Abstract
Hepatitis E virus (HEV) induces acute hepatitis in humans with a high fatality rate in pregnant women. There is a need for anti-HEV research to understand the assembly process of HEV native capsid. Here, we produced a large virion-sized and a small T=1 capsid by expressing the HEV capsid protein in insect cells with and without the N-terminal 111 residues, respectively, for comparative structural analysis. The virion-sized capsid demonstrates a T=3 icosahedral lattice and contains RNA fragment in contrast to the RNA-free T=1 capsid. However, both capsids shared common decameric organization. The in vitro assembly further demonstrated that HEV capsid protein had the intrinsic ability to form decameric intermediate. Our data suggest that RNA binding is the extrinsic factor essential for the assembly of HEV native capsids.
Keywords: Crystallography, Microscopic Imaging, Protein Structure, RNA-Protein Interaction, Virus Assembly, Hepatitis E Virus, Infectious Disease
Introduction
Hepatitis E virus (HEV),3 the causative agent of acute hepatitis in humans, is primarily transmitted through contaminated water and generally results in epidemic outbreaks in many developing countries. Sporadic cases have also been reported between outbreaks in HEV-endemic regions as well as in non-endemic areas, and these cases are transmitted through zoonotic route. The overall mortality rates of HEV during outbreaks range from 1 to 15% in general, and the highest mortality occurs in pregnant women, with fatality rates of up to 30% (1).
HEV consists of a non-enveloped icosahedral capsid and a single-stranded, positive-strand RNA genome of ∼7.2 kb that encodes three open reading frames (ORFs) (2). The capsid protein, encoded by the ORF2, is composed of 660 amino acids and responsible for most capsid-related functions, such as virion assembly, host interaction, and immunogenicity. Like other hepatitis viruses, HEV is unable to propagate in currently available cell culture systems, and the research of HEV relies largely on the recombinant HEV capsid proteins (3–6). Virus-like particle (VLP) was obtained when the truncated HEV capsid protein was expressed in insect Tn5 cells with deletion of 52 residues from the C terminus and 111 residues from the N terminus (PORF2) (7). Our previous structural analysis of this HEV-VLP by cryo-electron microscopy (cryo-EM) provided a basic understanding of the quaternary arrangement of PORF2, where the reconstructed VLP displayed a T=1 icosahedral particle composed of 60 copies of PORF2 (8). The essential element of PORF2 protein for T=1 VLP assembly includes amino acids 125–600 (9). Recently, the structural information was further refined by the crystal structures of genotype-3 T=1 VLP (10) and genotype-4 T=1 VLP (11), which revealed the tertiary structure of PORF2 to the level of amino acids. However, the T=1 VLPs used in these experiments were much smaller than that of the native virion, which has a diameter of 320–340 Å, as determined by immuno-EM (12). There is still a need to investigate the assembly pathway of HEV capsid.
HEV virion is hypothesized to be made of 180 copies of the capsid protein (8, 11). To test this hypothesis, we screened for HEV genotype expression and successfully produced a virion-sized VLP from the HEV genotype-3 ORF2 protein after deleting 52 residues from the C terminus. This VLP allowed us to investigate the molecular interactions that govern HEV virion assembly.
EXPERIMENTAL PROCEDURES
Production of HEV-VLPs and in Vitro Disassembly and Reassembly
HEV-VLPs were produced and purified according to the protocol described previously (7, 13). Briefly, the recombinant baculovirus Ac(G3n13ORF2), which harbored the genome of the N-terminal 13 amino-acid-deleted genotype-3 HEV ORF2, was infected into Tn5 cells with recombinant baculovirus at a multiplicity of infection of 10. The recombinant baculovirus-infected Tn5 cells were harvested at 7 days after infection. The medium and cells were separated by centrifugation at 3,000 rpm for 15 min at 4 °C. The cells were treated with a denaturation buffer containing 50 mm sodium borate, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 5% 2-mercaptoethanol and gently rocked at room temperature for 2 h. The lysate was diluted with EX-CELL 405, and centrifuged at 32,000 rpm for 3 h in a Beckman SW32Ti rotor, and the pellet was resuspended in EX-CELL 405. The VLP was purified by multiple ultracentrifugations followed by separation on a CsCl density gradient. The final pellet was resuspended in 10 mm potassium MES buffer, pH 6.2. A homemade dialysis device was used in the disassembly and reassembly experiments because it allowed dialysis with a small amount of sample (20–40 μl). Purified VLP was disrupted by dialysis against buffer containing EDTA (10 mm) and DTT (20 mm) at different pHs. After VLP dissociation, 150 mm NaCl in Tris-HCl buffer (pH 7.5) was added, and sample was examined under the electron microscope after a 1-h incubation in the presence of the divalent ion Ca2+ (20 mm).
Scanning Transmission Electron Microscopy Analysis of HEV-VLPs
Scanning TEM (STEM) was performed at the Brookhaven National Laboratory STEM facility, with tobacco mosaic virus (TMV) as an internal control. The mixture of VLP and TMV was quickly frozen in liquid nitrogen and then maintained at −150 °C during data collection to eliminate contamination and reduce mass loss. The specimen was scanned by a 40-keV electron beam of 0.25 nm in size, and images were collected with a preamp gain of 10 for both large and small angle detectors (14). The image was recorded with a pixel size of 10 Å and was analyzed with the PCMass29 program. After normalizing the background, the mass of the VLPs was selected with the murine sarcoma virus (MSV) shell model provided by the program. Mass measurements for TMV and HEV-VLPs were always performed from the same image. The HEV-VLP mass was measured in MDa (mass per particle), and the TMV mass was measured in KDa/Å (mass per unit length) (15).
Cryo-electron Microscopic Structure Determination of HEV T=3 VLP
The collection of cryo-EM data for image reconstruction was performed on a JEOL JEM-2100F TEM operating at 200 kV according to the procedure described in detail previously (8). Briefly, a 3-μl solution containing VLP or reassembled ORF2 complex was placed on holey carbon film-coated copper grids and then quickly plunged into liquid ethane after the removal of excess solution. The VLPs were embedded into a thin layer of vitrified ice and transferred into the EM using a Gatan 626 cryo-transferring system. The specimen was observed under 50,000× magnification, and the area of interest was recorded on a TVIPS CCD camera (TemCam-F415). The micrographs were recorded with a pixel size of 2.0 Å at a specimen space and defocus level of 0.7–3.5 Å (supplemental Fig. 1A). Digital images with no stigmatism or drift were selected for later image processing. Images of individual HEV T=3 VLPs were then boxed out and processed with an established software package for icosahedral particles (16, 17). In total, 7,720 individual images were included in the process, and their defocus levels were distributed mainly within 1.0–2.5 Å.
To correct contrast transfer function effect, we applied phase flipping on each image with an in-house program. The density maps were initially reconstructed by combining 1,812 individual images to an effective resolution of 14 Å. Next, amplitude correction was applied during map reconstruction while new data were added. The final density map was reconstructed by combining images of 4,348 individual particles, and the final resolution was assessed as 10.6 Å by Fourier shell correlation with a cutoff of 0.5 (supplemental Fig. 1B).
Docking of the T=1 crystal structure into the T=3 cryo-EM density map was first done manually with the program O (18) and then refined with the Situs software package (19). The PORF2 monomer was treated as a rigid body during the initial fitting and refinement processes.
X-ray Crystallographic Structure Determination of T=1 HEV-VLP
Crystallization of the VLPs was performed according to a previously described method (20). Crystals were directly flash-frozen in liquid nitrogen, and x-ray diffraction experiments were performed. All x-ray experiments of the HEV-VLP crystals were performed at SPring-8 in Hyogo, Japan. Particle orientation in the unit cell was determined with a self-rotation function (21), and the particle position was determined by a translation search with the cryo-EM structure as the model. The asymmetric crystal unit contains one particle; as a result, 60-fold non-crystallographic symmetry averaging was enforced. The cryo-EM structure (8) was used to obtain the initial phases of Data I (supplemental Table 1) and generated the envelope (mask) used for non-crystallographic symmetry averaging. The phases were refined by real space electron density averaging with icosahedral symmetry elements and solvent flattening. The resolution was gradually extended to 8.3 Å (R-factor = 0.21, correlation coefficient = 0.92). This structure was used for the phasing of Data II (supplemental Table 1), and the phases were refined and extended to a 3.8 Å resolution (the overall R-factor and correlation coefficient were 0.18 and 0.97, respectively). The electron density map revealed a clear main chain structure. Thus, we built an atomic model into this electron density map using the program O (18) without difficulty.
Sixty icosahedrally related S-subunits were treated as identical, and strict non-crystallographic symmetry constraints were applied during refinement. The data with the resolution range of 20–3.8 Å were included in the refinement (supplemental Table 1 and supplemental Fig. 2) using the CNS program (22). The subunit position and orientation were again clarified by rigid body refinement (R-factor of 0.391 (Rfree = 0.395)). After the first cycle of simulated annealing refinement, positional and B-factor refinement followed, and the model was enhanced to an R-factor of 0.261 (Rfree = 0.264). Further positional and B-factor refinement, followed by manual revision of the model, resulted in an R-factor of 0.242 (Rfree = 0.245) with reasonable stereochemistry (root mean square deviations in bond lengths and bond angles were 0.010 Å and 1.68°, respectively). Because of the high non-crystallographic symmetry, the R-factor and Rfree factor were almost identical. After refinement, the stereochemistry of the structure was checked with Procheck (23). 98.1% of the non-glycine residues were within the most favored and the additional allowed regions of the Ramachandran plot, and none of the residues were in the additional regions. Atomic structure representations were generated using MolScript (24) and Raster3D (25).
Nucleic Acid Extraction and Characterization
100 μg of purified large and small VLPs were treated with DNase I (final concentration; 0.01 mg/ml) (Sigma) and RNase A (final concentration; 0.5 μg/ml) at 37 °C for 1 h and then followed by centrifugation at 50,000 rpm for 2 h at 4 °C in a Beckman TLA55Ti rotor. After removal of the supernatant, the pellets were resuspended in 200 μl of PBS minus buffer. The nucleic acids were extracted with RNAzol-LS reagent (Tel-test, Inc., Friendswood, TX) and analyzed on 1% agarose gels.
For detection of HEV RNA in VLPs, the extracted RNAs from VLPs were mixed with sample buffer (final concentration: 2% formaldehyde, 50% formamide in MOPS buffer containing 20 mm MOPS, 5 mm sodium acetate, and 1 mm EDTA (pH 7.0), Sigma-Aldrich, Tokyo, Japan). The solution was denatured at 65 °C for 10 min and cooled immediately in ice-cold water and then mixed with 6× loading buffer (80% formamide, 0.25% bromphenol blue, 0.25% xylene cyanol, 6 mm EDTA, Sigma-Aldrich). After separation on the formaldehyde-denatured agarose gel (18% formaldehyde, 1% agarose, Sigma-Aldrich) in 1× MOPS buffer, the gel was washed twice for 15 min with 20× SSC (3 m NaCl, 0.3 m trisodium citrate dehydrate (pH 7.0), Roche Applied Science, Tokyo, Japan) and transferred onto a Hybond N+ membrane filter (GE Healthcare, Tokyo, Japan) by the capillary method. RNAs were fixed on the membrane by irradiation with UV light (Stratalinker UV crosslinker, Stratagene, Tokyo, Japan) and dried in the air. RNAs on the membrane were hybridized with DIG-labeled probe. DIG-labeled RNAs were detected by using a DIG Northern starter kit (Roche Applied Science) according to the manufacturer's protocol and visualized by the LAS-3000 imaging analyzer, Fujifilm, Tokyo, Japan). The purified template DNA for the probe RNA was prepared by digesting the plasmid carrying 584-base cDNA of HEV ORF2 (5,903–6,486 nucleotides) under the T7 promoter with BamHI. The probe RNA was prepared using the DIG Northern starter kit according to the manufacturer's manual (incorporation of DIG-UTP during RNA transcription). The norovirus-like particles that were produced by the recombinant baculovirus were used as a control, and the same procedure was used with the HEV-VLPs.
RESULTS
STEM
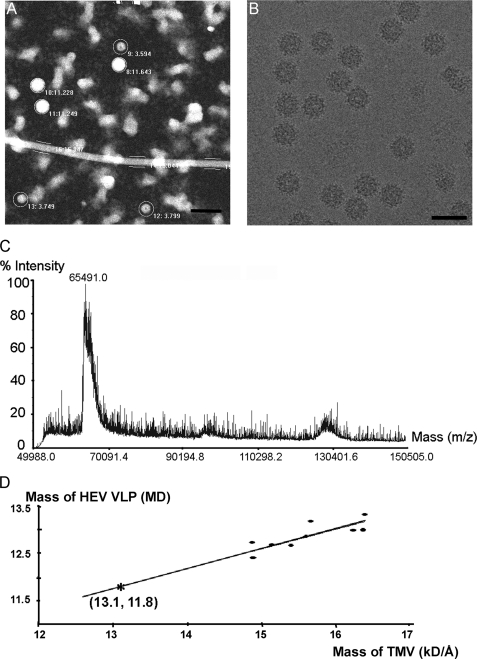
The virion-sized HEV-VLP was recovered when the genotype-3 ORF2 sequence was expressed in insect cells. This VLP projected as a spherical image with a diameter of ∼40 nm, larger than the T=1 VLP (27 nm in diameter) (Fig. 1A). In cryo-electron micrographs, the images of HEV-VLP are decorated with spike-like features and are homogeneous in contrast (Fig. 1B).
FIGURE 1.
The large HEV-VLP is composed of 180 copies of ORF2 protein. A, STEM micrograph of HEV-VLPs. Both large and small T=1 HEV-VLPs are projected as spherical images, and their corresponding particle mass was calculated. The long straight rod is TMV, which was added as an internal mass standard. (Bar = 1,000 Å.) B, the large HEV-VLP appeared as an intact particle decorated with a dot-like pattern on the surface on cryo-EM of the large HEV-VLP. (Bar = 500 Å.) C, mass spectrum of the large HEV-VLP showing that the molecular mass of the ORF2 protein is 65.5 kDa. D, plot showing the observed mass/length of TMV against the observed mass of the large HEV-VLP from different image conditions. For a known TMV mass/length of 13.1 kDa/Å, the mass of the large HEV-VLP was calculated as 11.8 MDa (asterisk). MD, megadalton.
To determine the composition of the large HEV-VLP, we performed mass measurements by using STEM, a technique that measures the amount of electrons scattered from the objects, such as VLPs, on an EM grid. A mixture of purified large and small HEV-VLPs was freeze-dried onto EM grids for STEM mass measurement. TMV with a known mass-to-length ratio was used as an internal standard. The HEV-VLPs appeared as spherical projections with white contrast on the dark field STEM images (Fig. 1A). White cloud-like objects were present in the background, which might be the VLPs broken during sample preservation. The mean mass of large VLP and TMV in the images was measured to generate a plot of the mean TMV mass per unit length versus mean VLP mass per particle (Fig. 1A). A first-order fit was calculated, and the mass of the large HEV-VLP was determined to be 11.8 MDa (Fig. 1D). The mass of the genotype-3 ORF2 protein, which was recovered from the large VLP, was measured as 65.5 KDa by mass spectrometry (Fig. 1C). Therefore, the large HEV-VLP contains 180 copies of ORF2 proteins, suggesting that the large HEV-VLP is a T=3 icosahedral particle (T=3 VLP). We further performed element analysis with x-ray photoelectron spectroscopy. X-ray photoelectron spectroscopy, also known as electron spectroscopy for chemical analysis, determines the chemical composition of the sample with a depth of 50–70 nm. Phosphorus element, as a characteristic element of nucleic acid, was detected from the G3-VLP that was applied to the carbon-coated copper grid (supplemental Fig. 1C). Although the signal of phosphorus element is weak when compared with that of carbon, the phosphorus peak is not detectable in the control grid without VLP materials. This result suggests the co-existence of nucleic acid within the large G3-VLPs.
Three-dimensional Reconstruction of the HEV Virion-sized Particle
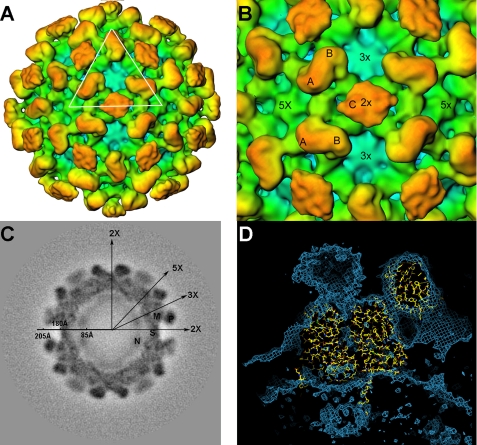
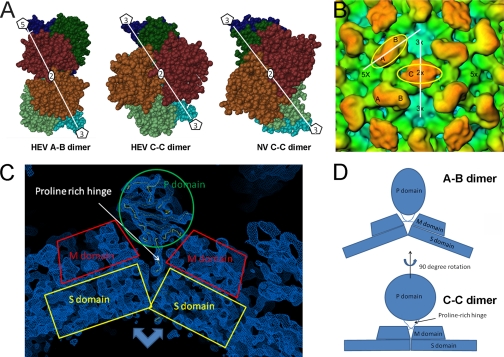
The cryo-EM structure of the large HEV-VLP revealed 90 protruding spikes on a complete icosahedral shell (Fig. 2A), which is consistent with the T=3 icosahedral symmetry and the results of the STEM mass measurements. The VLP had an overall diameter of 410 Å and a central cavity of 170 Å in radius as measured from the three-dimensional density map (Fig. 2C). The single-layer capsid contained 180 copies of the ORF2 protein, which were grouped into three unique monomers according to their geometric environments. Although monomers A and B formed dimeric spikes (A-B dimers) around each of the five-fold axes, two two-fold related C monomers formed a spike (C-C dimer) at each of the icosahedral two-fold axes (Fig. 2B). The surface lattices of ORF2 proteins in HEV T=3 VLP were similar to the capsid arrangement of caliciviruses. When compared with the A-B dimer, the morphology of the HEV C-C dimer was less well defined, perhaps due to flexibility in the angle of the protruding domain toward the icosahedral shell.
FIGURE 2.
Three-dimensional structure of HEV T=3 VLP. A, overall structure of the large HEV-VLP reveals the T=3 icosahedral lattice of the ORF2 proteins. One icosahedral facet is defined as the triangular area within the three adjacent five-fold axes. B, there are two unique dimeric ORF2 spikes on the HEV T=3 VLP surface. The A-B dimer is located around the five-fold axis, and the C-C dimer is located at the two-fold axis. C, HEV T=3 VLP has a radius of 205 Å and contains a low density cavity with a radius of 85 Å in the particle center. The distribution of the cryo-EM density revealed four ORF2 domains, P, M, S, and N, at 50 Å from the equatorial section. D, the crystal structure of the HEV subunit from T=1 VLP docks well with the cryo-EM density in the shell region of HEV T=3 VLP, with the N-terminal loop pointing toward the center.
The density map of the T=3 VLP displayed four discrete domains, designated from the outside inward as P, M, S, and N, on a section 52 Å from the equatorial plane (Fig. 2C). The density profile of the P, M, and S domains displayed less variation from that observed in T=1 HEV-VLP, and the docking of the crystal structure of the T=1 PORF2 protein to the density map of T=3 HEV-VLP showed a very good agreement between the two structures (Fig. 2D). The docking positioned N-terminal tail of the PORF2 protein at the capsid inner surface aligned well with the density linker in T=3 VLP (Fig. 2D). The linker density served as a tag to connect the N domain with the icosahedral capsid, indicating the location of the N-terminal 111 amino acids of the ORF2 protein in T=3 HEV-VLP.
Crystal Structure of the Genotype-1 T=1 HEV-VLP
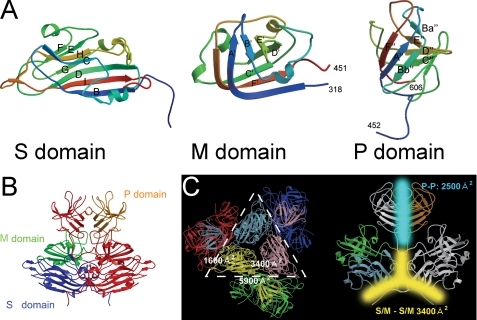
The crystal structure of the truncated genotype-1 capsid protein (PORF2, containing residues 112–608) can be separated into three domains, S, M, and P, with a less resolved region covering residues 555–560. The S domain formed by residues 118–317 folds into a classical eight-stranded β-barrel with a jelly roll motif (Fig. 3A), as observed in many T=3 viral capsid proteins (26, 27). Uniquely, three additional short α-helixes were observed in the S domain between strands E and F and strands G and H. The capsid shell was mainly stabilized by intersubunit interactions between the S domains. The folded M domain, consisting of residues 318–451, was a twisted antiparallel β-sheet with an α-helix between the B′ and C′ strands (Fig. 3A). The P domain, composed of residues 452–606, folded into a β-barrel composed of antiparallel β-sheets, F"A"Bb" and Ba"E"D"C" (Fig. 3A), and was connected with the M domain through a long proline-rich hinge (PTPSPAPSRP of residues 452–461) (Fig. 3A). Although both the M and the P domains existed above the S domain, the protruding spikes in the HEV cryo-EM map contain only the P domain density, which is a clear difference to those caliciviruses (supplemental Fig. 3). The PORF2 dimers have the largest buried surface area between monomers (5,900 Å2) mainly due to the interface between P domains (Fig. 3B). The buried surface area is 3,400 and 1,600 Å2 for the two adjacent PORF2 subunits around a three-fold axis and a five-fold axis, respectively (Fig. 3B). Moreover, the buried surface area of three molecules around a three-fold axis (9,500 Å2) is much wider than that around a five-fold axis (4,700 Å2).
FIGURE 3.
The structure of genotype-1 PORF2 protein. A, ribbon representations of S, M, and P. The structure is covered from blue (N terminus of the domain) to red (C terminus of the domain). B, dimer structure of the capsid protein. One subunit is colored red, and the other subunit is colored according to its domain structure (blue, S domain; green, M domain; orange, P domain). C, surface areas that buried at the interfaces between two adjacent subunits are overlapped with a PORF2 hexamer (left) and at the PORF2 dimeric interface (right). One icosahedral facet is defined as the triangular area within the three adjacent five-fold axes.
Sequence alignment of genotype-1 PORF2 with the sequences of genotype-3 (10) and genotype-4 (11) revealed that the S domain is the most conserved region among HEV genotypes, whereas greater divergence was seen in the N-terminal region (supplemental Fig. 4A). Among the solved structures, genotype-3 appeared flexible at the N-terminal end and was 11 amino acids shorter than the others, whereas the structure deviation from genotype-1 PORF2 is very small (total root mean square deviation is 0.62 for the 472 equivalent amino acids) (supplemental Fig. 4B). Because amino acids 118–129 play an important role in bridging the N-domain to the S-domain in T=3 VLP and serve as a docking register, we used the crystal structure of genotype-1 to decipher the T=3 cryo-EM density map.
Consistent Interdimeric Interactions between T=3 and T=1 HEV-VLPs
To understand the mechanism of ORF2 protein transition between T=1 and T=3 assemblies, we docked the T=1 decamer and hexamer into the T=3 cryo-EM density map. The decamer of T=1 VLP consisted of 10 adjacent PORF2 monomers corresponding to five dimers around a five-fold axis, whereas the T=1 hexamer corresponded to three adjacent dimers around a three-fold axis. Unlike the hexamer, the coordinates of the PORF2 decamer fitted very well with the curvature of the T=3 density map at the five-fold vertex (Fig. 4A) and with the domain separation (Fig. 4B). The curvature of T=3 capsid at the three-fold axis did not agree with the coordinates of the PORF2 hexamer as one of the dimers appeared to be sticking out of the cryo-EM density map (data not shown). Besides, the orientation of the P domain of the C-C dimer relative to its M/S domains was 90° different from that of the A-B dimer (Fig. 5). This suggests that the molecular interactions among A-B dimers in the T=3 icosahedron are consistent with the dimer-dimer interactions in the T=1 icosahedral assembly, whereas the interaction between the A-B dimer and C-C dimer is unique to the T=3 assembly.
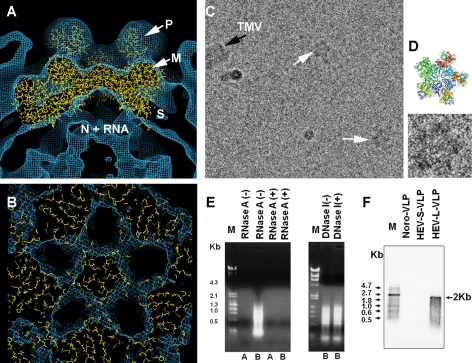
FIGURE 4.
Structure of ORF2 decamer is consistent in both T=1 and T=3 VLPs. A, the cryo-EM density of HEV T=3 VLP agrees well with the coordinates of T=1 VLP at the region around the five-fold axis. The P, M, S, and N + RNA mark the corresponding density layer. B, consistent features between the decamer in HEV T=3 and T=1 VLP were revealed by the crystal structure docking of PORF2 dimers into the cryo-EM density map of HEV T=3 VLP. C, reassembly of the ORF2 protein in vitro led to ORF2 star-shaped complex formation (white arrows). The size of this complex fits well into that of the ORF2 decamer after calibration with TMV (black arrow) as an internal standard. D, one ORF2 complex was zoomed in twice and is displayed in comparison with the crystal structure of PORF2 decamer (ribbon drawing). E, nucleic acids were extracted from HEV T=1VLP (lane A) and T=3 VLP (lane B) and degraded in the presence of RNase A but remained as intact when incubated with DNase I. No nucleic acid was detected from T=1 VLP (lane A). DNA maker (lane M): λ-HindIII + φ-174 HincII. F, Northern blotting to detect HEV RNA. RNA marker (lane M): 0.5–10-kb RNA ladder. RNA extract from large HEV T=3 VLP, small HEV T=1 VLP, and noravirus VLP was loaded in lane marked with HEV-L-VLP, HEV-S-VLP, and nora-VLP, respectively.
FIGURE 5.
The orientation of P domain relative to the M and S domain in the C-C dimer appears different from that in A-B dimer. A, the dimeric interactions in the HEV A-B dimer, the HEV C/C dimers, and the Norwalk virus C-C dimer are shown in space-filling models as observed from the outside of the particles. S, M (P1) and P (P2) domains in a subunit are highlighted in dark/light blue, dark/light green, and brown/chocolate, respectively. B, in the T=3 cryo-EM density map, the orientation of the P domain is shown as the angle between the platform of the spike (yellow circles) relative to the M/S domains in the C-C dimer (white line passing through the two adjacent three-fold axes) or the M/S domains in the A-B dimer (white line passing through a five-fold axis and the neighboring three-fold axis). C, the crystal density map showing the position of the proline-rich hinge within the cleft of the M domains. D, the cleft in between the M/S domains provides sufficient room to accommodate the proline-rich hinge in the A-B dimer, where the domains take a bent conformation. The cleft is narrowed down in the C-C dimer due to the flat conformation between the M/S domains, thus pushing the hinge up and out of the cleft.
In Vitro Reassembly of the ORF2 Protein
To understand the role of ORF2 decamer in T=3 VLP assembly, we analyzed the self-assembly process of HEV-VLP in vitro. A combination of chelating (EDTA) and reducing (DTT) agents was found to disassemble T=3 VLP in a high alkaline environment (pH 10) without denaturating the ORF2 protein (data not shown). The addition of 20 mm CaCl2 into the disassembly solution led to the association of the ORF2 dimers into star-shaped complexes, and no refolded VLP was found (Fig. 4C). When we examined the star-shaped complexes, we found that the distance between two opposite vertices was ∼18 nm, close to the diameter of TMV (Fig. 4D). This size was consistent with that measured from PORF2 decamers. Thus, the star-shaped complexes resembled not only the appearance but also the size of the ORF2 decamer (a pentamer of dimers). Although the overall buried surface area around the three-fold axis is larger than that around the five-fold axis, we did not find any complexes that could fit with the PORF2 hexamer.
The in vitro disassembly and reassembly suggested that other factors than ORF2 protein contribute to T=3 VLP assembly. Considering the electropositivity of the ORF2 N-terminal 111 amino acids, we performed nucleic acid extraction from both the T=3 and the T=1 VLPs. Electrophoresis results demonstrated the presence of nucleic acids in the T=3 extract, whereas the T=1 VLP extract was negative for nucleic acids (Fig. 4E). The extracted nucleic acids were sensitive to RNase treatment and resistant to DNase treatment. To further characterize the nucleic acid, we performed Northern blotting according to the protocol described under “Experimental Procedures.” An RNA band about 2 kb in size was detected from large particles, whereas no band was detected from the small particles and the control norovirus-like particles (Fig. 4F). This result is consistent with the VLP profiles observed from the cryo-electron micrographs and further indicated that the T=3 particles encapsulated the HEV gene encoding the ORF2 protein.
DISCUSSION
Hepatitis E virus is a human pathogen that causes acute liver failure. Like other hepatitis viruses, HEV cannot be propagated with currently available cell culture techniques. The capsid protein of genotype-3 HEV can be expressed in insect cells as PORF2 protein, including amino acids 112–608 that self-assemble into T=1 VLP, and as ORF2 protein, including amino acids 14–608 that form T=3 VLP.
The crystal structures of PORF2 revealed three functional domains, S, M, and P, and the function of each domain constrained its sequence flexibility. The S domain formed an icosahedral shell that served as the base for arranging M and P domains; hence, the subunit surface should be highly conserved among genotypes. Sequence alignment agreed very well with this function, identifying the S domain as the most conserved region among HEV genotypes (28). The P domain serves as the putative binding site for both neutralizing antibody and cellular receptor (29) and contains 19 divergent amino acids across four genotypes (supplemental Fig. 4). Only nine of these amino acids were exposed at the surface of the P domain. Inspection of the binding footprint of antibodies on the cryo-EM density map indicated that only one amino acid was buried within the antibody-binding interface4. This explains why the HEV serotype is non-divergent despite sequence variation among HEV genotypes. The direct correlation between sequence variability and domain functionality may be necessary for the HEV capsid to carry multiple functions and to ensure error-free assembly. It also explains why the transition of HEV-VLP from the T=3 to the T=1 lattice does not disturb its antigenicity and why T=1 VLP can be disassembled and reassembled in vitro to carry foreign antigenic epitopes (30) or DNA plasmids (31).
The T=3 HEV-VLP has a similar morphology to that of calicivirus; however, the crystal structures of PORF2 revealed a distinctive M domain arrangement, although the folding of the HEV M domain is similar to the folding of the P1 domain in caliciviruses (supplemental Fig. 3). In HEV, the P domain is located at the C-terminal end of the M domain, whereas the P2 domain of caliciviruses is inserted into the P1 domain at the region between the A′ and B′ strands (32, 33). Furthermore, the M domain of HEV interacts strongly with the S domain and connects to the P domain via a long proline-rich hinge, whereas the P1 domain in caliciviruses is a subdomain of the protrusion spike (supplemental Fig. 3). This seems to have an impact on VLP stability; the spike of the HEV C-subunits appeared weakly defined when compared with that in the A-B dimer, whereas the spike of the Norwalk virus C-subunit appeared rigid and similar to that in the A-B dimer in the cryo-EM structure (34). Additionally, deletion of the N-terminal positively charged amino acids from the Norwalk virus capsid protein does not induce T=1 VLP because the Norwalk virus capsid protein only contains a short N-terminal tail of 20 amino acids (35).The HEV C-C dimer is profoundly different from the HEV A-B dimer in the orientation of the P domain relative to the M/S domain (Fig. 5). Conformational difference between the A-B dimer and C-C dimer has been reported earlier T=3 viruses. In tomato bushy stunt virus, binding of RNA plays an important role to differentiate the C-C dimer from the A-B dimer. The N-terminal arm of the C-C dimer is well ordered and interacts with the RNA genome, whereas the A-B dimer is disordered and free from RNA interactions (36). In the flock house virus, the C-C dimeric contact acquires a flat conformation to accommodate the RNA duplex, whereas the A-B dimer is in a bent conformation and involves no RNA (37). The different orientation observed between the HEV C-C dimer and A-B dimer may result from the difference in RNA occupancy. The A-B dimers do not interact with RNA and have a bent conformation. As a result, the angled contact of the M/S domains accommodated the proline-rich hinge within the V-shaped cleft, similar to that in the T=1 VLP, thus solidifying the orientation of the P domain (Fig. 5C). In contrast, the contact with the RNA led the C-C dimer to a flat conformation that pushes the hinge out of the cleft. Thus, the P domain in the C-C dimer is flexible and could take a 90° rotation from the orientation in the A-B dimer (Fig. 5D).
The native HEV capsid was predicted to possess T=3 icosahedral symmetry (8, 11), and Guu et al. (11) suggested that such T=3 capsids require the dimer to have both flat and inwardly bent conformation at quasi-two-fold and icosahedral two-fold positions, respectively. Here, with detailed features, our cryo-EM structure of T=3 VLP provided direct observation of the two unique conformations between the A/B and C/C dimer at the foothold of S-domain contact but also in the arrangement of the protruding P-domain.
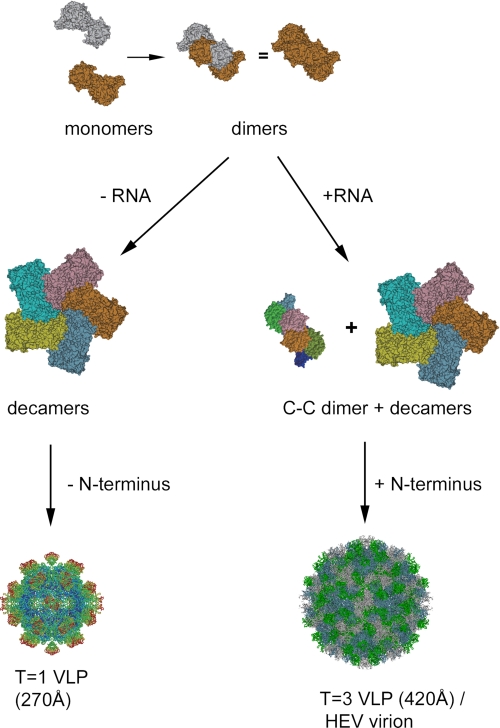
It is suggested by molecular simulation that T=3 icosahedral capsid assembly utilizes a mechanism in which preformed aggregates of intermediates combine in contrast to the formation of the T=1 icosahedral capsid that includes the addition of predominately monomers (38). The ORF2 decamer is therefore the assembly intermediate of T=3 HEV capsid and is located at each of the five-fold vertexes. The appearance of a hexameric ring at icosahedral three-fold positions is the critical step in T=3 capsid assembly and depends on the C-C dimer. The in vitro disassembly and reassembly also indicates the involvement of an extrinsic factor other than the ORF2 protein in the assembly of T=3 VLP and the C-C dimer is in a flat conformation that is concomitant with RNA binding. The induction of C-C conformation has been reported with bacteriophage MS2, where the complete assembly of capsid requires the presence of synthetic RNA fragment (39). Therefore, the pentamer of dimer (Fig. 4C) may be the assembly intermediate that is common for the T=3 virus. The interaction of RNA with the N-terminal end of ORF2 is the driving force leading the C-C dimer to the flat formation and ultimately full capsid formation through the integration of 30 copies of C-C dimers with 12 copies of A-B decamers (Fig. 6).
FIGURE 6.
Diagram showing the putative assembly process of HEV T=1 and T=3 VLP. The ORF2 subunit encodes information that governs the assembly of decamers. Interaction with the RNA fragment induces flat dimeric contact and the formation of C-C dimers, which guides the assembly of the complete icosahedral capsid.
The existence of the N-terminal amino acids 14–111 prevents ORF2 proteins from forming T=1 VLP. The capsid of T=1 VLP encloses a central cavity with a volume allowing a maximum of 55 additional residues on each copy of PORF2 protein, if the average protein density is considered to be 1.30 g/ml. The maximum diameter of the central cavity is about 340 Å for the T=3 VLP and thus is sufficient to accommodate both the HEV genome and the ORF2 N-terminal domains. By characterizing the size and the sequence of the encapsidated RNA, we found that the T=3 HEV VLP selectively encapsidated the RNA fragment that encodes the ORF2 protein. Thus, it is very possible that the native HEV capsid is the T=3 icosahedron. There, the encapsulated genomic RNA may play a direct role in the assembly of the HEV infectious virion. Our data demonstrated here that HEV was different from caliciviruses in its assembly pathway, protein domain arrangement, and genome organization, although both viruses are T=3 icosahedral particles with dimeric spikes. Hepatitis E virus showed a high similarity to some plant viruses in its assembly pathway: the utilization of a long electropositive N-terminal domain to interact with genomic RNA. Although the evolutionary origin for such similarities requires further investigation, our data place the HEV structure in a unique position, deviating from that of human caliciviruses and approaching that of T=3 small plant viruses.
Supplementary Material
Acknowledgments
We are grateful to Masaaki Kawano and Carlos G. Moscoso for fruitful discussion on the technical aspects of the manuscript and to Akifumi Higashiura and Atsushi Nakagawa for technical support. We thank Kai Sun from the University of Michigan for technical assistance on x-ray photoelectron spectroscopy analysis. The Brookhaven National Laboratory STEM facility is supported by the Department of Energy, Office of Health and Environmental Research.
This work was supported, in whole or in part, by a grant from the National Institutes of Health Roadmap Project on Nanomedicine (to R. H. C.). This work was also supported by grants from the U. S. Department of Agriculture Hatch Fund, STINT Foundation, and Strategic Research Foundation (to R. H. C.). This study was also partly funded by a grant from the Swedish Research Council (to L. X.) and grants for Research on Emerging and Re-emerging Infectious Diseases, Research on Hepatitis, and Research on Food Safety from the Ministry of Health, Labor, and Welfare, Japan (to T.- C. L.).
The atomic coordinates and structure factors (codes 2ZZQ and 3IYO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The EM data reported in this paper have been submitted to the Electron Microscopy Data Bank (Electron Microscopy Data Bank) with accession number EMD-5173.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and supplemental Table 1.
L. Xing, C.-Y. Wang, T.-C. Li, Y. Yasutomi, J. Lara, Y. Khudyakov, D. Schofield, S. Emerson, R. Purcell, N. Takeda, T. Miyamura, and R. H. Cheng, manuscript in preparation.
- HEV
- hepatitis E virus
- VLP
- virus-like particle
- TEM
- transmission electron microscopy
- STEM
- scanning TEM
- TMV
- tobacco mosaic virus
- DIG
- digoxigenin.
REFERENCES
- 1.Naik S. R., Aggarwal R., Salunke P. N., Mehrotra N. N. (1992) Bull. World Health Organ 70, 597–604 [PMC free article] [PubMed] [Google Scholar]
- 2.Tam A. W., Smith M. M., Guerra M. E., Huang C. C., Bradley D. W., Fry K. E., Reyes G. R. (1991) Virology 185, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schofield D. J., Purcell R. H., Nguyen H. T., Emerson S. U. (2003) Vaccine 22, 257–267 [DOI] [PubMed] [Google Scholar]
- 4.Li T. C., Suzaki Y., Ami Y., Dhole T. N., Miyamura T., Takeda N. (2004) Vaccine 22, 370–377 [DOI] [PubMed] [Google Scholar]
- 5.Purdy M. A., McCaustland K. A., Krawczynski K., Spelbring J., Reyes G. R., Bradley D. W. (1993) J. Med. Virol. 41, 90–94 [DOI] [PubMed] [Google Scholar]
- 6.Riddell M. A., Li F., Anderson D. A. (2000) J. Virol. 74, 8011–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li T. C., Yamakawa Y., Suzuki K., Tatsumi M., Razak M. A., Uchida T., Takeda N., Miyamura T. (1997) J. Virol. 71, 7207–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing L., Kato K., Li T., Takeda N., Miyamura T., Hammar L., Cheng R. H. (1999) Virology 265, 35–45 [DOI] [PubMed] [Google Scholar]
- 9.Li T. C., Takeda N., Miyamura T., Matsuura Y., Wang J. C., Engvall H., Hammar L., Xing L., Cheng R. H. (2005) J. Virol. 79, 12999–13006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita T., Mori Y., Miyazaki N., Cheng R. H., Yoshimura M., Unno H., Shima R., Moriishi K., Tsukihara T., Li T. C., Takeda N., Miyamura T., Matsuura Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12986–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guu T. S., Liu Z., Ye Q., Mata D. A., Li K., Yin C., Zhang J., Tao Y. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12992–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balayan M. S., Andjaparidze A. G., Savinskaya S. S., Ketiladze E. S., Braginsky D. M., Savinov A. P., Poleschuk V. F. (1983) Intervirology 20, 23–31 [DOI] [PubMed] [Google Scholar]
- 13.Li T. C., Scotti P. D., Miyamura T., Takeda N. (2007) J. Virol. 81, 10890–10896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall J. S., Hainfeld J. F., Simon M. N. (1998) Methods Cell Biol. 53, 139–164 [DOI] [PubMed] [Google Scholar]
- 15.Wall J. S., Simon M. N. (2001) Methods Mol. Biol. 148, 589–601 [DOI] [PubMed] [Google Scholar]
- 16.Baker T. S., Cheng R. H. (1996) J. Struct. Biol. 116, 120–130 [DOI] [PubMed] [Google Scholar]
- 17.Ji Y., Marinescu D. C., Zhang W., Zhang X., Yan X., Baker T. S. (2006) J. Struct. Biol. 154, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 19.Chacón P., Wriggers W. (2002) J. Mol. Biol. 317, 375–384 [DOI] [PubMed] [Google Scholar]
- 20.Wang C. Y., Miyazaki N., Yamashita T., Higashiura A., Nakagawa A., Li T. C., Takeda N., Xing L., Hjalmarsson E., Friberg C., Liou D. M., Sung Y. J., Tsukihara T., Matsuura Y., Miyamura T., Cheng R. H. (2008) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blow D. M., Rossmann M. G., Jeffery B. A. (1964) J. Mol. Biol. 8, 65–78 [DOI] [PubMed] [Google Scholar]
- 22.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 23.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR. 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 24.Kraulis P. (1991) J. Appl. Cryst. 24, 946–950 [Google Scholar]
- 25.Merritt E. A., Murphy M. E. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 869–873 [DOI] [PubMed] [Google Scholar]
- 26.Rossmann M. G., Johnson J. E. (1989) Ann. Rev. Biochem. 58, 533–573 [DOI] [PubMed] [Google Scholar]
- 27.Harrison S. C. (2001) Curr. Opin. Struct. Biol. 11, 195–199 [DOI] [PubMed] [Google Scholar]
- 28.Zhai L., Dai X., Meng J. (2006) Virus Res. 120, 57–69 [DOI] [PubMed] [Google Scholar]
- 29.He S., Miao J., Zheng Z., Wu T., Xie M., Tang M., Zhang J., Ng M. H., Xia N. S. (2008) J. Gen. Virol. 89, 245–249 [DOI] [PubMed] [Google Scholar]
- 30.Niikura M., Takamura S., Kim G., Kawai S., Saijo M., Morikawa S., Kurane I., Li T. C., Takeda N., Yasutomi Y. (2002) Virology 293, 273–280 [DOI] [PubMed] [Google Scholar]
- 31.Takamura S., Niikura M., Li T. C., Takeda N., Kusagawa S., Takebe Y., Miyamura T., Yasutomi Y. (2004) Gene Ther. 11, 628–635 [DOI] [PubMed] [Google Scholar]
- 32.Chen R., Neill J. D., Estes M. K., Prasad B. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prasad B. V., Hardy M. E., Dokland T., Bella J., Rossmann M. G., Estes M. K. (1999) Science 286, 287–290 [DOI] [PubMed] [Google Scholar]
- 34.Prasad B. V., Matson D. O., Smith A. W. (1994) J. Mol. Biol. 240, 256–264 [DOI] [PubMed] [Google Scholar]
- 35.Bertolotti-Ciarlet A., White L. J., Chen R., Prasad B. V., Estes M. K. (2002) J. Virol. 76, 4044–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmins P. A., Wild D., Witz J. (1994) Structure 2, 1191–1201 [DOI] [PubMed] [Google Scholar]
- 37.Fisher A. J., Johnson J. E. (1993) Nature 361, 176–179 [DOI] [PubMed] [Google Scholar]
- 38.Nguyen H. D., Reddy V. S., Brooks C. L., 3rd (2009) J. Am. Chem. Soc. 131, 2606–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockley P. G., Rolfsson O., Thompson G. S., Basnak G., Francese S., Stonehouse N. J., Homans S. W., Ashcroft A. E. (2007) J. Mol. Biol. 369, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.