Abstract
Tumor suppressor p53 plays the central role in regulating apoptosis in response to genotoxic stress. From an evolutionary perspective, the activity of p53 has to be backed up by other protein(s) in case of any functional impairment of this protein, to trigger DNA damage-induced apoptosis in cancer cells. We adopted multiple experimental approaches to demonstrate that in p53-impaired cancer cells, DNA damage caused accumulation of p53 paralogue p73 via Chk-1 that strongly impacted Bax expression and p53-independent apoptosis. On the contrary, when p53 function was restored by ectopic expression, Chk-2 induced p53 accumulation that in turn overshadowed p73 activity, suggesting an antagonistic interaction between p53 family members. To understand such interaction better, p53-expressing cells were impaired differentially for p53 activity. In wild-type p53-expressing cancer cells that were silenced for p53 for several generations, p73 was activated, whereas no such trend was observed when p53 was transiently silenced. Prolonged p53 interference, even in functional p53 settings, therefore, leads to the “gain of cellular adaptation” in a way that alters the cellular microenvironment in favor of p73 activation by altering p73-regulatory proteins, e.g. Chk1 activation and dominant negative p73 down-regulation. These findings not only unveil a hitherto unexplained mechanism underlying the functional switchover from p53 to p73, but also validate p73 as a promising and potential target for cancer therapy in the absence of functional p53.
Keywords: Apoptosis, Cancer Therapy, Leukemia, Signal Transduction, Tumor Suppressor, Acute Myeloid Leukemia
Introduction
The transcription factor p53 responds to diverse stresses to regulate many target genes that induce cell cycle arrest, apoptosis, and DNA repair (1, 2). Moreover, during cancer treatment, genotoxic drugs generally target p53 to exert apoptogenic effects (3). However, mutation or deletion of p53 lies at the heart of the events leading to cancer (4). In fact, mutation in p53 gene or deletion of p53 allele that compromises p53 function occurs in 50% of all human cancers (5), and the alteration of regulators of p53 occurs in many of the remainder (6). Nonetheless, the alternative signaling mechanism regulating apoptotic processes in functional p53-deficient cancer cells is still the Cinderella of investigation.
Until 1997, scientists working on p53 assumed that this gene was unique. Therefore, the discovery of p53-related genes, p73 and p63, was both challenging and confounding (7). Both genes encode proteins with transactivation, DNA binding, and tetramerization domains, and they share considerable homology with p53 (8). A role for p73 in DNA damage-induced apoptosis was recently discovered in cell-based and knock-out mice studies (9, 10). p73 functions in a manner analogous to p53 by inducing tumor cell apoptosis and participating in cell cycle checkpoint control through trans-activation of an overlapping set of p53/p73 target genes (11). Hence, the idea is that certain cellular response, previously assumed to be “p53-independent,” might be attributed to this particular relative of p53. Interestingly, in sharp contrast to p53, p73 is expressed as two N-terminally distinct isoforms: transcriptionally active TAp732 or p73 and transcriptionally inactive ΔNp73 forms (12). ΔNp73 has oncogenic potential, acts in a dominant negative manner against TAp73, and is very much related to tumor development (13). However, although the role of p53 in apoptotic responses is well established (14), there is a need to seek out the mechanism underlying the functional switchover from p53 to p73 in relation to the execution of genotoxic drug-induced death of cancer cells with differential p53 status.
Innumerable types of assaults can cause DNA damage, and multiple signals might be triggered inducing apoptosis. Although many signaling pathways are poorly understood, there has been significant advancement in elucidating pathways involving the checkpoint kinases (Chk-1/Chk-2) (15). Both kinases have important and differential roles, especially in the coordination of checkpoint and repair control, but the potential apoptotic correlations remain to be fully elucidated. Some published data show that under certain circumstances, a full complement of both Chk-1 and Chk-2 is essential, implying not only functional commonality but also synergism (16). Other reports indicate that Chk-1 alone can activate p73 to induce apoptosis (17). Many interesting questions surface from these results and await future investigation. Therefore, it is of immense interest to determine the specific functions of Chks in p53 to p73 switchover.
We introduce “binary switches” as new concepts in p53 biology, elucidating mechanisms that might govern either p53 or p73 to induce apoptosis in cancer cells depending on distinct modification patterns of p53/p73-regulatory proteins. Specifically, our study provides missing models for the apoptosis of cancer cells with nonfunctional p53. We provide substantial evidence that p53 paralogue p73 can induce Bax in absence of functional p53; however, when p53 is present in its functional form it overpowers p73. To understand better the antagonistic interactions between the p53 family members, we silenced p53-expressing cancer cells differentially for p53 expression by genetic engineering and noticed that unlike transient p53 silencing, prolonged p53 silencing makes the cellular milieu favorable for p73 activation. In addition, using curcumin, a member of the flavonoid class of phytochemicals (18, 19) that acts as a genotoxic stress-inducing agent to induce apoptosis (20, 21), we searched for the signaling mechanisms regulating the functional switchover from p53 to p73 in p53-impaired cancer cells. Such progress in understanding the molecular changes that underlie cancer biology offers the prospect of specifically targeting malfunctioning molecules and pathways to achieve more effective and rational cancer therapy.
EXPERIMENTAL PROCEDURES
Cell Culture
The acute myelogenous leukemia cells HL-60, U937, KG1, breast cancer cells MCF-7/MDAMB-231, fibrocircoma cells HT-1080, colon cancer cells Colo-205/HT-29, and Chk2−/− mutant p53-expressing (SW-620)/Chk2−/− wild-type p53-expressing (HCT-15) cells were obtained from National Centre for Cell Science, India. Human peripheral blood monocytes as well as patients' acute myeloid leukemia cells were purified from total leukocytes by cell sorting (FACS Aria; BD Biosciences) using FITC-CD14/-CD33 antibodies. Cells were routinely maintained in complete DMEM/RPMI 1640 medium at 37 °C in a humidified incubator containing 5% CO2. Viable cell numbers were determined by the Trypan blue exclusion test.
Flow Cytometry
For the determination of cell death, cells were stained with 7AAD and Annexin-V-FITC and analyzed on flow cytometer (FACS Calibur, BD Biosciences). Electronic compensation of the instrument was done to exclude overlapping of the emission spectra. A total of 10,000 events were acquired for analysis using CellQuest software. For the determination of cell cycle phase distribution of nuclear DNA Cycle TEST PLUS DNA reagent kit (BD Biosciences) was used. Histogram display of DNA content (x axis, PI-fluorescence) versus counts has been displayed. CellQuest statistics was employed to quantitate the data at different phases of the cell cycle (15, 16). For the assessment of mitochondrial transmembrane potential, cells were incubated with curcumin for 24 h and then for an additional 15 min at 37 °C in the dark with 40 nm DiOC6. The cells were analyzed flow cytometrically for DiOC6 fluorescence.
Co-immunoprecipitation and Immunoblotting
Cells were lysed in buffer (10 mm Hepes, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, and 0.5 mm DTT) and spun at 105,000 × g to get a cytosolic fraction. For whole cell lysates, cells were homogenized in buffer (20 mm Hepes, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm Na-EDTA, 1 mm Na-EGTA, and 1 mm DTT). All buffers were supplemented with protease and phosphatase inhibitor mixtures (15, 16). For direct Western blot analysis, the cell lysates or the particular fractions were separated by SDS-PAGE, transferred to nitrocellulose membrane, and visualized by chemiluminescence. For the determination of direct interaction between two proteins, the co-immunoprecipitation technique was employed. The immunopurified proteins were then detected by Western blot using specific antibody (Santa Cruz Biotechnology). Equal protein loading was confirmed with α-actin/MnSOD antibody (Santa Cruz Biotechnology).
RT-PCR Assay
Two μg of the total RNAs extracted from cells was reverse transcribed and then subjected to PCR. The cDNAs were amplified with primers specific for Bax (5′-TTTGCTTCAGGGTTTCATCC-3′/5′-CAGTTGAAGTTGCCGTCAGA-3′), p53 (5′-GGCCCACTTCACCGTACTAA-3′/5′-GTGGTTTCAAGGCCAGATGT-3′), p73 (5′-CAGACAGCACCTACTTCGACCTT-3′/5′-CCGCCCACCACCTCATTA-3′), ΔNp73 (5′-TTCAGCCAGTTGACAGAACTAAGG-3′/5′-GCGTTTGTTGGCATTT-3′), and GAPDH (internal control: 5′-CAGAACATCATCCCTGCCTCT-3′/5′-GCTTGACAAAGTGGTCGTTGAG-3′).
Plasmids, siRNA, and Transfections
Wild-type (WT) p53-HL-60 was derived from cells through an adenoviral vector expression system expressing WT p53 under the control of the cytomegalovirus promoter (Clontech). Plasmid or the control vector was introduced separately into each cell using Lipofectamine-2000 (Invitrogen). Stably expressing clones were isolated by limiting dilution and selection with G418 sulfate (1 μg/ml; Cellgro), and cells surviving this treatment were cloned and screened by immunofluorescence and Western blotting with specific antibodies. For double transfection experiments, ΔNp53 (in Prk5 vector) was introduced into WT p53-HL-60 cells, and stably expressing clones were isolated by limiting dilution and selection with hygromycin B (800 μg/ml). Cells surviving this treatment were cloned and screened by immunofluorescence and Western blotting. Cells were transfected with 300 pmol of p73-/p53-/Bax-/Chk-1-/Chk-2-/control-ds-siRNA (Santa Cruz Biotechnology) and Lipofectamine-2000 separately for 12 h. The mRNA and protein levels were determined by RT-PCR and Western blotting.
Statistical Analyses
Values are shown as standard error (S.E.). Data were analyzed, and, when appropriate, significance of the differences between mean values was determined by Student's t test. Results were considered significant at p < 0.05.
RESULTS
Genotoxic Stress Kills Functional p53-deficient Cancer Cells through the Activation of Intrinsic Apoptotic Pathway
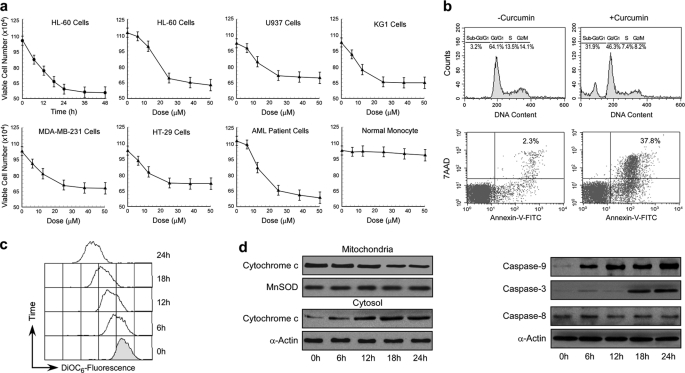
A battery of p53-null/-mutated human cancer cells from various origins such as acute myeloid leukemia (HL-60/U937/KG1), breast cancer (MDA-MB-231), and colon cancer (HT-29) was selected to study the effect of genotoxic drug, curcumin. Curcumin treatment resulted loss in these p53-null/-mutated cancer cell viability without showing any toxicity to peripheral circulatory monocytes (Fig. 1a), indicating that the cytotoxic effect is specific for malignant cells. Augmentation of sub-G0/G1 phase and Annexin-V/7AAD-positive cell populations by this genotoxic drug indicated an apoptotic mode of cell death (Fig. 1b). This effect is not selective to genotoxic drug such as curcumin because genotoxic stress like ionization radiation also furnished similar effects (Fig. 1a).
FIGURE 1.
Genotoxic stress kills functional p53-deficient cancer cells through the activation of intrinsic apoptotic pathway. a, functional p53-deficient cancer cells, AML patient cells, and peripheral monocytes were treated with a dose range of curcumin, and viable cell numbers were scored. In a parallel experiment ionizing radiation was used as a positive control for such genotoxic stress. b, curcumin (25 μm)-treated HL-60 cell cycle phase distribution of nuclear DNA was determined flow cytometrically (upper panel). 7AAD/Annexin-V-FITC-positive HL-60 cells (regarded as apoptotic cells) were analyzed flow cytometrically (lower panel). c, curcumin-treated HL-60 cells were flow cytometrically assessed for MTP loss by decreasing DiOC6 fluorescence. d, curcumin-treated HL-60 cells were analyzed by Western blotting for cytochrome c release (left panel) and caspase-9/-3/-8 activation (right panel). α-Actin/MnSOD was used as internal loading control. Values are mean ± S.E. (error bars) of five independent experiments in each case or representative of typical experiment.
Further study demonstrating genotoxic drug-induced mitochondrial transmembrane potential loss and cytochrome c release (Fig. 1, c and d) indicated the involvement of the mitochondria-mediated apoptotic pathway in these p53-impaired cancer cells. Existence of the intrinsic apoptosis pathway was further confirmed by examining the activation of caspase-9/-3 but not caspase-8 in these cells as was evident from the Western blot analysis with antibodies of active caspases (Fig. 1d). These results together suggested that even in absence of functional p53, curcumin induces cancer cell apoptosis by activation of the intrinsic apoptotic pathway.
p53-independent Bax Expression Plays Important Role in DNA Damage-induced Apoptosis
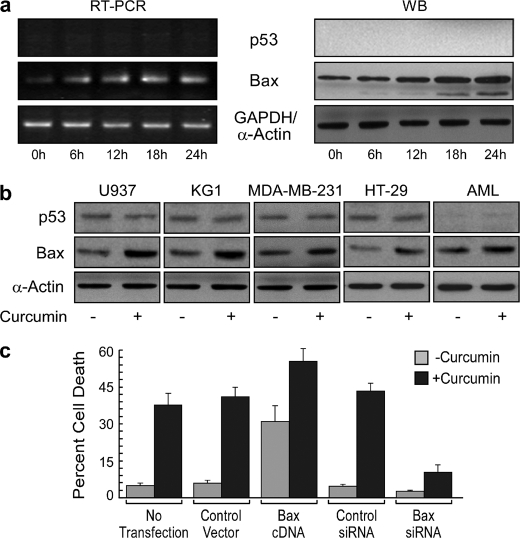
It is well reported that in HL-60/U937/KG1/MDA-MB-231/HT-29 cancer cells p53 is either absent or has lost the ability to activate its target gene Bax (22). Western blots and RT-PCR analysis reconfirmed that p53 was absent/low in these cells and also in AML patients and remained unchanged after genotoxic stress (Fig. 2, a and b). Interestingly, this genotoxic stress brought about a significant increase in Bax expression both at mRNA as well as at protein levels in these functional p53-deficient cells (Fig. 2, a and b). To confirm the involvement of Bax in DNA damage-induced apoptosis, we extraneously modulated its expression at the mRNA level. Introduction of Bax gene into HL-60 cells rendered these engineered cells susceptible to death that could be further augmented by curcumin. On the other hand, unlike control siRNA-transfected cells, Bax siRNA-transfected cells were only minimally affected, with an average of only 10–12% of the transfected cells succumbing to DNA damage-induced apoptotic death (Fig. 2c). These results establish the contribution of Bax in genotoxic stress-induced apoptosis in a p53-independent manner.
FIGURE 2.
p53-independent Bax expression plays an important role in DNA damage-induced apoptosis. a, HL-60 cells were treated with 25 μm curcumin, and p53/Bax was determined at the mRNA level by RT-PCR (left panel) and at the protein level by Western blotting (WB; middle panel). Ionizing radiation (UV) was used as positive a control for genotoxic stress. Wild-type p53-expressing HT-1080 cells were used as a positive control (right panel). b, lysates of untreated or curcumin-treated U937/KG1/MDA-MB-231/HT-29/patients' AML cells were subjected to Western blot analysis for the determination of changes in p53 and Bax expression. c, untransfected, control vector-/Bax-transfected and control/Bax siRNA-transfected HL-60 cells were incubated with or without curcumin for 24 h and were scored for percent apoptosis by Annexin-V-/7AAD positivity. α-Actin and GAPDH were used as internal controls. Values are mean ± S.E. (error bars) of five independent experiments in each case or representative of typical experiment.
p73 Activation Is Crucial for Apoptosis in Functional p53-deficient Cancer Cells
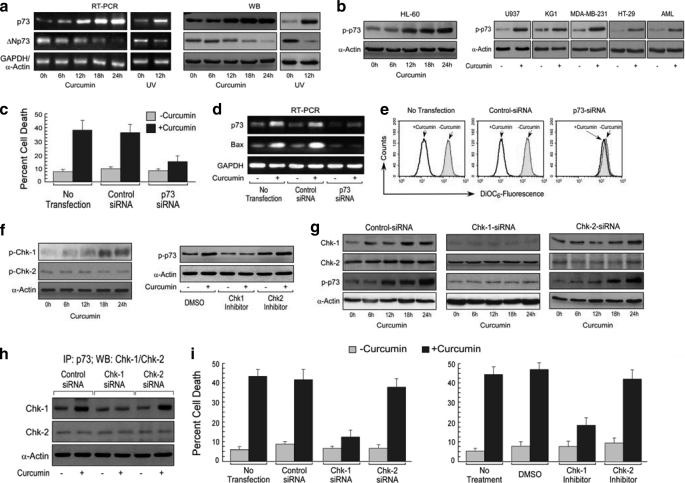
Involvement of Bax in functional p53-deficient cell death prompted us to examine whether p73, a paralogue of p53, is involved in Bax transactivation and apoptosis. Interestingly, we observed that genotoxic stress augmented p73 both at mRNA and protein levels in p53-null HL-60 cells (Fig. 3a) and simultaneously induced p73 Ser phosphorylation (23) in all of the p53-null/-mutated cells (Fig. 3b). Because, ΔNp73 is dominant negative toward p73 (24), in the second approach we investigated the influence of curcumin on ΔNp73. Our data revealed that curcumin efficiently blocked ΔNp73 expression in these cells (Fig. 3a), indicating that during functional impairment of p53, the apoptotic role is conferred probably by disabling ΔNp73 expression and thus increasing the activity of p73. To validate this hypothesis, endogenous p73 was knocked down with siRNA and was exposed to curcumin. Multiple experiments showed that p73 siRNA could efficiently prevent curcumin-induced apoptosis (Fig. 3c), implicating the crucial role of p73 in the induction of apoptotic signaling in functional p53-impaired cells.
FIGURE 3.
Chk-1-mediated p73 activation is crucial for apoptosis in functional p53-deficient cancer cells. a, expression of p73 and ΔNp73 at mRNA (left panel) and protein (right panel) levels was determined by RT-PCR and Western blotting (WB) from curcumin-treated HL-60 cells. Ionizing radiation (UV) was used as a positive control for genotoxic stress. b, phosphorylation (Ser) status of p73 in curcumin-exposed HL-60 cells (left panel) and U937/THP1/KG1/MCF-7/HT-29 cells/patients' AML cells (right panels) was detected by Western blot analysis. Untransfected and control/p73 siRNA-transfected HL-60 cells were treated with or without curcumin for 24 h. c, cells were then flow cytometrically assayed for percent apoptosis. d, expressions of p73 and Bax were determined at the mRNA level. e, in parallel, MTP loss was measured flow cytometrically. f, left panel, HL-60 cells were treated with curcumin, and cell lysates were Western blotted for the determination of phosphorylation status of Chk-1 and Chk-2. Right panel, control vector (DMSO)/Chk-1 inhibitor (PF-477736; 0.5 nm)/Chk-2 inhibitor (Chk-2 inhibitor ii; 10 nm)-pretreated HL-60 cells were treated with curcumin, and cell lysates were Western blotted for the determination of phosphorylation status of p73. g, HL-60 cells were transfected with control/Chk-1/Chk-2 siRNA, and expression level of Chk-1/Chk-2 and phospho-p73 pattern as a result of curcumin treatment was determined by Western blot analysis. h, control/Chk-1/Chk-2 siRNA-transfected HL-60 cells were incubated with curcumin, and p73-associated proteins were co-immunoprecipitated with anti-p73 antibody. Western blot analysis was done with specific antibody to check the specific association of Chk-1/Chk-2 with p73. i, in parallel experiments, after 24 h of curcumin treatment, percent control/Chk-1/Chk-2 siRNA-transfected or Chk-1/Chk-2 inhibitor-pretreated HL-60 cell death was scored by Annexin-V-/7AAD positivity. α-Actin/GAPDH was used as an internal control. Values are mean ± S.E. of five independent experiments in each case or representative of a typical experiment.
p73 Transactivates Bax to Induce Mitochondrial Transmembrane Potential Loss
Previous results indicating genotoxic stress-induced elevation of p73 as well as induction of Bax, in the absence of functional p53, led us to investigate as to whether p73 transactivates Bax. To achieve this goal, endogenous p73 was knocked down by siRNA, and the cells were challenged with the genotoxic drug curcumin. Fig. 3d shows that p73 siRNA could efficiently prevent Bax up-regulation, thereby indicating that curcumin-mediated p73 induction augments apoptotic signaling by transactivating Bax. The change in MTP was also suppressed dramatically in p73 siRNA-transfected cells (Fig. 3e), validating that the apoptosis induced by curcumin is mediated by p73-induced Bax expression and MTP loss. These findings strongly support our hypothesis that p73 is the major regulator of Bax in absence of functional p53.
Chk-1 Accumulates p73 in Functional p53-deficient Cancer Cells
Next we checked whether Chk-1/Chk-2 activation has any role in genotoxic stress-induced p73 activation. Of two isoforms only Chk-1 is phosphorylated and activated in curcumin-exposed p53-null HL-60 cells (Fig. 3f). The Chk-1 inhibitor (PF-477736; 0.5 nm), but not the Chk-2 inhibitor (Chk-2 inhibitor ii), selectively blocked p73 phosphorylation in presence of curcumin (Fig. 3f), thereby indicating that Chk-1 is involved in p73 phosphorylation. To validate these results, we transfected these cells with siRNA targeting Chk-1 or Chk-2, and the phosphorylation status of p73 was analyzed in presence of curcumin. Knocking down Chk-1 diminished p73 Ser phosphorylation efficiently, whereas control/Chk-2-siRNA had no effect (Fig. 3g), suggesting that Chk-1 plays key role in phosphorylation and accumulation of p73. Our co-immunoprecipitation study revealed direct association of p73 with Chk-1 but not with Chk-2 (Fig. 3h). To correlate the involvement of the isoforms of Chks to DNA damage-induced cancer cell death, HL-60 cells were transfected with Chk-1-/Chk-2-siRNA. It was observed that unlike control/Chk-2-siRNA-transfected cells, Chk-1-siRNA-transfected cells were only minimally affected by curcumin (Fig. 3i). Similar trends were observed with Chk-1/Chk-2 inhibitor. Therefore, our studies suggested that p73 plays as the downstream effector of Chk-1 in mediating DNA damage-induced apoptosis in functional p53-deficient cells.
Functional p53 Overshadows p73 to Induce Bax-mediated Apoptosis
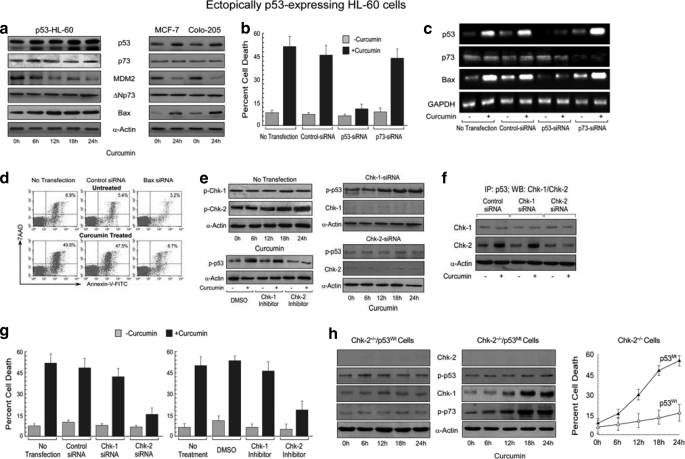
It is acknowledged that the chemistry between p53 and p73 has remained poorly understood. To establish the relationship between these two tumor suppressors we first investigated the role of p73 in functional p53-containing cells. For the same, we ectopically expressed the wild-type p53 gene in p53-null HL-60 cells and assessed the ability of curcumin to influence the expression levels of p53/MDM2/p73/ΔNp73/Bax in these cells. It was observed that there was an augmentation in the Bax expression with concomitant induction of p53 in ectopically p53-expressing HL-60 cells. Surprisingly, in these cells curcumin could neither induce p73 accumulation nor suppress ΔNp73 expression (Fig. 4a). Moreover, MDM2, which plays important role in p53 destabilization, exhibited pronounced declination after genotoxic stress (Fig. 4a). In accordance with the engineered WT p53-HL-60 cells, functional p53-expressing breast cancer MCF-7 and colon cancer Colo-205 cells also exhibited similar trends in response to curcumin (Fig. 4a), suggesting that functional p53 has an advantage over p73 in executing DNA damage-induced apoptosis in cancer cells.
FIGURE 4.
Functional p53 overshadows p73 to induce Bax-mediated apoptosis. a, ectopically p53-expressing HL-60 cells and functional p53-expressing breast cancer MCF-7 and colon cancer Colo-205 cells were incubated with 25 μm curcumin for different time points, and expression levels of p53/p73/MDM2/ΔNp73/Bax were analyzed by Western blotting. b, untransfected or control/p53/p73 siRNA-transfected p53-HL-60 cells were incubated with or without curcumin for 24 h and were scored for percent apoptosis. c, in parallel, the siRNA-transfected p53-HL-60 cells were treated with or without curcumin, and p53/p73/Bax at mRNA level was determined by RT-PCR. d, untransfected, control/Bax siRNA-transfected cells were assessed for curcumin-induced apoptosis. e, left upper panel, p53-expressing HL-60 cells were incubated with curcumin for different time points, and lysates were Western blotted to assess the phosphorylation status of Chk-1/Chk-2. Left lower panel, dimethyl sulfoxide (DMSO)/PF-477736 (0.5 nm)/Chk-2 inhibitor ii (10 nm)-pretreated HL-60 cells were incubated with curcumin, and cell lysates were Western blotted for the determination of phosphorylation status of p53. Right panels, p53-HL-60 cells were transfected with Chk-1/Chk-2 siRNA then treated with curcumin. Cell lysates were Western blotted to assess the level of Chk-1/Chk-2 proteins as well as Ser15 phosphorylation status of p53. g, control/Chk-1/Chk-2 siRNA-transfected HL-60 cells were incubated with curcumin, and p53-associated proteins were co-immunoprecipitated with anti-p73 antibody. Western blot analysis was done with specific antibody to check the specific association of Chk-1/Chk-2 with p53. In parallel experiments, percent siRNA-transfected or inhibitor-treated cell death was analyzed by Annexin-V-/7AAD positivity. GAPDH and α-actin were used as internal control. h, Chk-2−/− wild-type p53-expressing HCT-15 cells or Chk-2−/− mutant p53-expressing SW-620 cells were incubated with curcumin for different time periods, and the status of Chk-1/Chk-2/phospho-p53/phospho-p73 was assessed by Western blot analysis (left panels). Percent cell death as a result of curcumin treatment was analyzed by Annexin-V-/7AAD positivity (right panel). Values are mean ± S.E. (error bars) of five independent experiments in each case or representative of a typical experiment.
To reconfirm the above results, WT p53-HL-60 cells were transfected with p53/p73 siRNA, and the apoptosis index was checked in response to DNA damage. Interestingly, p53 silencing significantly abrogated curcumin-induced apoptosis (Fig. 4b). However, there was no pronounced difference in the percent of apoptosis between control and p73 siRNA-transfected cells, thereby reconfirming that in functional p53-expressing cells, DNA damage-induced apoptotic pathway prefers p53 but not p73.
These results were further validated by knocking out p53/p73 in WT p53-HL-60 cells with their specific siRNAs and then checking the status of Bax in response to DNA damage. Results in Fig. 4c depict that the expression level of Bax in these cells is directly related to the induction of p53 and not p73. Moreover, silencing of Bax decreased both the basal and curcumin-induced apoptosis significantly in WT p53-HL-60 cells (Fig. 4d). Therefore, unlike in p53-impaired cancer cells, there is an apoptotic pathway that involves p53-dependent but p73-independent Bax transactivation in functional p53-expressing cancer cells.
Chk-2 Activation Is Required for p53 Accumulation in Functional p53-expressing Cancer Cells
Next, in an attempt to identify the isoform of Chks responsible for p53 activation, we investigated the status of Chk-1/Chk-2 phosphorylation in response to genotoxic stress in engineered WT p53-HL-60 cells. It was revealed by the increase in phospho-Chk-2 content that in these cells only Chk-2 was activated (Fig. 4e). The Chk-2 inhibitor (Chk-2 inhibitor ii; 10 nm), but not the Chk-1 inhibitor (PF-477736; 0.5 nm), selectively blocked p53 phosphorylation in presence of curcumin (Fig. 4e), thereby indicating that Chk-2 is involved in p53 phosphorylation. To reconfirm the role of Chks in p53 phosphorylation, we next silenced Chk-1/Chk-2 expression in WT p53-HL-60 through RNA interference. Interestingly, curcumin failed to induce p53 Ser15 phosphorylation in Chk-2 siRNA-transfected cells, whereas, in Chk-1 siRNA-transfected cells it could do so efficiently (Fig. 4e). Our co-immunoprecipitation study revealed direct association of p53 with Chk-2 but not with Chk-1 (Fig. 4f). In parallel, when these cells were quantitated for curcumin-induced apoptosis, the percent cell death was effectively abrogated following Chk-2-silencing only (Fig. 4g). Similar trends were observed with Chk-1/Chk-2 inhibitor. Taken together, these data demonstrated that the Chk-2 pathway is involved in DNA damage-induced p53 accumulation and apoptosis in cells where p53 is present in its functional form.
To validate the above findings, we exploited two Chk-2−/− cell lines with different p53 status. In Chk-2−/− mutant p53-expressing cells we observed that p73 was elevated through Chk-1 but not in Chk-2−/− wild-type p53-expressing cells, in which Chk-1 was not activated, in response to genotoxic stress (Fig. 4h). These results not only indicate that wild-type p53 suppresses p73 induction via Chk-1 suppression and that Chk-2 is not required for p73 activation but also convincingly proves the interrelationship of Chk-1/Chk-2 and p53/p73. Further study revealed that in Chk-2−/− mutant p53-containing cells, due to activation of Chk-1, p73 is activated to induce apoptosis (Fig. 4h). However, in Chk-2−/− wild-type p53-expressing cells, due to impairment of Chk-1 by functional p53, p73 could not be phosphorylated (Fig. 4h). Moreover, due to absence of Chk-2, genotoxic stress failed to phosphorylate p53. These cells, therefore, escape from genotoxic stress-induced apoptosis (Fig. 4h). These results further confirm that (i) Chk-2 is required for p53 activation, and (ii) Chk-1 plays a major role in functional switchover from p53 to p73.
Unlike Prolonged ΔNp53 Overexpression, Transient Silencing of p53 Cannot Induce p73-mediated Apoptosis in p53-expressing Cancer Cells
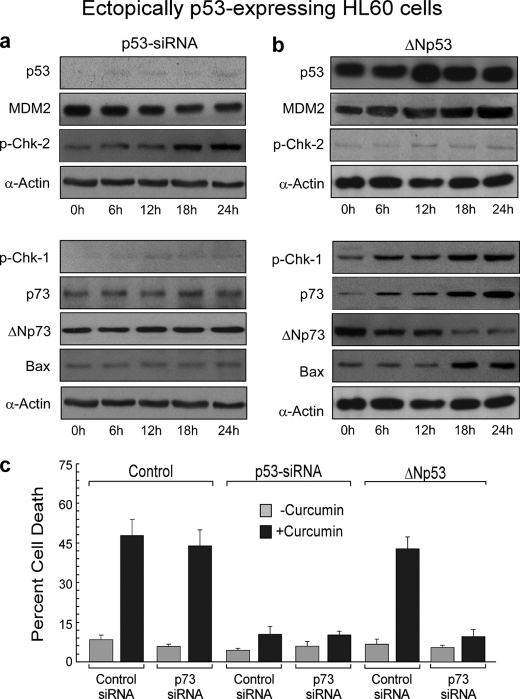
To further establish the relationship between p53 and p73, next we verified our previous findings that p73 was activated only in the absence of functional p53. At this end, activity of p53 was blocked either by siRNA or by a dominant negative clone in WT p53-HL-60 cells. These engineered cells were then exposed to curcumin, and cell lysates were subjected to Western blot analysis. To our surprise, we observed that when functional p53-expressing HL-60 cells were transfected with p53 siRNA that only transiently down-modulated p53 expression, despite moderate MDM2 down-regulation or Chk-2 activation, curcumin could neither reduce ΔNp73 nor induce Chk-1-mediated p73 accumulation as well as Bax expression (Fig. 5a). On the other hand, when these WT p53-HL-60 cells were stably transfected with dominant negative clone of p53 and a single clone was obtained after several generations, curcumin efficiently repressed ΔNp73 and induced Chk-1 activation and p73 accumulation followed by Bax induction (Fig. 5b). Consistent with these results, when functional p53-expressing MCF-7 or Colo-205 cells were transfected with p53 siRNA or p53 dominant negative clone, similar trends were observed (data not shown), indicating that this phenomenon is not cell- or tissue-specific but is universal.
FIGURE 5.
Unlike prolonged ΔNp53 overexpression, transient silencing of p53 in p53-expressing cells cannot induce p73-mediated apoptosis following DNA damage. a and b, p53 siRNA-transfected (a) and ΔNp53 overexpressed p53-expressing HL-60 cells (b) were challenged with 25 μm curcumin for different time periods, and expression levels of p53, MDM2, p73, Chk-1, Chk-2, ΔNp73, and Bax were analyzed by Western blotting. c, untransfected, p53 siRNA-transfected, and ΔNp53 overexpressed ectopically p53-expressing HL-60 cells were further transfected with control/p73 siRNA and then incubated with or without curcumin for 24 h, and the percent cell death was analyzed by Annexin-V-/7AAD positivity. α-Actin was used as internal loading control. Values are mean ± S.E. (error bars) of five independent experiments in each case or representative of a typical experiment.
The findings that (i) in functional p53-expressing cells, genotoxic drugs prefer a p53-dependent pathway to induce Bax-mediated apoptosis, and (ii) in cells where p53 function is naturally impaired, DNA damage induces p73-mediated Bax expression, prompted us to investigate the involvement of p73 in genotoxic stress-induced apoptosis in both of these systems. In WT p53-HL-60 cells, transfection of p73 siRNA imposed no effect on curcumin-induced apoptosis (Fig. 5c). On the contrary, although silencing of p53 completely abrogated curcumin-induced apoptosis, co-transfection of p73 siRNA failed to show any further change (Fig. 5c), indicating that p73 plays no significant role in these transiently p53-silenced cells. Interestingly, in ΔNp53-overexpressing WT p53-HL-60 cells, where p73 was augmented along with Bax, curcumin-induced apoptosis could be blocked successfully by p73 siRNA (Fig. 5c).
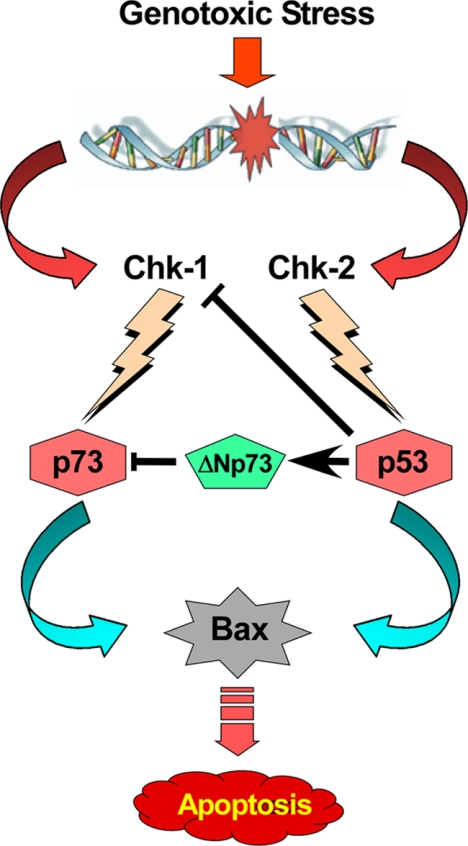
All of these data lead to the conclusion that when p53 is functional, apoptotic signals modify p53/p73-regulatory proteins in a way that DNA-damaging agents induce p53 but not p73 to transactivate Bax for executing apoptotic death (Fig. 6). On the other hand, in functional p53-impaired cancer cells, genotoxic stress induces Bax through p73-mediated transcriptional activation. Our results also signify that prolonged p53 interference, but not transient one, shifts the cellular microenvironment in favor of p73 activation by altering p73-regulatory proteins, e.g. Chk1 activation and ΔNp73 down-regulation.
FIGURE 6.
Schematic diagram for DNA damage-triggered Chk-1/Chk-2-induced p73/p53 activation.
DISCUSSION
From an evolutionary perspective, p53 activity has to be tightly regulated and backed up by other protein(s) if one fails for any reason. The majority of research with respect to p53 family members has focused on defining the individual activities of p53, p63, and p73 in apoptosis induction and have demonstrated that p73, and to a lesser extent p63, share similar activities with p53 (25). Moreover, mice heterozygous for both p53 and p73 display a more aggressive cancer phenotype compared with heterozygous p53 mice (26). A few research studies that examined the interplay between p53 family members in apoptosis induction and tumorigenesis have unveiled either cooperative or antagonistic interactions. Here, we have established the antagonistic interrelationship between p73 and p53 and delineated the mechanism involving the checkpoint kinases and ΔNp73, underlying the functional switchover from p53 to p73. These results have implications for cancers with impaired p53.
Discovery of the p73 gene that functions in a manner analogous to p53 through transactivation of an overlapping set of target genes has added complexity to the current view of p53-signaling. Because p73, unlike p53, is rarely mutated in human cancer (27, 28), we propose that p73 itself or agents that can activate the proapoptotic p73 pathway can be used as potential therapeutic agents against p53-defective cancers. In this study, an alternative mechanism of induction of apoptosis through p73 in functional p53-deficient cancer cells has been unveiled. Here, we provide substantial evidence that p53 paralogue p73 can be transformed into an inducer of Bax by genotoxic stress in the absence of functional p53. However, when p53 is present in its functional form, it overpowers p73 to induce Bax-mediated apoptosis because the transactivation and apoptotic potential of p73 are generally considered to be weaker than those of the corresponding functional p53 (29).
Chk-1 and Chk-2 are structurally unrelated yet functionally overlapping serine/threonine kinases activated after DNA damage to transduce checkpoint signals from the proximal kinases (ATM/ATR) (30, 31). Many researchers reported that Chk-2 is relatively stable protein whereas Chk1 is inducible (32–34). Our study suggests that in functional p53-expressing cells, Chk-2-activated p53 represses transcription of Chk-1, resulting in suppression of p73 phosphorylation and hence accumulation. On the other hand, when p53 is functionally impaired, DNA damage-induced Chk-1 channels the apoptotic signal through p73 accumulation.
Similar to p53, the p73 gene has two promoters (P1 and P2), and their transcripts undergo extensive splicing at the N terminus. In sharp contrast to p53, however, p73 is expressed as two N-terminally distinct isoforms: transcriptionally active TAp73 and a truncated transcriptionally inactive ΔNp73 (11). ΔNp73 acts in a dominant negative manner against p73 probably by two independent mechanisms: promoter competition and heterocomplex formation (35, 36). Interestingly, functional p53 can induce ΔNp73 by directly activating its transcription from the P2 promoter of the p73 gene (37, 38) and accumulated ΔNp73, in turn, suppresses p73 activity. Interestingly, our study revealed that following genotoxic stress the degree of p73 induction differs with the nature of p53-impairing transfection. When p53 is transiently silenced by double-stranded siRNA, the genotoxic drug fails to activate p73 because in these cells ΔNp73 expression is not repressed, and therefore ΔNp73 still acts in a dominant negative manner against p73. Interestingly, when p53 function is prolong-impaired by ΔNp53 overexpression and single clone is obtained after several generations, the genotoxic stress activates p73 and induces p73-dependent Bax as well as apoptosis.
Next, we sought other means to prolong the suppressive effect of siRNA and therefore transfected the wt-p53-HL-60 cells with ΔNp53. It is noteworthy that such suppression is long lasting and can be passed across generations in cell system. In these engineered cells, prolong p53 suppression leads to the “gain of cellular adaptation” in a way that changes the cellular microenvironment in favor of p73 activation by differentially altering p53/p73-regulatory proteins, e.g. Chk1, Chk2, MDM2 and ΔNp73. The genotoxic drug curcumin exploits such cellular adaptation to induce p73-dependent apoptosis. In fact, this phenomenon may be explained by the fact that nonfunctional p53 failed to suppress Chk-1 and/or to induce ΔNp73 expressions. All these findings together suggest that prolonged gene silencing might have advantage in enhancing the efficacy of treatment modalities.
In summary, our novel findings indicate that functional p53 overshadows p73 to trigger DNA damage-induced apoptosis in cancer cells by transactivating Bax. However, in p53-impaired cells, DNA-damage utilizes alternative pathway involving p73-dependent Bax-mediated intrinsic death cascade. Another fact came out from the above discussion is that, different cells have different constellation of signaling pathways that are activated under different situations to finally determine which Chk and p53 family member will take the lead role for inducing apoptosis in cancer cells. Because, functions of the tumor suppressor protein p53 are commonly lost in cancer, the above finding that p53 functions can be allotted to an alternative tumor suppressor, p73, is of potential therapeutic significance for designing novel strategies to improve efficacy in cancer treatment.
Acknowledgments
We thank R. Sarkar for valuable suggestions during the preparation of this paper and U. Ghosh and R. Dutta for technical help.
This work was supported by research grants from the Department of Science and Technology, Government of India.
- TA
- transcriptionally active
- AML
- acute myeloid leukemia
- Chk
- checkpoint kinase
- ΔN
- dominant negative.
REFERENCES
- 1.Green D. R., Kroemer G. (2009) Nature 458, 1127–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnstone R. W., Ruefli A. A., Lowe S. W. (2002) Cell 108, 153–164 [DOI] [PubMed] [Google Scholar]
- 3.Kravchenko J. E., Ilyinskaya G. V., Komarov P. G., Agapova L. S., Kochetkov D. V., Strom E., Frolova E. I., Kovriga I., Gudkov A. V., Feinstein E., Chumakov P. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6302–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whibley C., Pharoah P. D., Hollstein M. (2009) Nat. Rev. Cancer 9, 95–107 [DOI] [PubMed] [Google Scholar]
- 5.Yang A., McKeon F. (2000) Nat. Rev. Mol. Cell Biol. 1, 199–207 [DOI] [PubMed] [Google Scholar]
- 6.Johnson R. A., Shepard E. M., Scotto K. W. (2005) J. Biol. Chem. 280, 13213–13219 [DOI] [PubMed] [Google Scholar]
- 7.Aqeilan R. I., Pekarsky Y., Herrero J. J., Palamarchuk A., Letofsky J., Druck T., Trapasso F., Han S. Y., Melino G., Huebner K., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4401–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grob T. J., Fey M. F., Tobler A. (2002) Cell Death Differ. 9, 229–230 [DOI] [PubMed] [Google Scholar]
- 9.Blint E., Phillips A. C., Kozlov S., Stewart C. L., Vousden K. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3529–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ra H., Kim H. L., Lee H. W., Kim Y. H. (2009) FEBS Lett. 583, 1516–1520 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Chen X. (2007) Mol. Cell. Biol. 27, 3868–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tannapfel A., John K., Mise N., Schmidt A., Buhlmann S., Ibrahim S. M., Pützer B. M. (2008) Carcinogenesis 29, 211–218 [DOI] [PubMed] [Google Scholar]
- 13.Ehrhardt H., Häcker S., Wittmann S., Maurer M., Borkhardt A., Toloczko A., Debatin K. M., Fulda S., Jeremias I. (2008) Oncogene 27, 783–793 [DOI] [PubMed] [Google Scholar]
- 14.Seifert H., Mohr B., Thiede C., Oelschlägel U., Schäkel U., Illmer T., Soucek S., Ehninger G., Schaich M. (2009) Leukemia 23, 656–66319151774 [Google Scholar]
- 15.Bhattacharyya S., Mandal D., Saha B., Sen G. S., Das T., Sa G. (2007) J. Biol. Chem. 282, 15954–15964 [DOI] [PubMed] [Google Scholar]
- 16.Choudhuri T., Pal S., Das T., Sa G. (2005) J. Biol. Chem. 280, 20059–20068 [DOI] [PubMed] [Google Scholar]
- 17.Sa G., Das T. (2008) Cell Div. 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya S., Mandal D., Sen G. S., Pal S., Banerjee S., Lahiry L., Finke J. H., Tannenbaum C. S., Das T., Sa G. (2007) Cancer Res. 67, 362–370 [DOI] [PubMed] [Google Scholar]
- 19.Ng C. P., Lee H. C., Ho C. W., Arooz T., Siu W. Y., Lau A., Poon R. Y. (2004) J. Biol. Chem. 279, 8808–8819 [DOI] [PubMed] [Google Scholar]
- 20.Urist M., Tanaka T., Poyurovsky M. V., Prives C. (2004) Genes Dev. 18, 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez S., Prives C., Cordon-Cardo C. (2003) Mol. Cell. Biol. 23, 8161–8171 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Durland-Busbice S., Reisman D. (2002) Leukemia 16, 2165–2167 [DOI] [PubMed] [Google Scholar]
- 23.Kramer S., Ozaki T., Miyazaki K., Kato C., Hanamoto T., Nakagawara A. (2005) Oncogene 24, 938–944 [DOI] [PubMed] [Google Scholar]
- 24.Belloni L., Moretti F., Merlo P., Damalas A., Costanzo A., Blandino G., Levrero M. (2006) Oncogene 25, 3606–3612 [DOI] [PubMed] [Google Scholar]
- 25.Pietsch E. C., Sykes S. M., McMahon S. B., Murphy M. E. (2008) Oncogene 27, 6507–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flores E. R., Sengupta S., Miller J. B., Newman J. J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. (2005) Cancer Cell 7, 363–373 [DOI] [PubMed] [Google Scholar]
- 27.Hollstein M., Hergenhahn M., Yang Q., Bartsch H., Wang Z. Q., Hainaut P. (1999) Mutat. Res. 431, 199–209 [DOI] [PubMed] [Google Scholar]
- 28.Kaelin W. G., Jr. (1999) Oncogene 18, 7701–7705 [DOI] [PubMed] [Google Scholar]
- 29.Vilgelm A., El-Rifai W., Zaika A. (2008) Drug Resist. Updates 11, 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse A. N., Carvajal R., Schwartz G. K. (2007) Clin. Cancer Res. 13, 1955–1960 [DOI] [PubMed] [Google Scholar]
- 31.Bartek J., Lukas J. (2003) Cancer Cell 3, 421–429 [DOI] [PubMed] [Google Scholar]
- 32.Damia G., Broggini M. (2004) Cell Cycle 3, 46–50 [PubMed] [Google Scholar]
- 33.Damia G., Sanchez Y., Erba E., Broggini M. (2001) J. Biol. Chem. 276, 10641–10645 [DOI] [PubMed] [Google Scholar]
- 34.Gottifredi V., Karni-Schmidt O., Shieh S. S., Prives C. (2001) Mol. Cell. Biol. 21, 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagawa T., Takahashi M., Ozaki T., Watanabe, Ki K., Todo S., Mizuguchi H., Hayakawa T., Nakagawara A. (2002) Mol. Cell. Biol. 22, 2575–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaika A. I., Slade N., Erster S. H., Sansome C., Joseph T. W., Pearl M., Chalas E., Moll U. M. (2002) J. Exp. Med. 196, 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grob T. J., Novak U., Maisse C., Barcaroli D., Lüthi A. U., Pirnia F., Hügli B., Graber H. U., De Laurenzi V., Fey M. F., Melino G., Tobler A. (2001) Cell Death Differ. 8, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 38.Kartasheva N. N., Contente A., Lenz-Stöppler C., Roth J., Dobbelstein M. (2002) Oncogene 21, 4715–4727 [DOI] [PubMed] [Google Scholar]