Abstract
Increasing evidence suggests that adenosine monophosphate-activated protein kinase (AMPK) exerts protective effects for cardiovascular diseases apart from the regulation of energy homeostasis. However, the role of AMPK and its underlying mechanism on macrophage foam cell formation are poorly understood. In this study, we sought to investigate the potential effects of AMPK in modulating cholesterol deposition by using murine macrophage-derived foam cells. Incubation with 5-aminoimidazole-4-carboxyamide ribonucleoside (AICAR) markedly attenuated the cholesterol ester accumulation in oxidized low density lipoprotein-loaded macrophages. Notably, AICAR treatment significantly increased ATP-binding cassette transporters G1 (Abcg1) mRNA and protein levels without affecting mRNA and protein expression of ABCA1, scavenger receptors, including scavenger receptor-A, CD36, and scavenger receptor-BI (SR-BI), and cholesterol synthesis-related genes. The up-regulation of Abcg1 by AICAR was independent of the liver X receptor/retinoid X receptor pathway but dependent on ERK activation. AICAR elevates Abcg1 expression through a post-transcriptional mechanism that stabilizes the mRNA. Using a heterologous system with luciferase as a reporter, we further identify the Abcg1 mRNA 3′-UTR responsible for the regulatory effect of AICAR. Prevention of ABCG1 expression by small interfering RNA abolished the AICAR-mediated attenuation on foam cell formation. Furthermore, increased ABCG1 expression and reduced lipid accumulation were demonstrated in AICAR-treated macrophages isolated from apolipoprotein E-deficient mice (apoE−/− mice). AICAR treatment also inhibited atherosclerotic plaque formation in apoE−/− mice. Our findings elucidate a precise mechanism involved in the prevention of atherogenesis by AMPK.
Keywords: ABC Transporter, AMP-activated kinase (AMPK), Atherosclerosis, Cholesterol Metabolism, Macrophage
Introduction
Atherosclerosis is a leading cause of illness and death worldwide. The differentiation of macrophages into lipid-laden foam cells is a crucial process during the development of atherosclerosis (1–3). The transformation of foam cells is due mainly to dysregulated uptake of modified low density lipoprotein (LDL) by macrophages, resulting in excessive deposition of lipoprotein-derived cholesterol inside the cells (4, 5). Traditionally, it has been assumed that the uncontrolled uptake of oxidized low density lipoproteins (OxLDL)3 by macrophage scavenger receptors is largely responsible for this process. Several members of macrophage scavenger receptors, including class A SRs (SR-AI) and class B SRs (such as SR-BI and CD36), have been implicated in the formation of foam cells by its capacity to bind and endocytose OxLDL and shown to play a critical role in the development of atherosclerotic lesions (6, 7). On the contrary, cholesterol efflux is regarded as the most important key point with regard to maintenance of cholesterol homeostasis and atherosclerosis.
Two major potential cholesterol efflux pathways from macrophages have been described as follows: SR-BI-mediated cholesterol efflux and active cholesterol efflux mediated by the ATP-binding cassette transporters ABCA1 and ABCG1. ABCA1 promotes efflux of phospholipids and cholesterol to lipid-poor apoA-I in a process that involves the direct binding of apoA-I to the transporter. SR-BI and ABCG1 were identified as the key mediators of macrophage cholesterol efflux to mature HDL (8–10). The intracellular cholesterol homeostasis in macrophages is dynamically regulated by cholesterol uptake and cholesterol efflux, processes that are tightly controlled by these receptors.
Adenosine monophosphate-activated protein kinase (AMPK), which is expressed in most mammalian cell types, including the vascular cells such as endothelial cells, smooth muscle cells, and macrophages, plays a central role in the regulation of energy metabolism under stress conditions (11, 12). Activation of AMPK leads to the phosphorylation of a number of key enzymes in metabolic pathways, such as acetyl-CoA carboxylase or mammalian target of rapamycin, by modulating their activities and by regulating the activity of transcription factors and transcriptional cofactors (13, 14). Additionally, AMPK has been reported to exert multiple protective effects in vascular cells by inhibiting inflammation, oxidant production, vascular smooth muscle cell proliferation, and insulin resistance (15, 16). However, little is known about the influence of AMPK on foam cell formation. The purpose of this study was to investigate the impact of AMPK on macrophage foam cell formation and its molecular mechanisms.
EXPERIMENTAL PROCEDURES
Materials
AMPK agonist 5-aminoimidazole-4-carboxyamide ribonucleoside (AICAR), anti-phospho-AMPK Thr-172, and anti-β-actin were purchased from Cell Signaling Technology. AMPK inhibitor compound C and LXRα antagonist geranylgeranyl pyrophosphate ammonium salt were obtained from Calbiochem. A-769662, a cell-permeable drug that potently activates AMPK in cells, was provided by Symansis Pty Ltd. (Washdyke, New Zealand). Human 3,3′-dioctadecylindocarbocyanine-labeled fluorescent high density lipoprotein (DiI-HDL) was obtained from Biomedical Technologies, Inc. RPMI 1640 culture medium and fetal bovine serum (FBS) were provided by Invitrogen. GW3965, metformin, 1α,2α-[3H]cholesterol, actinomycin D, HDL, and penicillin/streptomycin were purchased from Sigma.
Cell Culture
Murine macrophage J774.A1 cells (ATCC, TIB-67) were cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin, and streptomycin.
LDL Modification
LDL was isolated from fresh plasma of healthy subjects as described previously (17). LDL was exposed to 5 μm CuSO4 for 24 h at 37 °C; then Cu2+ was removed by extensive dialysis with phosphate-buffered saline (PBS) overnight. The extent of modification was determined by measurement of thiobarbituric acid-reactive substances. The protein concentration was determined by a Lowry assay (17). OxLDL containing 20–50 nm thiobarbituric acid-reactive substances defined as malondialdehyde equivalents/mg of LDL protein was used for further experiments (18).
Oil Red O Staining
The macrophages were fixed with 4% paraformaldehyde and then stained by 0.5% Oil Red O. Hematoxylin was used as a counterstain. The density of lipid content was measured with a microplate reader (absorbance at 540 nm, BioTek Instruments, Inc.) after alcohol extraction (18).
Filipin Staining
Macrophages were pretreated with AICAR (1 mm) for 24 h. The cells were then loaded with OxLDL (50 μg/ml) in RPMI 1640 medium supplemented with 1% Nutridoma for 24 h. After incubation, cells were rinsed twice with ice-cold PBS and fixed with paraformaldehyde (4% in PBS) for 30 min at 4 °C before being incubating with filipin (50 μg/ml in PBS) for 30 min at room temperature. Labeling was then visualized by a fluorescent microscope (Nikon ECLIPSE TI-E), and fluorescent signals were quantified (19).
Intracellular Lipid Analysis in Macrophages
Intracellular lipids in macrophages were extracted with hexane/isopropyl alcohol (3:2) after 24 h of incubation with the vehicle or AICAR, evaporated, and dissolved in isopropyl alcohol containing 10% Triton X-100 for preparation of a sample solution. Free cholesterol and total cholesterol were determined by commercial assay systems (Wako Chemicals). Cholesterol ester was estimated by subtracting free cholesterol from total cholesterol. Data were normalized to cellular protein content (20).
Cholesterol Efflux Assays
Adherent macrophages were incubated in RPMI 1640 medium with OxLDL (50 μg/ml) at 37 °C for 24 h to induce macrophage foam cells. The cells were then washed in ice-cold PBS and incubated with AICAR in medium containing 0.2% low endotoxin fatty acid-free bovine serum albumin (BSA) for the indicated time. After that, cells were washed three times in PBS, and HDL-mediated cholesterol efflux studies were immediately performed by adding fresh medium in the presence or absence of HDL (10 μg/ml) for 24 h. At the end of this incubation, lipids of cells and media were separately extracted in chloroform and methanol, and the samples were then dried under nitrogen, and free cholesterol and total cholesterol were measured by enzymatic assays. Cholesterol ester was determined as the difference between total and free cholesterol. Cellular proteins were collected by digestion in NaOH and measured by using the Bradford method. The percent change of intracellular cholesterol amounts in the presence of HDL relative to HDL-free culture medium was determined as the percent counts in medium over counts in medium + cells. Each assay was performed in triplicate (20).
In the experiments with [3H]cholesterol, macrophages were incubated for 24 h in RPMI 1640 medium supplemented with 5% lipoprotein-deficient serum, 1.0 μCi/ml [3H]cholesterol, and 50 mg/ml OxLDL in the absence or presence of AICAR (0.5–2 mm) for 24 h. To equilibrate cholesterol pools, cells were washed twice with PBS and incubated for 24 h in RPMI 1640 medium containing 0.2% BSA with no lipoproteins. Cells were washed again and incubated in RPMI 1640 medium containing 0.2% BSA in the absence or presence of HDL (50 μg/ml) for 24 h. The percentage of cholesterol efflux was calculated by dividing the medium-derived radioactivity by the sum of the radioactivity in the media and the cells (21).
Cytotoxicity Tests
Cells were grown in microtiter plates and subjected to the experimental culture conditions and treatments as described for efflux experiments. 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide was added to each well and incubated for 4 h in the cell culture incubator. Solubilization buffer (10% SDS in 0.01 m HCl) was added to each well and incubated in a cell culture incubator overnight. Absorbance was measured at 550 nm on a microtiter plate reader. Percent 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cleavage was determined as follows: (treatment value − media with vehicle value)/(0.1% Triton X-100 value − media with vehicle value) × 100 (22). A lactate dehydrogenase release assay was performed according to the manufacturer's instructions (Biovision).
Quantitative Real Time RT-PCR
Total RNA was extracted from the cultured cells using TRIzol reagent according to the protocol provided by the manufacturer (Invitrogen). Real time quantitative PCR analysis was used to measure the relative levels of gene expression. The amplification was performed on ABI Prism 7900-HT sequence detection system and Universal MasterMix (Applied Biosystems). The relative levels of the mRNA were calculated with GAPDH mRNA levels as the invariant control (23).
Western Blot
Proteins were size-fractionated electrophoretically using SDS-polyacrylamide gel electrophoresis gels and transferred to polyvinylidene fluoride membranes. The membranes were incubated with the primary and secondary antibody and visualized using an enhanced chemiluminescence detection system (ECL, Cell Signaling Technology Inc). Anti-β-actin (Cell Signaling Technology Inc) was used for equal protein loading.
ABCG1 mRNA Stability
For mRNA stability measurements, actinomycin D (5 mg/ml) was used to inhibit gene transcription. At the indicated time points after the addition of actinomycin D, cells were harvested, and total RNA was extracted. The expression levels of Abcg1 at each time point were measured by quantitative RT-PCR and normalized to the GAPDH levels. The remaining mRNA was determined by comparison with the expression level of the relevant gene at the zero time point (designated 100%) when actinomycin D was added (24).
ABCG1 3′-Untranslated Region Constructs
Segments of the murine ABCG1 3′-UTR were PCR-amplified using oligonucleotide primers containing flanking SpeI recognition sequences and human genomic DNA as a template. The PCR products were gel-purified and ligated downstream of the firefly luciferase coding region of the pGL3-Control vector (Promega). The pGL3-Control vector was chosen because it contains the SV40 promoter without enhancers; changes in luciferase activity can be attributed to the effect of 3′-UTR inserts. The following primers were used to amplify the 3′-UTR of mouse Abcg1: forward, 5′-GACTAGTGTACAAAATCCGGGCAGAGA-3′; reverse, 5′-GACTAGTGCTTAAAATAAGAAGCACGTGGA-3′. To create the mutant Abcg1 3′-UTR (LUCΔ3′-UTR) that containing no 3′-UTR, we cut plasmids with Apa1 to remove the AU-rich element-containing region and religated the remaining vector with the 5′ proximal region of UTR. All constructs were sequenced, and the correct clones were further propagated to isolate plasmid DNA (25).
Adenoviruses
The adenoviral vector expressing a dominant-negative AMPKα mutant (DN-AMPK) and a constitutively active form of AMPK (CA-AMPK) were kindly donated as a gift by Dr J. Ha (Department of Molecular Biology, Kyung Hee University, College of Medicine, Seoul, Korea) (26).
LXR Reporter Gene Assay
LXR-response element (LXRE)-driven luciferase reporter vector (LXRE-tk-Luc) was kindly provided by David J. Mangelsdorf (27) (University of Texas Southwestern Medical Center). For LXR activation studies, 0.75 μg of LXRE-driven luciferase reporter vector (LXRE-tk-Luc) and 0.75 μg of β-galactosidase control vector (Promega) were used. Six hours after transfection, cells were treated with AICAR for 12 h. Luciferase and β-galactosidase (β-gal) activities were determined in cell lysates. The amount of luciferase activity was normalized for β-gal and reported as relative light units.
DiI-HDL Binding
The cultured cells were incubated with different concentrations of AICAR for 24 h. After that, the cells were incubated with 5 μg/ml DiI-HDL for another 4 h. The cells were detached using cell removal buffer containing EDTA and were washed, and the cells were resuspended in FACS solution (PBS with 0.5% BSA and 0.02% sodium azide) at a density of 1 × 106 cells/ml. The mean fluorescence intensity was analyzed by FACS (FACSort, BD Biosciences) (28).
Small Interfering RNA (siRNA)
cRNA oligonucleotides derived from mouse ABCG1 sequence (ONTARGETplus SMARTpool targeting mouse ABCG1) were obtained from Dharmacon (Chicago) and used to knock down ABCG1 expression in macrophages. Scrambled oligonucleotides (ONTARGETplus siCONTROL nontargeting pool) were used as control. Expression levels of ABCG1 in transfected cells were determined by real time PCR and Western blot.
AMPK Activity Assay
AMPK activity was actually measured by using a nonradioisotopic kit from MBL International Corp. according to the provided protocol (Woburn, MA, catalog no. CY-1182).
ERK Activation
Macrophages were lysed with phospho-Tyr protecting lysis buffer (1% Triton X-100, 10% glycerol, 50 mm NaCl, 50 mm HEPES, 2 mm EDTA, 1 mm Na3VO4, 10 mm NaF, 10 mm NaPO4, 10 mm p-nitrophenyl phosphate, 10 mm β-glycerol phosphate, and protease inhibitor mixture). Samples were subjected to SDS-PAGE and immunoblotted with anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies (Cell Signaling Technology).
Mice and Procedures
Animal experiments were conducted in accordance with institutional guidelines of Sun Yat-sen University. Male apoE-deficient (apoE−/−) mice at 6 weeks of age (C57BL/6J genetic background; The Jackson Laboratories) were used for this study. The animals were maintained in a 22 °C room with a 12-h light/dark cycle and received drinking water ad libitum. The mice were fed on a standard purified mouse diet (AIN-93G) for 2 weeks prior to the start of experiment. The animals were then switched to high fat diet for 12 weeks (containing 21% fat) receiving either AICAR (n = 10, 200 mg·kg−1·day−1) or vehicle (n = 10, 0.9% NaCl) via subcutaneous injection once daily.
Isolation of Mouse Peritoneal Macrophages
To harvest mouse peritoneal macrophages, the mice were sacrificed, and ice-cold PBS was injected into the peritoneal cavity of each mouse. This fluid was carefully collected and centrifuged at 3000 rpm. The supernatant was then withdrawn, and the cell pellet was resuspended in RPMI 1640 medium, allowed to adhere for 3 h, and then washed three times with pre-warmed PBS to remove nonadherent cells. The medium was then replaced with fresh RPMI 1640 medium for further analysis (29).
Plasma Measurements
Total cholesterol, triglycerides, and free fatty acids were determined with colorimetric assay systems (Wako chemicals) adapted for microtiter plate assay. Particle size distribution of the lipoproteins was determined by fast performance liquid chromatography (FPLC), using pooled samples of plasma as described previously (30). In brief, plasma from mice was subjected to FPLC analysis using a Superose 6 column (GE Healthcare) on an HPLC system model 600 (Waters chromatography). A 100-μl aliquot of plasma was injected onto the column and separated with a buffer containing 0.15 m NaCl, 0.01 m Na2HPO4, 0.1 mm EDTA (pH 7.4), at a flow rate of 0.5 ml/min. Forty 0.5-ml fractions were collected, and tubes 11–40 were analyzed for cholesterol. Fractions 14–17 contain VLDL and chylomicra; fractions 18–24 contain LDL and intermediate density lipoprotein; fractions 25–29 contain HDL; and fractions 30–40 contain nonlipoprotein-associated proteins.
Statistical Analysis
Continuous data were expressed as mean ± S.E. Comparison between groups was performed by Student's paired two-tailed t test. Two-way analysis of variance was used to examine differences in response to treatments between groups, with post hoc analysis performed by the method of Student-Newman-Keuls. p < 0.05 was considered significant.
RESULTS
AICAR Inhibits the Formation of Macrophage Foam Cells
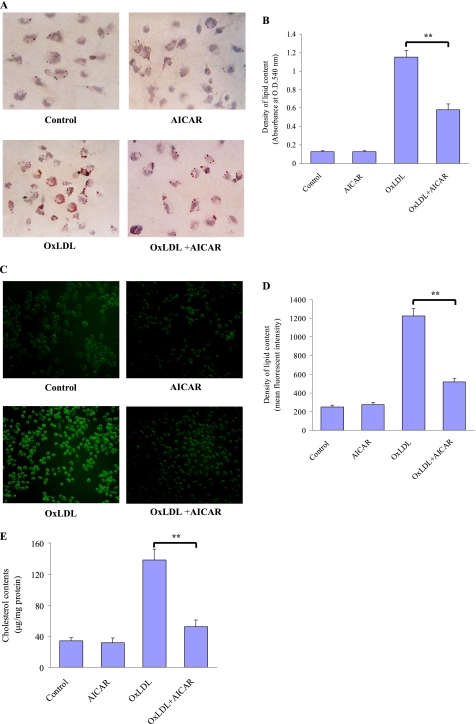
To examine the potential effect of AMPK on formation of foam cells, the mouse macrophages were loaded with OxLDL (50 μg/ml) for 24 h to promote cholesteryl ester accumulation in the absence or presence of the AMPK activator AICAR. Combined treatment with AICAR and OxLDL significantly ameliorated intracellular lipid accumulation in macrophages compared with the OxLDL-loaded cells as determined by Oil Red O (Fig. 1, A and B) and filipin staining (Fig. 1, C and D). This reduction in intracellular lipid accumulation was also evidenced by direct measurements. AICAR treatment significantly reduced cholesterol accumulation in OxLDL-loaded macrophages (Fig. 1E). These data suggest that AICAR confers a protective role in the formation of macrophage foam cells.
FIGURE 1.
AICAR attenuates OxLDL-induced lipid accumulation in macrophages. J774.A1 mouse macrophages were incubated with vehicle (PBS), OxLDL (50 μg/ml) alone, AICAR (1 mm) alone, or a combination of AICAR and OxLDL for 24 h. After fixation by 4% paraformaldehyde, cells were stained with Oil Red O (A and B) or filipin (C and D), respectively, to detect the lipid accumulation, and hematoxylin was used as counterstaining. Magnification was ×200. The intracellular lipid level (B and D) was measured by alcohol extraction after staining. E, intracellular levels of cholesterol were analyzed by colorimetric assay kits. **, p < 0.01.
We also assessed the AMPK activation in this cellular model. Compared with untreated control, AICAR treatment strikingly stimulated the Thr-172 phosphorylation of AMPK, which is an essential marker of AMPK activity (supplemental Fig. 1A). Consistent with this result, AICAR strongly stimulates AMPK enzymatic activity (∼2-fold) in macrophages (supplemental Fig. 1B).
AICAR Induces Expression of ABCG1 in Macrophages
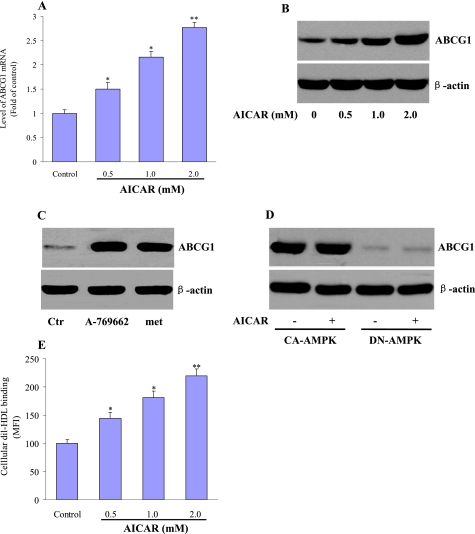
ABCG1, ABCA1, SR-A, CD36, and SR-BI have shown their crucial role in cholesterol homeostasis during foam cell formation (6–10, 31–33). We therefore delineated the mechanism of AMPK to attenuate lipid accumulation by examining the alterations of these receptors and transporters. Macrophages treated with AICAR (0.5, 1.0, or 2.0 mm) for 24 h showed a dose-dependent induction of Abcg1 mRNA levels as determined by quantitative RT-PCR (Fig. 2A). In accordance with this result, AICAR treatment also caused a concentration-dependent increase in ABCG1 protein levels (Fig. 2B). However, AICAR did not affect the mRNA (supplemental Fig. 2, A–D) and protein (supplemental Fig. 2E) levels of ABCA1, SR-A, CD36, and SR-BI.
FIGURE 2.
Effects of AICAR on the expression of ABCG1. J774.A1 macrophages were incubated with AICAR (0.5, 1.0, 2.0 mm) for 24 h. A, cell lysates were subjected to real time quantitative PCR to determine Abcg1 mRNA expression. The results are expressed as fold of control from at least three independent assays. B, Western blot was used to measure the protein levels of ABCG1. The representative blots from three independent experiments are shown. C, macrophages were treated with A769662 (100 μm) or metformin (met, 1 mm) for 24 h. Ctr, control. D, macrophages were infected with adenoviruses encoding CA- or DN-AMPKα in the presence of AICAR (1 mm) or vehicle. ABCG1 protein expression was measured by Western blot. The representative blots from three independent experiments are shown. E, J774.A1 macrophages were incubated with AICAR (0.5, 1.0, ad 2.0 mm) for 24 h. After then, the cells were incubated with 5 μg/ml DiI-HDL for 4 h, resuspended in FACS solution, and analyzed with the FACS flow cytometer. The mean fluorescence intensity (MFI) of untreated cells (control) is expressed as 100%. The data shown are expressed as a percentage of the control from three independent measurements. *, p < 0.05, or **, p < 0.01 compared with control.
Unrelated AMPK activators, such as metformin (34) and A-769662 (35), were also shown to increase ABCG1 expression in macrophages (Fig. 2C). Furthermore, overexpression of a CA-AMPKα led to a robust increase of ABCG1 expression that could not be further enhanced by AICAR. In contrast, when a DN-AMPKα was overexpressed, AICAR was unable to enhance ABCG1 expression (Fig. 2D).
We next tested the effects of AICAR on cellular surface binding of HDL to ABCG1, which could account for the higher cholesterol efflux. DiI-HDL binding was increased in a dose-dependent fashion by AICAR incubation (Fig. 2E). Taken together, these data show that AICAR increases the cellular expression of ABCG1 and its interactions with cholesterol acceptor HDL.
AICAR Did Not Affect Expression of Cholesterol Synthesis-related Genes
We then examined the impact of AICAR on principal target molecules that regulate cellular cholesterol ester synthesis and hydrolysis in macrophages. There was a modest but insignificant reduction in acyl-coenzyme A:cholesterol-acyltransferase 1 (Acat1), a key enzyme of cholesterol esterification, in the AICAR-treated macrophages (supplemental Fig. 3A), but no effect of AICAR was observed on the expression of hormone-sensitive lipase (Hsl) (supplemental Fig. 3B), which acts as the neutral cholesterol esterase, in macrophages.
We also investigated the effect of AICAR on endogenous cholesterol synthesis-related gene expression. Our data showed that AICAR exposure did not alter the mRNA expression of sterol regulatory element binding protein (Srebp) 1 (supplemental Fig. 3C) and Srebp2 (supplemental Fig. 3D), two key transcription factors regulating multiple genes involved in the synthesis and metabolism of cholesterol and fatty acids. Additionally, neither 3-hydroxy-3-methylglutaryl coenzyme A reductase (Hmgcr) (supplemental Fig. 3E) nor LDL receptor (Ldlr) (supplemental Fig. 3F) was influenced by AICAR treatment.
AICAR Induces HDL-mediated Cholesterol Efflux from Macrophage Foam Cells
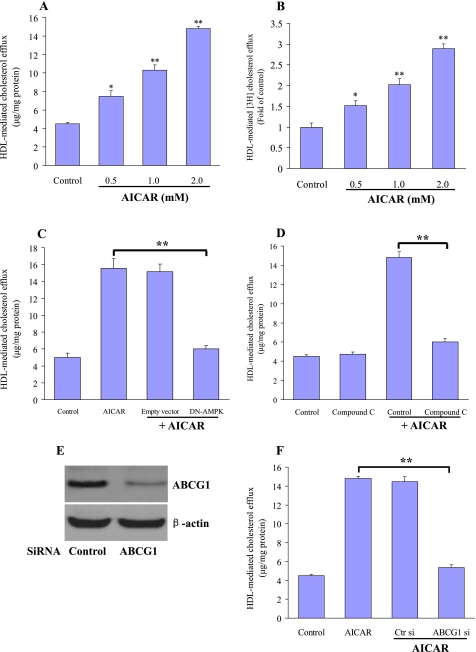
We next investigated the effects of AICAR on HDL-mediated cholesterol efflux in macrophage foam cells. The OxLDL-loaded macrophages were treated with AICAR for 24 h and subsequently exposed to HDL. After 24 h, the change of intracellular cholesterol levels was measured. Cholesterol efflux from macrophage foam cells was dose-dependently increased in the presence of AICAR (Fig. 3A). To demonstrate that the variation of intracellular lipids was not due to the action of AICAR on de novo cholesterol synthesis, we loaded macrophages with 250 μCi of [3H]cholesterol plus OxLDL (50 μg/ml) for 24 h and determined HDL-mediated efflux of cholesterol by measuring the appearance of cholesterol in the medium. AICAR treatment increased [3H]cholesterol release in a concentration-dependent fashion compared with untreated cells (Fig. 3B). Transfection with a dominant-negative AMPK mutant (Fig. 3C) or coincubation with compound C (Fig. 3D), a selective potent inhibitor of AMPK, largely abolished the positive effects of AICAR on cholesterol transport. These data suggest that AMPK confers a protective role in the formation of macrophage foam cells by promotion of cholesterol efflux. Treatment with AICAR did not elicit any release of lactate dehydrogenase (data not shown), ruling out that its effect on cholesterol efflux from foam cells were due to cellular toxicity.
FIGURE 3.
AICARs enhance HDL-mediated cholesterol efflux in macrophages. A, macrophages were loaded with OxLDL (50 μg/ml) alone or in the presence of different concentrations of AICAR for 24 h. These cells were then washed with PBS and incubated with HDL (20 μg/ml) as an acceptor for a further 16 h. Determination of cholesterol efflux from the cells was performed as described under “Experimental Procedures.” The results are represented as the mean ± S.E. of three individual experiments. B, [3H]cholesterol-loaded macrophages were treated with AICAR (0.5–2.0 mm) and subsequently incubated with RPMI 1640 medium with HDL (20 μg/ml) as indicated. HDL-induced [3H]cholesterol efflux was measured as described. Values are expressed relative to the control, set as 1. Results are the mean ± S.E. of triplicate determinations, representative of three independent experiments. *, p < 0.05, or **, p < 0.01 compared with control. C and D, cholesterol-loaded macrophages were transfected with DN-AMPKα (C) or pretreated with the compound C (D) (10 mm). The cells were then incubated with AICAR (1 mm) for a further 24 h. HDL-mediated cholesterol efflux was determined as under “Experimental Procedures.” Results are represented as mean ± S.E. of three individual experiments. **, p < 0.01. E and F, macrophages were transfected with ABCG1 siRNA or a nonspecific control siRNA. E, Western blot analyses using antibody against ABCG1 and β-actin were performed to test transfection efficiency. Representative blots from three independent assays are presented. F, 24 h after transfection, the cells were further incubated with OxLDL (50 μg/ml) alone or in the presence of AICAR (1 mm) for 24 h. Macrophages were then washed with PBS and incubated with or without HDL (20 μg/ml) for 16 h. Results are represented as mean ± S.E. of three individual experiments. **, p < 0.01.
To determine whether enhanced cholesterol efflux following AICAR treatment requires an increase in ABCG1, we transfected the specific ABCG1 siRNA in macrophages to knock down ABCG1 expression. Western blot analysis revealed that ABCG1 protein expression is effectively suppressed by siRNA, but control siRNA had no such effect (Fig. 3E). ABCG1 silencing significantly reduced cholesterol mass efflux induced by AICAR (Fig. 3F). This result strongly suggests that ABCG1 transporter is responsible for facilitating cholesterol efflux induced by AMPK activation.
AICAR Regulates Abcg1 mRNA Stability in Macrophages
To further determine the potential mechanism for the induction of ABCG1 expression, we determined the nuclear protein level of LXRα and RXR in AICAR-treated macrophages. However, AICAR treatment did not affect the nuclear level of LXRα and RXR (supplemental Fig. 4A). Furthermore, AICAR did not influence the LXRα activation by assessing LXRE-mediated luciferase activity (supplemental Fig. 4B). Moreover, coincubation with geranylgeranyl pyrophosphate (GGPP) a pharmacological LXR antagonist or LXR small interfering RNA (siRNA) did not affect the induction of ABCG1 by AICAR (supplemental Fig. 4C). These findings imply that LXRα activation did not play the essential role in AICAR-regulated gene expression of ABCG1 in macrophages.
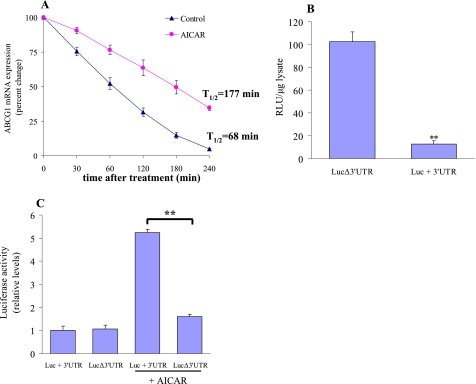
To further explore the mechanisms by which AICAR increased Abcg1 mRNA levels in macrophages, actinomycin D was added to both vehicle- and AICAR-treated macrophages for specified periods, and the half-life of ABCG1 mRNA was determined by quantitative RT-PCR in each case. Based on the observed decay values, we found that AICAR prolongs the turnover rate of ABCG1 transcripts by ∼3-fold (177 min versus 68 min; Fig. 4A).
FIGURE 4.
AICAR regulates Abcg1 mRNA decay through 3′-UTR. A, macrophages were left untreated or incubated with AICAR (1 mm) for 24 h, and actinomycin D (5 μg/ml) was added to the cells for different intervals. Total RNA was then isolated, and Abcg1 mRNA was assayed by quantitative RT-PCR, and the normalized Abcg1 mRNA signals were plotted as the percentage of the Abcg1 mRNA remaining. Decay curves were plotted versus time. B, LUC/3′-UTR-reporter constructs containing no 3′-UTR (LUCΔ3′-UTR) or Abcg1 3′-UTR (LUC+3′-UTR) were transfected into J774.1 macrophages. Luciferase activity (RLU) was normalized to total protein from the three independent experiments. **, p < 0.01 compared with LUCΔ3′-UTR. C, macrophages were transfected with LUC/3′-UTR or LUCΔ3′-UTR reporter constructs, respectively, and kept in culture medium or incubated with 1 mm AICAR. Relative luciferase activity was normalized to total protein, and values shown are based on luciferase expression from control transfections. **, p < 0.01.
A central mechanism that controls synthesis of ABCG1 protein involves rapid degradation of the ABCG1 transcript (42). Post-transcriptional regulation of ABCG1 expression has been shown to be mediated through the 3′-UTR of Abcg1 mRNA. To determine whether the 3′-UTR of Abcg1 regulates its expression in macrophages, we transfected J774.1 macrophages with LUC/3′-UTR reporter constructs containing the full-length ABCG1 3′-UTR (LUC+3′-UTR) or with ABCG1 3′-UTR deleted (LUCΔ3′-UTR), and luciferase (LUC) expression was measured. As in Fig. 4B, ABCG1 3′-UTR reduced luciferase expression ∼8-fold in macrophages, consistent with rapid decay of the mRNA. We next investigated the ability of AICAR to influence post-transcriptional regulation mediated through ABCG1 3′-UTR. Macrophages transfected with the LUC+3′-UTR construct or LUCΔ3′-UTR construct yielded similar levels of expression when the cells were incubated in culture medium. In contrast, addition of AICAR increased luciferase expression 7-fold in LUC+3′-UTR-transfected macrophages, but this response was not observed in LUCΔ3′-UTR-transfected cells (Fig. 4C). These findings indicate that AICAR promoted Abcg1 expression through inhibition of rapid mRNA decay mediated by the 3′-UTR of the ABCG1 transcript.
ERK Activation Is the Prerequisite for AICAR to Increase Abcg1 Expression
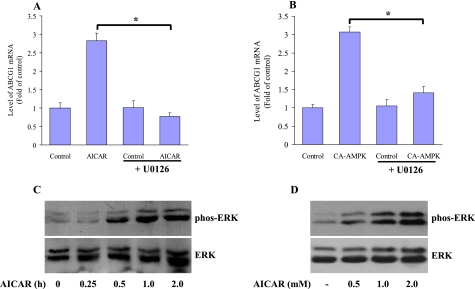
To further identify the potential signaling pathway involved in AMPK-mediated up-regulation of ABCG1 expression, the different kinase inhibitors were used, including the inhibitor of MEK1 U0126, the p38 kinase inhibitor SB203580, the c-Jun N-terminal kinase inhibitor curcumin, the PI3K inhibitor wortmannin, and the PKC inhibitor calphostin C. We found that the induction of ABCG1 mRNA expression by AICAR was most sensitive to the U0126. U0126 incubation almost completely abolished the activity of AICAR on ABCG1 mRNA expression, as confirmed by real time quantitative PCR (Fig. 5A). The effect of U0126 was also confirmed in CA-AMPK-transfected macrophages (Fig. 5B). However, SB203580, curcumin, wortmannin, and calphostin C could not influence the impact of AICAR on macrophage ABCG1 expression (data not shown).
FIGURE 5.
Blocking ERK activation abolished the regulatory effect of AICAR on ABCG1. A, relative amount of Abcg1 mRNA was measured by a quantitative real time PCR after macrophages were treated with 1.0 mm AICAR for 8 h in the presence or absence of 0.5 μm U0126. B, macrophages were infected with adenoviruses encoding CA-AMPK for 12 h. After that, the cells were incubated with medium or treated with 0.5 μm U0126 for 8 h. The amounts of ABCG1 transcript were expressed as fold in untreated control cells (defined as 1), and amounts of Abcg1 mRNA were plotted relative to that value. *, p < 0.05. C, cellular lysates were harvested from macrophages that were untreated or treated with AICAR at a dose of 1.0 mm for different intervals as indicated. Total cellular proteins (40 μg/lane) were subjected to SDS-PAGE and Western blot using antibodies specific for either the activated and phosphorylated forms of ERK1/2 or total ERK1/2. The representative blot from the three independent experiments is shown. D, J774 macrophages were treated with AICAR for 1 h at the indicated concentrations, and total cell lysates were used to detect phosphorylated (phos) and nonphosphorylated ERK by Western blot. The representative blot from the three independent experiments is shown.
To further determine whether AICAR directly activates the MEK1-ERK pathway, the cultured macrophages were treated with AICAR for different intervals, and we assessed the levels of activated ERK in control and AICAR-treated cells. AICAR rapidly activated ERK, and the kinetics of ERK activation preceded the up-regulation of ABCG1 expression by AICAR (Fig. 5C). Exposure to AICAR also dose-dependently activated ERK phosphorylation compared with untreated quiescent macrophages (Fig. 5D). These data indicate that activation of ERK pathway is a prerequisite event in AICAR-mediated stabilization of the ABCG1 transcript.
AICAR Induces ABCG1 Expression and Attenuates Atherogenesis in ApoE−/− Mice
Finally, we evaluated the impact of chronic AICAR treatment on ABCG1 expression, macrophage foam cell formation, and atherosclerotic lesion area. Six-week-old apoE−/− mice fed a high fat diet were randomized to receive either AICAR or vehicle for 12 weeks. We then isolated mouse peritoneal macrophages and examined the ABCG1 expression. The protein expression of ABCG1 was significantly higher in AICAR-treated macrophages compared with vehicle-treated macrophages (Fig. 6, A and B). Moreover, OxLDL-induced lipid accumulation was markedly attenuated in AICAR-treated macrophages, and this suppressive effect was abolished by pretreatment with compound C or ABCG1 siRNA (Fig. 6, C and D). Furthermore, we analyzed the development of atherosclerotic lesion in the aortic sinus. Treatment with AICAR markedly diminished the size of atherosclerotic plaques (Oil Red O staining) by 42.60 ± 9.53% (Fig. 6, E and F). All the biological parameters were not altered significantly by AICAR treatment in apoE−/− mice (Table 1). Examination of the distribution of cholesterol among the serum lipoprotein fractions revealed similar lipoprotein profiles between the groups (supplemental Fig. 5). These data showed clearly that chronic AMPK activation by AICAR significantly improved the efficiency of macrophage cholesterol transport and ameliorated atherosclerotic lesion formation.
FIGURE 6.
AICAR increases ABCG1 expression and inhibits atherogenesis in apoE−/− mice. ApoE−/− mice, fed a high fat cholesterol diet, were randomly administered with either AICAR (n = 8, 200 mg·kg−1·day−1) or vehicle (n = 8, 0.9% NaCl) for 12 weeks. A, peritoneal macrophages were isolated, lysed, and subjected to Western blot to evaluate the protein levels of ABCG1 and β-actin. B, quantification of ABCG1 protein relative levels. **, p < 0.01. C and D, peritoneal macrophages from vehicle- and AICAR-treated mice were preincubated with compound C or transfected with ABCG1 siRNA for 6 h, followed by the treatment of OxLDL (50 μg/ml) for an additional 24 h. After incubation, cells were fixed and stained with Oil Red O to detect lipid accumulation. Hematoxylin was used as counterstaining. **, p < 0.01. E, determination of atherosclerotic lesion size in apoE−/− mice. The aortic roots of the animals treated as described were analyzed for atherosclerotic lesion size with Oil Red O staining. Magnification was ×400. F, quantitative analysis of the atherosclerotic lesion areas in the aortic sinus (n = 8, **, p < 0.01).
TABLE 1.
Biochemical characteristics of apoE−/− mice treated with or without AICAR
Data are n or means ± S.E.
| Variables | Treatment | Control | AICAR |
|---|---|---|---|
| n | 8 | 8 | |
| Body weight | Before | 23.48 ± 0.32 | 22.84 ± 0.41 |
| After | 30.97 ± 0.25 | 31.15 ± 0.27 | |
| Total cholesterol, mg/dl | Before | 482.5 ± 48.2 | 474.8 ± 35.6 |
| After | 1469.1 ± 84.4 | 1531.8 ± 91.3 | |
| Triglycerides, mg/dl | Before | 181.0 ± 24.0 | 175.0 ± 15.6 |
| After | 208.2 ± 26.7 | 212.4 ± 22.9 | |
| Free fatty acid (mm) | Before | 0.85 ± 0.19 | 0.77 ± 0.03 |
| After | 1.78 ± 0.31 | 2.00 ± 0.26 |
DISCUSSION
Macrophage-derived foam cells are the prominent characteristic in all stages of atherosclerosis (36). These lipid-laden foam cells, present from the earliest discernable fatty streak lesions to advanced complicated lesion and plaque vulnerability, are key regulators of the pathological behavior of plaques (37). AMPK is a heterotrimeric enzyme that is expressed in most mammalian tissues. Regulation of fuel supply and energy-generating pathways in response to the metabolic needs of the organism is the fundamental function of this enzyme complex (38, 39). Although AMPK signaling pathway is traditionally thought of as an intracellular fuel gauge and regulator of energy metabolism, recent evidence indicates that it also has vasculoprotective effects such as maintenance of endothelial function (27, 40), regulation of the disturbed redox balance (41), and suppression of aberrant endoplasmic reticulum stress (42). However, the effect of AMPK in the transformation of macrophage foam cells, a crucial step for the initiation and progression of atherosclerosis, has never been established. Here, we demonstrated a novel effect of AMPK and its underlying molecular mechanism in suppressing macrophage foam cell formation. We first showed that AMPK activation by AICAR strongly ameliorated the OxLDL-induced foam cell formation. Using this experimental cell culture model, we then uncovered that the beneficial function of AMPK exerted on foam cell formation is dependent on ABCG1. AMPK activator AICAR induces ABCG1 expression in macrophages by regulating Abcg1 mRNA stability via its 3′-UTR. At the cellular level, increased ABCG1 expression by AMPK activation promotes cholesterol efflux to HDL from macrophage-derived foam cells, causing a marked reduction of cholesterol ester accumulation in macrophages. In vivo experiments using apoE−/− mice show that AICAR infusion augments ABCG1 expression and reduces the cholesterol content in peritoneal macrophages, thus reducing the area of atherosclerotic plaque. This study therefore sheds a new light into the potential anti-atherogenic properties of AMPK in addition to anti-oxidative/anti-inflammatory functions.
The intracellular lipid homeostasis of macrophage-derived foam cells is dynamically regulated by OxLDL internalization and cholesterol efflux. Macrophage scavenger receptors, particularly the SR-A and CD36, have been implicated in processes that contribute both to early foam cell formation and to the progression toward more complex vulnerable plaques (6, 7, 43). On the other hand, ABCA1, ABCG1, and SR-BI, the three major transporters for cholesterol efflux of foam cells, are pivotal in maintaining the cholesterol homeostasis in macrophages (8–10). Studies using gene-manipulated mice demonstrated that foam cell accumulation and atherosclerotic lesions are significantly promoted in individual transporter-deficient animals (9, 44, 45). Therefore, we examined the regulatory role of AICAR on these receptors. Interestingly, AICAR treatment selectively induced both mRNA and protein expression of ABCG1 but not ABCA1, SR-A, CD36, and SR-BI. AICAR treatment was also observed to increase the functional activity of ABCG1 by increasing DiI-labeled HDL binding with macrophages. Furthermore, we found that AICAR treatment did not affect the gene expression of Acat1, Hsl, Srebp1, Srebp2, Hmgcr, and Ldlr. Collectively, these data suggest the reduction of macrophage cholesterol accumulation by AICAR is not likely due to the cholesterol uptake and inhibition of endogenous lipid synthesis and hydrolysis but is specifically dependent on ABCG1-mediated cholesterol efflux.
Previous reports suggested that Abcg1 is the LXR target gene, and LXR-mediated transcriptional regulation is required for the induction of this transporter. Beyea et al. (46) and Joseph et al. (47) found that the endogenous LXR ligand 24-(S), 25-epoxycholesterol up-regulated Abca1 and Abcg1 and that synthetic LXR ligands inhibited the development of atherosclerosis in mice, respectively. However, in this study, neither the LXRα/RXR protein expression nor the LXRα transcriptional activity in macrophages was altered in the presence of AICAR. Furthermore, inhibition of LXR activation by preincubation of macrophages with geranylgeranyl pyrophosphate or LXR-specific siRNA did not reverse the induction of ABCG1 expression by AICAR, indicating that AICAR-mediated up-regulation of Abcg1 expression is independent on the activation of LXRα. Thus, the post-transcriptional regulation seems to be the major mechanism underlying the effects of AICAR upon ABCG1 expression. When in the presence of actinomycin D, a general inhibitor of gene transcription, we observed that the mRNA half-life of ABCG1 was considerably extended in AICAR-treated macrophages versus vehicle-treated cells, suggesting that AICAR increased the steady-state expression of ABCG1. We thus sought to understand how AMPK affects the stability of ABCG1 transcripts by examining the Abcg1 mRNA 3′-UTR. Based on recent evidence, 3′-UTRs play an important role in mRNA degradation by interacting with microRNA (48). We therefore performed a reporter gene assay using the luciferase constructs containing Abcg1 3′-UTRs to assess a possibility whether AICAR affects these regions. Exposure of macrophages to AICAR markedly restored the attenuated luciferase activity induced by the presence of 3′-UTR, indicating that this region of ABCG1 was actually involved in mRNA stabilization by AICAR. Because ERK activation is required for AICAR-elicited Abcg1 mRNA stabilization, interactions of mRNA-binding proteins with these motifs may be a direct downstream event of the ERK signaling cascade and merit further investigation.
Finally, similar data were observed in the in vivo experiment using atherosclerosis-prone apoE−/− mice, which again demonstrated the protective effects of AICAR on foam cells and atherogenesis. The peritoneal macrophages isolated from AICAR-treated apoE−/− mice showed significant increase in ABCG1 expression and reduced lipid accumulation by OxLDL compared with macrophages from vehicle-treated mice. Concomitantly, AICAR treatment strongly suppressed the atherosclerotic lesion area. It has recently been shown that AMPK activation suppresses OxLDL-induced proliferation of macrophages (49). It is therefore possible that AICAR inhibits atherosclerosis in part by inhibiting proliferation of lesion macrophages. However, we did not observed the evident effects of AICAR on macrophage proliferation in this study. This is largely due to the OxLDL concentration and incubation time. Ishii et al. (49) incubated the macrophages with a lower dose of OxLDL (20 μg/ml) for a long time (6 days), whereas we stimulated macrophages with 50 μg/ml OxLDL for 24 h, which was commonly used to induce cholesterol accumulation in macrophages (18, 50, 51).
In conclusion, this study demonstrates a novel mechanistic insight that AMPK exerts anti-atherogenic effects by increasing Abcg1 expression and enhancing HDL-mediated cholesterol efflux from the macrophages. Our findings suggest that AMPK may potentially be of therapeutic value in inhibiting the process of atherosclerosis-related cardiovascular disease.
Supplementary Material
This work was supported by National Natural Science Foundation Grants 30730079 and 30700665, Guangdong Natural Science Foundation Grant 9151012003000002, Foundation for the Author of National Excellent Doctoral Dissertation of China Grant 200978, Research Fund for the Doctoral Program of Higher Education of China Grant 20070558276, and the Fundamental Research Funds for the Central Universities of Sun Yat-Sen University 10ykzd04.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- OxLDL
- oxidized low density lipoprotein
- AMPK
- adenosine monophosphate activated protein kinase
- ABCG1
- ATP-binding cassette transporters G1
- SR-A
- scavenger receptor-A
- LXR
- liver X receptor
- RXR
- retinoid X receptor
- apoE−/− mice
- apolipoproteinE-deficient mice
- LXRE
- LXR-response element
- AICAR
- 5-aminoimidazole-4-carboxyamide ribonucleoside
- CA-AMPKα
- constitutively active form of AMPKα
- DN-AMPK
- dominant-negative AMPKα mutant
- DiI
- 3,3′-dioctadecylindocarbocyanine.
REFERENCES
- 1.Ross R. (1999) N. Engl. J. Med. 340, 115–126 [DOI] [PubMed] [Google Scholar]
- 2.Glass C. K., Witztum J. L. (2001) Cell 104, 503–516 [DOI] [PubMed] [Google Scholar]
- 3.Lusis A. J. (2000) Atherosclerosis 407, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berliner J. A., Navab M., Fogelman A. M., Frank J. S., Demer L. L., Edwards P. A., Watson A. D., Lusis A. J. (1995) Circulation 91, 2488–2496 [DOI] [PubMed] [Google Scholar]
- 5.Ross R. (1993) Nature 362, 801–809 [DOI] [PubMed] [Google Scholar]
- 6.Kunjathoor V. V., Febbraio M., Podrez E. A., Moore K. J., Andersson L., Koehn S., Rhee J. S., Silverstein R., Hoff H. F., Freeman M. W. (2002) J. Biol. Chem. 277, 49982–49988 [DOI] [PubMed] [Google Scholar]
- 7.Rahaman S. O., Lennon D. J., Febbraio M., Podrez E. A., Hazen S. L., Silverstein R. L. (2006) Cell Metab. 4, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang N., Silver D. L., Thiele C., Tall A. R. (2001) J. Biol. Chem. 276, 23742–23747 [DOI] [PubMed] [Google Scholar]
- 9.Kennedy M. A., Barrera G. C., Nakamura K., Baldán A., Tarr P., Fishbein M. C., Frank J., Francone O. L., Edwards P. A. (2005) Cell Metab. 1, 121–131 [DOI] [PubMed] [Google Scholar]
- 10.Tall A. R., Yvan-Charvet L., Terasaka N., Pagler T., Wang N. (2008) Cell Metab. 7, 365–375 [DOI] [PubMed] [Google Scholar]
- 11.Lage R., Diéguez C., Vidal-Puig A., López M. (2008) Trends Mol. Med. 14, 539–549 [DOI] [PubMed] [Google Scholar]
- 12.Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 13.Hahn-Windgassen A., Nogueira V., Chen C. C., Skeen J. E., Sonenberg N., Hay N. (2005) J. Biol. Chem. 280, 32081–32089 [DOI] [PubMed] [Google Scholar]
- 14.Gleason C. E., Lu D., Witters L. A., Newgard C. B., Birnbaum M. J. (2007) J. Biol. Chem. 282, 10341–10351 [DOI] [PubMed] [Google Scholar]
- 15.Fisslthaler B., Fleming I. (2009) Circ. Res. 105, 114–127 [DOI] [PubMed] [Google Scholar]
- 16.Kahn B. B., Alquier T., Carling D., Hardie D. G. (2005) Cell Metab. 1, 15–25 [DOI] [PubMed] [Google Scholar]
- 17.Lowry O. H., Rosebrough N. J., Farr A. L., Randall A. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 18.Lu K. Y., Ching L. C., Su K. H., Yu Y. B., Kou Y. R., Hsiao S. H., Huang Y. C., Chen C. Y., Cheng L. C., Pan C. C., Lee T. S. (2010) Circulation 121, 1828–1837 [DOI] [PubMed] [Google Scholar]
- 19.Rigamonti E., Helin L., Lestavel S., Mutka A. L., Lepore M., Fontaine C., Bouhlel M. A., Bultel S., Fruchart J. C., Ikonen E., Clavey V., Staels B., Chinetti-Gbaguidi G. (2005) Circ. Res. 97, 682–689 [DOI] [PubMed] [Google Scholar]
- 20.Furuhashi M., Tuncman G., Görgün C. Z., Makowski L., Atsumi G., Vaillancourt E., Kono K., Babaev V. R., Fazio S., Linton M. F., Sulsky R., Robl J. A., Parker R. A., Hotamisligil G. S. (2007) Nature 447, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia M., Hou M., Zhu H., Ma J., Tang Z., Wang Q., Li Y., Chi D., Yu X., Zhao T., Han P., Xia X., Ling W. (2005) J. Biol. Chem. 280, 36792–36801 [DOI] [PubMed] [Google Scholar]
- 22.Denizot F., Lang R. (1986) J. Immunol. Methods 89, 271–277 [DOI] [PubMed] [Google Scholar]
- 23.Pawar A., Botolin D., Mangelsdorf D. J., Jump D. B. (2003) J. Biol. Chem. 278, 40736–40743 [DOI] [PubMed] [Google Scholar]
- 24.Stefulj J., Panzenboeck U., Becker T., Hirschmugl B., Schweinzer C., Lang I., Marsche G., Sadjak A., Lang U., Desoye G., Wadsack C. (2009) Circ. Res. 104, 600–608 [DOI] [PubMed] [Google Scholar]
- 25.Dixon D. A., Tolley N. D., Bemis-Standoli K., Martinez M. L., Weyrich A. S., Morrow J. D., Prescott S. M., Zimmerman G. A. (2006) J. Clin. Invest. 116, 2727–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabol S. L., Brewer H. B., Jr., Santamarina-Fojo S. (2005) J. Lipid Res. 46, 2151–2167 [DOI] [PubMed] [Google Scholar]
- 27.Park K. G., Min A. K., Koh E. H., Kim H. S., Kim M. O., Park H. S., Kim Y. D., Yoon T. S., Jang B. K., Hwang J. S., Kim J. B., Choi H. S., Park J. Y., Lee I. K., Lee K. U. (2008) Hepatology 48, 1477–1486 [DOI] [PubMed] [Google Scholar]
- 28.Li D., Zhang Y., Ma J., Ling W., Xia M. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 1354–1362 [DOI] [PubMed] [Google Scholar]
- 29.Gu X., Trigatti B., Xu S., Acton S., Babitt J., Krieger M. (1998) J. Biol. Chem. 273, 26338–26348 [DOI] [PubMed] [Google Scholar]
- 30.Ottnad E., Parthasarathy S., Sambrano G. R., Ramprasad M. P., Quehenberger O., Kondratenko N., Green S., Steinberg D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1391–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babaev V. R., Patel M. B., Semenkovich C. F., Fazio S., Linton M. F. (2000) J. Biol. Chem. 275, 26293–26299 [DOI] [PubMed] [Google Scholar]
- 32.Mauldin J. P., Srinivasan S., Mulya A., Gebre A., Parks J. S., Daugherty A., Hedrick C. C. (2006) J. Biol. Chem. 281, 21216–21224 [DOI] [PubMed] [Google Scholar]
- 33.Argmann C. A., Edwards J. Y., Sawyez C. G., O'Neil C. H., Hegele R. A., Pickering J. G., Huff M. W. (2005) J. Biol. Chem. 280, 22212–22221 [DOI] [PubMed] [Google Scholar]
- 34.Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., Musi N., Hirshman M. F., Goodyear L. J., Moller D. E. (2001) J. Clin. Invest. 108, 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cool B., Zinker B., Chiou W., Kifle L., Cao N., Perham M., Dickinson R., Adler A., Gagne G., Iyengar R., Zhao G., Marsh K., Kym P., Jung P., Camp H. S., Frevert E. (2006) Cell Metab. 3, 403–416 [DOI] [PubMed] [Google Scholar]
- 36.Shyu A. B., Wilkinson M. F., van Hoof A. (2008) EMBO J. 27, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li A. C., Glass C. K. (2002) Nat. Med. 8, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 38.Osterud B., Bjorklid E. (2003) Physiol. Rev. 83, 1069–1112 [DOI] [PubMed] [Google Scholar]
- 39.Lim C. T., Kola B., Korbonits M. (2010) J. Mol. Endocrinol. 44, 87–97 [DOI] [PubMed] [Google Scholar]
- 40.Zhang B. B., Zhou G., Li C. (2009) Cell Metab. 9, 407–416 [DOI] [PubMed] [Google Scholar]
- 41.Schulz E., Anter E., Zou M. H., Keaney J. F., Jr. (2005) Circulation 111, 3473–3480 [DOI] [PubMed] [Google Scholar]
- 42.Schulz E., Dopheide J., Schuhmacher S., Thomas S. R., Chen K., Daiber A., Wenzel P., Münzel T., Keaney J. F., Jr. (2008) Circulation 118, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Y., Zhang M., Liang B., Xie Z., Zhao Z., Asfa S., Choi H. C., Zou M. H. (2010) Circulation 121, 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore K. J., Freeman M. W. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1702–1711 [DOI] [PubMed] [Google Scholar]
- 45.McNeish J., Aiello R. J., Guyot D., Turi T., Gabel C., Aldinger C., Hoppe K. L., Roach M. L., Royer L. J., de Wet J., Broccardo C., Chimini G., Francone O. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trigatti B., Rayburn H., Viñals M., Braun A., Miettinen H., Penman M., Hertz M., Schrenzel M., Amigo L., Rigotti A., Krieger M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beyea M. M., Heslop C. L., Sawyez C. G., Edwards J. Y., Markle J. G., Hegele R. A., Huff M. W. (2007) J. Biol. Chem. 282, 5207–5216 [DOI] [PubMed] [Google Scholar]
- 48.Joseph S. B., McKilligin E., Pei L., Watson M. A., Collins A. R., Laffitte B. A., Chen M., Noh G., Goodman J., Hagger G. N., Tran J., Tippin T. K., Wang X., Lusis A. J., Hsueh W. A., Law R. E., Collins J. L., Willson T. M., Tontonoz P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7604–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishii N., Matsumura T., Kinoshita H., Motoshima H., Kojima K., Tsutsumi A., Kawasaki S., Yano M., Senokuchi T., Asano T., Nishikawa T., Araki E. (2009) J. Biol. Chem. 284, 34561–34569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., Brewer H. B., Fruchart J. C., Clavey V., Staels B. (2001) Nat. Med. 7, 53–58 [DOI] [PubMed] [Google Scholar]
- 51.Conway J. P., Kinter M. (2006) J. Biol. Chem. 281, 27991–28001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.