax6 (mPax6) was systematically verified in each cotransfection experiment by Western blot analyses (data not shown). Quantification of luciferase assays was done with a Luminometer (Labsystem). Placental alkaline phosphatase was measured by spectrophotometry. A minimum of three independent transfections were performed, each of them carried out in duplicate.
Electrophoretic Mobility Shift Assays
EMSA reactions were performed as previously described (38). To assess for the presence of Pax6-DN306 in InR1G9 stable clones, EMSAs were performed using the −100 to −49 bp sequence of the 5′-flanking region of the rat glucagon gene promoter corresponding to the G1 element (15) with nuclear extracts from Pax6-DN306- or empty vector-expressing InR1G9 stable clones (C4 and A5). EMSAs were also performed using the 5′-flanking region sequence of rat cMaf (+362/+396 bp and +511/+545 bp) and mouse MafB (−357/−322 bp) and Beta2/NeuroD1 (−2133/−2096 bp, −1510/−1471 bp, and −775/−741 bp) genes corresponding to the promoter regions including conserved putative Pax6 binding sites, using in silico analyses of the binding site search with Genomatix software (Matinspector). Each probe was incubated with nuclear extracts from BHK-21 cells overexpressing mouse Pax6 (p46). An anti-Pax6 antibody was used to determine the specificity of Pax6 binding. Mutated Pax6 putative binding site oligonucleotides were also used for cold probe competition (supplemented Table S2).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assays were performed as previously described (15). Briefly, formaldehyde cross-linked chromatin extracts were prepared from rat islets, αTC1.9 and InR1G9 cell lines, and fragmented by enzymatic digestion (Enzymatic shearing kit, Active Motif Europe, Belgium). Fifty μg of chromatin extract were used for experiments and incubated with 4 μg of anti-Pax6 and α-acetyl-histone H4 antibodies (number 06-866, Upstate, Lake Placid, NY) as well as rabbit IgG (number sc-2027, Santa Cruz). The sets of PCR primers used for analysis of binding correspond to highly conserved regions of the rat and mouse Glucagon, MafB, cMaf, NeuroD1/Beta2, and PC2 gene promoters (containing the Pax6 binding site) (supplemented Table S3). PCR products were verified on ethidium bromide-stained 3% agarose gels and analyzed by real time PCR using a Light-Cycler (Roche Diagnostics).
Site-directed Mutagenesis
The promoter mutants were constructed using standard PCR. The procedure utilized the MafB, cMaf, and NeuroD1/Beta2 promoter DNA constructs as template and two synthetic oligonucleotide primers containing the desired mutation for the reaction (mutated oligonucleotides are listed in supplemented Table S2). PCR were performed with Pfu DNA polymerase (Stratagene, La Jolla, CA) during the temperature cycling of 95 °C for 30 s, 55 °C for 1 min, and 68 °C for 15 min for a total of 18 cycles. The mutant products were treated with DpnI endonuclease (Stratagene) to digest the parental DNA template and confirmed by DNA sequencing. The plasmids were then prepared using Midiprep kit (Qiagen) for transfection into αTC1.9 cells.
Data Analysis
Data are presented as mean ± S.E. and analyzed by Student's t test. A p value of less than 0.05 was considered statistically significant.
RESULTS
To better understand the role of Pax6 in differentiated glucagon-producing cells, we developed a Pax6-deficient model in adult primary rat α cells (Fig. 1) and extended our analyses in glucagon-producing cell lines. The aim of the present study was to determine Pax6 target genes involved in the differentiation of pancreatic α cells and more specifically on glucagon gene expression and glucagon biosynthesis.
Identification of Pax6 Target Genes with Specific Inhibition of the Pax6 Gene by siRNA in Primary Rat α Cells
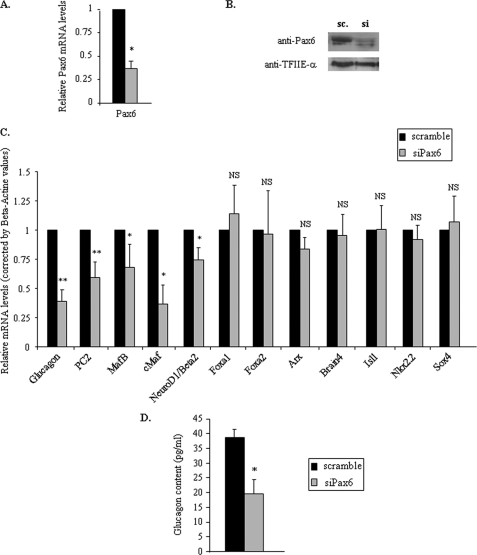
We first developed a Pax6-deficient model in adult rat primary α cells by using specific siRNAs directed against rat Pax6 mRNA. We performed these experiments with Pax6 or corresponding scramble siRNAs for 96 h. We first verified the efficiency of Pax6 silencing in our system by determining the cellular amounts of Pax6 mRNA and protein in each condition. Transient transfection of Pax6 siRNA in primary α cells led to a 70% decrease of Pax6 mRNA levels at 96 h (Fig. 2A), accompanied by a corresponding decrease in Pax6 protein content (Fig. 2B) thus achieving partial knockdown of Pax6 in differentiated α cells.
FIGURE 2.
Effects of specific inhibition of Pax6 gene expression by siRNA in primary rat α cells. Enriched primary rat α cells were transfected with 100 nm of the Pax6 siRNA mixture (siPax6–614 and siPax6–1007) or corresponding scramble during 96 h. A, effects of Pax6 siRNA on Pax6 gene expression by real time RT-PCR. Data are corrected by β-actin mRNA values. B, Western blot analyses of Pax6 protein content from transfected enriched primary rat α cells with scramble or specific Pax6 siRNAs after 96 h. TFIIE-α serves as a control for the specificity of siRNA effects. C, quantitative analyses of the expression of key pancreatic endocrine genes coding for glucagon and transcription factors involved in α cell differentiation or glucagon gene expression 96 h after scramble (sc) or Pax6 siRNA (si) transfection. Data are expressed relative to β-actin mRNA values and are presented as the mean ± S.E. for at least six different experiments. D, glucagon content after specific Pax6 gene silencing. Histograms represent glucagon content in rat primary enriched α cells 96 h after scramble or Pax6 siRNA transfection. Glucagon contents are corrected by total protein amounts. Data are presented as the mean ± S.D. (error bars) for at least two different experiments. * indicates statistical significance with p < 0.05 value using a Student's t test and NS indicates no significant effect.
We then measured the level of expression of key genes involved in α cell differentiation and function by real time RT-PCR through a candidate gene approach. We analyzed the mRNA levels for Glucagon, Arx, Brain4, and MafB (which represent specific markers of differentiated α cells) as well as for Isl1, cMaf, NeuroD1/Beta2, Nkx2.2, Sox4, Foxa1, and Foxa2 (which are all involved in α cell development/differentiation and glucagon gene expression) and PC2 (prohormone convertase 2), which are responsible for proglucagon processing in pancreatic α cells.
As expected, the specific silencing of Pax6 in primary α cells led to a significant decrease of Glucagon and PC2 mRNA levels (Fig. 2C). Furthermore, partial knockdown of Pax6 was accompanied by a 50% decrease of glucagon content (Fig. 2D). This observation illustrates the crucial role exerted by Pax6 on glucagon biosynthesis in primary α cells (transcription and processing) as we previously proposed in cultured α cells (39). Among the different mRNA levels measured in primary α cells, only those coding for MafB, cMaf, and NeuroD1/Beta2 were decreased significantly after 96 h of Pax6 silencing (Fig. 2C), indicating that MafB, cMaf, and NeuroD1/Beta2 are potential Pax6 target genes. By contrast Arx, Brain4, Foxa1, Foxa2, Isl1, Nkx2.2, and Sox4 mRNA levels were unaffected by Pax6 knockdown. These observations strongly suggest that Pax6 specifically affects these three genes among the different genes coding for transcription factors previously shown to be involved in α cell differentiation and function. Taken together, our data indicate that Pax6 controls the mRNA levels of key genes in adult pancreatic α cells particularly involved in Glucagon gene transcription (MafB, cMaf, and NeuroD1/Beta2) or glucagon biosynthesis (PC2) in addition to its direct control on the glucagon gene.
Effects of Pax6 Silencing in Glucagon-producing Cell Lines
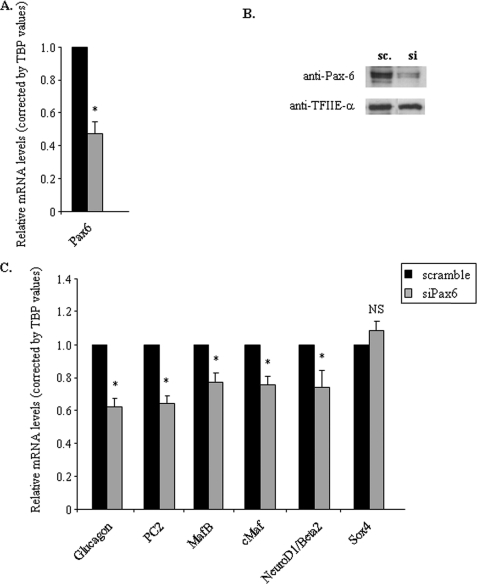
To confirm the effects of Pax6 on MafB, cMaf, and NeuroD1/Beta2 gene transcription, we generated partial knockdown of Pax6 in the glucagon-producing cell line αTC1.9. We thus performed experiments with specific anti-Pax6 or corresponding scramble siRNAs for 96 h in these cells. We obtained a 50% decrease of Pax6 content (Fig. 3, A and B). We next analyzed the mRNA levels of Glucagon, PC2, MafB, cMaf, and NeuroD1/Beta2 as well as the mRNAs that were unaffected in primary α cells. We confirmed that knockdown of Pax6 led to a significant and specific decrease of Glucagon, PC2, MafB, cMaf, and NeuroD1/Beta2 mRNA levels (Fig. 3C), whereas Sox4, Foxa2, Arx, Isl1, Brain4, and Nkx2.2 mRNA levels were unchanged (data not shown). Of note, the Foxa1 gene is not expressed in α-TC1.9 cells. The same results were also obtained in another α cell line, InR1G9, except for Isl1 mRNA levels, which were slightly but significantly decreased in these cells (data not shown).
FIGURE 3.
Effects of specific inhibition of the Pax6 gene by siRNA in αTC1.9 cells. Cells were transfected with 100 nm Pax6 siRNA (si) mixture (siPax6–614 and siPax6–1007) or scramble (sc) siRNA during 96 h. A, effects of Pax6 siRNA on Pax6 gene expression by real time RT-PCR. Data are corrected by TBP mRNA values. B, Western blot analyses of Pax6 protein content from transfected αTC1.9 cells with scramble or specific Pax6 siRNA after 96 h. TFIIE-α serves as a control for the specificity of siRNA effects. C, quantitative analyses of the expression of key pancreatic endocrine genes coding for glucagon and transcription factors involved in α cell differentiation or glucagon gene expression 96 h after scramble or Pax6 siRNA transfection. Only the regulated mRNA levels and a control mRNA (Sox4) are shown. Data are expressed relative to TBP mRNA values and are presented as the mean ± S.D. (error bars) for at least three different experiments. * indicates statistical significance with p < 0.05 value using a Student's t test and NS indicates no significant effect.
Identification of Functional Pax6 Binding Sites on MafB, cMaf, and NeuroD1/Beta2 Gene Promoter
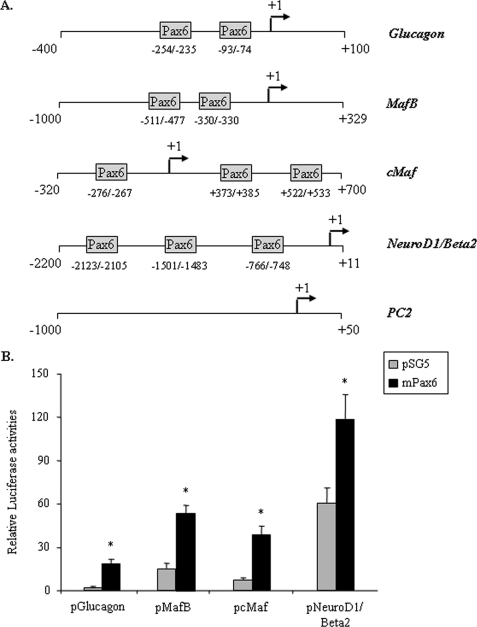
To further characterize the effects of Pax6 on MafB, cMaf, and NeuroD1/Beta2 gene expression, we searched for potential binding sites on their promoter regions. In silico analyses of the 5′-flanking region of the Glucagon, MafB, cMaf, and NeuroD1/Beta2 genes revealed at least one Pax6 putative binding site in each promoter (Fig. 4A). The PC2 gene promoter, which does not contain any Pax6 binding site was used as a control in ChIP experiments and the Glucagon promoter was used as a positive control in transactivation and binding experiments.
FIGURE 4.
Pax6 target gene promoter structure and effects of Pax6 overexpression on transcription in BHK-21 cells. A, schematic representation of the rat Glucagon and cMaf and mouse MafB, NeuroD1/Beta2, and PC2 gene promoters. Putative Pax6 binding sites are represented by boxes. B, the Glucagon, MafB, cMaf, and NeuroD1/Beta2 gene promoters were cotransfected in BHK-21 cells with pSG5 (gray bars) or mouse Pax6 cDNA (mPax6, black bars). Histograms represent normalized LUC activities of promoter constructs versus corresponding promoter-less LUC reporter genes 48 h after transfection. Data are presented as the mean ± S.D. (error bars) for three different transfection experiments. * indicates statistical significance with p < 0.05 using Student's t test and NS indicates no significant effect.
We first investigated the respective transcriptional effects of Pax6 on the MafB, cMaf, and NeuroD1/Beta2 genes. We performed transient transfections in BHK-21 cells with the mouse MafB and NeuroD1/Beta2 gene promoters as well as the rat cMaf and Glucagon gene promoters linked to the LUC reporter gene along with the expression plasmid containing the full-length mouse Pax6 (Fig. 4B). Pax6 overexpression led to a significant activation of the glucagon gene promoter as previously described (40, 41), as well as the promoters of the MafB, cMaf, and NeuroD1/Beta2 gene. These results suggest that Pax6 acts directly on the respective promoters to regulate gene expression.
To further analyze the respective binding sites of Pax6 on the MafB, cMaf, and NeuroD1/Beta2 gene promoters, we performed EMSA with probes containing each identified putative Pax6 binding sites (Fig. 5). DNA-proteins complexes were observed with extracts from BHK-21 cells overexpressing mPax6. To verify the nature of these complexes, an antibody raised against Pax6 was used and able to supershift the complexes, indicating that Pax6 can interact with each of these promoter regions. Cold wild-type and mutant oligonucleotides were also added at a 200-fold excess to further verify specificity; whereas wild-type Pax6-binding sites disrupted the complexes, mutant sites did not, indicating that Pax6 binds specifically the MafB (−357/−322 bp), cMaf (+362/+396 bp and +511/+545 bp), and NeuroD1/Beta2 (−2133/−2096 bp, −1510/−1471 bp, and −775/−741 bp) gene promoter regions. Other potential sites identified in the search of MafB (−511/−477 bp) and cMaf (−276/−267 bp) promoters were also studied in EMSA experiments but displayed no Pax6 binding (data not shown).
FIGURE 5.
Analyses of Pax6 binding on the MafB, cMaf, and NeuroD1/Beta2 gene promoters by electrophoretic mobility shift assays experiments. Representative gels for Pax6 binding on MafB, cMaf, and NeuroD1/Beta2 gene promoters. EMSA were performed with 5′ end-labeled MafB (−357/−322bp), cMaf (+362/+396 and +511/+545 bp), and NeuroD1/Beta2 (−2133/−2096, −1510/−1471, and −775/−741 bp) oligonucleotides (listed in supplemental Table S2) in the presence of 10 μg of nuclear extracts from BHK-21 cells (pSG5, negative control) or BHK-21 cells overexpressing mouse Pax6 (p46). An anti-Pax6 antibody was used to test the specificity of Pax6 binding as well as native and mutated cold probes in 200-fold excess. Arrows indicate specific shifts and supershifts for Pax6 proteins on probes. EMSA experiments were performed at least three times for each probe.
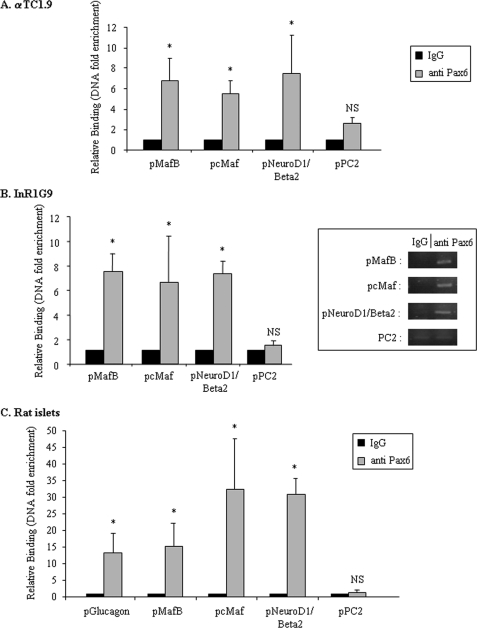
To determine in vivo binding of Pax6 on the promoter region of MafB, cMaf, and NeuroD1/Beta2, we performed ChIP assays on αTC1.9 and InR1G9 cell lines and rat islets cells (Fig. 6, A–C). Due to the limited number of α cells in islets preparations, we had to use total islets to assess Pax6 binding on target gene promoters, thus restricting the conclusions for the cMaf and NeuroD1/Beta2 promoters. We designed primers for PCR amplification corresponding to promoter regions including the Pax6 binding sites revealed by in silico analyses as well as EMSA experiments. We found that Pax6 binds the MafB, cMaf, and NeuroD1/Beta2 promoter regions in glucagon-producing cells as well as in rat islets confirming the involvement of Pax6 on the transactivation of these promoters. We also investigated Pax6 binding on the PC2 gene promoter and confirmed that the PC2 gene is not a direct target of Pax6 in rat islets as we previously reported in glucagon-producing cell lines (39). We also confirm that Pax6 binds the Glucagon gene promoter in rat islets (Fig. 6C). Taken together, our results indicate that Pax6 binds directly and activates the promoter region of MafB, cMaf, and NeuroD1/Beta2 genes in α cells.
FIGURE 6.
In vivo interactions of Pax6 on the MafB, cMaf, NeuroD1/Beta2, and PC2 gene promoters. Quantitative ChIP analyses in glucagon-producing cell lines (A, αTC1.9 and B, InR1G9) and rat islet cells (C). Histograms represent relative binding of Pax6 on the Glucagon (pGlucagon), MafB (pMafB), cMaf (pcMaf), NeuroD1/Beta2 (pNeuroD1/pBeta2), and PC2 (pPC2) gene promoters in different cell lines or islets. After cross-linking between chromatin and the interacting proteins, specific immunoprecipitations with anti-Pax6 antibody were performed as indicated under “Experimental Procedures.” Binding was analyzed by real time PCR with a Light-Cycler (Roche Diagnostics). Binding intensity data are expressed relative to IgG immunoprecipitation (nonspecific binding) and presented as the mean ± S.D. (error bars) for at least three independent experiments for cell lines and two independent experiments for rat islets cells. An anti-Histone H4 immunoprecipitation was also performed as a positive control for each promoter (data not shown). * indicates statistical significance with p < 0.05 value using a Student's t test and NS indicates no significant effect. Qualitative aspects of quantitative PCR are also presented in-frame with representative signals for IgG and anti-Pax6 interactions to target gene promoters in InR1G9 cells.
Generation and Characterization of Pax6-DN306 InR1G9 Stable Clones
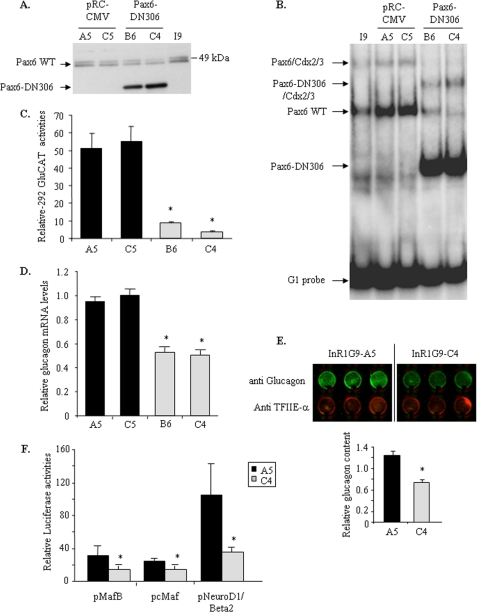
To further assess Pax6 involvement in transcriptional control of the MafB, cMaf, and NeuroD1/Beta2 genes, we generated InR1G9 stable clones expressing either a dominant-negative form of Pax6 (Pax6-DN306) or an empty expressing vector (pRC-CMV). We used a dominant-negative form of human Pax6 (Pax6-DN306) that had previously been shown to inhibit wild-type Pax6-induced transcription (35). We first verified Pax6-DN306 expression in different stable clones by Western blot analyses and selected two clones expressing either the empty vector (A5 and C5) or Pax6-DN306 (B6 and C4) (Fig. 7A). We also performed EMSA experiments with nuclear extracts from the four clones. Pax6-DN306 was able to bind G1, alone or in combination with Cdx-2/3 (Fig. 7B) as previously shown, and inhibited the binding of wild-type Pax6 (clones B6 and C4). The specificity of Pax6-DN306 binding on G1 was confirmed with anti-Pax6 antibodies (data not shown).
FIGURE 7.
Characterization of Pax6-DN306 InR1G9 stable clones. A, Pax6-DN306 expression in InR1G9 stable clones. Western blot analyses of nuclear extracts from native InR1G9, A5-C5 clones (pRC-CMV), and B6-C4 clones (Pax6-DN306). Wild-type Pax6 is visualized by two specific bands at around 49 kDa (corresponding to p46 and p48 isoforms) and Pax6-DN306 at ∼39 kDa. B, analyses of Pax6-DN306 binding to the G1 element of the rat glucagon gene promoter by EMSA experiments. 32P-Labeled G1 was incubated with native InR1G9 nuclear extract as well as nuclear extracts from A5-C5 and B6-C4 clones. Arrows indicate specific binding to the G1 element. C, analyses of −292 bp glucagon gene promoter activities in Pax6-DN306- and empty vector-InR1G9 clones. InR1G9-stable clones were transfected with the rat glucagon promoter (−292GluCAT) for 48 h. Data are expressed relative to the empty CAT reporter gene and presented as mean ± S.D. (error bars) for three different transfection experiments. D, quantitative analyses of glucagon mRNA levels in InR1G9-stable clones. Results were corrected by β-actin mRNA levels and are presented as the mean ± S.D. (error bars) for at least three independent experiments. * indicates statistical significance with p < 0.05 value using a Student's t test and NS indicates no significant effect. E, analyses of glucagon content in InR1G9-stable clones by the “In-cell Western blot” technique (described under “Experimental Procedures”). F, the rat cMaf and mouse MafB and NeuroD1/Beta2 gene promoters were transfected in InR1G9-stable clones. Histograms represent normalized LUC activities of promoter constructs versus corresponding promoter-less LUC reporter genes 48 h after transfection. Promoter activities were analyzed in A5 (black bars) and C4 (gray bars) InR1G9-stable clones after 48 h of transfection. Data are presented as the mean ± S.E. for three different transfection experiments. * indicates statistical significance with p < 0.05 using Student's t test and NS indicates no significant effect.
We then investigated the functional consequences of Pax6-DN306 expression in InR1G9 stable clones by assessing glucagon gene promoter activity. Transcriptional activities of −292GluCAT in the B6 and C4 stable clones were markedly lower compared with that observed in A5 and C5 stable clones (Fig. 7C). Moreover, quantification of Glucagon mRNA levels and protein contents in InR1G9 clones clearly confirmed that chronic expression of Pax6-DN306 decreased expression of the Glucagon gene (Fig. 7, D and E). These results indicate that Pax6-DN306 is able to inhibit Pax6 effects on the glucagon gene promoter acting as a dominant-negative protein by competition with the wild-type form.
We also investigated the potential effects of Pax6-DN306 overexpression on α cell proliferation and death. We found that constitutive expression of Pax6-DN306 did not affect cell proliferation and death rates of InR1G9 cells (supplemental Fig. S1).
Our results thus suggest that Pax6-DN306 affects glucagon gene expression by interfering with wild-type Pax6 transactivation allowing us to use these clones to investigate the functional role of Pax6 on the transcription of the newly identified target genes, MafB, cMaf, and NeuroD1/Beta2. We thus performed transient transfections of different promoter constructs in Pax6DN306 (C4) and empty clones (A5). The activities of each tested promoter construct were significantly lower in the Pax6-DN306 (C4) compared with the empty clone (A5) (Fig. 7F); similar results were obtained in the B6 compared with the C5 clones (data not shown). Our results thus show that disruption of Pax6 action in pancreatic α cells leads to a decrease of MafB, cMaf, and NeuroD1/Beta2 promoter activities, indicating that Pax6 is involved in the transcriptional control of these genes.
Mutations of Pax6 Binding Sites on the MafB, cMaf, and NeuroD1/Beta2 Gene Promoters Decreases Pax6 Transactivation
To further prove Pax6 effects on the identified target genes, we performed mutations of Pax6 binding sites on the MafB, cMaf, and NeuroD1/Beta2 gene promoters (Fig. 8). We verified the consequences of these mutations on Pax6 binding in EMSA experiments. An excess of the mutated probes was indeed unable to compete for Pax6 binding indicating that the mutated sites should be unable to significantly bind and thus transactivate the respective MafB, cMaf, and NeuroD1/Beta2 promoter constructs (Fig. 5). Mutations of the Pax6 binding sites located between −350 and −330 bp on the MafB gene promoter as well as between +373/+385 bp of the cMaf gene promoter and −2123/−2105 bp and −1501/−1483 bp of the NeuroD1/Beta2 gene promoter were all associated with decreased promoter activities (Fig. 8A).
FIGURE 8.
Effects of Pax6 binding site mutations on MafB, cMaf, and NeuroD1/Beta2 gene promoters. The rat cMaf and mouse MafB, NeuroD1/Beta2 gene promoters, and corresponding mutated promoter constructs were transfected in αTC1.9 cells. A, histograms represent normalized LUC activities of promoter constructs 48 h after transfection. Data are presented as the mean ± S.D. (error bars) for three different transfection experiments. * indicates statistical significance with p < 0.05 value using Student's t test and NS indicates no significant effect. B, schematic representation of MafB, cMaf, and NeuroD1/Beta2 gene promoters. Only Pax6 binding sites characterized by EMSA experiments are represented. Functional binding sites are represented by an oval sphere.
By contrast, mutations of the other Pax6 binding sites on the cMaf (+522/+533 bp) and the NeuroD1/Beta2 genes (−766/−748 bp) did not affect promoter activities. We also investigated the effects of mutations on multiple Pax6 sites of the cMaf and NeuroD1/Beta2 gene promoters but found no additional or synergistic effects (data not shown). These data indicate that Pax6 acts directly on one single site of the gene promoters of MafB and cMaf, and two of NeuroD1/Beta2 to control their activities in glucagon-producing cells (Fig. 8B).
DISCUSSION
α Cell differentiation and function result from a combinatorial network of transcription factors that remains poorly characterized although targeted disruption studies in mice have revealed the critical importance of Pax6, Arx, and Foxa2. More detailed studies on the transcriptional network of these three transcription factors and their inter-relationship have been prevented by the lack of an adequate system to study primary α cells mainly due to their low number and the difficulty to isolate them among endocrine cells.
Because Pax6 is crucial for endocrine cell development and function particularly for the differentiation of glucagon-producing cells, we investigated the transcriptional network of Pax6 in primary α cells. Several Pax6 target genes have been identified in the eye and the central nervous system (42–44); however, in the endocrine pancreas only Glucagon, Insulin, Somatostatin, Pdx1, PC2, and PC1/3 genes have been determined mostly through overexpression of Pax6 in transfection studies (39–41, 45, 46). Thus, our aim was to further define the Pax6 target genes involved in differentiation of the α cells and particularly in glucagon gene expression and glucagon biosynthesis to better understand the dramatic deleterious effects observed in the absence of Pax6 (3, 9).
We previously reported the critical role of Pax6 in transcription of the Proglucagon and PC2 genes in glucagon-producing cells (39, 40). We now report that Pax6 regulates directly the transcription of the MafB, cMaf, and NeuroD1/Beta2 genes in glucagon-producing cells and thus provide new evidence for the critical role of Pax6 in α cell differentiation and function.
Mice with a targeted disruption of Pax6 display a drastic decrease of all four types of differentiated endocrine cells without affecting endocrine cell mass, suggesting that Pax6 is required for endocrine cell differentiation (2, 3, 9). The finding that Pax6-deficient mice contain few or no glucagon-positive cells in the adult pancreas underlines the crucial role of Pax6 in α cell development (3). This role is not limited to the α cell because Pax6-deficient mice also present a strong reduction of insulin+ (70%), somatostatin+, and PP+ cells with a significant increase of ghrelin+ glucagon− ϵ-cell number (47). Pax6 deficiency is also accompanied by a strong decrease of the transcription factors MafB, Pdx1, GLUT2, and MafA proteins, which are implicated in the terminal differentiation process of endocrine cells and particularly β cells. By contrast, Pax6 deficiency does not affect the expression of Ngn3, Nkx2.2, and Nkx6.1 at embryonic day 15.5. Overall, these observations suggest that Pax6 plays a key role in determining the endocrine cell fate favoring α, β, and δ cells at the expense of ghrelin cells but is not required for initiation of endocrine cell differentiation (9, 47, 48).
To better understand how Pax6 specifically controls α cell differentiation and function, we investigated the consequences of Pax6 deficiency on the expression of genes coding for proteins known to play key roles in the differentiation of α cells and particularly in the expression of its main marker, glucagon. We show that in primary rat α cells Pax6 not only controls Proglucagon and PC2 gene transcription but also the MafB, cMaf, and NeuroD1/Beta2 genes. We found that Pax6 directly regulates MafB, cMaf, and NeuroD1/Beta2 genes through activation of their promoter regions. Although several Pax6 putative binding sites were identified in the promoter region of all three genes, we characterized only one functional Pax6 binding site on MafB and cMaf gene promoters and two in the NeuroD1/Beta2 by both ChIP and EMSA assays as well as site-directed mutagenesis. Indeed although three Pax6 binding sites were identified by EMSA for the NeuroD1/Beta2 gene, only the two distal elements −2123/−2105 and −1501/−1483 were found to be functional, at least for basal transcriptional activity. Furthermore, it is important to note that only the −1501/−1483 elements are perfectly conserved among species. Similarly, it has previously been reported that Pax6 activates cMaf gene expression in the eye through three potential Pax6 binding sites on its promoter (49); our results clearly show that only the +373/+385 proximal site is implicated in control of cMaf gene transcription, at least in α cells.
Overall, our results indicate that Pax6 directly regulates transcription of MafB, cMaf, and NeuroD1/Beta2 genes in glucagon-producing cells. It is relevant to indicate here that Pax6, glucagon, MafB, cMaf, and NeuroD1/Beta2 are expressed approximately at the same time during mouse pancreas development (embryonic kidney 9–9.5).
Our data indicate that Pax6 affects Glucagon gene expression directly and indirectly through the regulation of MafB, cMaf, and NeuroD1/Beta2, which have all been implicated by gene transfection or knockout studies to affect Glucagon gene transcription (15, 24, 25). We indeed previously showed that Pax6 is crucial for glucagon gene transcription through its binding to the G1 and G3 elements (12, 40); more recently we reported that cMaf and Pax6 act synergistically on G1 to activate glucagon gene expression in InR1G9 cells (15).
MafB was also proposed to activate the glucagon gene (25), and we previously showed by ChIP analyses that MafB and cMaf interact with the glucagon gene promoter in InR1G9 and αTC1.9 cells and that Pax6 can associate directly with MafB and cMaf independently (15). Furthermore, MafB and cMaf can form heterodimers (50). Interestingly, it was reported that MafB and cMaf can control their own gene expression (49, 51). Overall, these data indicate that Pax6 directly activates Glucagon gene expression in association with MafB and cMaf and indirectly through the direct control of MafB and cMaf gene expression in association with their encoded protein. The indirect effects of Pax6 on glucagon gene expression may also involve NeuroD1/Beta2, which has also been reported to activate the glucagon gene in combination with E47 transcription factor through the G4 element (24). Overall, the previously reported results and our present data indicate that Pax6 is critical for Glucagon gene transcription through direct and indirect mechanisms involving MafB, cMaf, and NeuroD1/Beta2 proteins.
Pax6, however, plays a much broader role than just the activation of Glucagon gene transcription in α cells. Indeed MafB, cMaf as well as NeuroD1/Beta2 are all involved in α cell development and differentiation (17, 18, 20). Functionally, MafB- or cMaf-deficient mice show an important reduction in both insulin- and glucagon-positive cells throughout development indicating a significant role of MafB and cMaf in differentiation, replication, and/or survival of endocrine precursor cells (17, 20).
Interestingly MafB deficiency does not affect endocrine specification but the lineage commitment of the endocrine cells and their maturation (48). Nishimura et al. (48) has compared MafB and Pax6 deficiency on the development of glucagon- and insulin-positive cells. They showed that, similar to Pax6-deficient mice, MafB-deficient mice displayed reductions both in insulin and glucagon expressing cells. Furthermore, Pax6-deficient mice had a strong reduction in the number of MafB-expressing cells, whereas MafB-deficient mice exhibited no effect on Pax6 expression suggesting that MafB may function as a downstream mediator of Pax6 in regulating the specification of insulin- and glucagon-expressing cells, in agreement with our present results.
cMaf is also involved in endocrine cell differentiation inasmuch as knock-out mice are characterized by a reduction of α (55%) and β cells (65%) (20). However, the transcriptional network upstream and downstream of cMaf in pancreatic endocrine cells has not been characterized. Our results show that Pax6 is an activator of cMaf.
NeuroD1/Beta2 knock-out mice present mature endocrine precursor cells with a reduction of insulin+ (74%) and glucagon+ (39%) cell number. Moreover, these mice are characterized by an abnormal islets structure and excessive programmed cell death. NeuroD1/Beta2 thus appears as a critical factor for endocrine cell maturation but is not required for initial islet cell formation (19).
Thus, our results suggest that Pax6, through the control of many factors, among them MafB, cMaf, and NeuroD1/Beta2, regulates the differentiation of α cells and particularly glucagon biosynthesis through the direct and indirect activation of its gene and through processing of its precursor, proglucagon. Indeed, we previously reported that Pax6 controls indirectly PC2 gene expression through cMaf and NeuroD1/Beta2 (39), illustrating again the major and multiple roles that Pax6, MafB, cMaf, and NeuroD1/Beta2 exert in glucagon biosynthesis.
Of importance, our data suggest that Pax6 deficiency does not affect Arx, Nkx2.2, Foxa1/Foxa2, and Isl1 mRNA levels, which are also involved in α cell development (4, 5, 16, 21, 22). Arx and Foxa2 knockout models present, respectively, an absence or a drastic decrease of glucagon-positive cells without alterations of endocrine progenitor cell numbers indicating their critical role in the acquisition of α cell fate by endocrine progenitors (4, 5). Furthermore, Arx and Foxa2 deficiencies do not affect the number of Pax6-positive cells. Therefore, our results and these observations strongly suggest a crucial role of Pax6 in α cell development through Arx- and Foxa2-independent pathways.
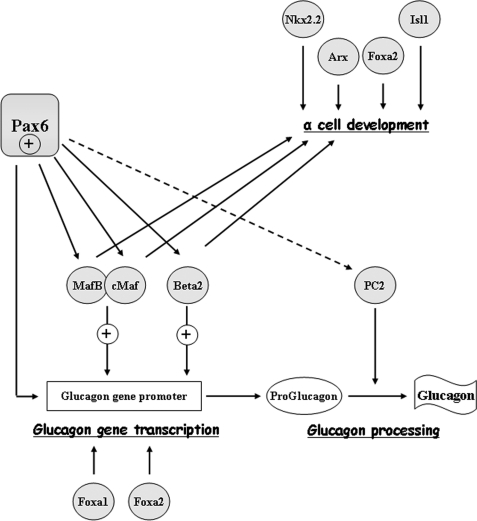
In conclusion, we have demonstrated that Pax6 acts as a crucial transcription factor in α cell differentiation and function through the control of glucagon biosynthesis. We propose that Pax6 activates directly and indirectly Proglucagon gene transcription through the control of MafB/cMaf and NeuroD1/Beta2 (Fig. 9).
FIGURE 9.
Schematic representation of Pax6 effects on α cell development and glucagon biosynthesis. Pax6 involvement on α cell differentiation, glucagon gene transcription, and processing is represented by black arrows for direct and the dotted arrow for indirect Pax6 effects.
Supplementary Material
Acknowledgments
We thank Prof. P. Halban and Dr. K. Bouzakri (Department of Genetic Medicine and Development, University of Geneva Medical School) for helpful advice and knowledge to develop primary rat islet models in our laboratory as well as for the generous gift of laminin-804G clone. We also thank Dr. S. Singh (Department of Biochemistry and Molecular Biology, The University of Texas M. D. Anderson Cancer Center, Houston, TX) for generously providing the dominant-negative form of human Pax6 (Pax6-DN306). We also thank Dr. S. Saule (CNRS/UMR 146, Institut Curie Section de Recherche, France) for generously providing Pax6 antibody and Dr. M. Sakai (Department of Biochemistry and Department of Ophthalmology, Hokkaido University School of Medicine, Japan) and Dr. M. J. Tsai (Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX) for the generous gift, respectively, of the rat cMaf and mouse MafB promoters and the mouse NeuroD1/Beta2 promoter constructs. We thank Dr. Richard James for critically reading the manuscript.
This work was supported by the Swiss National Science Foundation, Novo-Nordisk, Switzerland, and the Biology and Cancer Research Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S3.
- BHK
- baby hamster kidney
- CAT
- chloramphenicol acetyltransferase
- LUC
- luciferase
- CMV
- cytomegalovirus.
REFERENCES
- 1.Dunning B. E., Gerich J. E. (2007) Endocr. Rev. 28, 253–283 [DOI] [PubMed] [Google Scholar]
- 2.Sander M., Neubüser A., Kalamaras J., Ee H. C., Martin G. R., German M. S. (1997) Genes Dev. 11, 1662–1673 [DOI] [PubMed] [Google Scholar]
- 3.St-Onge L., Sosa-Pineda B., Chowdhury K., Mansouri A., Gruss P. (1997) Nature 387, 406–409 [DOI] [PubMed] [Google Scholar]
- 4.Collombat P., Mansouri A., Hecksher-Sorensen J., Serup P., Krull J., Gradwohl G., Gruss P. (2003) Genes Dev. 17, 2591–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C. S., Sund N. J., Behr R., Herrera P. L., Kaestner K. H. (2005) Dev. Biol. 278, 484–495 [DOI] [PubMed] [Google Scholar]
- 6.Callaerts P., Halder G., Gehring W. J. (1997) Annu. Rev. Neurosci. 20, 483–532 [DOI] [PubMed] [Google Scholar]
- 7.Walther C., Gruss P. (1991) Development 113, 1435–1449 [DOI] [PubMed] [Google Scholar]
- 8.Turque N., Plaza S., Radvanyi F., Carriere C., Saule S. (1994) Mol. Endocrinol. 8, 929–938 [DOI] [PubMed] [Google Scholar]
- 9.Ashery-Padan R., Zhou X., Marquardt T., Herrera P., Toube L., Berry A., Gruss P. (2004) Dev. Biol. 269, 479–488 [DOI] [PubMed] [Google Scholar]
- 10.Yasuda T., Kajimoto Y., Fujitani Y., Watada H., Yamamoto S., Watarai T., Umayahara Y., Matsuhisa M., Gorogawa S., Kuwayama Y., Tano Y., Yamasaki Y., Hori M. (2002) Diabetes 51, 224–230 [DOI] [PubMed] [Google Scholar]
- 11.Nishi M., Sasahara M., Shono T., Saika S., Yamamoto Y., Ohkawa K., Furuta H., Nakao T., Sasaki H., Nanjo K. (2005) Diabetic Med. 22, 641–644 [DOI] [PubMed] [Google Scholar]
- 12.Philippe J., Morel C., Cordier-Bussat M. (1995) J. Biol. Chem. 270, 3039–3045 [DOI] [PubMed] [Google Scholar]
- 13.Jin T., Drucker D. J. (1996) Mol. Cell. Biol. 16, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planque N., Leconte L., Coquelle F. M., Benkhelifa S., Martin P., Felder-Schmittbuhl M. P., Saule S. (2001) J. Biol. Chem. 276, 35751–35760 [DOI] [PubMed] [Google Scholar]
- 15.Gosmain Y., Avril I., Mamin A., Philippe J. (2007) J. Biol. Chem. 282, 35024–35034 [DOI] [PubMed] [Google Scholar]
- 16.Ahlgren U., Pfaff S. L., Jessell T. M., Edlund T., Edlund H. (1997) Nature 385, 257–260 [DOI] [PubMed] [Google Scholar]
- 17.Artner I., Blanchi B., Raum J. C., Guo M., Kaneko T., Cordes S., Sieweke M., Stein R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H. P., Chu K., Nemoz-Gaillard E., Elberg D., Tsai M. J. (2002) Mol. Endocrinol. 16, 541–551 [DOI] [PubMed] [Google Scholar]
- 19.Naya F. J., Huang H. P., Qiu Y., Mutoh H., DeMayo F. J., Leiter A. B., Tsai M. J. (1997) Genes Dev. 11, 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura W., Kondo T., Khattabi I., Dodge R., Ho I., Weir S., Sharma A. (2006) Diabetes: A Journal of the American Diabetes Association 55, Suppl. 1, A362 [Google Scholar]
- 21.Sussel L., Kalamaras J., Hartigan-O'Connor D. J., Meneses J. J., Pedersen R. A., Rubenstein J. L., German M. S. (1998) Development 125, 2213–2221 [DOI] [PubMed] [Google Scholar]
- 22.Shih D. Q., Navas M. A., Kuwajima S., Duncan S. A., Stoffel M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10152–10157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson M. E., Yang K. Y., Kalousova A., Lau J., Kosaka Y., Lynn F. C., Wang J., Mrejen C., Episkopou V., Clevers H. C., German M. S. (2005) Diabetes 54, 3402–3409 [DOI] [PubMed] [Google Scholar]
- 24.Dumonteil E., Laser B., Constant I., Philippe J. (1998) J. Biol. Chem. 273, 19945–19954 [DOI] [PubMed] [Google Scholar]
- 25.Artner I., Le Lay J., Hang Y., Elghazi L., Schisler J. C., Henderson E., Sosa-Pineda B., Stein R. (2006) Diabetes 55, 297–304 [DOI] [PubMed] [Google Scholar]
- 26.Gauthier B. R., Schwitzgebel V. M., Zaiko M., Mamin A., Ritz-Laser B., Philippe J. (2002) Mol. Endocrinol. 16, 170–183 [DOI] [PubMed] [Google Scholar]
- 27.Wang M., Drucker D. J. (1995) J. Biol. Chem. 270, 12646–12652 [DOI] [PubMed] [Google Scholar]
- 28.Sutton R., Peters M., McShane P., Gray D. W., Morris P. J. (1986) Transplantation 42, 689–691 [DOI] [PubMed] [Google Scholar]
- 29.Rouiller D. G., Cirulli V., Halban P. A. (1990) Exp. Cell Res. 191, 305–312 [DOI] [PubMed] [Google Scholar]
- 30.Van De Winkel M., Pipeleers D. (1983) Biochem. Biophys. Res. Commun. 114, 835–842 [DOI] [PubMed] [Google Scholar]
- 31.Meyer K., Irminger J. C., Moss L. G., de Vargas L. M., Oberholzer J., Bosco D., Morel P., Halban P. A. (1998) Diabetes 47, 1974–1977 [DOI] [PubMed] [Google Scholar]
- 32.Parnaud G., Hammar E., Ribaux P., Donath M. Y., Berney T., Halban P. A. (2009) Mol. Endocrinol. 23, 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takaki R., Ono J., Nakamura M., Yokogawa Y., Kumae S., Hiraoka T., Yamaguchi K., Hamaguchi K., Uchida S. (1986) In Vitro Cell Dev. Biol. 22, 120–126 [DOI] [PubMed] [Google Scholar]
- 34.Philippe J., Drucker D. J., Knepel W., Jepeal L., Misulovin Z., Habener J. F. (1988) Mol. Cell. Biol. 8, 4877–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S., Tang H. K., Lee J. Y., Saunders G. F. (1998) J. Biol. Chem. 273, 21531–21541 [DOI] [PubMed] [Google Scholar]
- 36.Kumar V., Chambon P. (1988) Cell 55, 145–156 [DOI] [PubMed] [Google Scholar]
- 37.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1988) EMBO J. 7, 4221–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordier-Bussat M., Morel C., Philippe J. (1995) Mol. Cell. Biol. 15, 3904–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katz L. S., Gosmain Y., Marthinet E., Philippe J. (2009) Mol. Cell. Biol. 29, 2322–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritz-Laser B., Estreicher A., Klages N., Saule S., Philippe J. (1999) J. Biol. Chem. 274, 4124–4132 [DOI] [PubMed] [Google Scholar]
- 41.Hussain M. A., Habener J. F. (1999) J. Biol. Chem. 274, 28950–28957 [DOI] [PubMed] [Google Scholar]
- 42.Bernier G., Vukovich W., Neidhardt L., Herrmann B. G., Gruss P. (2001) Development 128, 3987–3994 [DOI] [PubMed] [Google Scholar]
- 43.Chauhan B. K., Reed N. A., Zhang W., Duncan M. K., Kilimann M. W., Cvekl A. (2002) J. Biol. Chem. 277, 11539–11548 [DOI] [PubMed] [Google Scholar]
- 44.Yamada R., Mizutani-Koseki Y., Hasegawa T., Osumi N., Koseki H., Takahashi N. (2003) Development 130, 1759–1770 [DOI] [PubMed] [Google Scholar]
- 45.Samaras S. E., Cissell M. A., Gerrish K., Wright C. V., Gannon M., Stein R. (2002) Mol. Cell. Biol. 22, 4702–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen J. H., Chen Y. Y., Song S. J., Ding J., Gao Y., Hu Q. K., Feng R. P., Liu Y. Z., Ren G. C., Zhang C. Y., Hong T. P., Gao X., Li L. S. (2009) Diabetologia 52, 504–513 [DOI] [PubMed] [Google Scholar]
- 47.Heller R. S., Jenny M., Collombat P., Mansouri A., Tomasetto C., Madsen O. D., Mellitzer G., Gradwohl G., Serup P. (2005) Dev. Biol. 286, 217–224 [DOI] [PubMed] [Google Scholar]
- 48.Nishimura W., Rowan S., Salameh T., Maas R. L., Bonner-Weir S., Sell S. M., Sharma A. (2008) Dev. Biol. 314, 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakai M., Serria M. S., Ikeda H., Yoshida K., Imaki J., Nishi S. (2001) Nucleic Acids Res. 29, 1228–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kataoka K., Fujiwara K. T., Noda M., Nishizawa M. (1994) Mol. Cell. Biol. 14, 7581–7591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang K. (1999) Hokkaido Igaku Zasshi 74, 419–430 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.