Abstract
Caveolin-1 has a segment of hydrophobic amino acids comprising approximately residues 103–122. We have performed an in silico analysis of the conformational preference of this segment of caveolin-1 using PepLook. We find that there is one main group of stable conformations corresponding to a hydrophobic U bent model that would not traverse the membrane. Furthermore, the calculations predict that substituting the Pro110 residue with an Ala will change the conformation to a straight hydrophobic helix that would traverse the membrane. We have expressed the P110A mutant of caveolin-1, with a FLAG tag at the N terminus, in HEK 293 cells. We evaluate the topology of the proteins with confocal immunofluorescence microscopy in these cells. We find that FLAG tag at the N terminus of the wild type caveolin-1 is not reactive with antibodies unless the cell membrane is permeabilized with detergent. This indicates that in these cells, the hydrophobic segment of this protein is not transmembrane but takes up a bent conformation, making the protein monotopic. In contrast, the FLAG tag at the N terminus of the P110A mutant is equally exposed to antibodies, before and after membrane permeabilization. We also find that the P110A mutation causes a large reduction of endocytosis of caveolae, cellular lipid accumulation, and lipid droplet formulation. In addition, we find that this mutation markedly reduces the ability of caveolin-1 to form structures with the characteristic morphology of caveolae or to partition into the detergent-resistant membranes of these cells. Thus, the single Pro residue in the membrane-inserting segment of caveolin-1 plays an important role in both the membrane topology and localization of the protein as well as its functions.
Keywords: Caveolae, Endocytosis, Lipid Droplet, Membrane, Membrane Function, Membrane Trafficking, Trafficking, Caveolin-1, Transmembrane Helix
Introduction
Caveolae are specialized domains of the plasma membrane found in most cell types and particularly abundant in highly differentiated cells, such as endothelial cells, adipocytes, or muscle cells. They were described as invaginations of the plasma membrane (1, 2). Caveolae appear to have a number of functions, including roles in signal transduction, lipid exchange, cell entry, and intracellular delivery of bacterial toxins, viruses, and growth factors (3–13), but the molecular aspects of their formation and functions are still being unraveled (14). Electron microscopy allows the visualization of caveolae as 50–100-nm flask-shaped invaginations of the plasma membrane or as circularized single or clustered vesicles underneath the plasma membrane. Caveolae are specialized membrane microdomains enriched in sphingolipids, cholesterol, and receptor proteins. It is also the site of NO production and of cholesterol efflux from the cell. Several isoforms of caveolin are characteristic proteins of caveolae (15, 16).
Caveolin inserts into membranes of phosphatidylcholine in a cholesterol-dependent manner (17) and is anchored to the membrane with a hydrophobic segment comprising residues 105–125 as well as with three palmitoyl chains attached to Cys residues. Palmitoylated proteins are known to translocate into cholesterol-rich domains (18), and this may contribute to the partitioning of caveolin to the cholesterol-rich domain of caveolae. However, a mutant form of caveolin with the three palmitoylation sites removed still translocates to caveolae (19). Thus, the protein itself must promote the interaction of caveolin-1 with cholesterol-rich domains. There is a CRAC domain adjacent to the hydrophobic segment of caveolin. CRAC domains have been suggested to be important in facilitating the translocation of peptides and proteins to cholesterol-rich domains (20, 21), and there is evidence that the CRAC domain of caveolin facilitates translocation of the protein into caveolae (22, 23).
Another feature of the interaction of caveolin with membranes is that the hydrophobic segment is thought to form a U-shaped, re-entrant helix rather than a transmembrane helix (15, 24, 25). The finding that the sequence of this hydrophobic segment of caveolin-1 is well conserved in evolution (22) suggests that it plays an important functional role.
The protein caveolin-1 is the major isoform of the caveolin family and is expressed in most cell types. Caveolin-1 expression is related to caveolar formation. There are no caveolae in caveolin-1 null cells (26), and expression of caveolin-1 in such cells results in the de novo formation of caveolae (27).
In the current study, we chose to use HEK 293 cells that express virtually no endogenous caveolin-1 and lack the most common putative fatty acid transport protein, FAT/CD36 (28, 29). Expression of caveolin-1 in these cells caused lipid uptake (28). In addition, the uptake of BODIPY-labeled lactosylceramide (LacCer)4 and globoside are selectively internalized by a caveola-related process (30–34) in human skin fibroblasts through a mechanism that is dynamin-dependent and clathrin-independent. These labeled lipids as well as labeled albumin can be used as markers for the caveolar endocytic pathway (11, 12).
We have previously shown that the Pro residue in the hydrophobic segment of diacylglycerol kinase-ϵ is important in allowing this segment to form a bend in a membrane, resulting in a re-entrant helix (35). In addition, we found that substitution of the Pro residue in the hydrophobic segment of this enzyme with Ala resulted in it converting to a transmembrane helix. Caveolin-1 also has a Pro residue in the hydrophobic segment at position 110. We determined if substitution of this residue in caveolin with Ala would allow it to become a transmembrane helix and what effect this would have on caveolar structure and function. To determine the membrane topology of caveolin-1 and its P110A mutant, we expressed an N-terminal FLAG tag-labeled form of these proteins in HEK 293 cells. If the hydrophobic segment formed a transmembrane helix, the FLAG epitope would be exposed to the cell exterior. However, if the hydrophobic segment formed a re-entrant helix, the protein would remain monotopic and be located on the cytoplasmic face of the plasma membrane. It is conceivable that the presence of the polar FLAG tag at the N terminus inhibits the formation of a re-entrant helix after passage of the protein through the translocon. However, this would seem unlikely, a priori, because of the separation of the membrane inserting segment from the N terminus and because of our finding that the same FLAG tag does not prevent the wild type caveolin-1 from forming a re-entrant helix. In addition, to know if substitution of Pro110 with Ala influenced the ability of caveolae to be endocytosed, wild type and mutant caveolin-1 were expressed in cells and analyzed for the uptake of BODIPY-LacCer and BODIPY-BSA. Another of the multiple functions of caveolae is to mediate the uptake of long-chain fatty acids, resulting in the formation of lipid droplets (36, 37). We observed the formation of lipid droplets and their localization in cells transfected with either the wild type or the P110A mutant form of caveolin-1, to determine the importance of the topology of caveolin for this function. In addition, the effects of the mutation for caveolae formation were investigated with immunoelectron microscopy.
Our studies with caveolin-1 were supplemented with in silico calculations that are in accord with several of the experimental findings and provide a thermodynamic basis for the experimental observations.
EXPERIMENTAL PROCEDURES
Construction of FLAG Epitope-tagged Caveolin-1 Expression Vectors
Mouse caveolin-1 DNA fragment was amplified from pcDNA3.1 hygro vector constructed with the fragment of interest (we are grateful to Dr. Pilch's research group, Boston University (28) for generously providing this material) by PCR. The following primers were used: forward, 5′-CCAAGCTTATGTCTGGGGGCAAATA-3′; reverse, 5′-CGGGATCCTCATATCTCTTTCTGC-3′. The fragments of interest were subcloned into the corresponding site of a p3XFLAG-CMV-7.1 mammalian vector (Sigma-Aldrich), which attaches a FLAG epitope at the N terminus of the protein. The P110A mutant of the FLAG-tagged caveolin-1 was designed using the QuikChange protocol (Stratagene, La Jolla, CA). A mutated DNA plasmid was amplified from an N-terminal caveolin-1 by 18 cycles using Pfu DNA polymerase (Fermentas) and the following mutagenic primers: forward, 5′-ACGATCTTCGGCATCGCCATGGCACTCATCTGG-3′; reverse, 5′-CCAGATGAGTGCCATGGCGATGCCGAAGATCGT-3′. After digestion of the nonmutated parental DNA with DpnI restriction enzyme, the resulting PCR mix, containing the mutated DNA plasmid, was transformed into competent cells. DNA was purified from the bacterial culture using the QIAprep miniprep kit (Qiagen). The presence of the desired mutation was verified by sequencing analysis.
Cell Culture and Transfection of Caveolin-1 in HEK 293 Cells
HEK 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (Lonza) at 37 °C in an atmosphere of 5% CO2. The p3XFLAG constructs were transiently transfected into HEK 293 cells using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Cells were transfected in parallel with the p3XFLAGCMV-7.1 vector as a control.
Western Blotting and Antibodies
In confluent 6-well plates, HEK 293 cells transfected with FLAG-tagged caveolin-1 or with the P110A were rinsed with PBS and scraped into ice-cold cell lysis buffer (50 mm Hepes, 150 mm NaCl, 10% glycerol, 0.5% Triton X-100, 1 mm EDTA, and protease inhibitor mixture (1×) (Sigma-Aldrich)). Proteins were separated by SDS-PAGE (15% gel) and then transferred onto Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore). Blotting was performed according to the manufacturer's instructions (GE Healthcare), using mouse anti-FLAG antibody followed by anti-mouse IgG secondary antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Immune complexes were detected using an ECL solution detection system (GE Healthcare). The presence of FLAG-tagged caveolin-1 and the P110A proteins were detected by Western blotting using mouse anti-FLAG M2 antibody (Sigma-Aldrich). Anti-actin and anti-Gαi1 primary antibodies were obtained from Santa Cruz Biotechnology, Inc., and anti-transferrin receptor antibody was obtained from Sigma-Aldrich.
Indirect Immunofluorescence
HEK 293 cells were grown on coverslips in 6-well plates. The cells were grown to 60–70% confluence in DMEM with 10% FBS and 1% penicillin/streptomycin. The cells were then transiently transfected with Lipofectamine 2000 reagent from Invitrogen. The medium was replaced after 5 h, and the cells were left in the incubator for 24 h. The next day, the cells were fixed with 4% PFA in PBS, pH 7.4, and after several washes the cells were incubated with 5% BSA in PBS for 1.5 h, or they were treated with 0.1% Triton X-100 in PBS for 10 min, washed three times, and then incubated with 5% BSA in PBS for 1.5 h. The cells were then rinsed with PBS and incubated with the mouse monoclonal anti-FLAG antibody (Sigma-Aldrich) in PBS (1:200) for 1.5 h at 37 °C. After rinsing three times with PBS, the cells were incubated with Alexa Fluor 488-labeled goat anti-mouse IgG (Molecular Probes/Invitrogen) in PBS (1:500) for 1 h at 37 °C. After washing five times with PBS, the glass coverslips were mounted onto the glass slides and left to dry at room temperature in the dark overnight. The coverslip was then sealed onto the slide with nail polish and left to dry. The slides were visualized using the spinning disk confocal microscope (Leica DMI 6000 B).
Fluorescent Lipid and Other Reagents
BODIPY-LacCer was from Invitrogen. To prepare for the caveolar invagination assay, BODIPY-LacCer was complexed (1:1, mol/mol) with fatty acid-free bovine serum albumin (defatted BSA) (31, 32, 38, 39) as follows; BODIPY-LacCer 25 μg was dissolved in 500 μl of chloroform/ethanol (19:1, v/v) and put into a 5-ml glass tube. It was dried under nitrogen and then under vacuum for 1 h. Dried BODIPY-LacCer was dissolved in 200 μl of absolute ethanol. An ethanolic solution of BODIPY-LacCer was added dropwise to 300 μl of defatted BSA/PBS solution. The mixture was then dialyzed against PBS overnight at 4 °C with four changes of PBS to remove the ethanol. After dialysis, the mixture was centrifuged once or twice in an ultracentrifuge for 20 min at 100,000 × g, 4 °C, and the supernatant was collected. The concentration of the BODIPY-LacCer·BSA complex was determined by measurements of fluorescence intensity, relative to known standards of BODIPY-C5-sphingomyelin (Invitrogen) using a spectrofluorimeter (AMINCO/Bowman, Series 2). Bovine serum albumin conjugated with BODIPY (33, 34) (BODIPY-BSA) was purchased from Invitrogen. BODIPY 493/503 stock solution (1 mg/ml) was prepared by dissolving 5 mg of BODIPY 493/503 (Molecular Probes) in a 5-ml ethanol aliquot and stored at −20 °C. The defatted BSA/oleic acid (OlAc) medium was prepared as indicated (40).
Incubation of Caveolin-1-expressing HEK 293 Cells with Fluorescent Probes
Either caveolin-1 or the P110A mutant was expressed into HEK 293 cells. BODIPY-LacCer was complexed to defatted BSA as described above. Cells were typically incubated for 30 min at 10 °C with either 2 μm BODIPY-LacCer·BSA or 50 μg of BODIPY-BSA, washed twice with HMEM+G buffer (13.8 mm Hepes acid, 137.0 mm NaCl, 5.4 mm KCl, 5.5 mm glucose, 2.0 mm glutamine, 0.4 mm KH2PO4, 0.18 mm Na2HPO4, minimum essential medium vitamins (Invitrogen), minimum essential medium amino acids (Invitrogen), pH 7.4) (31, 32), and further incubated for 5 min at 37 °C. Fluorescent markers (BODIPY-LacCer·BSA or BODIPY-BSA) present at the plasma membrane were then removed by back-exchange using 5% defatted BSA in HMEM without glucose (HMEM−G buffer) (four times for 10 min each at 10 °C) (39). In some experiments, HEK 293 cells were pretreated with 80 μm genistein, an inhibitor of caveola-mediated endocytosis, prior to incubation with fluorescent markers (2 h at 37 °C) and during the above mentioned steps. Cell viability in the presence of 80 μm genistein was analyzed using a cell proliferation assay kit (Millipore) and was found to be higher than 90%. After removing the excess of fluorescent markers, the cells were then fixed with 4% PFA in PBS for 15 min at room temperature, quenched with 25 mm glycine in PBS for 10 min at room temperature, permeabilized with 0.1% Triton X-100 in PBS for 2 min at room temperature, and further incubated in blocking buffer (10% fetal bovine serum in PBS) for 30 min at room temperature. The specimens were then incubated for 1.5 h at 37 °C with mouse anti-FLAG antibody, washed three times in PBS, incubated with secondary antibody conjugated to Alexa Fluor 647 for 1 h at room temperature, washed five times in PBS, and mounted in Prolong (Molecular Probes). Using a spinning disk confocal microscope (Leica DMI 6000 B), BODIPY-labeled samples were excited at 450–490 nm and viewed as green fluorescence (λem ∼540 nm). Alexa Fluor 647-labeled proteins were excited at 650 nm and viewed as red (λem ∼692 nm). For double-labeled experiments, control samples were labeled identically with the individual fluorophores and measured identically to the dual labeled samples at each wavelength to verify that there was no cross-over between emission channels.
Oil Red O Staining
HEK 293 cells were seeded in 6-well plates and transfected with FLAG-tagged caveolin-1 or with the P110A mutant. Cells supplemented with OlAc for the indicated time (36) were washed with PBS and fixed with 10% formalin in PBS for 15 min. This reagent was discarded, and then fresh formalin was added, and the samples were incubated for 1 h. After two washes in PBS, cells were incubated with 60% isopropyl alcohol for 5 min at room temperature. Cells were dried completely at room temperature, stained for 10 min in freshly diluted Oil Red O (Sigma-Aldrich) solution (6 parts Oil Red O stock solution and 4 parts H2O; Oil Red O stock solution is 0.5% Oil Red O in isopropyl alcohol). The cells were stained at room temperature for 20 min, followed by passage through a 0.2-μm filter. The stain was removed, and cells were washed with water four times. The plates were measured at A490 nm in a plate reader.
Determination of Triglyceride Content
Because the HEK 293 cells lack the most common putative fatty acid transporter, FAT/CD36, we verified that caveolin-dependent fatty acid uptake led to the accumulation of triglyceride. Triglyceride content was determined in cell lysates using a colorimetric Triglyceride assay as previously described (36) according to the manufacturer's instructions (Point Scientific Inc.). Cell lysates were prepared using 1% Igepal CA-630 in PBS, and results were normalized to 100 μg of total cellular protein.
BODIPY 493/503 Staining
Cells were plated on 8-well chamber glass slides (Lab-Tek II, Nalge Nunc) and incubated overnight. Growth medium was changed to 400 μm defatted BSA/OlAc medium with serum and incubated for 24 h. Cells were transiently transfected in the same medium and incubated for 48 h. Transfected cells were washed twice with PBS, fixed by incubating with 3% PFA for 20 min at room temperature, and washed four times with PBS. The blocking buffer (0.2 m glycine, 0.1 mg/ml saponin, and 30 mg/ml BSA in PBS) was added for 45 min at room temperature to prevent nonspecific antibody binding. Cells were incubated with anti-FLAG antibody 1:200 in the antibody diluent (0.1 mg/ml saponin, 1 mg/ml BSA in PBS) for 1.5 h at 37 °C and washed four times (10 min each) with PBS on a gently rocking platform. The anti-mouse IgG Alexa Fluor 647 was diluted 1:150 in the antibody diluent containing the BODIPY 493/503 (final concentration 1 μg/ml), and the cells were incubated for 1 h at room temperature. Cells were washed four times as above, and the coverslips were mounted onto slides with 20–40 μl of SlowFade mounting medium (Invitrogen) (40–43). Four specimens, individually prepared, were observed with a multiphoton excitation microscope (Leica DMI 6000 B; scanner, Leica TCS SP5 with AOBS).
Successive Detergent Extraction of Lipid Rafts
Cells were plated on a 10-cm culture dish and transiently transfected. Cells were scraped in ice-cold PBS, 2 mm EDTA and pelleted by centrifugation at 2,506 × g for 7 min at 4 °C. Cells were resuspended in 50 mm HEPES, pH 7.4, 10 mm NaCl, 5 mm MgCl2, and 0.1 mm EDTA, supplemented with protease inhibitor mixture, and subjected to mechanical disruption with 12 strokes of a Dounce homogenizer. Following centrifugation at 14,000 × g for 20 min at 4 °C, the supernatant (cytosol) was removed. Cells were resuspended in buffer A (25 mm MES, 150 mm NaCl, pH 6.5). To this, an equal volume of the same buffer with 2% Triton X-100, 2 mm Na3VO4, and 2 mm phenylmethylsulfonyl fluoride was added, and the cells were incubated on ice for 30 min. Insoluble fractions were pelleted in a microcentrifuge (14,000 × g) for 20 min at 4 °C. The supernatant was removed (detergent soluble membrane fraction), and the insoluble pellet was resuspended in buffer B (1% Triton X-100, 10 mm Tris (pH 7.6), 500 mm NaCl, 2 mm Na3VO4, 60 mm β-octyl glucoside (Sigma-Aldrich), and 1 mm phenylmethylsulfonyl fluoride) for 30 min on ice. Debris was pelleted in a microcentrifuge (14,000 × g) for 20 min at 4 °C, and the supernatant was collected. This fraction is referred to as the detergent-resistant membrane (44–48). Immunoblotting was performed as described above.
Immunoelectron Microscopy
Either caveolin-1 or the P110A mutant was expressed in HEK 293 cells on 6-well plates. Cells were fixed with 4% (w/v) PFA and then dehydrated using increasing concentrations of ethanol. Propylene oxide was added to remove cell layers from plates. Cell layers were placed in vials, rinsed with propylene oxide, and then rinsed with 100% EtOH (3 × 10 min). The cell layers were infiltrated and embedded with LR White resin (Polysciences Inc.) and then polymerized at −20 °C under UV light. 100-nm ultrathin sections were prepared and mounted on Formvar-coated nickel grids. Ultrathin sections were incubated in PBS containing 5% normal goat serum (NGS) and 1% BSA (PBS/BSA/NGS) for 10 min and then incubated for 1 h with affinity-purified mouse anti-FLAG antibodies diluted 1:100 in PBS/BSA/NGS. The sections were rinsed in PBS and then incubated for 1 h in a 1:30 dilution of 6-nm colloidal gold-labeled anti-mouse IgG and IgM (Jackson ImmunoResearch Laboratories, Inc.) in PBS/BSA/NGS, followed by a rinse in PBS and distilled water. The 6-nm gold particles were enhanced using an R-Gent SE-EM silver enhancing kit (Aurion). Immunolabeled sections were counterstained with uranyl acetate and lead citrate and then observed using a transmission electron microscope (JEM-1200EX, JEOL, Tokyo, Japan).
Molecular Modeling
Sequences used for modeling are for the native fragments 103–122 (103LSALFGIPMALIWGIYFAIL122) and 94–122 (94VTKYWFYRLLSALFGIPMALIWGIYFAIL122); for the mutants, Pro110 is substituted by A.
PepLook
we used the Boltzmann-stochastic in silico procedure, PepLook, from Biosiris_RA, France (available on the World Wide Web) (49) to calculate the populations of three-dimensional models of caveolin sequence fragments. The N- and C-ends of every peptide were NH and CO to take into account their central positions in the protein sequence. The PepLook procedure implicates iterative runs of calculation of 10,000 different random models. At each run, 10,000 random models are generated from the sequence fragment and from a set of 64 pairs of Φ/Ψ of angles randomly sorted for every residue. Energies of all conformations are calculated and ranked using the force field described previously (35). In the first step, the probability of sorting any Φ/Ψ value for a residue is equal; in the next steps, probability varies according to whether tested angle values had contributed to exclusively poor or exclusively good structural solutions. The calculation steps are iterated up to when the sum of all Φ/Ψ angle probability remains constant. The process is stopped, and the 99 structures of lower energy are saved. The 99 models are compared on the basis of their root mean square (r.m.s.) deviations (r.m.s. deviation matrices). Three-dimensional models were calculated considering a hydrophobic environment. The insertion of the three-dimensional model in the membrane was tested using the slab model IMPALA as described previously (50).
RESULTS
Fluorescence Microscopy Immunodetection of the FLAG Epitope of FLAG-Caveolin-1 and the P110A Mutant
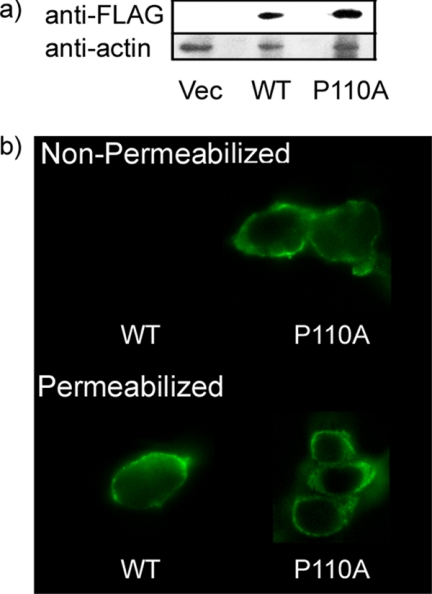
We expressed FLAG-tagged caveolin-1 and the P110A mutant form in HEK 293 cells that have little or no detectable endogenous caveolin-1 (28, 51) (Fig. 1a). The present study allowed detection of the topology of the N terminus of the FLAG-tagged proteins by comparing the exposure of the FLAG tag to antibody before and after detergent permeabilization of the cell membrane. The results show that for the wild type FLAG-tagged caveolin-1, more of the N-terminal FLAG tag is detected upon cell permeabilization (Fig. 1b) with much of the caveolin-1 label in the plasma membrane. We show a representative result. Empty vector control cells (not shown) exhibit only background fluorescence. In contrast, with the P110A mutant, the fluorescent signals from the permeabilized and non-permeabilized cells (Fig. 1b) are very similar, indicating that the major fraction of the P110A-caveolin-1 has a transmembrane helix. The observation that the exposure of the FLAG tag did not markedly increase upon detergent solubilization indicates that the N terminus of the mutant protein is oriented on the extracellular side of the membrane.
FIGURE 1.
Confocal fluorescence microscopy of HEK 293 cells transfected with either caveolin-1 or the P110A mutant. a, expression of caveolin-1 or the P110A mutant in HEK 293 cells. Shown (from the left) are transfectant of p3XFLAG vector (Vec), caveolin-1 (WT), and P110A mutant. b, confocal fluorescence microscopy of cells transfected with caveolin-1 (WT cells, left) or with the P110A mutant (right). Top, non-permeabilized cells. The cells were fixed with PFA and left non-permeabilized prior to indirect immunofluorescence using an antibody directed against the FLAG tag and an Alexa 488 secondary antibody. Bottom, permeabilized cells. The cells were fixed with PFA and permeabilized with 0.1% Triton X-100 prior to indirect immunofluorescence using an antibody directed against the FLAG tag and an Alexa 488 secondary antibody.
Caveolin Membrane Topology and Caveola-related Endocytosis
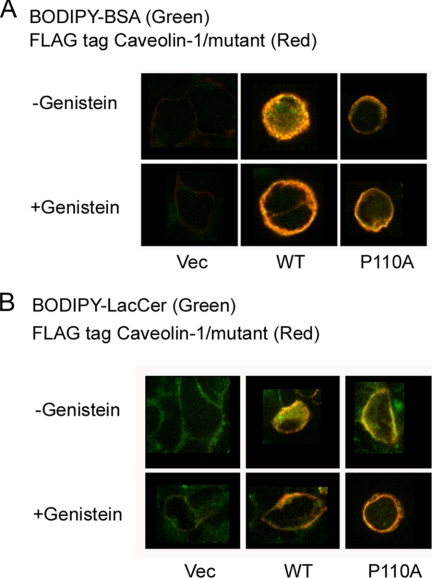
To investigate the structural importance of the topology of the hydrophobic segment of caveolin-1 for the endocytosis of caveolae, we compared HEK 293 cells expressing FLAG-tagged caveolin-1 or the P110A mutant. To induce caveola-related endocytosis, we used BODIPY-BSA and BODIPY-LacCer as markers under the conditions described under “Experimental Procedures” (31, 32). Briefly, HEK 293 cells expressing one of the FLAG-tagged proteins were incubated for 2 h either in HMEM+G medium (see “Experimental Procedures”) or in this same medium to which the caveolin-1 inhibitor, genistein, was added. Cells were labeled with BODIPY-BSA or BODIPY-LacCer and washed to remove excess BODIPY-labeled compound. After the cells were fixed with PFA, they were immunostained with anti-FLAG antibodies. We show a representative result. Untreated (not shown) and empty vector control cells exhibit only background fluorescence. The FLAG-tagged caveolin-1-expressing cells induce endocytosis of BODIPY-BSA and of BODIPY-LacCer, but endocytosis is inhibited in genistein-treated cells. This demonstrates that cells expressing FLAG-tagged caveolin-1 (WT) are capable of caveola-related endocytosis (Fig. 2). In contrast, endocytosis is not observed with cells expressing the P110A mutant with either BODIPY-BSA or with BODIPY-LacCer (Fig. 2). The different behavior in caveola-related endocytosis of the cells expressing the wild type versus the P110A mutant of caveolin-1 is probably related to the change in membrane topology of the protein as a consequence of the mutation.
FIGURE 2.
BODIPY-BSA and BODIPY-LacCer internalization in HEK 293 cells transfected with either caveolin-1 or the P110A mutant. A, top row, the cells were incubated with 50 μg of BODIPY-BSA for 30 min at 10 °C, washed, and warmed for 5 min at 37 °C before back-exchange. The cells were fixed with PFA and permeabilized with 0.1% Triton X-100 prior to indirect immunofluorescence using an antibody directed against the FLAG tag and an Alexa 647 secondary antibody. BODIPY-BSA was observed at green wavelengths, and Alexa 647 (FLAG) was observed at far red wavelengths (see “Experimental Procedures”). The figure shows the merged images. Bottom row, the cells were pretreated with 80 μm genistein, an inhibitor of caveola-mediated endocytosis, prior to incubation with fluorescent markers (2 h at 37 °C) and during the above mentioned steps. Columns show examples of different cells. B, top row, the cells were incubated with 2 μm BODIPY-LacCer·BSA for 30 min at 10 °C, washed, and warmed for 5 min at 37 °C before back-exchange. The cells were treated as for A. Bottom row, the cells were pretreated with 80 μm genistein as for A. There was background green fluorescence in the plasma membrane of control cells (the vector-transfected cells). Pictures show the midportion of cells using the confocal fluorescence microscopy. Columns show examples of different cells. Vec, vector.
Caveolin-1 Topology and the Formation of Lipid Droplets
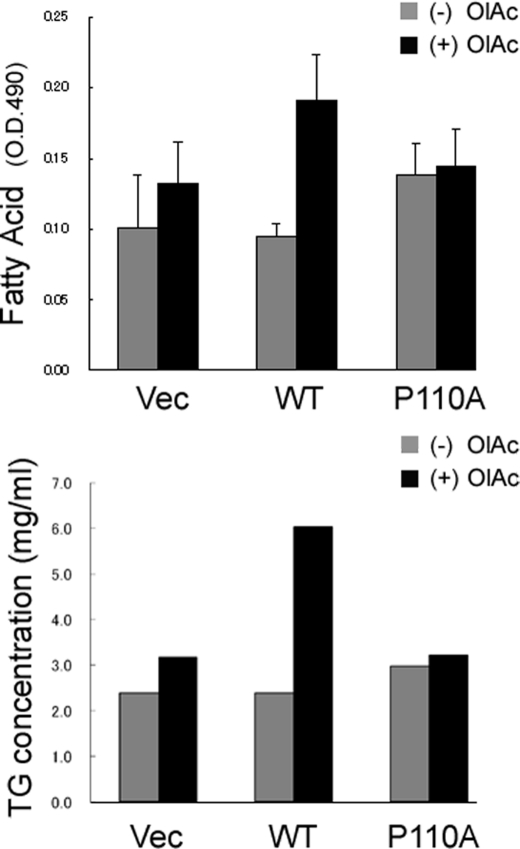
There are multiple functions of caveolae, and one of them is to mediate cell uptake of long-chain fatty acids (36, 37). The HEK 293 cells transiently transfected with either wild type or mutant caveolin-1 exhibited very different abilities to take up fatty acids and form lipid droplets. As displayed by Oil Red O staining of lipids (Fig. 3, top), expression of caveolin-1 results in substantially more lipid accumulation than does expression of the P110A mutant or with control cells. Quantitative analysis reveals an approximate doubling of triglyceride accumulation with the WT cells in the presence of OlAc (Fig. 3, bottom).
FIGURE 3.
Caveolin-1 expression enhances cellular lipid accumulation. Top, cells were incubated with 80 μm OlAc for 48 h and then fixed and stained with Oil Red O. Expression of wild type caveolin-1 (WT) upon the addition of OlAc results in substantially enhanced lipid accumulation compared with cells transfected with the empty vector (Vec) or with cells transfected with the P110A mutant (P110A). Bottom, triglyceride accumulation was determined and normalized to total cellular protein. The cells were incubated either with or without 80 μm OlAc for 48 h. Error bars, S.D. < 0.001.
Caveolin Association with Lipid Droplets
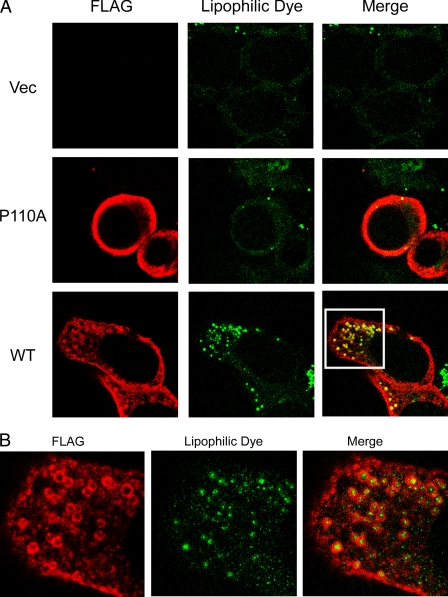
Lipid droplets consist of a core of neutral lipids, predominantly triacylglycerols and cholesteryl esters. This core is surrounded by a phospholipid monolayer and associated proteins. An especially important role for lipid droplets is the storage of cholesterol in the form of cholesteryl esters. The formation of lipid droplets contributes to regulating the level of intracellular free cholesterol, along with other homeostatic mechanisms (52, 53). Early studies showed that endogenous caveolin-1 localizes to lipid droplets (42). Caveolin-1 binds to cholesterol (54) and fatty acids (55) and can be internalized from the cell surface in response to stimulation by lipids, including glycosphingolipids and cholesterol. We investigated whether either FLAG-tagged caveolin-1 or the P110A mutant could associate with lipid droplets and whether this could occur in response to lipid loading of cells grown in the presence of elevated levels of the fatty acid (oleic acid). Treatment of either FLAG-tagged caveolin-1- or the P110A mutant-expressing HEK 293 cells for 24 h with OlAc caused redistribution of caveolin-1 to ring-shaped cytoplasmic structures (Fig. 4A, bottom row) identified as lipid droplets by the colocalization with the lipophilic stain, BODIPY 493/503 (Fig. 4B). Association of caveolins with the lipid droplets was accompanied by an apparent decrease in staining at the plasma membrane for caveolin-1 in comparison with the P110A mutant (Fig. 4A, middle row), whereas the empty vector-transfected cells showed little staining (Fig. 4A, top row).
FIGURE 4.
Caveolin-1 expression enhances the formation of lipid droplets. Cells were incubated with OlAc for 48 h and labeled with anti-FLAG antibodies. A, the addition of OlAc to the culture medium causes lipid storage droplets in caveolin-1-expressing cells and redistribution of caveolin-1 (red in merged pictures) to ring-shaped cytoplasmic structures identified as lipid droplets by BODIPY 493/503 (green in merged pictures). In contrast, the mutant did not form lipid droplets. B, an enlarged image of the section bounded by the white box in A to illustrate the location of the FLAG tag surrounding the lipid droplet.
Localization of FLAG-tagged Caveolin-1 and the P110A Mutant in the Detergent-resistant Membrane Fraction
Lipid rafts are cholesterol- and sphingolipid-enriched microdomains in cell membranes that regulate phosphorylation cascades originating from membrane-bound proteins (56, 57). Caveolae are a specialized type of raft that has a distinct morphology and protein composition but a lipid composition similar to other raft domains. Resistance to detergent solubilization has been frequently used to identify proteins that associate with membrane domains having this lipid composition. Although the basis of this criterion has been criticized (58), it does provide a phenomenological property that has often proven to be useful (18). We tested whether the alteration of the Pro110 residue affects partitioning of FLAG-tagged caveolin-1 into the detergent-resistant membrane fraction. Cytosolic, non-raft membrane (membrane) and lipid raft fractions were isolated from FLAG-tagged caveolin-1 or P110A mutant-expressing HEK 293 cells. In these preparations, the lipid raft fraction consists of detergent-resistant membranes that are insoluble in cold Triton X-100, from which cytoskeletal and nuclear structures are removed by pelleting of the insoluble material (see “Experimental Procedures”). Lipid raft material is solubilized in buffer containing octyl glucoside. These preparations are essentially identical to raft fractions obtained by density gradient centrifugation of Triton-insoluble material (44–48). We probed membrane and lipid raft fractions with antibodies for membrane markers shown in prior studies to be present or enriched in rafts. Heterotrimeric G proteins are enriched in rafts, and transferrin receptors are in non-raft membranes (Fig. 5). Caveolin was detected with anti-FLAG antibodies. As shown in Fig. 5, the P110A mutant protein localizes less to the lipid raft fraction of plasma membrane compared with the FLAG-tagged caveolin-1.
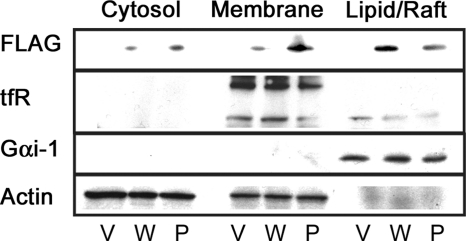
FIGURE 5.
Location in detergent-resistant membranes. Caveolin-1-expression (W) and the P110A mutant-expressing (P) cell lysates as well as an empty vector control (V) were separated by centrifugation into cytosolic (cytosol) and membrane fractions. The membrane fraction was partially solubilized with 1% Triton X-100. Equal amounts of protein from the 1% Triton-soluble (Membrane) and insoluble (Lipid/Raft) fractions were then analyzed by Western blotting. The mutant protein no longer localizes to the lipid raft fraction of plasma membrane as much as WT does. tfR, transferrin receptor.
Transmission Electron Microscopy Immunodetection of Caveolar Structure
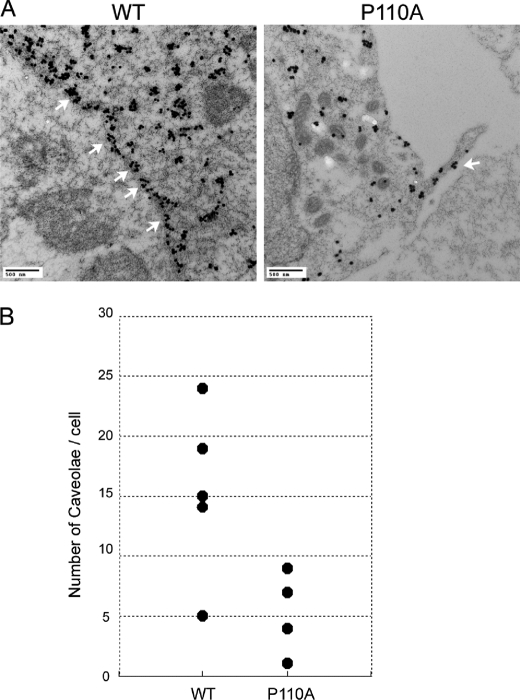
Caveolae are small, 50–100-nm invaginations of the plasma membrane defined on the basis of their morphological appearance in electron micrographs (invaginated caveolae). Caveolin-1 is an essential structural protein of caveolae. We investigated the influence of the single amino acid substitution at Pro110 on the ability of the mutant caveolin to form the morphologically distinct structure of caveolae. The number and distribution of caveolae were studied by immunoelectron microscopy of either FLAG-tagged caveolin-1 or P110A mutant-expressing HEK 293 cells. FLAG-tagged caveolin-1 and the P110A mutant were detected using anti-FLAG antibodies and 6-nm colloidal gold-labeled anti-mouse IgG and IgM. The term caveolae as used here is defined morphologically as invaginations of the plasma membrane that are readily identified by immunoelectron microscopy (Fig. 6A) (59–61). For quantification, we only included invaginated caveolae that clearly opened to the cell surface because it is clear that caveolae that may exist in a non-invaginated form cannot be distinguished from other portions of the plasma membrane and therefore are not included (Fig. 6B). As shown in Fig. 6, the plasma membrane formed typical invaginated caveolar structures in the FLAG-tagged caveolin-1-expressing cells, but there were very few such structures in the mutant-expressing cells.
FIGURE 6.
Transmission electron micrographs of thin sections of either caveolin-1- or the P110A mutant-expressing cells labeled with anti-FLAG-coated gold particles. A, the plasma membrane was invaginated by typical invaginated caveolar structures (marked by white arrows), and a greater amount of FLAG internalized in the cell on the left (WT) but not on the right (the P110A mutant). B, scatter plots of the number of caveolae in each cell. The P110A mutant-expressing cells do not form caveolae as much as WT do.
Modeling
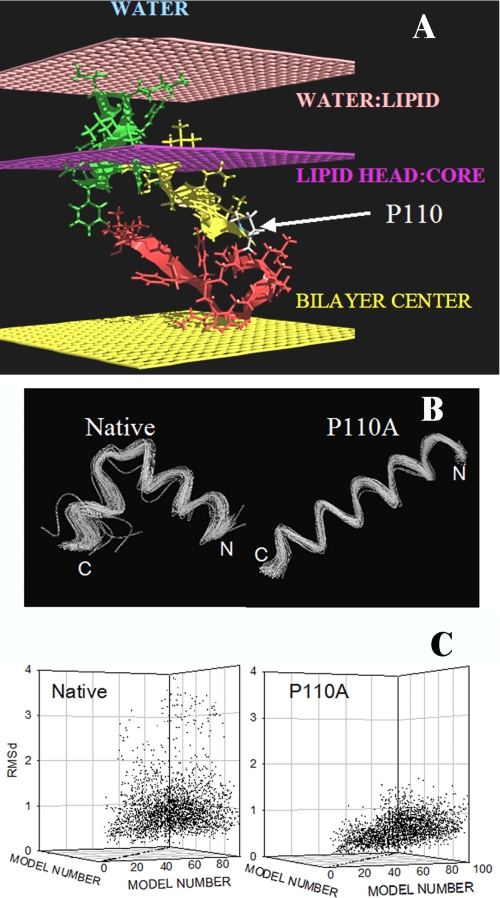
The 99 lower energy conformations of the caveolin segment 103–122 are bent conformations (Fig. 7B). This U conformation is helical on the N-side and more extended on the C-side. The U form is lost when Pro110 is replaced by Ala, resulting in the 99 lower energy conformations being straight helices.
FIGURE 7.
Modeling of the transmembrane segment of caveolin-1 using PepLook. A, best position of the caveolin 94–122 fragment in a membrane. All PepLook models were systematically pulled through a membrane slab bilayer (50), across the water/lipid interfaces (light pink layers), the lipid polar heads/acyl chain interfaces (dark pink layers), and the membrane center (yellow layer). The model structure and position corresponding to the lower restraint energy is here presented. This model has the U bent conformation; the N- side is yellow, and the C-side is red. The center is in the hydrophobic layer of membrane, and the 94VTKYWFYRL102 fragment (green) is in the membrane interface layer in connection with lipid polar heads. Note that only one monolayer of the membrane bilayer is shown in the figure. The position of the proline residue is indicated. B, spline image of the 99 three-dimensional models calculated using PepLook (49). All models are fitted by adjusting their backbone atoms (N–C–C=O) to those of the lowest energy structure (the Prime), the three-dimensional model of lower energy using Qmol. The N- and C-ends are indicated. Shown are conformations of the 99 PepLook three-dimensional models of the caveolin fragment 103–122. Left, native fragment; right, P110A mutant. C, r.m.s. deviation matrices of all pairs of three-dimensional models.
The Prime structure is defined as the lowest energy structure representing the most populated conformation. The 99 lower energy conformations of the caveolin segment 103–122 as well as the P110A mutant exhibit little deviation from the three-dimensional conformation of the Prime as demonstrated by the narrow dispersion of the r.m.s. deviation matrix of couples of calculated models (Fig. 7C). For the native fragment, the median r.m.s. deviation is 0.89 ± 0.59 Å; for the P110A mutant, the median deviation is even less, 0.61 ± 0.25 Å (Fig. 7C).
If native and mutant models are homogenous structures, they are also very hydrophobic with a hydrophobic/hydrophilic accessible surface area ratio of 4.3 and 4.7 for the best models of the native fragment and the P110A mutant, respectively. Accordingly, appropriate locations of those models are in the hydrophobic core or the interface of a membrane rather than in water. We demonstrated this by systematically “pulling” all calculated three-dimensional models across a slab membrane model made using IMPALA (50). The results show that an intramembrane orientation is preferred over an interfacial one, when and only when more residues on the N-side are taken into consideration. We used the segment 94–122 (VTKYWFYRLLSALFGIPMALIWGIYFAIL). Interestingly, the added residues carry a CRAC motif (VTKYWFYR). With this fragment, the U bent conformation is inserted in the region of the membrane acyl chains with the N- and C-ends turned toward the lipid polar head interface (Fig. 7A). The additional N-end fragment (residues 94–103) anchors the native 103–122 U model within one monolayer of the membrane. In the same conditions, the P110A mutant crosses both monolayers of the bilayer (i.e. it is a transmembrane helix).
DISCUSSION
Caveolin inserts into membranes of phosphatidylcholine in a cholesterol-dependent manner (17). Many aspects of the hydrophobic segment of caveolin-1 have been investigated (22, 62, 63). Caveolin-1 is anchored to the membrane with a hydrophobic segment that promotes interaction with cholesterol-rich domains independent of palmitoylation of the Cys residues (19, 23). Studies with caveolin-1 mutants indicate that the segment comprising residues 82–101 (human) is necessary and sufficient for membrane binding and has been termed the scaffolding domain (24). Studies using model peptides indicated that a segment comprising residues 94–101, with the sequence VTKYWFYR that corresponds to a CRAC motif, promotes the formation of cholesterol-rich domains (22).
In addition to the CRAC motif, caveolin-1 also has a hydrophobic segment that is believed not to be a transmembrane helix but rather has both N- and C-terminal ends protruding from the cytoplasmic side of the membrane. This is supported by the results of the present study. In addition, we show that the membrane topology of the hydrophobic segment is altered in the P110A mutant to become transmembrane (Fig. 1).
The modeling studies support our experimental observations and provide a mechanistic rationale for our observations on the topology of caveolin-1 and the P110A mutant. For caveolin-1, analysis using PepLook of the segment 103–122 detects almost no tendency to structural polymorphism. The 99 models of lower energy give one major conformation: a U-bent helix for WT and a long helix for the P110A mutant. These two models (WT and the mutant) support the conclusion that the mutation should alter the membrane topology, in support of the experimental findings.
One of the major functions of caveolin-1 is to contribute to the formation of caveolae in the plasma membrane. The protein caveolin-1 has particular importance in cholesterol efflux (64) and binding to intracellular lipid droplets (65). Caveolae are cholesterol- and sphingomyelin-rich invaginations (50–100 nm) in the plasma membrane and have been suggested to be involved in the transport of macromolecules across endothelial cells through a process called transcytosis. Transcytosis of caveolae involves the internalization at the luminal surface of the endothelium, budding through the cell and releasing at the abluminal apical surface of the endothelium (66). The mechanism of internalization of caveolae is still the object of intense investigation. The involvement of caveolae in constitutive endocytosis is not completely established but is a likely contributing pathway. Most of what has been discovered about caveolar endocytosis comes from experiments carried out with labeled albumin internalization in endothelial cells, simian virus (SV40) entry into cells, and internalization of the ganglioside GM1-binding cholera toxin (11, 67). The clearest evidence comes from electron microscopy that shows labeled serum albumin endocytosed through caveolae and traversing endothelial cells. Albumin binds to its caveola-localized receptor GP60 and induces a signaling cascade, including caveolin-1 phosphorylation (68–72).
Recently, Dr. Pagano's group (31, 32) has developed methods to study the endocytosis of fluorescent glycosphingolipid analogs in various cell types using pathway-specific inhibitors and colocalization studies with endocytic markers and caveolin-1. They describe how the internalization mechanism for these glycosphingolipids was unaffected by varying the carbohydrate headgroup or sphingosine backbone chain length, and additionally their evidence suggested that the ceramide core of the glycosphingolipids may be important for caveolar uptake (34). Caveolar internalization was observed using the fluorescent glycosphingolipid analog, BODIPY-LacCer, in caveolin-1-transfected HeLa cells. However, cancer cell lines have low levels of caveolin-1 mRNA and low caveolin-1 expression relative to some normal terminally differentiated cell types (73, 74). The HEK cells have virtually no endogenous caveolin-1 and lack the most common putative fatty acid transport protein, FAT/CD36, which is found in caveolae (28). This cell line does not take up lipids through caveolae. However, when these cells are transfected with caveolin-1, they exhibit caveolin-dependent lipid uptake and caveolar internalization unlike untreated cells or genistein-treated caveolin-1-expressing cells (Fig. 2). One of the multiple functions of caveolae is cellular lipid accumulation. We used oleic acid as a representative long-chain fatty acid. As shown in Fig. 3, upon the addition of oleic acid, there is a marked increase in fatty acid accumulation in the cells expressing wild type caveolin but not in the non-transfected cells or in the cells expressing the mutant caveolin. This indicated the possibility that Pro110 could have an important role in determining the curvature of caveolae. It has been shown that the wild type caveolin-1 facilitates caveolae attaining its characteristic shape (75). This has been suggested to be a consequence of the hairpin membrane insertion of caveolin that would promote positive membrane curvature in the monolayer in which the re-entrant helix of caveolin was inserted. The curvature modulation would contribute both to the positive curvature found in the cytoplasmic monolayer of caveolae and the positive curvature of the phospholipid monolayer surrounding lipid droplets. This curvature modulation would not occur with the P110A mutant that forms a transmembrane helix. This difference can explain why the P110A mutant was not able to internalize fatty acids (Fig. 3) or form lipid droplets (Fig. 4) as efficiently as the wild type protein can.
The results above clearly indicate that the topology of caveolin-1 is changed by the P110A mutation from a re-entrant helix to a transmembrane helix and that this has marked effects on several functional properties. We also addressed the question of whether this change in membrane topology of caveolin-1 also affected its ability to partition into cholesterol-rich domains and if the mutant protein would still promote the formation of membrane structures having the typical morphology of caveolae. The P110A mutant protein still contains the CRAC segment adjacent to a transmembrane helix. It should be noted that in the P110A mutant, this CRAC domain would be on the external side of the plasma membrane, whereas with wild type protein, the CRAC domain interacts with the cytoplasmic leaflet. By the indirect criterion of partitioning into the detergent-resistant membrane fraction, we observe that the mutant protein does not partition with cholesterol-rich “raft” domains (Fig. 5). This is confirmed by our observation that the P110A protein does not form structures typical of caveolae (Fig. 6).
In summary, the conformational state of the caveolin-1 can be shifted toward the transmembrane arrangement by a single amino acid mutation, changing the Pro110 residue to Ala. This amino acid substitution prevents not only the localization of the protein into lipid rafts but also caveolar formulation. As a result, caveola-related endocytosis as well as lipid accumulation and caveolin-1 lipid droplet formulation are markedly reduced in cells expressing the P110A mutant of caveolin-1 in comparison with the wild type. The multiple activities of caveolin-1 are a consequence of the topology- and compartment-specific cellular localization of this protein.
Acknowledgments
We are grateful to Dr. P. F. Pilch and Dr. T. Meshulam (Boston University School of Medicine) for providing the caveolin-1 plasmid and the HEK 293 cell line. We are also grateful to Marnie Timleck for assistance with the electron microscopy study.
This work was supported by the Government of Canada, by Canadian Post-Doctoral Research Fellowships 2008-09 and by Natural Sciences and Engineering Research Council of Canada Grant 9848.
- LacCer
- lactosylceramide
- FLAG
- an epitope tag with the sequence DYKDDDDK
- OlAc
- oleic acid
- PFA
- paraformaldehyde
- EtOH
- ethanol
- NGS
- normal goat serum
- 6 m1
- monosialotetrahexosylganglioside.
REFERENCES
- 1.Palade G. E. (1953) J. Histochem. Cytochem. 1, 188–211 [DOI] [PubMed] [Google Scholar]
- 2.Yamada E. (1955) J. Biophys. Biochem. Cytol. 1, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson R. G. (1998) Annu. Rev. Biochem. 67, 199–225 [DOI] [PubMed] [Google Scholar]
- 4.Fielding C. J., Fielding P. E. (2003) Biochim. Biophys. Acta 1610, 219–228 [DOI] [PubMed] [Google Scholar]
- 5.Gleizes P. E., Noaillac-Depeyre J., Dupont M. A., Gas N. (1996) Eur. J. Cell Biol. 71, 144–153 [PubMed] [Google Scholar]
- 6.Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. (2004) J. Biol. Chem. 279, 3787–3792 [DOI] [PubMed] [Google Scholar]
- 7.Lobie P. E., Sadir R., Graichen R., Mertani H. C., Morel G. (1999) Exp. Cell Res. 246, 47–55 [DOI] [PubMed] [Google Scholar]
- 8.Marjomäki V., Pietiäinen V., Matilainen H., Upla P., Ivaska J., Nissinen L., Reunanen H., Huttunen P., Hyypiä T., Heino J. (2002) J. Virol. 76, 1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto Y., Ninomiya H., Miwa S., Masaki T. (2000) J. Biol. Chem. 275, 6439–6446 [DOI] [PubMed] [Google Scholar]
- 10.Pelkmans L., Kartenbeck J., Helenius A. (2001) Nat. Cell Biol. 3, 473–483 [DOI] [PubMed] [Google Scholar]
- 11.Schnitzer J. E., Oh P., Pinney E., Allard J. (1994) J. Cell Biol. 127, 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert W., Frank P. G., Razani B., Park D. S., Chow C. W., Lisanti M. P. (2001) J. Biol. Chem. 276, 48619–48622 [DOI] [PubMed] [Google Scholar]
- 13.Shin J. S., Gao Z., Abraham S. N. (2000) Science 289, 785–788 [DOI] [PubMed] [Google Scholar]
- 14.Parat M. O. (2009) Int. Rev. Cell Mol. Biol. 273, 117–162 [DOI] [PubMed] [Google Scholar]
- 15.Okamoto T., Schlegel A., Scherer P. E., Lisanti M. P. (1998) J. Biol. Chem. 273, 5419–5422 [DOI] [PubMed] [Google Scholar]
- 16.Williams T. M., Lisanti M. P. (2004) Genome Biol. 5, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Song K. S., Lisanti M. P. (1996) J. Biol. Chem. 271, 568–573 [PubMed] [Google Scholar]
- 18.Brown D. A., London E. (2000) J. Biol. Chem. 275, 17221–17224 [DOI] [PubMed] [Google Scholar]
- 19.Dietzen D. J., Hastings W. R., Lublin D. M. (1995) J. Biol. Chem. 270, 6838–6842 [DOI] [PubMed] [Google Scholar]
- 20.Epand R. M. (2006) Prog. Lipid Res. 45, 279–294 [DOI] [PubMed] [Google Scholar]
- 21.Li H., Papadopoulos V. (1998) Endocrinology 139, 4991–4997 [DOI] [PubMed] [Google Scholar]
- 22.Epand R. M., Sayer B. G., Epand R. F. (2005) J. Mol. Biol. 345, 339–350 [DOI] [PubMed] [Google Scholar]
- 23.Uittenbogaard A., Smart E. J. (2000) J. Biol. Chem. 275, 25595–25599 [DOI] [PubMed] [Google Scholar]
- 24.Schlegel A., Schwab R. B., Scherer P. E., Lisanti M. P. (1999) J. Biol. Chem. 274, 22660–22667 [DOI] [PubMed] [Google Scholar]
- 25.Sargiacomo M., Scherer P. E., Tang Z., Kübler E., Song K. S., Sanders M. C., Lisanti M. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9407–9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drab M., Verkade P., Elger M., Kasper M., Lohn M., Lauterbach B., Menne J., Lindschau C., Mende F., Luft F. C., Schedl A., Haller H., Kurzchalia T. V. (2001) Science 293, 2449–2452 [DOI] [PubMed] [Google Scholar]
- 27.Fra A. M., Williamson E., Simons K., Parton R. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8655–8659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meshulam T., Simard J. R., Wharton J., Hamilton J. A., Pilch P. F. (2006) Biochemistry 45, 2882–2893 [DOI] [PubMed] [Google Scholar]
- 29.Ibrahimi A., Abumrad N. A. (2002) Curr. Opin. Clin. Nutr. Metab. Care 5, 139–145 [DOI] [PubMed] [Google Scholar]
- 30.Puri V., Watanabe R., Singh R. D., Dominguez M., Brown J. C., Wheatley C. L., Marks D. L., Pagano R. E. (2001) J. Cell Biol. 154, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marks D. L., Singh R. D., Choudhury A., Wheatley C. L., Pagano R. E. (2005) Methods 36, 186–195 [DOI] [PubMed] [Google Scholar]
- 32.Singh R. D., Marks D. L., Pagano R. E. (2007) Curr. Protoc. Cell Biol. 24, Unit 24.1 [DOI] [PubMed] [Google Scholar]
- 33.Sharma D. K., Choudhury A., Singh R. D., Wheatley C. L., Marks D. L., Pagano R. E. (2003) J. Biol. Chem. 278, 7564–7572 [DOI] [PubMed] [Google Scholar]
- 34.Singh R. D., Puri V., Valiyaveettil J. T., Marks D. L., Bittman R., Pagano R. E. (2003) Mol. Biol. Cell 14, 3254–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decaffmeyer M., Shulga Y. V., Dicu A. O., Thomas A., Truant R., Topham M. K., Brasseur R., Epand R. M. (2008) J. Mol. Biol. 383, 797–809 [DOI] [PubMed] [Google Scholar]
- 36.Simard J. R., Meshulam T., Pillai B. K., Kirber M. T., Brunaldi K., Xu S., Pilch P. F., Hamilton J. A. (2010) J. Lipid Res. 51, 914–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pohl J., Ring A., Stremmel W. (2002) J. Lipid Res. 43, 1390–1399 [DOI] [PubMed] [Google Scholar]
- 38.Marks D. L., Bittman R., Pagano R. E. (2008) Histochem. Cell Biol. 130, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin O. C., Pagano R. E. (1994) J. Cell Biol. 125, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Listenberger L. L., Brown D. A. (2007) Curr. Protoc. Cell Biol. 24, Unit 24.2 [DOI] [PubMed] [Google Scholar]
- 41.Brasaemle D. L., Wolins N. E. (2006) Curr. Protoc. Cell Biol. 3, Unit 3.15 [DOI] [PubMed] [Google Scholar]
- 42.Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. (2004) Mol. Biol. Cell 15, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pol A., Luetterforst R., Lindsay M., Heino S., Ikonen E., Parton R. G. (2001) J. Cell Biol. 152, 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Vizio D., Adam R. M., Kim J., Kim R., Sotgia F., Williams T., Demichelis F., Solomon K. R., Loda M., Rubin M. A., Lisanti M. P., Freeman M. R. (2008) Cell Cycle 7, 2257–2267 [DOI] [PubMed] [Google Scholar]
- 45.Zhuang L., Kim J., Adam R. M., Solomon K. R., Freeman M. R. (2005) J. Clin. Investig. 115, 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacLellan D. L., Steen H., Adam R. M., Garlick M., Zurakowski D., Gygi S. P., Freeman M. R., Solomon K. R. (2005) Proteomics 5, 4733–4742 [DOI] [PubMed] [Google Scholar]
- 47.Zhuang L., Lin J., Lu M. L., Solomon K. R., Freeman M. R. (2002) Cancer Res. 62, 2227–2231 [PubMed] [Google Scholar]
- 48.Solomon K. R., Mallory M. A., Finberg R. W. (1998) Biochem. J. 334, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas A., Deshayes S., Decaffmeyer M., Van Eyck M. H., Charloteaux B., Brasseur R. (2006) Proteins 65, 889–897 [DOI] [PubMed] [Google Scholar]
- 50.Ducarme P., Rahman M., Brasseur R. (1998) Proteins 30, 357–371 [PubMed] [Google Scholar]
- 51.Wharton J., Meshulam T., Vallega G., Pilch P. (2005) J. Biol. Chem. 280, 13483–13486 [DOI] [PubMed] [Google Scholar]
- 52.Listenberger L. L., Brown D. A. (2008) Curr. Biol. 18, R237–R238 [DOI] [PubMed] [Google Scholar]
- 53.Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 54.Murata M., Peränen J., Schreiner R., Wieland F., Kurzchalia T. V., Simons K. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10339–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trigatti B. L., Anderson R. G., Gerber G. E. (1999) Biochem. Biophys. Res. Commun. 255, 34–39 [DOI] [PubMed] [Google Scholar]
- 56.van Deurs B., Roepstorff K., Hommelgaard A. M., Sandvig K. (2003) Trends Cell Biol. 13, 92–100 [DOI] [PubMed] [Google Scholar]
- 57.Pike L. J. (2003) J. Lipid Res. 44, 655–667 [DOI] [PubMed] [Google Scholar]
- 58.Lichtenberg D., Goñi F. M., Heerklotz H. (2005) Trends Biochem. Sci. 30, 430–436 [DOI] [PubMed] [Google Scholar]
- 59.Scheiffele P., Verkade P., Fra A. M., Virta H., Simons K., Ikonen E. (1998) J. Cell Biol. 140, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lahtinen U., Honsho M., Parton R. G., Simons K., Verkade P. (2003) FEBS Lett. 538, 85–88 [DOI] [PubMed] [Google Scholar]
- 61.Uawithya P., Tuntitippawan T., Katzenmeier G., Panyim S., Angsuthanasombat C. (1998) Biochem. Mol. Biol. Int. 44, 825–832 [DOI] [PubMed] [Google Scholar]
- 62.Spisni E., Tomasi V., Cestaro A., Tosatto S. C. (2005) Biochem. Biophys. Res. Commun. 338, 1383–1390 [DOI] [PubMed] [Google Scholar]
- 63.Karsan A., Blonder J., Law J., Yaquian E., Lucas D. A., Conrads T. P., Veenstra T. (2005) J. Proteome Res. 4, 349–357 [DOI] [PubMed] [Google Scholar]
- 64.Fu Y., Hoang A., Escher G., Parton R. G., Krozowski Z., Sviridov D. (2004) J. Biol. Chem. 279, 14140–14146 [DOI] [PubMed] [Google Scholar]
- 65.Robenek M. J., Severs N. J., Schlattmann K., Plenz G., Zimmer K. P., Troyer D., Robenek H. (2004) FASEB J. 18, 866–868 [DOI] [PubMed] [Google Scholar]
- 66.Stan R. V. (2002) Microsc. Res. Tech. 57, 350–364 [DOI] [PubMed] [Google Scholar]
- 67.Ghitescu L., Bendayan M. (1992) J. Cell Biol. 117, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Minshall R. D., Tiruppathi C., Vogel S. M., Malik A. B. (2002) Histochem. Cell Biol. 117, 105–112 [DOI] [PubMed] [Google Scholar]
- 69.Minshall R. D., Tiruppathi C., Vogel S. M., Niles W. D., Gilchrist A., Hamm H. E., Malik A. B. (2000) J. Cell Biol. 150, 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shajahan A. N., Timblin B. K., Sandoval R., Tiruppathi C., Malik A. B., Minshall R. D. (2004) J. Biol. Chem. 279, 20392–20400 [DOI] [PubMed] [Google Scholar]
- 71.Shajahan A. N., Tiruppathi C., Smrcka A. V., Malik A. B., Minshall R. D. (2004) J. Biol. Chem. 279, 48055–48062 [DOI] [PubMed] [Google Scholar]
- 72.Tiruppathi C., Song W., Bergenfeldt M., Sass P., Malik A. B. (1997) J. Biol. Chem. 272, 25968–25975 [DOI] [PubMed] [Google Scholar]
- 73.Racine C., Bélanger M., Hirabayashi H., Boucher M., Chakir J., Couet J. (1999) Biochem. Biophys. Res. Commun. 255, 580–586 [DOI] [PubMed] [Google Scholar]
- 74.Skretting G., Torgersen M. L., van Deurs B., Sandvig K. (1999) J. Cell Sci. 112, 3899–3909 [DOI] [PubMed] [Google Scholar]
- 75.Shibata Y., Hu J., Kozlov M. M., Rapoport T. A. (2009) Annu. Rev. Cell Dev. Biol. 25, 329–354 [DOI] [PubMed] [Google Scholar]