Abstract
Infectious diseases that cause hemolysis are among the most threatening human diseases, because of severity and/or global distribution. In these conditions, hemeproteins and heme are released, but whether heme affects the inflammatory response to microorganism molecules remains to be characterized. Here, we show that heme increased the lethality and cytokine secretion induced by LPS in vivo and enhanced the secretion of cytokines by macrophages stimulated with various agonists of innate immune receptors. Activation of nuclear factor κB (NF-κB) and MAPKs and the generation of reactive oxygen species were essential to the increase in cytokine production induced by heme plus LPS. This synergistic effect of heme and LPS was blocked by a selective inhibitor of spleen tyrosine kinase (Syk) and was abrogated in dendritic cells deficient in Syk. Moreover, inhibition of Syk and the downstream molecules PKC and PI3K reduced the reactive oxygen species generation by heme. Our results highlight a mechanism by which heme amplifies the secretion of cytokines triggered by microbial molecule activation and indicates possible pathways for therapeutic intervention during hemolytic infectious diseases.
Keywords: Cytokine Induction, Heme, Inflammation, Innate Immunity, Toll-like Receptors (TLR), Hemolysis
Introduction
A general consequence of infectious diseases that cause hemolysis, internal hemorrhage, or extensive cell damage is the release of hemeproteins. Upon oxidation, hemeproteins release heme, a potentially harmful molecule (1). Heme-binding plasma proteins, such as hemopexin or albumin, remove the intravascular free heme, subsequently degraded by heme oxygenase-1 (HO-1), generating equimolar amounts of biliverdin, carbon monoxide, and free iron (2, 3). HO-1-deficient mice (Hmox−/−) have high plasma concentrations of heme and show increased susceptibility to LPS-induced lethality, associated with inflammation and oxidative damage (4). Accumulation of large amounts of heme might overwhelm the capacity of heme scavengers and degrading system, thus causing oxidative stress and inflammation (5, 6). In fact, recent studies suggest that heme, in combination with ROS3 and inflammatory mediators, increase blood brain barrier leakage and hepatocyte necrosis in models of malarial infection (7, 8).
Hemolysis or hemoglobinemia are associated with increased mortality in septic patients (9, 10). Hemoglobin increases the secretion of TNF triggered by LPS, whereas globin has an inhibitory effect (11), suggesting that heme is responsible for the cytokine amplification. Heme has several pro-inflammatory activities, including leukocyte activation and migration, up-regulation of adhesion molecules, ROS production, and induction of cytokine expression (12–14). Recently, we have shown that heme is able to activate Toll-like receptor 4 (TLR4) inducing TNF on macrophages and dendritic cells (DC) (15).
Mammalian pattern recognition receptors (PRRs) recognize conserved microbial molecules from all classes of microorganisms (16, 17). The activation of these receptors elicits selective intracellular signaling cascades that result in the production of cytokines, chemokines, lipid mediators, and reactive oxygen/nitrogen species. The synergistic effect of certain associations of PRR agonists on cytokine secretion by macrophages and DC promotes the inflammatory response and the activation of the adaptive immune system (18). The increased secretion of cytokines is also achieved by combinations of microbial molecules with molecules from host origin such as cytokines, ATP, and ROS (19, 20). This activation of the immune system is considered essential for pathogen killing but is also critically involved in tissue damage and sepsis (19). In this study, we investigated if heme would be able to affect the inflammatory response triggered by microbial molecules. Heme caused a remarkable increase in the production of cytokines induced by microbial molecules and the lethality induced by LPS. We demonstrate that the heme-LPS synergistic effect on cytokine secretion and the generation of ROS by heme were both dependent on Syk activation.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 mice were supplied by the UFRJ breeding facilities (Rio de Janeiro, Brazil) or bred at Cancer Research UK in specific pathogen-free conditions. Fetal liver cells from WT embryos or Syk−/− embryos were used to generate radiation chimeras in C57BL/6 hosts, as described previously (22). Tlr4−/−, Myd88−/−, Asc−/−, Rip2−/−, and Syk−/− mice were kindly provided by S. Akira, A. Coyle, V. Tybulewicz, D. Zamboni, and N. Camera. The animals were kept at a constant temperature (25 °C) with free access to chow and water in a room with a 12-h light/dark cycle. The animal protocols were approved by the Centro de Ciências da Saúde, Universidade Federal do Rio de Janeiro Animal Ethics Committee and by the London Research Institute Animal Ethics Committee and were performed under the authority of the Animal Scientific Procedures Act of 1986 (United Kingdom).
Reagents
LPS 0111:B4 from Escherichia coli, CpG, synthetic bacterial lipopeptide (Pan3Cys-Ser-(Lys)4), poly(I-C), were obtained from InvivoGen; FK565 was from Astellas Pharmaceutical, and MDP was from Calbiochem. Heme was purchased from Frontier Scientific, Logan, UT. Heme was dissolved in NaOH (0.1 m), diluted in RPMI 1640 medium, and filtered. Stock solutions of heme were prepared immediately before use in the dark to avoid the generation of free radicals. Diphenylene iodonium (DPI), N-acetyl-l-cysteine (NAC), rotenone, allopurinol, l-NAME, piceatannol, paraquat, and SP600125 were obtained from Sigma. CM-H2DCFDA was obtained from Invitrogen. PD98059, SB203580, calphostin C, wortmannin, and wedelolactone were from Calbiochem. RPMI 1640 medium for macrophage culture was obtained from Sigma and was supplemented with penicillin/streptomycin and fetal calf serum (FCS) from Invitrogen.
Hemolysis and Heme in Endotoxemic Mice
Mice (n = 10) C57Bl/6 received an intraperitoneal injection of phenylhydrazine (0.15 mg/g in a volume of 200 μl) and were injected with LPS (10 mg/kg) 4 h later. Control groups received an intraperitoneal injection of LPS or phenylhydrazine alone. Mice were monitored for 72 h. Mice (n = 10) C57Bl/6 were injected intraperitoneally with heme (50 mg/kg) and LPS (10 mg/kg). Control groups received an intraperitoneal injection of LPS or heme alone. Mice were monitored for 72 h. C67Bl/6 mice were injected intraperitoneally with saline solution, heme (50 mg/kg), LPS (1 mg/kg), or heme plus LPS. Three and 6 h after injection, animals were sacrificed, and their blood (to isolate the plasma) was collected and stored at −20 °C for cytokine determinations.
Cell Culture and Stimulation
Peritoneal macrophages were obtained 4 days after intraperitoneal inoculation of 2 ml of 3% thioglycollate by peritoneal washes with chilled RPMI 1640 medium. Cells were seeded at 2 × 105/well in 96-well plates in RPMI 1640 medium with 10% FCS. Nonadherent cells were removed by washing, and adherent cells were stimulated as indicated in the figure legends. Supernatants were collected at the intervals indicated in the figure legends and frozen until cytokine determination. Bone marrow was harvested from the tibias and femurs from WT mice and differentiated into bone marrow-derived macrophages as described previously (21). Briefly, the cells were resuspended at 4 × 106/10 ml in RPMI 1640 medium supplemented with 20% of FCS and 30% of supernatant from L929 cells cultures. After 4 days, fresh medium was added to the cell cultures. Macrophages were collected at day 7, and 2 × 105/well were plated in 96-well plates in RPMI 1640 medium with 10% of FCS. Nonadherent cells were removed, and adherent cells were stimulated as indicated in the figure legends. Supernatants were collected and frozen for cytokine determinations. Bone marrow-derived dendritic cells (DC) were prepared as described previously (22). Briefly, bone marrow was harvested from the tibias and femora from WT or chimeric mice and differentiated into DC. The cells were resuspended in a 24-ml final volume of RPMI 1640 medium containing 10% FCS, 50 μm β-mercaptoethanol, 100 units/ml penicillin/streptomycin, 1 mm sodium pyruvate, 10 mm HEPES, nonessential amino acids, and 200 units/ml GM-CSF. Four milliliters of cell suspension were plated per well into a 6-well plate (1× plate/mouse). Cells were incubated at 37 °C and 5% CO2. A partial medium change was performed after 2 days of culture. On day 3 of culture, all nonadherent cells were removed, and 4 ml of a new complete medium and 200 units/ml rmGM-CSF were added to each well. On day 5, DC were harvested in PBS, 2 mm EDTA and enriched using CD11c-microbeads (Miltenyi Biotec). DC were plated at 2 × 105/well in a 96-well plate for 2 h in RPMI 1640 medium with 10% of FCS. Cells were stimulated as indicated in the figure legends. Supernatants were collected and frozen for cytokine determination. Cell viability was analyzed by LDH measurement in supernatants collected after the 4-h stimulation period for all treatments with selective inhibitors in combination with heme and LPS. Cell viability of treated groups was similar to the nonstimulated controls.
Cytokine Measurements
The concentrations of TNF, IL-6, and IP-10 were determined using ELISA (PeproTech and R & D Systems). All the measurements were performed in duplicate according to the manufacturer's instructions.
Western Blot Analysis for Syk, MAPKs Phosphorylation, and IκBα Degradation
Elicited peritoneal macrophages were plated in 6-well plates at a density of 2.0 × 106 cells/well. Nonadherent cells were removed by washing with medium, and adherent cells were stimulated as indicated in the figure legends. After 30 min, cells were lysed in a buffer consisting of Tris-HCl (50 mm), NaCl (150 mm), 1% Nonidet P-40, 0.25% sodium deoxycholate, EDTA (1 mm), aprotinin (5 mg/ml), leupeptin (5 mg/ml), pepstatin (5 mg/ml), PMSF (1 mm), sodium orthovanadate (1 mm), and NaF (1 mm), pH 7.5. Cell lysates were centrifuged, and 20 μg of protein contained in the supernatants were boiled and subjected to electrophoresis in SDS-polyacrylamide gel in reducing conditions. Proteins were transferred to a nitrocellulose membrane at 4 °C for 2 h. Membranes were blocked with Tris-buffered saline solution with 0.05% of Tween 20 (TBS-T) and 5% of fat-free milk, incubated overnight with anti-phospho (p)-ERK1/2 (1:1000), anti-p-JNK (1:1000) (Santa Cruz Biotechnology), anti-p-p38 (1:1000), or anti-p-Syk (1:1000) (Cell Signaling Technology) diluted in blocking solution. Membranes were washed in TBS-T and incubated for 2 h with horseradish peroxidase-conjugated goat anti-rabbit (1:10,000) or goat anti-mouse (1:10,000) IgG polyclonal antibodies (Santa Cruz Biotechnology). The bands were revealed by chemiluminescence, using ECL substrate (Santa Cruz Biotechnology). The normalization of Syk and MAPKs phosphorylation was performed by stripping membranes during 30 min at 50 °C in stripping buffer (100 mm β-mercaptoethanol, 2% SDS, 62.5 mm Tris-HCl, pH 6.7). After stripping, membranes were washed with TBS-T, blocked with 5% fat-free milk TBS-T, incubated overnight with rabbit anti-β-actin (1:1000) (Sigma) for p-Syk membranes or rabbit anti-ERK2 (1:1000) (Santa Cruz Biotechnology) for MAPK membranes. Detection was performed as described above. The degradation of IκB protein was analyzed by Western blot with polyclonal rabbit anti-IκB (Sigma) diluted (1:1000) in block solution. Detection of nonphosphorylated ERK2 was used as loading control.
Quantitative Real Time RT-PCR
Total RNA was extracted using TRIzol (Invitrogen), and 1 μg was reverse-transcribed using M-MLV reverse transcriptase and random hexamer primers (Invitrogen). Subsequent quantitative real time PCR analysis was performed on an ABI 7500 (Applied Biosystems) using SYBR Green master mix (Applied Biosystems). Amplification conditions were as follows: 95 °C (10 min), 40 cycles of 95 °C (15 s), and 60 °C (60 s). All data were normalized to the corresponding HPRT expression, and the fold difference relative to the control level was shown. The analyses of relative gene expression data were performed by the 2−ΔΔCT method. TNF primers are forward 5′-GGTCCCCAAAGGGATGAGAAGTTC-3′ and reverse 5′-CCACTTGGTGGTTTACTACGACG-3′; HPRT primers are forward 5′-GCTGGTGAAAAGGACCTCT-3′ and reverse 5′-CACAGGACTAGAACACCTGC-3′.
Electrophoretic Mobility Shift Assay
Nuclear extracts were obtained and analyzed for NF-κB activation by EMSA, as described previously (23). Binding reactions were performed using 1 μg of nuclear protein in the presence of 40,000 cpm of 32P-end-labeled double-stranded consensus NF-κB oligonucleotide (sequence 5′-AGT TGA GGG GAC TTT CCC AGG C-3′, Santa Cruz Biotechnology) and 1 μg of poly(dI-dC)·(dI-dC) (Amersham Biosciences) in binding buffer (50 mm HEPES, pH 7.9, 20% glycerol, 5 mm DTT, 5 mm EDTA, and 0.5 μg of BSA) for 30 min at room temperature. The DNA-protein complex formed was separated from free oligonucleotide on 4% native polyacrylamide gel.
Reactive Oxygen Species Generation
Macrophages (106 cells/well) were stimulated in the presence of 10% FCS with heme during 1 h after the treatment with deferoxamine, piceatannol, wortmannin, or calphostin C. After the stimulus, cells were incubated with 2 μm 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA) in RPMI 1640 medium for 30 min at 37 °C under 5% CO2 atmosphere. ROS production was analyzed by flow cytometry using FACScan flow cytometer (BD Biosciences).
Statistical Analysis
Data are presented as means ± S.E. Results were analyzed using a statistical software package (GraphPad Prism 4). Statistical analyses were performed by one-way analysis of variance and Newman-Keuls post test. A significant difference between groups was considered if p < 0.05. Comparisons of survival curves were analyzed by log-rank (Mantel-Cox test).
RESULTS
Hemolysis and Heme Increased Lethality Induced by a Sublethal Dose of LPS
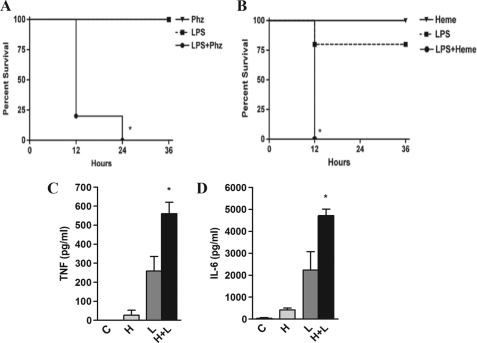
Previous studies have shown that hemolysis or hemoglobinemia is associated with increased mortality in patients with sepsis (9, 10). To investigate whether endogenous heme affects the host response to microbial products without interfering with bacterial growth, we used an established model of hemolysis caused by phenylhydrazine injection and challenged mice with a sublethal dose of LPS. This protocol induced a striking increase in lethality compared with LPS or phenylhydrazine alone (Fig. 1A). To define if exogenous heme could synergize with LPS in vivo, to cause mortality, we treated mice with heme and challenged them with a sublethal dose of LPS. This combination also caused a significant increase in lethality compared with the groups receiving LPS or heme alone (Fig. 1B). The combined treatment with heme and LPS caused a significant increase in plasma TNF and IL-6 compared with LPS or heme alone at 3 and 6 h after challenge, respectively (Fig. 1, C and D). These results indicate that heme increases both the lethality and cytokine concentration induced by LPS in vivo.
FIGURE 1.
Hemolysis and heme potentiated the lethal effect of LPS. A, mice (n = 10) C57Bl/6 received an intraperitoneal injection of phenylhydrazine (Phz) (0.15 mg/g) in a volume of 200 μl. Four hours later the animals were injected with LPS (10 mg/kg). Control (C) groups received an intraperitoneal injection of LPS or phenylhydrazine alone. Mice were monitored for 72 h. *, p < 0.0001 compared with LPS injection. B, mice (n = 10) C57Bl/6 were injected intraperitoneally with heme (50 mg/kg) and LPS (10 mg/kg). Control groups received an intraperitoneal injection of LPS or heme alone. Mice were monitored for 72 h. *, p < 0.001 compared with LPS injection. C and D, mice received heme (50 mg/kg) and/or LPS (1 mg/kg), and plasma concentrations of TNF and IL-6 were evaluated 3 and 6 h after the challenge, respectively. n = 4 *, p < 0.05 compared with LPS injection. H, heme; L, LPS.
Heme Increased the Secretion of Cytokines Induced by LPS in Macrophages
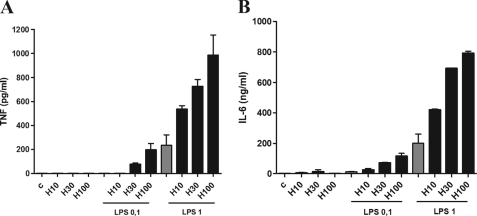
Macrophages are important sources of inflammatory cytokines during infection and inflammation. To define the mechanism by which heme enhances cytokine secretion induced by LPS, macrophages were stimulated with increasing concentrations of heme and LPS. In the presence of serum, stimulation with heme or low concentrations of LPS caused a negligible secretion of TNF or IL-6, whereas the combination of treatments induced a robust secretion of these cytokines (Fig. 2, A and B). This potentiating effect of heme upon LPS stimulation was dose-dependent and was observed at concentrations of 30 and 100 μm. Heme also potentiated the secretion of IP-10 induced by LPS (data not shown). These results indicate that heme increases the secretion of MyD88- and TIR domain-containing adaptor-inducing interferon β-dependent cytokines induced by LPS on macrophages.
FIGURE 2.
Heme increased the secretion of cytokines induced by LPS in macrophages. A, peritoneal macrophages from C57Bl/6 mice were stimulated with heme (10, 30, or 100 μm) 1 h before the stimulus with LPS (0.1 or 1 ng/ml). After 4 h of stimulus with LPS, the supernatants were collected for TNF (A) and IL-6 (B) determinations by ELISA. Results represents mean ± S.E. for cytokines determinations and are representative of three different experiments. C, control.
Heme Increased the Cytokine Secretion Induced by Different TLR and NOD Agonists
To define whether the effect of heme was exclusive to LPS stimulation, we used agonists of TLR2 (Pan3-Cys), TLR3 (poly(I-C)), and TLR9 (CpG). Heme significantly enhanced the secretion of TNF and IL-6 induced by all tested TLR agonists (supplemental Fig. 1, A–C, and data not shown). NOD-like receptors are cytosolic receptors involved in the recognition of microorganisms and viral molecules present in the cytosol (17). Treatment of macrophages with heme also increased the production of TNF induced by NOD1 and NOD2 agonists (supplemental Fig. 1, D and E). Together, these results indicate that heme increases the cytokine secretion by macrophages activated with TLR and NOD agonists.
Heme Advanced and Increased the Activation of NF-κB and MAPKs Induced by LPS
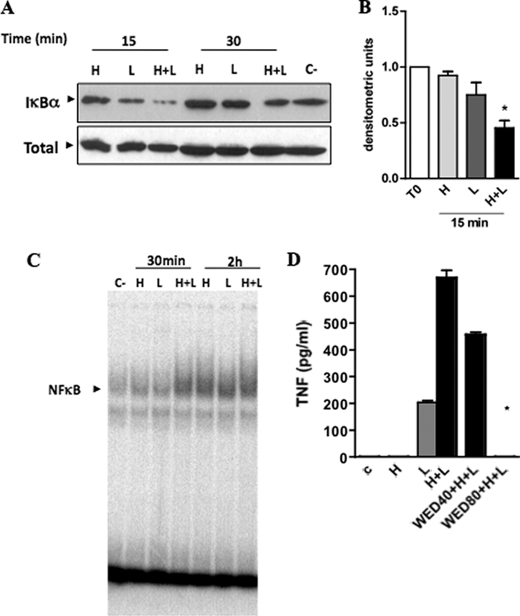
Activation of NF-κB is essential for cytokine production induced by several PRR agonists (16, 17). To determine the involvement of this pathway on macrophage activation induced by heme plus LPS, we initially characterized the content of IκBα by Western blot. Treatment of macrophages with heme plus LPS caused an early destruction of IκBα compared with the effect of heme or LPS alone (Fig. 3, A and B). Next, we determined the binding of NF-κB by EMSA in nuclear extracts of macrophages stimulated with heme, LPS, or heme plus LPS. As early as 30 min of stimulation, heme and LPS induced a strong activation of NF-κB compared with single stimulations (Fig. 3C). Treatment with an inhibitor of IKK inhibited the secretion of TNF in a concentration-dependent fashion (Fig. 3D).
FIGURE 3.
Heme increased the activation of NF-κB induced by LPS. A, macrophages from C57Bl/6 mice were stimulated with heme (H) (100 μm) and/or LPS (L) (0.1 ng/ml) in the time intervals indicated. Cell extracts were submitted to SDS-PAGE. IκB degradation was detected by immunoblotting. Detection of nonphosphorylated ERK2 was used as loading control (C). B, densitometric analysis of IκBα degradation relative to negative control in the 15-min interval. Values are mean ± S.E. of three different experiments. *, p < 0.05. C, NF-κB activation was analyzed by EMSA from nuclear extracts in the time intervals indicated. The figures are representatives of two different experiments with similar results. D, production of TNF was evaluated in peritoneal macrophages treated with wedelolactone (40 or 80 μm) for 30 min before the stimuli with heme (100 μm) and LPS (0.1 ng/ml). The supernatants were collected 4 h after the stimuli. Results represent mean ± S.E. for TNF determination and are representatives of two different experiments. *, p < 0.05. H, heme; L, LPS; WED, wedelolactone.
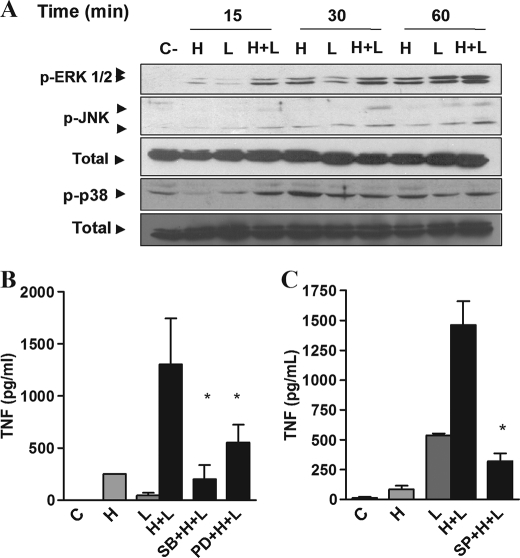
Activation of macrophages with LPS causes the phosphorylation of MAPKs, considered essential for the production of several cytokines, including TNF (16, 17). Thus, we investigated the effect of heme on the phosphorylation of MAPKs stimulated by LPS. At 15 and 30 min after stimulation with heme plus LPS, we observed an increased phosphorylation of ERK and p38 compared with controls receiving heme or LPS alone (Fig. 4A). An increase in JNK was observed at 30 and 60 min after treatment with heme and LPS (Fig. 4A). Treatment with ERK, p38, or JNK inhibitors blocked the potentiating effect of heme on LPS-induced cytokine secretion (Fig. 4, B and C). As expected, we also observed an increase in TNF mRNA on macrophages treated with heme plus LPS compared with single treatments (supplemental Fig. 2). These results indicate that NF-κB and MAPKs activation are essential to the cooperative effect of heme and LPS on cytokine production.
FIGURE 4.
Heme anticipates and increases the activation of MAPKs induced by LPS. A, macrophages from C57Bl/6 mice were stimulated with heme (100 μm) and/or LPS (0.1 ng/ml) in the time intervals indicated. Cell extracts were submitted to SDS-PAGE. ERK1/2, JNK, and p38 phosphorylations were detected by immunoblotting. Detection of nonphosphorylated ERK2 was used as loading control. The figures are representatives of three different experiments with similar results. B and C, production of TNF was evaluated in peritoneal macrophages treated with SB203580 (10 μm), PD98059 (30 μm) (B), or SP600125 (30 μm) (C) for 30 min before the stimuli with heme (100 μm) and LPS (0.1 ng/ml). The supernatants were collected 4 h after the stimuli. Results represent means ± S.E. for TNF determination and are representative of two different experiments. *, p < 0.05. H, heme; L, LPS; SB, SB203580; PD, PD98059; SP, SP600125.
Induction of ROS Was Involved in the Potentiating Effect of Heme upon LPS Stimulation
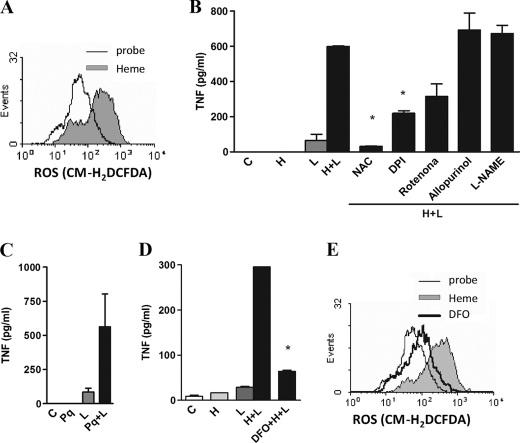
ROS participate as second messengers in several signaling pathways, affecting the activation of downstream molecules such as NF-κB and MAPKs and priming macrophages to subsequent activation by LPS (20, 24, 25). Heme induced a significant generation of ROS in macrophages (Fig. 5A). Thus, we tested the role of ROS on heme-induced cytokine amplification triggered by LPS. Pretreatment with the antioxidant NAC blocked the cytokine secretion induced by heme and LPS, whereas DPI, a flavoprotein inhibitor, or rotenone, a mitochondrial inhibitor, partially inhibited the enhancing effect of heme on LPS-induced TNF and IL-6 (Fig. 5B and data not shown). Similarly, apocynin, a NADPH inhibitor, also blocked the synergistic effect of heme and LPS (data not shown). Treatment with allopurinol, a xanthine oxidase inhibitor, or with l-NAME, a NO synthase inhibitor, did not affect the ability of heme to increase cytokine production induced by LPS (Fig. 5B). The treatment of macrophages with paraquat, a drug that causes the generation of superoxide, also induced an increase in TNF secretion triggered by LPS (Fig. 5C). Next, we analyzed the role of the coordinated iron of heme on cytokine production and ROS generation. The use of an iron chelator, deferoxamine, suppressed the effect of heme on cytokine secretion and ROS generation (Fig. 5, D and E). These results suggest that induction of ROS by heme is involved in the increased cytokine secretion triggered by LPS.
FIGURE 5.
Heme amplifies the production of cytokines dependently of ROS. A, macrophages were stimulated for 1 h with heme (100 μm), and ROS generation was evaluated by flow cytometry using the probe CM-H2DCFDA (2 μm). B, macrophages were preincubated for 30 min with NAC (10 mm), DPI (10 μm), rotenone (1 μm), allopurinol (10 μm), or l-NAME (100 μm) and stimulated with heme (100 μm) and LPS (0.1 ng/ml), and after 4 h the supernatants were collected for posterior determination by ELISA. C, macrophages were stimulated with paraquat (100 μm) and LPS (0.1 ng/ml), and after 4 h the supernatants were collected for TNF determination by ELISA. D, macrophages were preincubated for 30 min with deferoxamine (2 μm) and stimulated with heme (100 μm) and LPS (0.1 ng/ml), and after 4 h the supernatants were collected for posterior determination by ELISA. E, macrophages were pretreated with deferoxamine (2 μm) and stimulated for 1 h with heme (100 μm). The ROS generation was evaluated by flow cytometry using the probe CM-H2DCFDA (2 μm). All stimuli were performed in the presence of 10% FCS. Results represent mean ± S.E. for cytokines and are representative of two or three different experiments. *, p < 0.05. H, heme; L, LPS; Pq, paraquat; DFO, deferoxamine.
Synergistic Effect of Heme Combined with Microbial Molecules on TNF Secretion Required Syk
Macrophages from Tlr4−/− mice stimulated with heme plus CpG or from Myd88−/− mice stimulated with heme plus poly(I-C) showed a marked increase in the TNF secretion compared with macrophages stimulated with CpG or poly(I-C) alone (supplemental Fig. 3, A and B). Heme also potentiated the TNF production by Asc−/− and Rick−/− macrophages activated with LPS (supplemental Fig. 3, C and D). Together, these data indicate that TLR4 and the adaptor proteins MyD88, ASC, or RICK/Rip2 are not essential to the potentiating effect of heme upon LPS stimulation.
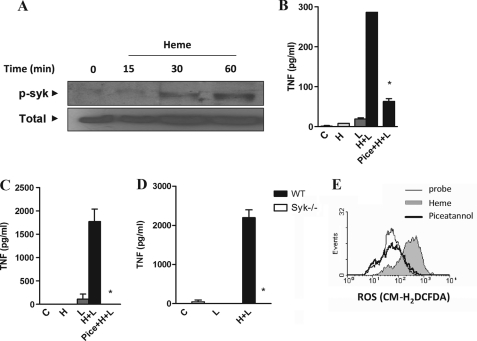
Recent studies indicate that receptors that signal through immunoreceptor tyrosine-based activation motifs (ITAMs) regulate the signaling by heterologous receptors such as TLRs, modulating cytokine production by macrophages and DC (18, 26–28). This effect of ITAM-associated receptors requires Syk, an essential molecule in downstream activation of NF-κB via the CARD9 adaptor, and of PKC and calcium signaling via activation of phospholipase Cγ. These pathways participate on activation of MAPKs, and Syk also activates PI3K and ROS production (29). The stimulation of macrophages with heme caused the phosphorylation of Syk, suggesting a putative role of this pathway on the heme potentiating effect upon LPS stimulation (Fig. 6A). To directly evaluate the role of Syk, we used piceatannol, a selective Syk inhibitor (29). Treatment of macrophages with piceatannol abolished the amplifying effect of heme on cytokine secretion induced by LPS, CpG, and poly(I-C) (Fig. 6B and supplemental Fig. 4). The synergistic induction of TNF by heme and LPS was also observed using DC and was abolished after treatment with piceatannol (Fig. 6C). More important, the synergism of heme and LPS was absent in Syk-deficient DC (Fig. 6D). Considering the critical role of ROS and Syk on heme-induced cytokine amplification, we analyzed the involvement of Syk in ROS generation induced by heme. Treatment of macrophages with piceatannol abrogated the ROS generation by heme (Fig. 6E). Together, these results indicated that activation of Syk is essential to ROS generation by heme affecting the cytokine production induced by microbial ligands.
FIGURE 6.
Syk is involved in the synergistic effect of heme on cytokine secretion. A, macrophages from C57Bl/6 mice were stimulated with heme (100 μm) in the time intervals indicated. Cell extracts were submitted to SDS-PAGE and Syk phosphorylation was detected by immunoblotting. Detection of β-actin was used as loading control. The figure is representative of three different experiments with similar results. B, peritoneal macrophages; C, dendritic cells from C57Bl/6 were stimulated with heme (100 μm) and/or LPS (0.1 ng/ml) 30 min after the treatment with 10 μm piceatannol. *, p < 0.05. D, DC from Syk−/− and WT mice were stimulated with heme (100 μm) and LPS (1 ng/ml), and supernatants were collected 4 h after treatment. TNF was quantified by ELISA. The stimuli were performed in the presence of 10% FCS. Results represent means ± S.E. and are representative of two different experiments. *, p < 0.05. E, peritoneal macrophages from C57Bl/6 were treated with 10 μm of the Syk inhibitor piceatannol for 30 min and stimulated with heme (100 μm) for 1 h. The ROS generation was evaluated by flow cytometry using the probe CM-H2DCFDA (2 μm). Results are representative of three different experiments. H, heme; L, LPS; Pice, piceatannol.
Involvement of Downstream Effectors of Syk on Heme-induced ROS
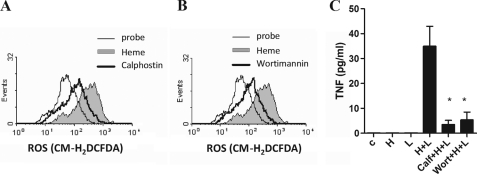
PKC and PI3K are downstream effectors in the Syk signaling pathway. On neutrophils, we previously observed a critical role of these molecules on ROS generation (14). Similarly, PKC and PI3K inhibitors reduced ROS generation by heme and blocked the synergistic effect of heme and LPS on cytokine production (Fig. 7, A–C). Together, these results indicate that Syk, PKC, and PI3K activation and the subsequent ROS generation are involved in the potentiating effect of heme upon LPS stimulation.
FIGURE 7.
PKC and PI3K are important to ROS generation induced by heme. Peritoneal macrophages from C57Bl/6 were treated with 10 μm of the PKC inhibitor calphostin (A) or 50 μm of the PI3K inhibitor wortmannin (B) for 30 min and stimulated with heme (100 μm) for 1 h. The ROS generation was evaluated by flow cytometry using the probe CM-H2DCFDA (2 μm). Inhibitors plus heme (thick line), heme (filled area curve), probe (fine line). The results are representative of three different experiments. C, macrophages were pretreated for 30 min with calphostin (10 μm) or wortmannin (50 μm) and were stimulated with heme, LPS, or heme plus LPS. TNF was quantified by ELISA in supernatants collected 4 h after treatment with heme and LPS. All experiments were performed with medium supplemented with 10% FCS. Results represent mean ± S.E. and are representative of three different experiments. *, p < 0.05.
DISCUSSION
In malaria, hemorrhagic fevers, leptospirosis, and septic shock, high quantities of hemoproteins are released, probably affecting the inflammatory response triggered by microbial molecules. Here, we showed that heme synergized with microbial products increased lethality and cytokine production in vivo and in vitro. Hemolysis induced by phenylhydrazine or treatment with heme caused an increased lethality when combined with a sublethal dose of LPS. The effect of heme on cytokine secretion was not restricted to LPS, because heme also synergizes with agonists of TLR2, TLR3, TLR9, NOD1, and NOD2. The increased cytokine production correlated with an earlier and heightened activation of NF-κB and with phosphorylation of MAPKs on macrophages treated with heme and LPS. Moreover, inhibition of these pathways abrogated the increased secretion of TNF. Furthermore, we demonstrated that generation of oxidative stress by heme was essential for its potentiating effects upon LPS stimulation. The upstream mechanism of ROS generation by heme and heme-LPS synergism to increase cytokine production involved the activation of Syk, PI3K, and PKC. Together, these results revealed a critical and previously unrecognized role of heme in enhancing innate immune response by affecting the signaling of PRRs through Syk activation and subsequent change of the redox state of macrophages.
A low dose of LPS alone resulted in insignificant cytokine secretion, whereas the same dose of LPS together with heme resulted in substantial production of cytokines, thus suggesting that this synergism may be particularly important under conditions of low agonist concentrations. Heme affected MyD88 and TIR domain-containing adaptor-inducing interferon β (TRIF) pathways induced by LPS upon activation of TLR4, increasing the secretion of TNF, IL-6, and IP-10. Heme also enhanced cytokine secretion induced by TLR2, TLR9, and TLR3 agonists. These results indicate that heme synergizes with those TLRs that use exclusively MyD88 (TLR2 and -9), with the TLR that activates TIR domain-containing adaptor-inducing interferon β only (TLR3), or with that able to induce both pathways simultaneously (TLR4). Moreover, heme increased cytokine secretion induced by NOD agonists. These receptors, in contrast to TLRs, are localized to the cytosol, instead of being associated to cell membranes, and use RICK/RIP2 as adaptor protein (30, 31). Together, these results establish that heme amplifies cytokine secretion by PRRs situated on distinct locations and using distinct signal transduction pathways.
The combination of heme and LPS caused an enhanced IκBα degradation, NF-κB activation, and MAPKs phosphorylation. The activation of these pathways was essential to the effect of heme, because treatment with selective inhibitors abrogated the increase in cytokine secretion. The redox state severely affects the response of leukocytes to microbial molecules and cytokines. ROS affect the activation of NF-κB, a key component in TLR and NLR signaling pathways, and are required for cytokine production upon receptor activation (16–18). Exogenously added H2O2 causes the activation of NF-κB and increases the secretion of cytokines by macrophages stimulated with LPS, whereas antioxidants have an inhibitory effect (20). The generation of ROS by heme was involved in the potentiating effect on LPS-induced cytokine secretion. NAC, DPI, and rotenone impaired this enhanced response, whereas inhibition of xanthine oxidase or NOS had no effect. These results suggest that ROS generated by NADPH and mitochondria are involved on heme-LPS synergism to produce cytokines.
Recent studies demonstrated an intense cooperation of several PRRs and ITAM-associated receptors, demonstrating both positive and negative regulation (22, 26–29, 32–34). It has been suggested that activation of ITAM-associated receptors with high affinity ligands causes cross-inhibition of TLR receptors, whereas low affinity ligands potentiate the response (34). The mechanism of the synergistic induction of cytokine production by TLRs and ITAM-associated receptors seems to be an enhanced activation of NF-κB and MAPKs (29). We observed that heme caused the phosphorylation of Syk and that treatment with a selective inhibitor of Syk caused the abrogation of heme-LPS synergistic effects on macrophages and DC. Using Syk-deficient DC, we confirmed the critical role of Syk on heme-LPS synergistic effects. The inhibitors of Syk and downstream effectors, including PKC and PI3K, all affected the generation of ROS and the heme-LPS synergism on cytokine production by macrophages. These results resemble the ROS production induced by Dectin-1 and TGF-β-activated kinase (TAK1)-mediated signal in cooperation with TLR2, which enhances activation of NF-κB (26).
The mechanism of ROS generation by heme is considered to be at least partially dependent on the Fenton reaction (13). However, we recently observed that PKC and PI3K are also involved on ROS generation induced by heme on neutrophils, thus also suggesting a receptor-mediated process (14). A recent report indicates that monosodium urate crystals activate Syk in DC by a mechanism independent of protein receptors in the plasma membrane and dependent on cholesterol in lipid rafts (35). H2O2 also activates NF-κB, a process dependent on Syk (36), thus suggesting a possible involvement of this pathway on H2O2-induced synergism on cytokine production induced by LPS. At present there is no evidence indicating that H2O2 uses a receptor to activate this pathway. The involvement of a putative specific ITAM-associated receptor activated by heme or, alternatively, Syk activation by heme in a membrane receptor-independent effect requires further investigation.
In conclusion, our results show that heme is capable of potentiating cytokine production induced by different innate receptors irrespective of their subcellular localization. This phenomenon ascribes to heme a general role on modulation of innate responses to pathogens and/or NF-κB activation. The molecular mechanism behind this effect of heme is the activation of Syk and subsequent ROS generation.
Supplementary Material
Acknowledgments
We thank Dr. Shizuo Akira, Dr. Anthony Coyle, Dr. Victor Tybulewicz, Dr. Dario Zamboni, Dr. Patricia Bozza, and Dr. Niels Camera for kindly providing mice strains used in this study and Miguel Rodriguez for the critical review of the manuscript.
This work was supported by Conselho de Desenvolvimento Científico e Tecnológico, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro, Fundação José Bonifácio, and Programa de Núcleos de Excelência, Secretaria Nacional de Ciencia Tecnologia e Innovacion de Panama.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- ROS
- reactive oxygen species
- DC
- dendritic cell
- DPI
- diphenyleneiodonium
- NAC
- N-acetyl-l-cysteine
- TLR
- Toll-like receptors
- PRR
- pattern recognition receptor
- CM-H2DCFDA
- 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester
- ITAM
- immunoreceptor tyrosine-based activation motif
- NOD
- nucleotide-binding oligomerization domain.
REFERENCES
- 1.Ferreira A., Balla J., Jeney V., Balla G., Soares M. P. (2008) J. Mol. Med. 86, 1097–1111 [DOI] [PubMed] [Google Scholar]
- 2.Muller-Eberhard U., Fraig M. (1993) Am. J. Hematol. 42, 59–62 [DOI] [PubMed] [Google Scholar]
- 3.Morse D., Choi A. M. (2002) Am. J. Respir. Cell Mol. Biol. 27, 8–16 [DOI] [PubMed] [Google Scholar]
- 4.Poss K. D., Tonegawa S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10925–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller-Eberhard U., Javid J., Liem H. H., Hanstein A., Hanna M. (1968) Blood 32, 811–815 [PubMed] [Google Scholar]
- 6.Jeney V., Balla J., Yachie A., Varga Z., Vercellotti G. M., Eaton J. W., Balla G. (2002) Blood 100, 879–887 [DOI] [PubMed] [Google Scholar]
- 7.Pamplona A., Ferreira A., Balla J., Jeney V., Balla G., Epiphanio S., Chora A., Rodrigues C. D., Gregoire I. P., Cunha-Rodrigues M., Portugal S., Soares M. P., Mota M. M. (2007) Nat. Med. 13, 703–710 [DOI] [PubMed] [Google Scholar]
- 8.Seixas E., Gozzelino R., Chora A., Ferreira A., Silva G., Larsen R., Rebelo S., Penido C., Smith N. R., Coutinho A., Soares M. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths E., Cortes A., Gilbert N., Stevenson P., MacDonald S., Pepper D. (1995) Lancet 345, 158–160 [DOI] [PubMed] [Google Scholar]
- 10.Su D., Roth R. I., Yoshida M., Levin J. (1997) Infect. Immun. 65, 1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H., Wang H., Bernik T. R., Ivanova S., Wang H., Ulloa L., Roth J., Eaton J. W., Tracey K. J. (2002) Shock 17, 485–490 [DOI] [PubMed] [Google Scholar]
- 12.Graça-Souza A. V., Arruda M. A., de Freitas M. S., Barja-Fidalgo C., Oliveira P. L. (2002) Blood 99, 4160–4165 [DOI] [PubMed] [Google Scholar]
- 13.Wagener F. A., Volk H. D., Willis D., Abraham N. G., Soares M. P., Adema G. J., Figdor C. G. (2003) Pharmacol. Rev. 55, 551–571 [DOI] [PubMed] [Google Scholar]
- 14.Porto B. N., Alves L. S., Fernández P. L., Dutra T. P., Figueiredo R. T., Graça-Souza A. V., Bozza M. T. (2007) J. Biol. Chem. 282, 24430–24436 [DOI] [PubMed] [Google Scholar]
- 15.Figueiredo R. T., Fernandez P. L., Mourao-Sa D. S., Porto B. N., Dutra F. F., Alves L. S., Oliveira M. F., Oliveira P. L., Graça-Souza A. V., Bozza M. T. (2007) J. Biol. Chem. 282, 20221–20229 [DOI] [PubMed] [Google Scholar]
- 16.Medzhitov R. (2007) Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 17.Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 18.Trinchieri G., Sher A. (2007) Nat. Rev. Immunol. 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 19.Nathan C. (2002) Nature 420, 846–852 [DOI] [PubMed] [Google Scholar]
- 20.Powers K. A., Szászi K., Khadaroo R. G., Tawadros P. S., Marshall J. C., Kapus A., Rotstein O. D. (2006) J. Exp. Med. 8, 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bittencourt V. C., Figueiredo R. T., da Silva R. B., Mourão-Sá D. S., Fernandez P. L., Sassaki G. L., Mulloy B., Bozza M. T., Barreto-Bergter E. (2006) J. Biol. Chem. 281, 22614–22623 [DOI] [PubMed] [Google Scholar]
- 22.Rogers N. C., Slack E. C., Edwards A. D., Nolte M. A., Schulz O., Schweighoffer E., Williams D. L., Gordon S., Tybulewicz V. L., Brown G. D., Reis e Sousa C. (2005) Immunity 22, 507–517 [DOI] [PubMed] [Google Scholar]
- 23.Scheinman R. I., Gualberto A., Jewell C. M., Cidlowski J. A., Baldwin A. S., Jr. (1995) Mol. Cell. Biol. 15, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathan C. (2003) J. Clin. Invest. 111, 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 26.Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M. (2003) J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. (2003) J. Exp. Med. 197, 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbull I. R., McDunn J. E., Takai T., Townsend R. R., Cobb J. P., Colonna M. (2005) J. Exp. Med. 202, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivashkiv L. B. (2009) Nat. Immunol. 10, 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carneiro L. A., Magalhaes J. G., Tattoli I., Philpott D. J., Travassos L. H. (2008) J. Pathol. 214, 136–148 [DOI] [PubMed] [Google Scholar]
- 31.Shaw M. H., Reimer T., Kim Y. G., Nuñez G. (2008) Curr. Opin. Immunol. 20, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross O., Poeck H., Bscheider M., Dostert C., Hannesschläger N., Endres S., Hartmann G., Tardivel A., Schweighoffer E., Tybulewicz V., Mocsai A., Tschopp J., Ruland J. (2009) Nature 459, 433–436 [DOI] [PubMed] [Google Scholar]
- 33.Hise A. G., Tomalka J., Ganesan S., Patel K., Hall B. A., Brown G. D., Fitzgerald K. A. (2009) Cell Host Microbe 5, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Gordon R. A., Huynh L., Su X., Park Min K. H., Han J., Arthur J. S., Kalliolias G. D., Ivashkiv L. B. (2010) Immunity 32, 518–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng G., Sharma K., Ward S. M., Desrosiers M. D., Stephens L. A., Schoel W. M., Li T., Lowell C. A., Ling C. C., Amrein M. W., Shi Y. (2008) Immunity 29, 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takada Y., Mukhopadhyay A., Kundu G. C., Mahabeleshwar G. H., Singh S., Aggarwal B. B. (2003) J. Biol. Chem. 278, 24233–24241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.