Abstract
In response to different environmental stresses, phosphorylation of eIF2 (eIF2∼P) represses global translation coincident with preferential translation of ATF4. ATF4 is a transcriptional activator of the integrated stress response, a program of gene expression involved in metabolism, nutrient uptake, anti-oxidation, and the activation of additional transcription factors, such as CHOP/GADD153, that can induce apoptosis. Although eIF2-P elicits translational control in response to many different stress arrangements, there are selected stresses, such as exposure to UV irradiation, that do not increase ATF4 expression despite robust eIF2∼P. In this study we addressed the underlying mechanism for variable expression of ATF4 in response to eIF2∼P during different stress conditions and the biological significance of omission of enhanced ATF4 function. We show that in addition to translational control, ATF4 expression is subject to transcriptional regulation. Stress conditions such as endoplasmic reticulum stress induce both transcription and translation of ATF4, which together enhance expression of ATF4 and its target genes in response to eIF2∼P. By contrast, UV irradiation represses ATF4 transcription, which diminishes ATF4 mRNA available for translation during eIF2∼P. eIF2∼P enhances cell survival in response to UV irradiation. However, forced expression of ATF4 and its target gene CHOP leads to increased sensitivity to UV irradiation. This combination of transcriptional regulation and translational control allows the eIF2 kinase pathway to selectively repress or activate key regulatory genes subject to preferential translation, providing the integrated stress response versatility to direct the transcriptome that is essential for maintaining the balance between stress remediation and apoptosis.
Keywords: ER Stress, Gene Expression, Protein Synthesis, Translation, Translation Control, Translation Initiation Factors, Translation Regulation, Integrated Stress Response, eIF2 Kinase
Introduction
Protein synthesis is dynamic, with rapid reductions in translation in response to many different cellular stresses. An important contributor to this stress adaptation is a family of protein kinases that phosphorylates eIF2 (eIF2∼P), a translation initiation factor that associates with initiator Met-tRNAiMet and GTP and facilitates 40 S ribosome binding to mRNA and subsequent recognition of the start codon (1, 2). In response to disruption of protein folding and assembly in the endoplasmic reticulum (ER),2 PERK (EIF2AK3/PEK) phosphorylation of the α subunit of eIF2 at serine 51 blocks the exchange of eIF2-GDP to eIF2-GTP required for binding and delivery of the initiator tRNA to the ribosomal machinery. The ensuing block in global protein synthesis reduces the influx of nascent polypeptides into the overloaded ER secretory pathway (3). Accompanying this global translational control, eIF2∼P selectively enhances the translation of ATF4 mRNA, encoding a transcription activator of genes important for essential adaptive functions (2, 4–6).
Preferential translation of ATF4 involves two upstream ORFs (uORFs) located in the 5′-leader of the ATF4 mRNA. After translation of uORF1, eIF2∼P delays ribosomal reinitiation, facilitating the bypass of the inhibitory uORF2, leading to enhanced translation of the ATF4 coding region (6). This leads to increased levels of the ATF4 transcription factor that then functions in conjunction with additional ER stress regulators, ATF6 and IRE1, to induce the transcription of genes involved in the unfolded protein response. Collectively, expression of the unfolded protein response genes lead to an enhanced processing capacity of the secretory pathway (3). In addition to PERK, there are other eIF2 kinases, including GCN2 that directs translational control in response to nutrient starvation and UV irradiation (7–9), PKR which participates in an anti-viral defense mechanism mediated by interferon (10–12), and HRI that is activated by heme deprivation in erythroid tissues (13, 14).
The idea that ATF4 is a common downstream target that integrates signaling from different eIF2 kinases has led to the eIF2∼P/ATF4 pathway being collectively referred to as the integrated stress response (ISR) (15). The ATF4 target genes are involved in protein folding and assembly, metabolism, nutrient uptake, gene expression, alleviation of oxidative stress, and the regulation of apoptosis (15, 16). Although the ISR serves essential adaptive functions, perturbations in or unabated induction of this stress response can contribute to morbidity. In this sense, the ISR, which ameliorates cellular damage in response to environmental stresses, becomes maladaptive (16–18). The processes by which the ISR can adversely affect cells is not well understood, but central to this process is the ATF4-target gene, CHOP/GADD153, a transcriptional regulator whose extended expression during stress can trigger apoptosis (1, 16, 17, 19).
A range of different environmental stresses has been reported to elicit the ISR. That is not to say that activation of the eIF2∼P/ATF4 pathway and its target genes is indistinguishable between various stress arrangements. Clearly there can be important differences in gene expression that are required for optimal alleviation of each stress condition. The underlying reason for the differences in gene expression elicited by eIF2∼P during various stress arrangements can involve the combined actions between the ISR and other stress response pathways. For example, PERK functions in combination with the unfolded protein response regulators, and GCN2 is integrated with TORC1 during nutrient stress (3, 20–23). A second reason for the distinct gene expression profiles is that there are some stress conditions, such as UV irradiation, that elicit robust eIF2∼P that is required for cell survival yet do not enhance ATF4 expression (8). Furthermore, eIF2∼P in the absence of induced ATF4 was also reported in brain ischemia and non-alcoholic steatohepatitis (24, 25).
In this study we addressed the underlying mechanism for variable expression of ATF4 in response to eIF2∼P induced by different stress conditions and the biological significance of omission of enhanced ATF4 function. We show that in addition to translational control, ATF4 expression is subject to transcriptional regulation. Stress conditions, such as ER stress, show significant induction of both transcription and translation of ATF4, which together enhance expression of ATF4 and its target genes in response to eIF2∼P. By comparison, UV irradiation represses ATF4 transcription, which diminishes mRNA available for translation during eIF2∼P. This combination of transcriptional regulation and translational control allows the eIF2 kinase pathway to selectively repress or activate key regulatory genes subject to preferential translation, providing the ISR versatility to direct the transcriptome and cell survival during different environmental stresses.
EXPERIMENTAL PROCEDURES
Cell Culture and Stress Conditions
ATF4−/−, CHOP−/−, and A/A (eIF2α-S51A) mouse embryonic fibroblast (MEF) cells along with their wild-type counterparts were described previously (26–31). MEF cells were cultured in Dulbecco's modified Eagle's media (DMEM), which was supplemented with 1 mm nonessential amino acids, 10% (v/v) fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C. ATF4−/− cells require additional essential amino acids along with 55 μm β-mercaptoethanol, and thus, all cell lines were cultured with the same enriched media when compared in the described experiments. Human glioblastoma M059K cells were cultured in medium containing a 1:1 mixture of DMEM and Ham's F-12 medium supplemented with 0.05 mm nonessential amino acids and 10% fetal bovine serum (32). Primary normal human keratinocyte cells were isolated from foreskin tissue and cultured in EpiLife medium (Cascade Biologics) supplemented with human keratinocyte growth supplement, Cascade Biologics) and 1000 units/ml penicillin and 1000 μg/ml streptomycin (Roche Applied Science) (33). Cells were cultured to 70% confluence and irradiated with the indicated dose of UV-C or UV-B followed by further incubation for the indicated number of hours (8, 33). Alternatively, cells were treated with up to 200 μm methyl methane sulfonate (MMS) or 1 μm thapsigargin for up to 6 h as indicated. In some cases MEF cells were pretreated with 10 μm salubrinal-003 for the indicated number of hours.
To measure the ATF4 mRNA half-life, wild-type MEF cells were treated with 1 μm thapsigargin, 40 J/m2 UV-C irradiation, or no stress. One hour later the cells were treated with 20 μm actinomycin D, the cells were cultured for 1, 2, or 4 h, and ATF4 mRNA levels were measured by qRT-PCR as described below. To determine whether protein synthesis is required for transcriptional regulation of ATF4 in response to ER stress or UV irradiation, wild-type MEF cells were treated with 50 μg/ml cycloheximide for 30 min. Cells were then treated with 1 μg/ml thapsigargin, 40 J/m2 UV-C irradiation, or no stress and then cultured for an additional 3 or 6 h. ATF4 mRNA levels were measured by qRT-PCR following the experimental details highlighted below.
Preparation of Protein Lysates and Immunoblot Analysis
Cultured cells treated with or without stress agents were washed twice with ice-cold phosphate saline buffer (pH 7.4) and lysed in a solution containing 50 mm Tris-HCl (pH 7.9), 150 mm NaCl, 0.1% SDS, 100 mm NaF, 17.5 mm glycerol phosphate, and 10% glycerol supplemented with protease inhibitors (100 μm phenylmethylsulfonyl fluoride, 0.15 μm aprotinin, 1 μm leupeptin and 1 μm pepstatin). Lysates were subjected to sonication for 30 s and precleared by centrifugation. Protein concentrations were measured by the Bio-Rad protein quantification kit for detergent lysis, and equal amounts of protein were subjected to electrophoresis by SDS/PAGE along with low or high molecular weight markers (Bio-Rad). Proteins were transferred to nitrocellulose filters and subsequently incubated with TBS-T solution (20 mm Tris/HCl (pH 7.9), 150 mm NaCl, and 0.2% Tween 20) supplemented with 4% (w/v) nonfat dried milk for 1 h followed by an overnight incubation with antibodies specific for phosphorylated eIF2α at serine 51, cleaved PARP, or cleaved caspase-3 (Cell Signaling Technologies). ATF4 antibody was prepared against recombinant protein (34). CHOP and β-actin antibodies were obtained from Santa Cruz Biotechnology (sc-7351) and Sigma (A5441), respectively. Monoclonal antibody that recognizes either phosphorylated or nonphosphorylated forms of eIF2α was provided by Dr. Scot Kimball (Pennsylvania State University College of Medicine, Hershey, PA). Filters were washed in TBS-Tween 3 times and subsequently incubated with secondary antibody tagged with horseradish peroxidase and chemiluminescent solution. Proteins bound to antibody in the immunoblots were visualized by exposure to x-ray film and by imaging using the LI-COR Odyssey system. Images shown in the figures are representative of three independent experiments.
Sucrose Gradient Centrifugation and Polysome Analysis
Cultured wild-type and A/A cells were treated with 1 μm thapsigargin for 6 h or subjected to 40 J/m2 UV-C irradiation and cultured for 6 h. Polysome analyses were carried out as described previously (34). Before lysis, cells were treated with 50 μg/ml cycloheximide to inhibit translation elongation. Cells were washed twice with an ice-chilled phosphate saline solution containing 50 μg/ml cycloheximide, and cellular lysates were prepared in a solution of 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 100 mm NaCl, and 0.4% Nonidet P-40 supplemented with 50 μg/ml cycloheximide. Lysates were gently passed through a 23-gauge needle followed by a 10-min incubation on ice to ensure proper lysis. The preparation was then cleared by microcentrifugation (10,000 × g for 10 min) at 4 °C and quantified for RNA concentration using a UV spectrophotometer (Nanodrop). Lysates were loaded onto 10–50% sucrose gradients (20 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 100 mm NaCl, and 50 μg/ml cycloheximide) and subjected to centrifugation in a Beckman SW-41Ti rotor for 2 h at 40,000 rpm. Gradients were fractionated using a Biocomp Gradient Station. Absorbance of RNA at 254 nm was recorded by an in-line UV monitor, and polysome to monosome ratios were quantitated by comparing the areas under the recorded peaks.
RNA Isolation and Real Time PCR
MEF cells were treated with the indicated stress conditions and harvested, and total cellular RNA was prepared using TRIzol reagent (Invitrogen). Single-stranded cDNAs were synthesized using the Taqman reverse transcriptase kit (Applied Biosystems) according to the manufacturer's protocol. Quantitative PCR reactions were carried out with 400 ng of cDNA sample from each reaction using Taqman probes (Applied Biosystems) specific for detection of ATF4, p21, CHOP, GADD45av and β-actin (u) genes in a Roche LightCycler real-time PCR system. Quantitation of the target genes was normalized using the reference 18 S rRNA to compensate for inter-PCR variations. Quantification was carried out using the Light Cycler 480 software (Version 1.2.9.11) to generate Cp values. Values are a representation of three independent experiments, with standard deviations as indicated. Statistical significance was calculated by using the two-tailed Student's t test.
Plasmid Constructions and Luciferase Assays
ATF4 translational control was measured using a previously constructed PTK-ATF4-Luc plasmid containing the cDNA for the full-length 5′-leader of the ATF4 mRNA along with the ATF4 start codon, inserted between the TK promoter and firefly luciferase gene in plasmid pGL3 (6). In parallel, mutant versions of the PTK-ATF4-Luc plasmid, with mutations in the start codon of uORF1 or both uORF1 and uORF2, were analyzed. ATF4 transcription was measured by fusing a 2.5-kb mouse ATF4 promoter to the firefly luciferase gene in plasmid pGL3, generating PATF4-Luc. Transient co-transfections were carried out in triplicate using the wild-type or mutant versions of the PTK-ATF4-Luc plasmid or the PATF4-Luc reporter along with the Renilla luciferase plasmid for normalization. Plasmid transfections were performed in the MEF cells using the FuGENE 6 reagent (Roche Applied Science). 24 h after transfection, the MEF cells were exposed to vehicle alone, 1 μm thapsigargin, or 40 J/m2 UV-C irradiation and cultured for 6 h. Dual luciferase assays were carried out as described by the Promega instruction manual in triplicates, and statistical significance was calculated with a Standard t test.
Cell Survival Assays
Cells were plated in 96-well plates and treated with UV irradiation followed by culture incubation for the indicated times. Selected cultures were pretreated with salubrinal-003 for 6 h followed by UV irradiation and then incubated for the indicated times. The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed by using the CellTitre 96 nonradioactive cell proliferation assay kit (Promega) to measure the number of viable cells as described in the manufacturer's protocol. Clonogenic assays involved plating cells at a density of 500 cells/60-mm plate. 48 h after seeding, cells were then treated with UV-C irradiation and Sal-003 as indicated, and the cells were cultured for a period of 7–10 days. Cells were fixed with a solution containing 10% methanol and 10% acetic acid and stained with 0.4% crystal violet. Colonies were counted by using the AlphaImager system from Innotech and plotted from three independent experiments. Statistical significance was calculated using the Student's t test.
RESULTS
UV Irradiation Induces eIF2α Phosphorylation without Activation of ATF4 and CHOP
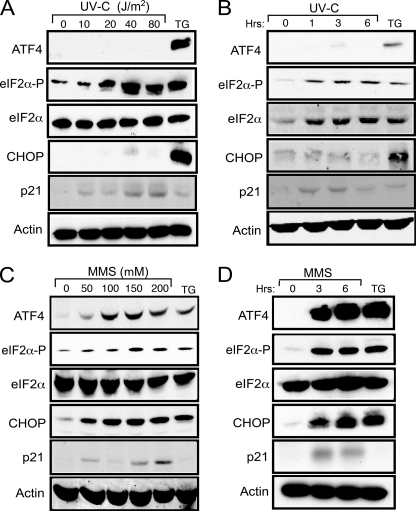
UV irradiation has been reported to induce translation control by eIF2∼P without activating its target genes, ATF4 and CHOP (8). To test this model, we measured the levels of eIF2α phosphorylation in MEF cells in response to increasing doses of UV-C irradiation (Fig. 1A). The levels of eIF2α phosphorylation were determined by immunoblot analysis using a polyclonal antibody that specifically recognizes the translation initiation factor phosphorylated at serine 51. eIF2∼P was detected with a dose range between 20 and 80 J/m2 of UV-C, with a maximum at 40 J/m2. Levels of eIF2∼P at 40 J/m2 UV-C irradiation were comparable with that measured in cells treated with 1 μm thapsigargin, a well established inducer of ER stress (Fig. 1A). Phosphorylated eIF2α was detected by 1 h after exposure to 40 J/m2 UV-C and was sustained for 6 h (Fig. 1B). Importantly, neither ISR regulators, ATF4, nor CHOP was appreciably induced in response to UV irradiation, whereas both were highly expressed during ER stress (Fig. 1, A and B). By comparison, expression of the p53 target gene, p21, was enhanced in response to UV irradiation but not during ER stress. We also carried out a dose- and time-dependent analysis of the ISR during treatment of the MEF cells with MMS, an alkylating agent that can damage DNA, and found induction of both ATF4 and CHOP (Fig. 1, C and D).
FIGURE 1.
UV irradiation elicits eIF2∼P in the absence of induced ATF4 and CHOP. A, wild-type MEF cells were treated with the indicated dosage of UV-C irradiation and then incubated in the culture medium for 6 h. B, alternatively, the MEF cells were treated with 40 J/m2 UV-C irradiation and then cultured for up to 6 h as indicated. In each case lysates were prepared from the UV-C irradiated cells, and the levels of ATF4, phosphorylated eIF2α, total eIF2α, p21, and β-actin were measured by immunoblot analysis using antibody specific to each protein. As a control, cells were subjected to ER stress elicited by 1 μm thapsigargin for 6 h, and immunoblot analyses were carried out using the cell lysates. C, MEF cells were treated with up to 200 μm MMS for 6 h as indicated. D, alternatively, the cells were exposed to 100 μm MMS for up to 6 h. Lysates were prepared from the treated cells, and the levels of the indicated proteins were measured by immunoblot analysis. Results shown in each panel are representative of three independent experiments.
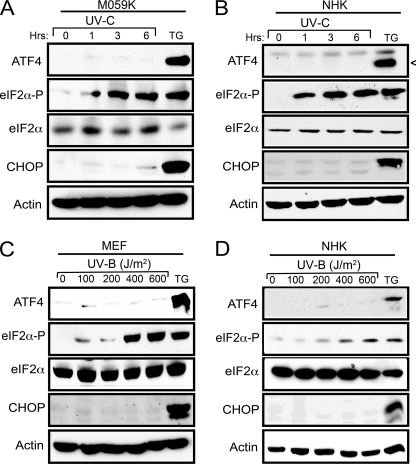
We also observed increased eIF2∼P without induced expression of the ISR genes in two different cultured human cell types (glioblastoma cell line M059K and primary human keratinocytes) treated with UV-C irradiation (Fig. 2, A and B). By contrast, in response to thapsigargin treatment, both of the human cells showed increased ATF4 and CHOP levels accompanied by eIF2∼P. Finally, we addressed whether UV-B irradiation elicits a similar discordant induction of the ISR. MEF and normal human keratinocyte cells were treated with increasing doses of UV-B irradiation, and although there were significant levels of eIF2∼P, there was no induction of ATF4 and CHOP (Fig. 2, C and D). These results indicate that both UV-C and UV-B irradiation significantly increase eIF2∼P without activation of the central ISR regulators, ATF4 and CHOP.
FIGURE 2.
UV-C and UV-B irradiation induces eIF2∼P in different cell types. Human glioblastoma cells M059K (A) and normal human keratinocytes (NHK) cells (B) were treated with 40 J/m2 UV-C irradiation and then cultured for up to 6 h. As a control, cells were exposed to 1 μm TG for 6 h. Lysates were prepared from the stressed human cells, and the levels of the indicated proteins were measured by immunoblot analysis. Wild-type MEF cells (C) and normal human keratinocyte cells (D) were treated with increasing dosages of UV-B irradiation and cultured for up to 6 h, and the indicated proteins were measured by immunoblot analyses. Each panel is representative of three independent experiments.
eIF2∼P by UV Irradiation Reduces Global Protein Synthesis
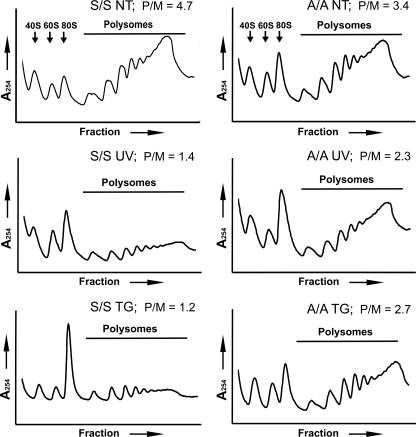
eIF2∼P is a potent repressor of translation initiation. We examined the effects of UV-induced eIF2∼P on global protein synthesis by sucrose gradient analyses of polysomes. UV irradiation of MEF cells significantly reduced polysomes, coincident with increased free ribosomes and monosomes, indicating repressed translation initiation (Fig. 3). By comparison, there was a substantial restoration of polysomes in UV-irradiated A/A MEF cells expressing a mutant form of eIF2α with alanine substituted for the phosphorylated serine 51. This result supports the model that eIF2∼P is the principal mediator of global translation repression in response to UV irradiation. This central idea is also true for ER stress, with eIF2∼P being required for a robust reduction in translation initiation (Fig. 3).
FIGURE 3.
Phosphorylation of eIF2α reduces translation initiation in response to UV irradiation or ER stress. Wild-type MEF cells (WT) or mutant cells expressing the nonphosphorylated eIF2α-S51A (A/A) were treated with 40 J/m2 UV-C irradiation (UV) followed by incubation for 6 h or to no stress treatment (NT). Alternatively, the MEF cells were subjected to ER stress by treatment with 1 μm TG for 6 h. Lysates were prepared and then subjected to centrifugation in a 10–50% sucrose gradient. Polysome profiles were generated, and absorbance was measured at 254 nm. Arrows indicate peaks corresponding to 40 S and 60 S ribosomal subunits and 80 S monosomes; the line highlights polysome fractions. The polysome to monosome (P/M) ratios are indicated in each panel.
ATF4 mRNA Is Lowered in Response to UV Irradiation
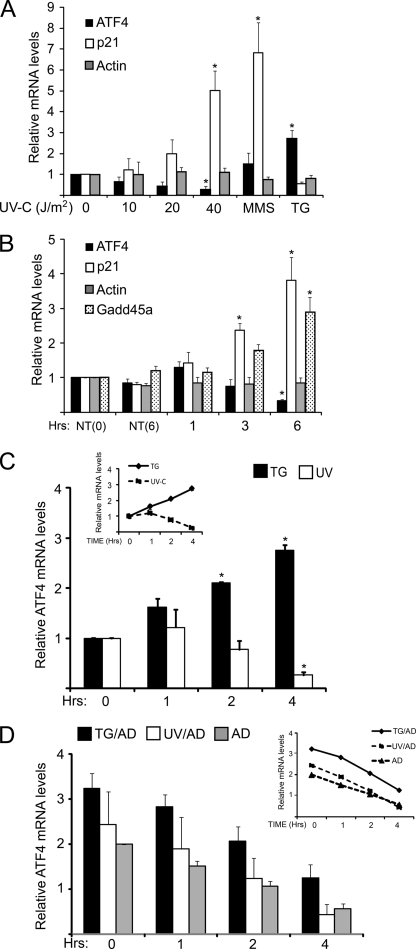
Both UV and ER stresses elicited eIF2∼P and repressed translation initiation, yet there is differential regulation of ATF4, with ER stress triggering enhanced ATF4 protein levels, whereas in response to UV irradiation, ATF4 is absent. The loss of ATF4 protein in response to UV irradiation may be due to altered regulation of the synthesis and/or turnover of ATF4 mRNA or protein. ATF4 mRNA levels were measured in MEF cells during the ER and UV stress conditions. Treatment with thapsigargin led to almost a 3-fold increase in ATF4 mRNA levels, whereas UV irradiation led to a lowering of ATF4 mRNA in a dose-dependent fashion, with significant reductions in the transcript levels in response to exposure to 40 J/m2 UV irradiation (Fig. 4A). Treatment with MMS, a condition that led to elevated ATF4 protein levels along with eIF2∼P, elicited a modest increase in ATF4 mRNA levels. UV irradiation did not lead to a general reduction in mRNA levels, as the amount of actin mRNA remained unchanged for up to 6 h after exposure to 40 J/m2 UV-C irradiation (Fig. 4, A and B). The amount of p21 and GADD45a transcripts, both known to be induced by genotoxic stress, were increased 3-fold or greater (Fig. 4B). The kinetics of the ATF4 mRNA changes are gradual, with lowered ATF4 transcript levels 2 h after UV irradiation and a further reduction 4 h after the UV stress (Fig. 4, B and C). By contrast, in MEF cells exposed to thapsigargin there was a continuous enhancement in ATF4 mRNA levels, with a 3-fold increase after 4 h of ER stress (Fig. 4C). These results indicate that changes in ATF4 transcript levels are an important reason for differential expression of ATF4 in response to UV and ER stress. UV-C irradiation leads to selective reduction in ATF4 mRNA levels, coincident with low ATF4 protein levels despite a robust eIF2∼P.
FIGURE 4.
Levels of ATF4 mRNA are reduced in response to UV irradiation. A, MEF cells were treated with the indicated doses of UV-C irradiation and subsequently incubated in the culture medium for 6 h. Alternatively cells were treated with either 100 μm MMS or 1 μm TG for 6 h. Total RNA was isolated from the samples, and the levels of ATF4, p21, and β-actin mRNAs were measured by qRT-PCR. Values were plotted as -fold change compared with the no treatment control (0). B, MEF cells were treated with 40 J/m2 UV-C and then incubated in the culture medium for up to 6 h as indicated. NT (0) indicates cells not treated with UV-C, and NT (6) indicates a mock-treated cell preparation that was followed by a 6-h incubation period. Transcript levels were measured by qRT-PCR for ATF4, p21, GADD45a, and β-actin as indicated. C, levels of ATF4 mRNA were measured in MEF cells treated with 40 J/m2 UV-C irradiation and then incubated in culture medium for 1, 2, or 4 h as indicated. Cells not subjected to stress are indicated as 0. Additionally, ATF4 transcript levels were measured in cells that were exposed to 1 μm TG for up to 4 h. D, measurement of the half-life of ATF4 mRNA was carried out by first treating MEF cells with 1 μm TG or to 40 J/m2 UV-C UV. 1 h later the cells were then treated with 20 μm actinomycin D (UV+AD or TG+AD) to halt transcription and then cultured for up to 4 h. Alternatively, cells were treated with actinomycin D alone (AD). ATF4 mRNA levels were measured by qRT-PCR at the indicated times, and the panels are presented as the averages ± S.D. of three independent experiments, with each measurement performed in triplicate (*, p < 0.05).
ATF4 mRNA Is Short-lived Independent of Stress
Reduced ATF4 mRNA in response to UV irradiation can be due to an increase in mRNA decay or repressed transcription. To address whether the half-life of ATF4 mRNA is decreased in response to UV irradiation, we utilized the transcription blocking capabilities of actinomycin D. MEF cells were treated with UV-C irradiation, thapsigargin, or no stress, and 1 h later 20 μm actinomycin D was added to the cells, which were then incubated for an additional period of 1, 2, or 4 h. Total RNA was isolated, and subsequently ATF4 mRNA was measured by qRT-PCR (Fig. 4, C and D). We anticipated that if the ATF4 mRNA half-life is a significant factor in the lowered levels of ATF4 transcript in response to UV irradiation that the turnover of ATF4 mRNA would be considerably greater than that measured in cells treated with thapsigargin or no stress. Although ATF4 mRNA was short-lived, with a half-life of ∼3 h, there was not a significant difference in ATF4 transcript turnover in the MEF cells treated with UV irradiation, thapsigargin, or no stress (Fig. 4D). These results indicate that the decay of ATF4 mRNA does not change between different stress arrangements and is not the regulatory switch for reduced mRNA levels in response to UV irradiation.
ATF4 Transcription Is Repressed in Response to UV Irradiation
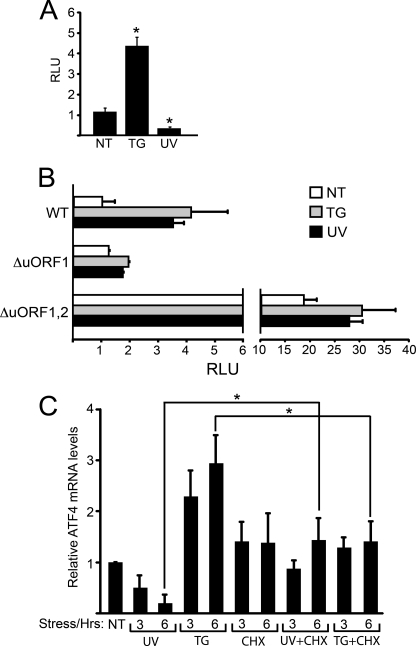
We propose that ATF4 transcription is repressed in response to UV irradiation, leading to low levels of ATF4 mRNA available for preferential translation in response to eIF2∼P. To test this model, a 2.5-kb insert encompassing the ATF4 promoter was fused to a firefly luciferase reporter gene. The resulting PATF4-Luc reporter, which did not encode the 5′-leader region of ATF4 mRNA required for translational control in response to eIF2∼P, was introduced into MEF cells and subjected to ER or UV stress. After thapsigargin treatment there was about a 4-fold increase of luciferase activity, consistent with the idea that enhanced ATF4 mRNA in response to ER stress was due to increased transcription (Fig. 5A). Exposure to 40 J/m2 UV-C resulted in a 3-fold reduction of luciferase activity, indicating that UV irradiation leads to transcriptional repression of ATF4.
FIGURE 5.
ATF4 transcription is regulated during stress. A, a 2.5-kb segment of the ATF4 promoter was fused to a firefly luciferase reporter and assayed for expression in MEF cells treated with 1 μm TG for 6 h, 40 J/m2 UV-C followed by culture incubation for 6 h or to no stress treatment (NT). The firefly luciferase activity was assayed as described under “Experimental Procedures,” and the relative light units (RLU) are presented relative to the non-stressed cells. B, wild-type and mutated versions of the 5′-leader sequences of the ATF4 mRNA, which mediate translational control, were inserted between the constitutive thymidine kinase promoter and the firefly luciferase reporter gene. MEF cells were co-transfected with the PTK-ATF4-Luc plasmid and a control Renilla luciferase plasmid. The transfected cells were treated with 1 μm thapsigargin, 40 J/m2 UV-C or no stress agent as indicated. The mutant versions of the 5′-leader of the ATF4 transcript include a mutation in the initiation codon of uORF1 (ΔuORF1), abolishing the positive-acting element for translational control. Alternatively the mutations were present in the initiation codons for both uORFs (ΔuORF1,2). For clarity, the histograms are represented in two different scales. C, protein synthesis in wild-type MEF cells was blocked by treatment with 50 μg/ml of cycloheximide (CHX) for 30 min. Cells were then exposed to 40 J/m2 UV-C (UV+CHX) or thapsigargin (TG+CHX) stress for 3 or 6 h. Control experiments were carried out by treating cells only with 40 J/m2 UV-C (UV), thapsigargin (TG), cycloheximide, or no stress (NT). Levels of ATF4 mRNA were measured by qRT-PCR. Panels A, B, and C illustrate experimental averages ± S.D. from three independent experiments (*, p < 0.05).
We next addressed whether ATF4 translational control can occur during UV irradiation and eIF2∼P if the ATF4 transcript is available. We analyzed an ATF4-Luc fusion reporter, which contained the 5′-leader of the ATF4 mRNA expressed from a constitutive thymidine kinase (TK) promoter (6). MEF cells transfected with the PTK-ATF4-Luc plasmid were treated with thapsigargin or UV irradiation. In response to either stress conditions, there was a significant increase in luciferase activity compared with nontreated cells (Fig. 5B). By comparison a similar reporter with mutations in the uORF1 that block preferential translation of ATF4 led to low levels of luciferase expression independent of stress. Finally, mutations in both uORF1 and uORF2 in the 5′-leader of the ATF4 mRNA remove the underlying translation control in response to eIF2∼P (6). In this case, there were high levels of luciferase activity in response to the stress and non-stressed conditions, consistent with the idea that the luciferase was subject to the constitutive transcription from the thymidine kinase promoter. These results indicate that if ATF4 mRNA is present in response to UV irradiation that the transcript is subject to preferential translation.
Our results indicate that ATF4 is transcriptionally regulated in response to UV-C and ER stress, suggesting that there is a transcriptional repressor(s) and activator(s) that contributes to ATF4 expression. To address the nature of these transcriptional regulators, we stressed the MEF cells with thapsigargin or UV irradiation in the presence or absence of cycloheximide. We reasoned that if the transcriptional regulators were present before stress and were subject to allosteric regulation or signaling that cycloheximide would not affect the changes in ATF4 mRNA levels in response to ER or UV stress. Alternatively, if the activities of the proposed transcriptional repressor(s) and activator(s) relied directly or indirectly on synthesis for their regulation in response to the stress conditions, cycloheximide would block the changes in ATF4 mRNA in response to these stress conditions. In both stress conditions, treatment with cycloheximide significantly blocked the changes in ATF4 mRNA levels (Fig. 5C). There was a 3-fold increase in ATF4 transcripts after 6 h of the thapsigargin exposure, whereas there were minimal changes in cells with the combined thapsigargin and cycloheximide treatment. Similarly, 6 h after the UV insult there was a 3-fold decrease in the levels of ATF4 mRNA. By contrast, this reduction was blocked in cells when cycloheximide was combined with UV irradiation (Fig. 5C). As a control we measured p21 mRNA levels 6 h after UV irradiation, and similar to the qRT-PCR measurements in Fig. 4B, there was a 5-fold increase in p21 mRNA levels compared with no treatment. This increase in p21 transcript levels after UV exposure was not significantly changed when cycloheximide was combined with UV irradiation (data not shown), which is consistent with the idea that induction of p21 transcription by genotoxic stress involves signaling events activating the p53 transcription factor (35). These results suggest that there are transcription factors that are synthesized after stress or are controlled by co-regulators expressed during stress, which are central to the transcriptional control of ATF4.
eIF2∼P Is Important for Cellular Survival in Response to UV-C
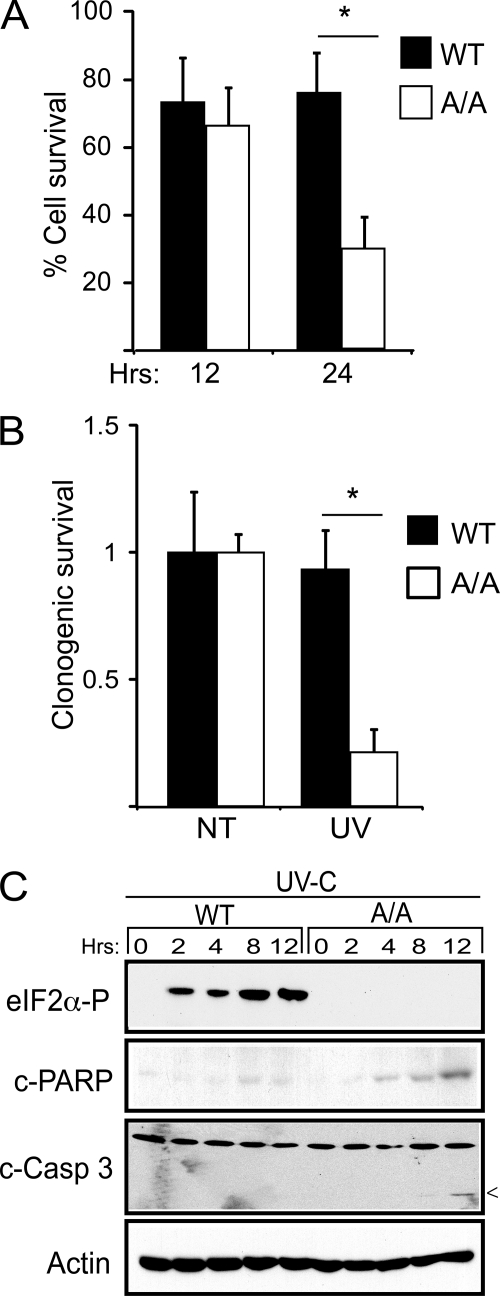
We next addressed the role of eIF2∼P in the resistance to UV irradiation and the functional consequences of the absence of induced ATF4 expression. The A/A MEF expressing the nonphosphorylated version of eIF2α (S51A) showed lowered cell survival in response to UV irradiation. There was a decrease in the number of surviving A/A cells as judged by the MTT assay, with a 2.5-fold reduction 24 h after exposure to 40 J/m2 UV-C (Fig. 6A). By comparison, wild-type MEF cells showed only a modest ∼20% reduction in cell count after the UV insult. Long term clonogenic survival assays also showed over a 3-fold decrease in surviving A/A cells after UV irradiation, whereas the wild type did not show significant reductions (Fig. 6B). Finally, A/A cells showed measureable cleavage of PARP and caspase-3, markers of apoptosis, after 8 h of exposure to UV-C (Fig. 6C). These studies indicate that eIF2∼P significantly contributes to cell survival after UV irradiation.
FIGURE 6.
Phosphorylation of eIF2α provides for resistance to UV irradiation. A, WT and eIF2α-S51A (A/A) MEF cells were treated with 40 J/m2 UV-C irradiation and cultured for 12 or 24 h. The number of viable cells was then determined by the MTT assay. B, the percentage of surviving cells after exposure to 40 J/m2 UV-C was determined by the clonogenic survival assay. NT indicates cells not treated with UV stress. The results in panels A and B correspond to the mean ± S.D. derived from three independent experiments and are normalized to the no treatment control. C, cells were subjected to the UV stress, cultured for up to 12 h as indicated, and phosphorylated eIF2α, β-actin, and apoptotic markers; cleaved caspase 3 and PARP were measured by immunoblot. 0 represents lysates not subjected to the UV stress.
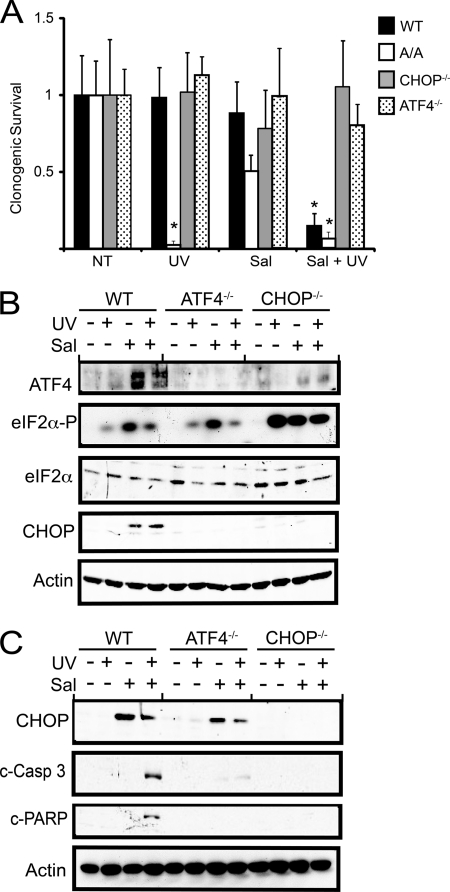
We next wished to address whether expression of ATF4, and its downstream target CHOP, have negative consequences after UV stress. To address this question, we used a derivative of the drug salubrinal, a selective inhibitor of eIF2α dephosphorylation (36), to induce ATF4 and CHOP along with eIF2α phosphorylation without adding any cellular stress. This drug has been used to precondition cells in response to oxidizing stress, providing for a heightened ISR gene expression that provides for increased resistance to stress conditions. Wild-type MEF cells were pretreated with salubrinal-003, a derivative of salubrinal that is more potent and soluble (37), for 6 h, which is sufficient for induced eIF2∼P and its downstream targets ATF4 and CHOP. After this pretreatment, salubrinal was removed from the media, and the cells were then exposed to 40 J/m2 UV- C. We found that the combined salubrinal and UV-irradiation significantly reduced survival of the wild-type cells as judged by the clonogenic assay (Fig. 7A). Salubrinal alone did not have any negative consequence on wild-type cells, although A/A cells showed a partial lowering in cell survival, suggesting that this drug may have consequences beyond eIF2∼P. Importantly, deletion of either ATF4 or CHOP restored cell survival in response to the combined salubrinal and UV treatment (Fig. 7A). These results indicate that although eIF2∼P provides for resistance to UV-C irradiation, activation of ATF4 and the downstream CHOP is detrimental to survival.
FIGURE 7.
Expression of ATF4 and CHOP elicited by pretreatment with salubrinal reduces viability of cells during UV stress. A, WT, eIF2α-S51A, ATF4−/−, and CHOP−/− MEF cells were treated with 10 μm salubrinal-003 (Sal) for 6 h. After pretreatment with salubrinal, the cells were washed and then treated with 40 J/m2 UV-C (UV) irradiation. Alternatively, cells were subjected to only the salubrinal-003 pretreatment, UV irradiation, or no stress treatment (NT). Survival of the stressed wild-type and mutant MEF was measured by clonogenic assays, which are represented as the mean ± S.D. derived from three experiments. Values for each are normalized to the no-treatment controls. B, induction of the ISR in the wild-type, ATF4−/−, and CHOP−/− MEF cells after 3 h of the stress treatments was validated by immunoblot analysis by using antibodies specific for phosphorylated eIF2α, total eIF2α, ATF4, CHOP, and β-actin. C, CHOP, β-actin, and two apoptotic markers (cleaved caspase 3 and PARP) were measured by immunoblot in the wild-type, ATF4−/−v and CHOP−/− after 24 h of the stress regimen.
We also carried out immunoblot analyses of the wild-type, ATF4−/−, and CHOP−/− cells after 3 and 24 h after UV irradiation. Early in the UV stress response, there were measurable increases in ATF4 and CHOP protein in the wild-type MEF cells after the combined salubrinal and UV treatments, as compared with UV-C irradiation alone, which yielded induced eIF2∼P but no detectable ATF4 and CHOP proteins (Fig. 7B). As expected, ATF4−/− cells displayed no expression of ATF4 or its target CHOP early in the salubrinal and UV treatment regimen. CHOP-deficient cells displayed elevated ATF4 protein but no CHOP. After an extended period after the salubrinal and UV treatment, there was measureable cleavage of PARP and caspase 3 in the wild-type cells, supporting a role for apoptosis in the reduced cell death (Fig. 7C). These apoptotic markers were not detectable in the ATF4−/− and CHOP−/− cells subjected to salubrinal and UV irradiation. It is noted that CHOP expression is robust in the wild-type cells 24 h after the treatment with salubrinal and UV or with salubrinal alone. This suggests that CHOP expression alone is not sufficient to trigger cell death but, rather, the timing and duration of CHOP expression may be critical for the sensitivity of cells to UV irradiation. Furthermore, ATF4−/− cells expressed measureable CHOP levels 24 h after treatment with salubrinal alone or the combined salubrinal and UV irradiation. This indicates that during extended stress conditions, CHOP can be expressed independent of ATF4.
DISCUSSION
This study addressed the regulatory mechanisms governing the variable ATF4 expression in response to eIF2∼P and different stress conditions. From our experimental results, we draw four central conclusions. First, eIF2∼P is induced by UV-B and UV-C irradiation in many different mammalian cell types (Figs. 1 and 2), and this phosphorylation event leads to a reduction in global translation initiation (Fig. 3). This finding is consistent with earlier studies that showed that UV irradiation can enhance GCN2 phosphorylation of eIF2α (8, 9).
The second conclusion is that expression of ATF4 in response to environmental stress involves changes in ATF4 mRNA levels. For example, ATF4 mRNA levels are lowered 3-fold in response to UV stress, whereas ATF4 transcript levels are significantly increased during ER stress (Fig. 4). ATF4 transcriptional regulation is the key switch for the changes in ATF4 mRNA levels in response to UV and ER stress conditions (Fig. 5A). ATF4 mRNA is also subject to rapid turnover, with a half-life of about 3 h, but this is independent of stress conditions (Fig. 4D). The lability of ATF4 mRNA renders it more sensitive to changes in transcriptional regulation elicited by stress conditions. For example, transcriptional repression in response to UV irradiation coupled with the constitutive short half-life facilitates reduced levels of ATF4 mRNA during this stress condition. These central ideas are further supported by earlier reports that observed increased ATF4 transcript levels in response to ER stress (4, 5, 38). Additionally, it was reported that ATF4 mRNA levels are elevated in response to amino acid starvation (39). These results indicate that expression of ATF4 is subject to both transcriptional regulation and translational control.
The third conclusion is that the combined transcriptional regulation and translational control of ATF4 provides for versatility in regulating the ISR gene expression. In response to UV irradiation, the lowered availability of the ATF4 transcript significantly reduces the amount of ATF4 synthesized, thus blocking ATF4 induction of the ISR. Therefore, there does not appear to be an inherent inability to elicit preferential ATF4 mRNA translation in response to eIF2∼P and UV stress. This was illustrated by our finding that constitutive transcription of the ATF4-luciferase reporter from the thymidine kinase promoter led to preferential translation during UV irradiation (Fig. 5B). Consistent with this idea, mutation of uORF1 in the 5′-leader of the ATF4 mRNA, which is required for reintiation of translating ribosomes and preferential translation, blocked expression of the luciferase reporter. During ER stress, there is elevated transcription of ATF4, which enhances the amount of ATF4 transcript available for preferential translation by eIF2∼P, therefore, amplifying expression of ATF4 and the ISR. These results indicate that the combination of transcriptional regulation with translational control allows for genes marked for preferential translation by eIF2∼P to be selectively induced in response to a range of environmental stresses. The ISR is not constricted to a specific program of gene expression but, rather, can tailor it for a given stress condition.
The fourth central conclusion is that the absence of ATF4 expression appears to be advantageous for cells during UV stress (Fig. 7). Clearly eIF2∼P and its reduction in translation initiation facilitates resistance to UV irradiation (Fig. 6). However, the versatility of the ISR has provided for selective loss of expression of ATF4 and its ISR target genes. Pretreatment with salubrinal, an inhibitor of eIF2∼P dephosphorylation that enhances the eIF2∼P/ATF4 pathway, typically provides for heightened resistance to stress conditions, such as those triggered by oxidizing agents (5, 36, 40–42). However, this pretreatment regimen renders cells much more susceptible to UV irradiation (Fig. 7). Importantly, this UV sensitivity is alleviated by deletion of either ATF4 or CHOP (Fig. 7). During the ISR, elevated CHOP levels for an extended period are thought to elicit gene expression that triggers apoptosis (17, 18, 43, 44). These findings suggest that cells encountering UV stress are hypersensitive to expression of CHOP.
The Combination of Transcriptional Regulation and Translational Control Allows for Differential Expression of ISR Target Genes
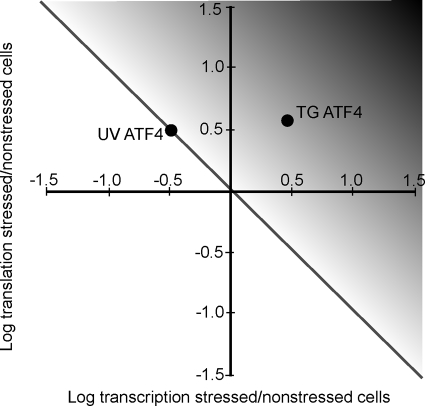
We propose that a combination of transcriptional regulation and translational control of ATF4 underlies the ability of the ISR to differentially express ATF4 depending on the precise stress condition and that this process is central to cell survival (Fig. 8). Although this model is specifically highlighted for ATF4, it would apply to other genes subject to translational control in response to eIF2∼P. Expression of genes slated for preferential translation in the ISR can be enhanced or blocked depending on their transcription status. Enhanced ATF4 transcript availability would ensure a greater level of ATF4 protein synthesis and activity, as illustrated by the degree of black color in the gradient depiction in Fig. 8. A range of ATF4 expression appears to be central for alleviation of different stress conditions. In most cases, including nutrient and ER stress, ATF4 activity provides for resistance to the stress insults, whereas in others, such as UV irradiation, ATF4 is suggested to be harmful. The detrimental properties of ATF4 are due at least in part to its ability to induce CHOP, which is suggested to elicit apoptosis through induction of pro-apoptotic genes, such as BCL2, DR5, and those tied to autophagy (43, 45, 46). In addition to UV irradiation, lowered ATF4 expression during heightened eIF2∼P has been reported during brain ischemia and non-alcoholic steatohepatitis (24, 25), suggesting that the dampened expression of ATF4 in the gradient model can be applied to a number of stress arrangements.
FIGURE 8.
Model depicting proposed transcriptional regulation and translational control of ATF4 expression and the ISR. The y axis represents the levels of mRNA translation in stressed cells compared with non-stressed, whereas the x axis represents changes in transcript levels of a given gene in stressed cells relative to non-stressed. The diagonal line represents the levels of transcription and translation that are proposed to yield no change in ATF4 protein levels. The black gradient depicts the levels of transcriptional regulation and translational control that would enhance ATF4 protein levels and induce its target ISR genes. The solid circles represent the outcome of the transcriptional regulation and translational control of ATF4 in response to UV irradiation (UV) or ER stress elicited by TG treatment.
The idea of a combination of transcriptional regulation and translational control of ATF4 redefines some features of the ISR. ATF4 was defined as a common downstream target, which integrates signaling from different eIF2 kinases (3). In this respect the mammalian ISR builds on and elaborates upon the earlier concept of the general amino acid control in yeast. This yeast pathway features the ability of different nutritional stresses to activate GCN2 phosphorylation of eIF2α and preferential translation of GCN4, a “master regulator” of genes involved in amino acid metabolism and the salvaging and uptake of nutrients (7, 47). It is noted that UV irradiation has been reported to enhance GCN4 translation in yeast, possibly in a GCN2-independent manner (48). Therefore, regulation of GCN4 by UV irradiation in yeast may differ from the ISR in mammalian cells.
The model featuring combined transcriptional regulation and translation control indicates that eIF2∼P in response to various stress arrangements does not lead to default activation of ATF4. Rather, there are additional target genes activated by eIF2∼P that play a major role in alleviating stress damage. In some cases these additional target genes may function in conjunction with ATF4, whereas in others they may function in the absence of the ATF4 transcriptional activator. These additional target genes could include those subject to preferential translation in response to eIF2∼P through 5′-leader configurations in their mRNAs. For example, several DNA repair enzymes were reported to be subject to preferential translation by eIF2∼P after UV stress (32). These preferentially translated genes include ERCC1, ERCC5, and DDB1, and the uORFs in the 5′-leaders of these encoded transcripts were suggested to be important to maintain elevated expression after UV irradiation. However, in these cases eIF2∼P in the absence of UV irradiation did not appear to be sufficient for translational control, suggesting that additional signaling pathways and proteins may be involved. It is also noted that in this earlier study that UV irradiation did not enhance expression of luciferase activity expressed from a transfected vector that included the 5′-leader of ATF4 (32). This result appears to differ from our findings in Fig. 5B. We are currently uncertain as to the underlying reason for this difference, but it may reflect the different cell type (HeLa cells) or the UV-B irradiation used in the earlier study.
Repressed translation by UV irradiation also reduces the synthesis of key labile regulatory proteins, such as IκBα (8), which can relieve its repression of NF-κB. Activation of NF-κB would then enhance the transcription of diverse target genes, such as those involved in inflammation and the regulation of apoptosis. In this respect, the ISR can be viewed as a collection of eIF2 kinases that recognize various stress arrangements activating different combinations of target gene modules that can provide for stress resistance.
Regulation of ATF4 Transcription in Response to Environmental Stresses
We do not yet know the regulators of ATF4 transcription during the different stress arrangements. Our experiments with UV and ER stress combined with cycloheximide suggest that the proposed repressor(s) and activator(s) of ATF4 transcription need to be synthesized during the stress conditions (Fig. 5C). Our preliminary studies using A/A MEF cells suggests that eIF2∼P is required at least in part for the changes in ATF4 mRNA levels in response to UV and ER stresses. Furthermore, analysis of the PATF4-Luc reporter in ATF4−/− cells suggests that ATF4 is not involved in autoregulation. Therefore, there is involvement of additional ISR regulatory genes in the transcriptional regulation of ATF4. Future studies are needed to define these transcription regulators and their control of the ATF4 promoter.
Acknowledgments
We thank Dr. Hua Lu and Latha Ramalingam for experimental assistance. We also acknowledge the Biochemistry Biotechnology Facility at Indiana University for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM49164 and GM64350 (to R. C. W.).
- ER
- endoplasmic reticulum
- uORF
- upstream ORF
- ISR
- integrated stress response
- MEF
- mouse embryonic fibroblast
- MMS
- methyl methane sulfonate
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TG
- thapsigargin
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1.Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 3.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 4.Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 5.Lu P. D., Jousse C., Marciniak S. J., Zhang Y., Novoa I., Scheuner D., Kaufman R. J., Ron D., Harding H. P. (2004) EMBO J. 23, 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vattem K. M., Wek R. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 8.Jiang H. Y., Wek R. C. (2005) Biochem. J. 385, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng J., Harding H. P., Raught B., Gingras A. C., Berlanga J. J., Scheuner D., Kaufman R. J., Ron D., Sonenberg N. (2002) Curr. Biol. 12, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 10.Gale M., Jr., Blakely C. M., Kwieciszewski B., Tan S. L., Dossett M., Tang N. M., Korth M. J., Polyak S. J., Gretch D. R., Katze M. G. (1998) Mol. Cell. Biol. 18, 5208–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadler A. J., Williams B. R. (2007) Curr. Top Microbiol. Immunol. 316, 253–292 [DOI] [PubMed] [Google Scholar]
- 12.García M. A., Meurs E. F., Esteban M. (2007) Biochimie 89, 799–811 [DOI] [PubMed] [Google Scholar]
- 13.Han A. P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G., Fleming M., Leboulch P., Orkin S. H., Chen J. J. (2001) EMBO J. 20, 6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J. J. (2007) Blood 109, 2693–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) Mol. Cell. 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 16.Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 17.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marciniak S. J., Ron D. (2006) Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 19.Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) PLoS Biol. 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherkasova V. A., Hinnebusch A. G. (2003) Genes Dev. 17, 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staschke K. A., Dey S., Zaborske J. M., Palam L. R., McClintick J. N., Pan T., Edenberg H. J., Wek R. C. (2010) J. Biol. Chem. 285, 16893–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Proud C. G. (2009) Trends Cell Biol. 19, 260–267 [DOI] [PubMed] [Google Scholar]
- 23.Anthony T. G., McDaniel B. J., Byerley R. L., McGrath B. C., Cavener D. R., McNurlan M. A., Wek R. C. (2004) J. Biol. Chem. 279, 36553–36561 [DOI] [PubMed] [Google Scholar]
- 24.Puri P., Mirshahi F., Cheung O., Natarajan R., Maher J. W., Kellum J. M., Sanyal A. J. (2008) Gastroenterology 134, 568–576 [DOI] [PubMed] [Google Scholar]
- 25.Kumar R., Krause G. S., Yoshida H., Mori K., DeGracia D. J. (2003) J. Cereb. Blood Flow Metab. 23, 462–471 [DOI] [PubMed] [Google Scholar]
- 26.Jiang H. Y., Wek S. A., McGrath B. C., Scheuner D., Kaufman R. J., Cavener D. R., Wek R. C. (2003) Mol. Cell. Biol. 23, 5651–5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. (2004) Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. (2001) Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 30.Zhang P., McGrath B., Li S., Frank A., Zambito F., Reinert J., Gannon M., Ma K., McNaughton K., Cavener D. R. (2002) Mol. Cell. Biol. 22, 3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P., McGrath B. C., Reinert J., Olsen D. S., Lei L., Gill S., Wek S. A., Vattem K. M., Wek R. C., Kimball S. R., Jefferson L. S., Cavener D. R. (2002) Mol. Cell. Biol. 22, 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powley I. R., Kondrashov A., Young L. A., Dobbyn H. C., Hill K., Cannell I. G., Stoneley M., Kong Y. W., Cotes J. A., Smith G. C., Wek R., Hayes C., Gant T. W., Spriggs K. A., Bushell M., Willis A. E. (2009) Genes Dev. 23, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Lewis D. A., Yi Q., Travers J. B., Spandau D. F. (2008) Mol. Biol. Cell. 19, 1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou D., Palam L. R., Jiang L., Narasimhan J., Staschke K. A., Wek R. C. (2008) J. Biol. Chem. 283, 7064–7073 [DOI] [PubMed] [Google Scholar]
- 35.Vousden K. H. (2002) Biochim. Biophys. Acta. 1602, 47–59 [DOI] [PubMed] [Google Scholar]
- 36.Boyce M., Bryant K. F., Jousse C., Long K., Harding H. P., Scheuner D., Kaufman R. J., Ma D., Coen D. M., Ron D., Yuan J. (2005) Science 307, 935–939 [DOI] [PubMed] [Google Scholar]
- 37.Robert F., Kapp L. D., Khan S. N., Acker M. G., Kolitz S., Kazemi S., Kaufman R. J., Merrick W. C., Koromilas A. E., Lorsch J. R., Pelletier J. (2006) Mol. Biol. Cell. 17, 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008) Cell. Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 39.Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S. (2002) J. Biol. Chem. 277, 24120–24127 [DOI] [PubMed] [Google Scholar]
- 40.Oh Y. K., Shin K. S., Yuan J., Kang S. J. (2008) J. Neurochem. 104, 993–1005 [DOI] [PubMed] [Google Scholar]
- 41.Lewerenz J., Maher P. (2009) J. Biol. Chem. 284, 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakka V. P., Gusain A., Raghubir R. (2010) Neurotox. Res. 17, 189–202 [DOI] [PubMed] [Google Scholar]
- 43.McCullough K. D., Martindale J. L., Klotz L. O., Aw T. Y., Holbrook N. J. (2001) Mol. Cell. Biol. 21, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song B., Scheuner D., Ron D., Pennathur S., Kaufman R. J. (2008) J. Clin. Invest. 118, 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi H., Wang H. G. (2004) J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 46.Rouschop K. M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J. W., Lambin P., van der Kogel A. J., Koritzinsky M., Wouters B. G. (2010) J. Clin. Invest. 120, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., Hinnebusch A. G., Marton M. J. (2001) Mol. Cell. Biol. 21, 4347–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelberg D., Klein C., Martinetto H., Struhl K., Karin M. (1994) Cell 77, 381–390 [DOI] [PubMed] [Google Scholar]