Abstract
Methionine sulfoxide reductases protect cells by repairing oxidatively damaged methionine residues in proteins. Here, we report the first three-dimensional structure of the mammalian selenoprotein methionine sulfoxide reductase B1 (MsrB1), determined by high resolution NMR spectroscopy. Heteronuclear multidimensional spectra yielded NMR spectral assignments for the reduced form of MsrB1 in which catalytic selenocysteine (Sec) was replaced with cysteine (Cys). MsrB1 consists of a central structured core of two β-sheets and a highly flexible, disordered N-terminal region. Analysis of pH dependence of NMR signals of catalytically relevant residues, comparison with the data for bacterial MsrBs, and NMR-based structural analysis of methionine sulfoxide (substrate) and methionine sulfone (inhibitor) binding to MsrB1 at the atomic level reveal a mechanism involving catalytic Sec95 and resolving Cys4 residues in catalysis. The MsrB1 structure differs from the structures of Cys-containing MsrBs in the use of distal selenenylsulfide, residues needed for catalysis, and the mode in which the active form of the enzyme is regenerated. In addition, this is the first structure of a eukaryotic zinc-containing MsrB, which highlights the structural role of this metal ion bound to four conserved Cys. We integrated this information into a structural model of evolution of MsrB superfamily.
Keywords: Enzyme Mechanisms, Evolution, Protein Folding, Proteomics, Reductase, BioNMR
Introduction
Methionine sulfoxide (MetSO)3 is readily formed by oxidation of methionine in cells, especially under conditions of oxidative stress, but can be enzymatically reduced back to methionine by MetSO reductases (Msrs). Msrs are thiol-based oxidoreductases in which cysteine (Cys) functions as a catalytic residue. These enzymes are classified into two families, MsrA and MsrB, according to their substrate specificity. MsrA catalyzes the reduction of the s-form of MetSO (Met-s-SO), whereas MsrB can only reduce the r-form (Met-r-SO). These two enzyme families reduce MetSO residues in proteins, but MsrA can also reduce free Met-s-SO. Msrs are crucial repair proteins that protect cells against oxidative stress and have been implicated delaying the aging process and protecting against neurodegeneration (1, 2). In addition, a new class of Msr, called fRMsr, has recently been identified (3) and characterized from Escherichia coli and Saccharomyces cerevisiae (3, 4). This enzyme is specific for free Met-r-SO but is not active with protein-based Met-r-SO. fRMsr also functions as an antioxidant protein (4).
Selenocysteine (Sec) is a rare amino acid that is co-translationally incorporated into proteins at UGA codons. Selenoproteins (i.e. Sec-containing proteins) are found in all three domains of life. Among the functionally characterized selenoproteins, most are oxidoreductases in which Sec replaces Cys normally found in the active site (e.g. glutathione peroxidase and thioredoxin reductase). The most profound characteristic provided by Sec in selenoenzymes is higher catalytic efficiency of these enzymes compared with their Cys mutants or Cys homologs (5–7). In mammals, there are three MsrBs, of which MsrB1 is a selenoprotein and MsrB2 and MsrB3 are Cys-containing isozymes (7).
In both MsrA and MsrB, the catalytic mechanism is characterized by sulfenic acid chemistry and consists of three steps (8–10). First, a catalytic Cys (or Sec) attacks the sulfoxide moiety of MetSO and is oxidized to a sulfenic acid (selenenic acid) intermediate, accompanied by the release of product, Met. Second, another Cys (called resolving Cys) interacts with the sulfenic acid (or selenenic acid) intermediate to form an intramolecular disulfide (selenenylsulfide) bond. Third, the disulfide (selenenylsulfide) is reduced by thioredoxin in vivo or by other reductants (e.g. dithiothreitol) in vitro to regenerate the active enzyme. In MsrBs lacking resolving Cys, the sulfenic acid intermediate can be directly reduced by thioredoxin (11, 12), but the selenenic acid intermediate, at least in two tested proteins, could not (12, 13). Cys4 in selenoprotein MsrB1 is required for the thioredoxin-dependent reaction and was found to serve as a resolving Cys. The resolving Cys are absent in MsrB2 and MsrB3 (13).
NMR spectroscopy and x-ray crystallography are the most powerful experimental techniques capable of providing information on three-dimensional structures of biomolecules at atomic resolution. In addition, NMR is a useful technique for monitoring interactions between molecules in solution due to sensitivity of the nuclear chemical shifts to the change of chemical environment of the nucleus, thus allowing one to obtain additional functional information. Both structural and functional features of NMR spectroscopy have been exploited in this work which characterized selenoprotein MsrB1.
Crystal structures of MsrBs from Neisseria gonorrhoeae, Streptococcus pneumoniae, Bacillus subtilis, and Xanthomonas campestris have previously been reported (8, 14, 36), and catalytic mechanisms for these Cys-containing proteins have been proposed. Two additional unpublished MsrB structures from B. subtilis and Burkholderia pseudomallei have been deposited in the Protein Data Bank (PDB) (1XM0/3E0O and 3CEZ). However, structures of selenoprotein MsrBs, which deviate sufficiently from other MsrBs (13), are not known. In this work, we report NMR solution structures of mouse selenoprotein MsrB1, in which Sec was replaced with Cys. Our NMR structural analysis provides insights into the catalytic mechanism of this selenoprotein and reveals novel structural features of this protein family. We integrate this information into a structural model of evolution of MsrB superfamily.
EXPERIMENTAL PROCEDURES
Sample Preparation
Cloning, expression, and purification of uniformly 15N/13C-labeled Sec95→Cys mutant of mouse MsrB1 (116 amino acids) and conditions for NMR measurements were described previously (13, 15). Samples of the reduced protein form had 1–1.5 mm MsrB1 in 20 mm sodium phosphate buffer, pH 5.5, containing 10 mm NaCl and 5 mm dithiothreitol in 90% H2O/10% D2O or 99.9% D2O.
NMR Spectroscopy
NMR spectra were recorded at 298 K on a Bruker Avance 600-MHz spectrometer (at the NT faculty NMR center at Norwegian University of Science and Technology) equipped with a 5-mm z-gradient CP-TCI(H/C/N) cryoprobe or Avance 800-MHz spectrometer (at the NMR center of the University of Florence) equipped with a 5-mm z-gradient CP-TCI(H/C/N) cryoprobe.
Three-dimensional 13C- and 15N-edited 1H-1H NOESY spectra were recorded in D2O and H2O, respectively. NMR data were processed using BRUKER XWinNMR, version 3.5. NMR spectral analysis was performed using CARA version 1.4.1/1.5.3 (16). 15N-{1H} heteronuclear NOEs were calculated as the ratio of corresponding cross-peak volumes, from two independently measured and integrated 15N-1H heteronuclear correlated spectra recorded using enhanced sensitivity sequence employing pulsed field gradient (17) with and without 1H saturation. Nuclear magnetic relaxation (T1 and T2) measurements of 15N nuclei were obtained by exponential fitting of peak intensities in 15N HSQC spectra acquired with different relaxation delays (17, 18).
Structure Calculation
NOE cross-peaks were identified, assigned, and integrated in the aforementioned NOESY spectra using the program NEASY in CARA suite (16). CALIBA (19) subroutine in CYANA 2.1 was used to convert cross-peak intensities from NOESY spectra into distance constraints. Dihedral angle constraints were derived from secondary chemical shifts using the TALOS program (20). Zinc ion was incorporated into the calculations using the binding geometry from other tetrahedral Cys-zinc-coordinating proteins available at the PDB with the following codes: 1QF8 (human casein kinase) (21), 8ADH (horse alcohol dehydrogenase) (22), and 1QQT (E. coli methionyl-tRNA synthetase) (23). The position of the Zn2+ ion within the protein was restricted by upper distance limit constrains set to 2.4 Å for Sγ-Zn2+ and 3.6 Å for Cβ-Zn2+and the lower distance limits set to 2.2 Å for Sγ-Zn2+ and 3.0 Å for Cβ-Zn2+ based on average distances found in the x-ray structures. The Zn2+ van der Waals radius was set to 1.39 Å (Cambridge Structural database). Based on the input described, the structure was calculated using the torsion angle dynamics program CYANA 2.1 (19). Structure calculations started from 96 conformers with random torsion angle values. 20 conformers with the lowest final CYANA target function values were energy-minimized in explicit water using the AMBER force field with the aid of AMBER 9.0 program (24). The distance and torsion angle constraints were applied with force constants of 50 kcal mol−1Å−2 and 32 kcal mol−1 rad−2, respectively.
Interaction of MsrB1 with MetSO and Methionine Sulfone (MetSO2)
The samples for NMR-based analysis of interaction of MsrB1 with MetSO or MetSO2 were prepared as described above for the reduced protein. Single buffer exchange of protein preparation, with ∼10-fold excess of reactants (10 mm Met-(r,s)-SO or l-MetSO2 in 20 mm phosphate buffer, pH 5.5, containing 10 mm NaCl in 90% H2O/10% D2O), was used to assure that any residual DTT is removed. Subsequently, three buffer exchanges of samples with a solution containing 2 mm MetSO or MetSO2, 20 mm phosphate buffer, pH 5.5, in the presence of 10 mm NaCl in 90% H2O/10% D2O were performed. Changes in the reduced MsrB1 upon addition of the compounds were detected through three-dimensional HNCA, CBCANH, 13C-edited 1H-1H NOESY, and 15N HSQC spectra.
pH Dependence
pH titration of MsrB1 was performed twice on uniformly labeled 13C/15N ∼1.0 mm MsrB1 in 20 mm phosphate buffer containing 10 mm NaCl and 5 mm DTT in 90% H2O/10% D2O in the pH range 4.5–9.7. The samples were titrated by transferring, with a glass rod, small amounts of 0.1 m NaOH or HCl, ensuring minimal changes in pH values and ionic strength. The pH of the sample was measured with a mini pH probe. For each titration step, one 15N HSQC and two 13C HSQCs were recorded, one for the aromatic (13Caro HSQC) region and the other for the aliphatic (13Cali HSQC) region of the NMR spectrum.
Activity Measurements
The reaction mixture for activity measurements (100 μl) contained 100 mm sodium phosphate at different pH values (5.7, 6.5, 7.0, or 7.5), 200 μm dabsyl-Met-r-SO, 20 mm DTT, and 20 μg of MsrB1(Sec95→Cys). The reaction was carried out at 37 °C for 30 min and the product, dabsyl-Met, was analyzed by the established HPLC procedure (9).
RESULTS AND DISCUSSION
Structure Description
MsrB1 solution structure was calculated based on the NOE-derived geometrical constraints, dihedral angles obtained from TALOS (20), and distance constraints for the tetrahedral Zn2+ ion derived from x-ray structures of proteins containing zinc coordinated by Cys residues (21–23). The geometrical constraints used in the calculations are summarized in Table 1. In total, 1155 NOE-based upper limit distances, 88 torsion angle restraints, and 8 upper limit and 8 lower limit distances for Zn2+ ion coordination were used to derive the MsrB1 structure. The resulting MsrB1 family was further energy-minimized. The geometrical constraint and coordinate files of the minimized MsrB1 family were deposited in the PDB under code 2KV1.
TABLE 1.
Structural statistics and geometrical constraints derived from NMR for the reduced form of mouse MsrB1
| Parameter | Number |
|---|---|
| Restraints used in structure calculation | |
| Total number of NOE distance restraints | 1155 |
| Intraresidual | 528 |
| Short range | 446 |
| Medium range | 36 |
| Long range | 145 |
| Number of lower limits for zinc ion | 8 |
| Number of upper limits for zinc ion | 8 |
| Torsion angle constraints | 88 |
| Structure statistics, 20 conformers | |
| CYANA target function value (Å2) | 5.24 ± 1.24 |
| Maximal distance constraint violation | 0.22 ± 0.06 |
| Maximal torsion angle constraint violation | 3.15 ± 0.06 |
| AMBER energies in water (kcal/mol) | −2.19 E+5 |
| PROCKECK-NMR Ramachandran statistics | |
| Residues in favorable regions (%) | 73.4 |
| Residues in additional allowed regions (%) | 24.0 |
| Residues in generously allowed regions (%) | 2.2 |
| Residues in disallowed regions (%) | 0.4 |
| Root mean square deviation to average coordinates (Å) | |
| N, Cα, C′ (18–108) | 1.59 ± 0.47 |
| N, Cα, C′ (secondary structure) | 0.84 ± 0.29 |
| Heavy atoms (18–108) | 2.64 ± 0.49 |
| Heavy atoms (secondary structure) | 1.70 ± 0.31 |
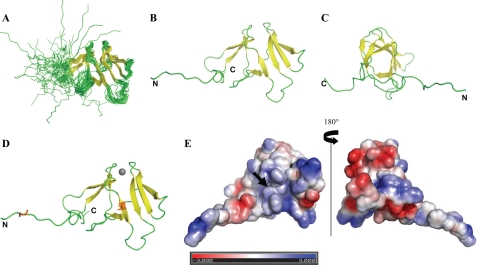
The family of 20 energy-refined MsrB1 conformers is shown in Fig. 1A, and the average family structures with the observed elements of the protein secondary structure are shown in Fig. 1, B and C (without Zn2+ coordination), and Fig. 1D (with Zn2+). Structural statistics data are shown in Table 1. The resulting MsrB1 structure represents the conformation of low energy in accordance with the geometrical constraints experimentally determined.
FIGURE 1.
Three-dimensional NMR structure of reduced mouse MsrB1. A, MsrB1 structural family consisting of 20 conformers with the lowest target function. The secondary structure elements, including strands β1 (residues 19–23), β8 (100–104), and β2 (28–30) in the listed order, form the first β-sheet. Another β-sheet is connected in the following order: β3 (45–48), β7 (93–96), β6 (77–82), β5 (66–72), and β4 (55–60). These β-sheets form an ellipsoidal β-sandwich. B and C, front and side orientations of the structure with the lowest target function. D, frontal view of MsrB1 structure with zinc ion indicated as gray sphere and the catalytic and resolving cysteines highlighted as orange sticks. E, electrostatic surface potential of MsrB1 front and back side views, where the position of the catalytic cysteine is indicated by a black arrow. The program PDB 2PQR, version 1.5 (33), was used to prepare the structures for electrostatic potential distribution calculations, performed with the programs APBS (34). Visualization of the potential plotted on the solvent accessible surface was made with the program PyMOL, version 0.99 (35). Red and blue denote regions of negative (i.e. less than −3 kT/|e|) and positive (i.e. more than +3 kT/|e|) potential on the MsrB1 surface, respectively. Structural models were visualized with PyMOL version 0.99.
The MsrB1 structure is characterized by an overall β-fold protein consisting of eight all antiparallel β-strands. Both N-terminal (amino acids 1–18) and C-terminal (amino acids 105–116) regions are flexible (Fig. 1, B and C). The overall MsrB1 structure is of ellipsoidal shape consisting of two antiparallel β-sheets. The first β-sheet is three-stranded, forming the back side of the structure, whereas the second sheet has five strands forming the front side. The active site is situated in the second β-sheet. The N-terminal region contains two hinge sections defined by Gly8-Gly9 and Pro18-Gly19 pairs followed by a back side β-sheet consisting of strand β1 (residues 19–23), β8 (residues 100–104), and β2 (residues 28–30) in the listed order. The C-terminal flexible region comes out of the middle of the back side β-sheet. The front side β-sheet is connected in the following order: β3 (45–48), β7 (93–96), β6 (77–82), β5 (66–72), and β4 (55–60). The front side β-sheet forms the protein hydrophobic core through residues Leu67, Val69, Phe94, and Ile96 linking to the back side β-sheet hydrophobic amino acids Tyr21, Phe31, and Phe103 at the bottom of the structure. The top part of MsrB1 is held together through tetrahedral structural zinc, in which the metal ion is bound to the protein matrix by Cys23, Cys26, Cys71, and Cys74. A more disordered region, consisting of 13 residues (amino acids 31–44) between β2- and β3-strands, connects front side and back side β-sheets of the protein.
Zinc-binding Site
MsrB1 was recognized as a zinc-containing protein in an earlier study (25). It was also suggested that the zinc has a structural role in this protein (9, 26, 31). The presence of the zinc ion within MsrB1 was clearly confirmed by our NMR assignment of MsrB1. Indeed, an average chemical shift value of 32.29 ± 0.94 ppm for 13Cβ of four of seven Cys was observed. This observation is in good agreement with the previous data (27), showing that the 13Cβ chemical shift of a reduced Cys that does not bind zinc is on average 28.92 ± 2.11 ppm, whereas 13Cβ chemical shifts for the zinc-coordinating Cys are 30.89 ± 1.01 ppm. This analysis demonstrates that Cys23, Cys26, Cys71, and Cys74 coordinate the zinc ion in MsrB1, validating the presence of the metal and its structural role in the enzyme (Fig. 1D).
Mobility Studies
To obtain information about the intramolecular dynamics of MsrB1, both 15N-{H} NOE and 15N backbone relaxation data were recorded and analyzed. As seen in Fig. 1A, analysis of MsrB1 structure revealed that N-terminal (residues 1–18) and C-terminal (residues 105–116) regions have highly flexible or ill-defined structures. This observation is consistent with the 15N-{H} NOE and 15N backbone relaxation data, even if the latter are not very well defined for some protein regions (supplemental Fig. S1). On the basis of the mobility data, we conclude that neither the structured protein regions nor loops within the protein core (amino acids 18–105) show increased mobility. Together, these data indicate that N- and C-terminal regions are largely unstructured because they are intrinsically flexible. As described below, the increased mobility of these regions is needed in catalysis to form an intermediate selenenylsulfide. It is also possible that flexibility of these regions may promote interactions of MsrB1 with its protein substrates.
Catalytic Site
Sec-to-Cys mutants of selenoproteins are commonly used for structural studies because of the difficulties in obtaining sufficient amounts of selenoproteins. Although the Sec95→Cys form has a significantly decreased activity, the protein is catalytically competent (13, 25), suggesting that the overall environment of the active site is preserved. Thus, the Cys mutant of MsrB1, which can be conventionally expressed in bacteria in amounts sufficient for NMR studies, can be used as a model to investigate functional and structural properties of the native selenoprotein form.
The catalytic Cys/Sec is located in the middle of the β7-strand on the lower end of the front side β-sheet, along with Arg93, Asp83, and His80, which were also suggested to participate in catalysis (13). In close vicinity of Cys95 and surrounding this residue, the side chains of three aromatic residues, Trp43, Phe82, and Phe97, are located. All of these residues constitute a binding cleft with hydrophobic side walls. The aromatic amino acids may stabilize the bound substrate. In particular, Trp43, a conserved residue in mammalian MsrBs, was proposed to stabilize and correctly orient the substrate sulfoxide group through interaction with its ϵ-methyl group (8, 28).
The MsrB reaction mechanism (8) involves formation of oxidized MsrB1 through an intramolecular disulfide bridge between the catalytic Cys95 and another Cys residue, acting as a resolving residue. Of the seven Cys residues in MsrB1, one is the catalytic (Cys95), and four are involved in zinc coordination (Cys23, Cys26, Cys71, and Cys74). Thus, only two remaining Cys, Cys4 and Cys58, may be candidates for the role of the resolving Cys. Cys58 is unlikely to serve this function because this residue is not conserved among mammalian MsrB1s. Moreover, our MsrB1 structure revealed that Cys58 is located at the opposite end of front side β-sheet in β4-strand in relation to Cys95. The remaining Cys, Cys4, is situated in the mobile N-terminal region, which is flexible enough to flip in and form the proposed selenenylsulfide bond with the catalytically active Sec95 in the native MsrB1 (or the disulfide bond with Cys95 in the Cys mutant form).
pH Dependence of Catalytic Residues
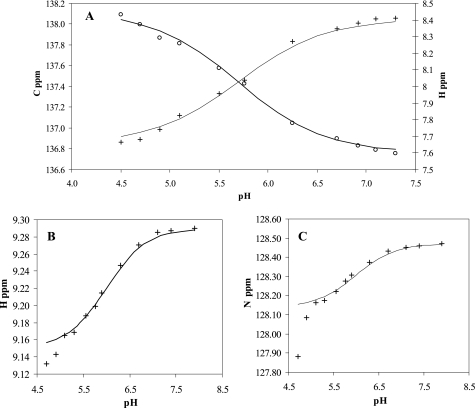
We employed NMR spectroscopy to monitor pH dependence of signals corresponding to the residues constituting the catalytic active site of MsrB1. Both 15N and 13C HSQC spectra were recorded at different pH values in the range from 5.0 to 8.0, and the data were analyzed by plotting the chemical shift values as a function of pH (Fig. 2). It was previously shown (30) that pKa values of catalytic Cys117 and His103 (corresponding to Cys95 and His80, respectively, in MsrB1) in the resting state of MsrB from Neisseria meningitidis are 9.3 and 6.6, respectively, being only slightly altered from the corresponding values for these free amino acids (Cys pKa ∼9.0–9.5; His pKa ∼7.0) (29). We carried out the pH titration for MsrB1 to examine how the use of Sec is accommodated with regard to these residues. To determine the pKa values of His80 and Cys95, the changes in chemical shift values for Cϵ1-Hϵ1 nuclei of His80 and the NH-H nuclei of Cys95 were followed during the titration.
FIGURE 2.
Transition curves for titration of His80 and Cys95 in mouse MsrB1. A, chemical shifts of Cϵ1 and Hϵ1 of His80 are plotted as a function of pH. B, proton chemical shift of HN of Cys95 is plotted as a function of pH. C, backbone nitrogen chemical shift of Cys95 is plotted as a function of pH.
The titration profile for His80 shows a sigmoidal transition (Fig. 2A), and its pKa was determined to be 5.7 ± 0.2 by fitting the experimental data to the Henderson-Hasselbach equation. This pKa value is close to an unperturbed histidine pKa value and is almost 1 unit lower than the corresponding value of the catalytically relevant His103 of N. meningitidis MsrB.
The titration profile for Cys95 (Fig. 2, B and C) appears to have two sigmoidal transitions. Although the first transition with the pKa of 6.0 ± 0.2 should correspond to acid-base equilibrium involving Cys95, the lower pKa around 4.3 corresponds most probably to protonation-deprotonation of another acidic group in the vicinity to Cys95. A plausible candidate acidic residue is Glu81; the change chemical shift obtained for Glu81 from titration data supports this conclusion. However, precise assignment of the observed acidic pKa value to a specific residue should await further investigation.
Thus, the transition in the pH interval 5.35–7.80 presumably reflects the protonation-deprotonation of Cys95. The pKa value for this transition was 6.0 ± 0.2 by fitting the data to the Henderson-Hasselbach equation. The comparison with the corresponding pKa value for the active site Cys in N. meningitidis MsrB reveals the difference of more than 3 units, thus indicating that MsrB1 at the neutral pH is ionized and therefore a significantly stronger nucleophile. Because the wild-type MsrB1 contains Sec, it is plausible that the protein active site has a structure/geometry that tunes the pKa of the attacking residue close to that of Sec even when the catalytic Sec is replaced with Cys. The observed Cys95 pKa value shift may, for example, be caused by the interaction of Sec/Cys95 and Arg93 side chains.
We also examined pH dependence of MsrB1 (Sec95→Cys) activity in the range from pH 5.7 to 7.5 and found an increase in activity upon with the increase in pH (supplemental Table S1). At pH 5.7, the MsrB activity was 24% of the activity at pH 7.5. This pH dependence correlates with the lower pKa value of Cys95 obtained from our NMR titration data.
The observed pH dependence indicates that catalytically relevant residues have different properties compared with the bacterial MsrBs (30). Altogether, comparative analysis of pH dependences observed for a bacterial MsrB by Neiers et al. (30) and for the Sec95→Cys variant of mammalian MsrB1 in this work indicates that, as judged from the acid-base properties of the catalytic residue, Sec-containing MsrBs have different catalytic features compared with Cys-containing MsrBs. This finding is also consistent with earlier biochemical data on the differences in catalytic features of MetSO reduction between Cys- and Sec-containing MsrBs (13).
Interaction of MsrB1 with Substrate and Inhibitor
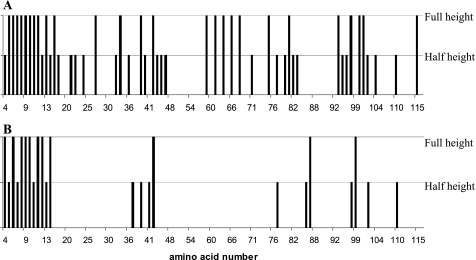
To obtain insights into the mechanism of the MsrB1-catalyzed reaction, we examined NMR spectra of this enzyme after incubation with its substrate (MetSO) or substrate analog/inhibitor (MetSO2). Because the nuclear chemical shift is an extremely sensitive indicator of the chemical environment of nucleus, this property can be used efficiently to detect structural alterations occurring upon interaction with its ligand. To follow the spectral changes occurring in MsrB1 upon its oxidation by MetSO or binding MetSO2 (9, 26), 15N HSQC of the perturbed protein was compared with the corresponding spectrum of the original MsrB1. The observed spectral differences were then confirmed through three-dimensional HNCA spectra recorded for each protein derivative. Fig. 3 shows variations (unaltered position and intensity, unaltered position and decreased intensity, or altered position and intensity) in the 15N HSQC cross-peak appearance for both oxidized and substrate analog-bound MsrB1 compared with the original protein. The comparison of panels in Fig. 3, A and B, immediately shows that oxidation of the protein causes structural perturbations in MsrB1. Indeed, the percentage of amino acids showing changes in the corresponding cross-peaks appearance is significantly higher for MetSO than for MetSO2. The calculated MsrB1 structure shows that the amino acids with spectral changes upon interaction with MetSO2 are those located in close vicinity to the catalytic Cys95, consistent with the fact that MetSO2 does not oxidize the enzyme.
FIGURE 3.
Observed changes in 15N HSQC spectra for mouse MsrB1 upon addition of the substrate, MetSO (A), or a substrate analog/inhibitor, MetSO2 (B). Half-height bars indicate reduction of peak intensity for the N-HN cross-peak; full-height bars show the appearance of a new N-HN cross-peak in the spectrum. Residue identity (with respect to mouse MsrB1 amino acid sequence) is provided below each corresponding peak in the spectrum.
It was previously suggested (13) that oxidation of MsrB1 results in the formation of an intramolecular selenenylsulfide bond between Cys4 and Sec/Cys95. However, the structure of reduced MsrB1 shows these two residues situated far away from each other (>15 Å). If Cys4 is the actual resolving Cys, formation of the intramolecular selenenylsulfide would require significant structural changes. Moreover, this structural reorganization should occur upon protein reduction-oxidation because in the reduced form, the two residues never come close enough. Indeed, our data (Fig. 3, A and B) qualitatively confirm the proposed mechanism of MsrB1 oxidation through the formation of intramolecular selenenylsulfide bridge. To monitor structural changes upon formation of selenenylsulfide bond between Cys4 and Sec/Cys95, we recorded 13C-edited NOESY and two-dimensional NOESY in 99.9% D2O. In the two-dimensional NOESY of the oxidized protein, two new cross-peaks compared with that of the reduced protein were observed. These peaks reveal dipolar interactions between His80 Hδ2 proton and supposedly the protons of the aromatic ring of phenylalanine. The only unassigned phenylalanine in the reduced MsrB1 is Phe3. The appearance of the previously undetected Phe3 aromatic signals (i.e. the new cross-peaks in the NOESY spectrum of the oxidized MsrB1), together with the data in Fig. 3A, indicate structural changes occurring in the N-terminal region of the protein upon its reduction-oxidation. The data suggest that the N-terminal tail is positioned over the protein active site. To understand the structure of the oxidized protein better, a structure calculation was set up based on the structure of the reduced form of MsrB1 with two new tentatively assigned NOE constrains (His80 Hδ2 to Phe3 Hδ/ϵ). The resulting structure (supplemental Fig. S2) shows that the N-terminal tail is positioned over the active site and suggests that a selenenylsulfide bond between Cys4 and Sec/Cys95 can indeed be formed. This tentative structure is also supported by the chemical shift changes observed in Fig. 3A.
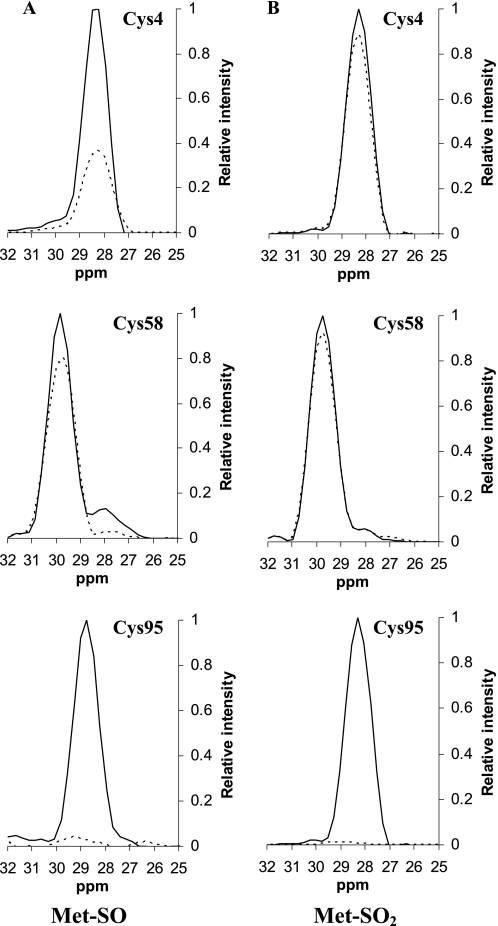
To examine this observation in a more quantitative manner, one-dimensional slice profiles for all Cys residues, as the most catalytically relevant residues, were monitored before and after incubation of MsrB1 substrate or substrate analog in 13C-edited NOESY spectrum (Fig. 4, A and B). Such analysis (instead of the analysis of whole spectra) significantly simplifies the procedure and makes it more reliable. Cys residues that bind Zn (Cys23, Cys26, Cys71, and Cys74) did not show spectral changes upon addition of either MetSO or MetSO2 (data not shown). Thus, Zn remains coordinated by the same amino acids in both oxidized and inhibited forms of the protein. A surface residue Cys58 also remains unaffected as shown in Fig. 4, and therefore, it is not directly involved in the catalytic reaction.
FIGURE 4.
Observed changes in cysteine Cβ chemical shifts for surface-exposed residues in mouse MsrB1 upon addition of the substrate, MetSO (A) or the substrate analog/inhibitor, MetSO2 (B). Solid line shows the peak intensity of cysteine Cβ before addition of substrate or inhibitor. Dotted line shows the peak intensity after addition of substrate or inhibitor.
However, the intensity of the cross-peak of Cys4, the resolving residue, is significantly decreased in the case of the oxidized protein (dashed line) compared with that of the reduced form (solid line) (Fig. 4A), whereas interaction with MetSO2 results only in minor changes in the intensity and the position of the corresponding peak (Fig. 4B). The cross-peak corresponding to catalytic Cys95 completely changed its position in the 13C-edited NOESY spectra of oxidized and MetSO2-bound forms of the protein. Thus, as expected, Cys95 is directly involved in the catalytic reaction. Therefore, the treatment of MsrB1 with substrate and substrate analog provided critical insights into structural and functional peculiarities of MsrB1 at the atomic level.
Additionally, we provide structural evidence that the MsrB1 structure is remarkably sensitive to the binding of the natural substrate, MetSO, to the active site. In particular, the unstructured N-terminal part and its high mobility play a crucial role in allowing the resolving Cys to approach the catalytic residue and also in determining the substrate specificity of the protein. The latter is a novel feature of selenoprotein MsrBs described in our study. A previous study (36) of X. campestris MsrB showed considerable structural flexibility in a bacterial MsrB, including (i) an N-terminal region containing a resolving Cys31 and (ii) a β-stranded region where a considerable rearrangement of secondary structure was registered. The resolving Cys of the bacterial MsrB (37) does not correspond, on sequence or structural levels, to that of the selenocysteine containing MsrBs, in which Cys4 appears to have evolved in metazoa. Cys4 is located in a highly unstructured N-terminal part of the protein, which allows significant movement of the resolving Cys without loss or rearrangement of preexisting secondary structures. Given the importance of the N-terminal segment, we discuss unique structural, functional and evolutionary features of this region in MsrB1 below.
Evolution of the N-terminal Region of MsrB1 and Structural Comparison of Mouse MsrB1 and Bacterial MsrB Proteins
Several structures of bacterial MsrBs have been determined previously (supplemental Fig. S5). Relative to MsrB1, the crystal structures of MsrBs from N. gonorhoeae (PDB code 1L1D) (8), X. campestris (PDB code 3HCI) (36), B. pseudomallei (PDB code 3CEZ/3CXK), S. pneumoniae (PDB code 3E0M) (31), and B. subtilis (3E0O) (31), as well as the solution structure of B. subtilis MsrB (PDB code 1XM0), have a highly similar β-fold consisting of two β-sheets, one with three strands and the other with five, which are the same as β-sheets found in mouse MsrB1. However, the bacterial MsrBs also have several α-helices on the exterior part of the protein. Another common feature of these α-helices is that they are not rigidly fixed to the β-strand core structure and seem to be flexible/adaptable based on the NMR structure of B. subtilis MsrB. Because MsrBs encounter many different protein substrates, it is plausible that the α-helices have adapted for different protein surfaces. However, this structural feature is absent in MsrB1. Based on sequence alignment of selenoprotein MsrBs (12) and our MsrB1 structure analyses, the selenoprotein MsrB family appears to be the smallest and the most compact among MsrBs.
Thus, an interesting aspect of the evolution of the MsrB superfamily is the dramatic variability with regard to length and secondary structure composition of N-terminal and C-terminal regions in these proteins. This is evident from superimposition of MsrB structures: whereas the β-core is well conserved (supplemental Fig. S3, A and C), the terminal parts of the proteins show significant differences (supplemental Fig. S3, B and D). This feature is further supported by bacterial MsrBs: although their structures are very similar (supplemental Fig. S3A), their N-terminal and C-terminal regions are not superimposable (supplemental Fig. S3B).
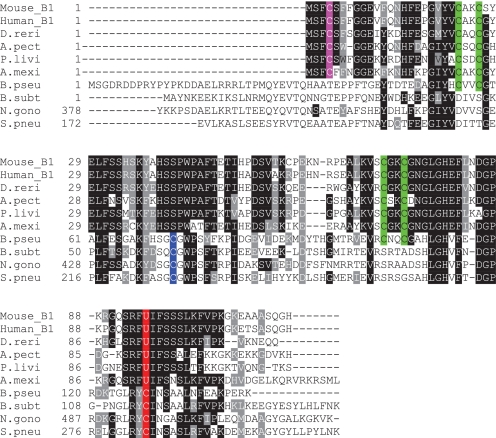
The data indicate that the N-terminal region may play an important role in the evolution of MsrB1. As discussed above, compared with other MsrBs, MsrB1 salient features include (i) a catalytic Sec residue and (ii) a short N-terminal region containing a resolving Cys in position 4 (the “classical” resolving Cys is present in the middle of MsrB sequences as shown in Fig. 5 and supplemental Fig. S4). The occurrence of the novel resolving Cys is tightly linked to the presence in the active site Sec (rather than Cys). The presence of highly homologous Sec-containing MsrBs can be traced back to early aquatic animals, such as Porifera and Antozoa. For example, the sponge Suberites sp. S29 selenoprotein MsrB has 67% sequence identity with mouse MsrB1 and 70% with Xenopus MsrB1 (supplemental Fig. S4) and retains all features distinguishing MsrB1s from ancestral MsrBs. Thus, the evolution of MsrB1 appears to tightly couple the shorter N-terminal region with the new resolving Cys and the use of catalytic Sec. In the present work (Fig. 4) we provide evidence that could explain the functional relevance behind this coupling: in Sec-containing MsrBs, the short and mobile N-terminal part is needed to resolve the catalytic residue, after the switching has occurred (i.e. after the reaction with the natural substrate, promoting the relocation of the N terminus).
FIGURE 5.
Multiple sequence alignment of selenoprotein MsrBs and structurally characterized bacterial Cys-containing MsrBs. Catalytic Sec (U) and Cys residues are highlighted in red. Conserved resolving Cys residues in the Cys-containing forms are highlighted in blue, and those residues in the N-terminal region of selenoprotein forms are highlighted in pink. Zinc-binding Cys residues in the two CXXC motifs are shown in green. GI numbers are as follows: Mus musculus, 7305479; Homo sapiens, 7706511; Danio rerio, 45501380; Asterina pectinifera, 93344816; Paracentrotus lividus, 89448082; Ambystoma mexicanum, 51000422; B. pseudomallei, 53719057; B. subtilis, 2634588; N. gonorrhoeae, 59802366; S. pneumoniae, 221232101.
Secondary structure prediction using PSI-PRED and the experimental evidence from our study indicate that one consequence of shortening the N-terminal region in MsrB1 is the loss of secondary structure elements (a helix in bacteria and a β-strand mixed with a helix in archaea, supplemental Fig. S3, C and D) in favor of a mainly unstructured N terminus. This loss seems to be specific for MsrB1 because N-terminal secondary structures are found in other MsrBs ranging from bacteria and archaea to animal Cys-containing MsrBs.
A crucial role in the interaction between the mobile N-terminal and the active site appears to be governed by hydrophobic interactions. The analysis of the N-terminal region of mouse MsrB1 revealed that this region is rich in aromatic amino acids; other selenoprotein MsrBs also have aromatic amino acids in their N-terminal parts (Fig. 5) (12). Of 19 N-terminal residues in MsrB1, 5 are phenylalanines, including Phe3, Phe6, and Phe7, which surround the resolving Cys4. Two other Phe residues, Phe82 and Phe97, are in the active site. In addition, as discussed above, Phe3 (a fully conserved residue in selenoprotein MsrBs) (Fig. 5 and supplemental Fig. S5) specifically approaches the catalytic site upon binding of MetSO. Furthermore, the role of hydrophobic residues is supported by the analysis of the electrostatic surface potential distribution for MsrB1, calculated at neutral pH (Fig. 1E). It shows that the front side of MsrB1 around the active site is mainly neutral and the charges are equally distributed on the rest of the frontal surface.
Bacterial MsrBs have a single Phe (corresponding to Phe82 for MsrB1) in their active sites (supplemental Fig. S5, lower panel). Another Phe, Phe97, is only present in selenoprotein MsrBs and is replaced with Asn in Cys-containing forms (12, 13). Thus, it appears that hydrophobic interactions between aromatic residues in the N-terminal region and the active site play an important role during catalysis, assisting the movement of the N-terminal region upon formation of the intramolecular selenenylsulfide (or disulfide). Previously, Phe97 was found to be crucial for the activity of selenoprotein MsrB1 (13). Furthermore, the aromatic amino acids in the N-terminal region and the extra Phe97 in mouse MsrB1 may have a dual role: stabilizing protein-substrate interactions in a way similar to the α-helices in bacterial MsrBs and promoting the intramolecular selenenylsulfide formation through hydrophobic interaction.
Glu81 is also a unique residue present in selenoprotein forms, which is replaced with Val or Ile in Cys-containing MsrBs. Our previous biochemical analysis has shown that this residue is important for MsrB1 activity (i.e. E81V mutant of selenoprotein MsrB1 had a >100-fold decreased activity compared with that of wild-type protein) (13). Our structure reveals that this Glu81 can form a salt bridge with Lys57 on the backside of the active site, contributing to the structural stability and/or water solubility.
As shown in the lower panel of supplemental Fig. S5, the resolving Cys in all reported bacterial MsrBs is located close to the active site. The distances between the catalytic and resolving Cys residues are 3.0 Å in N. gonorhoeae MsrB, 3.7 Å in B. pseudomallei MsrB, 4.1 Å in S. pneumoniae MsrB, and 3.5–4.1 Å in B. subtilis MsrB (3EOO). Thus, their catalytic mechanism involves formation of a disulfide bond between proximal catalytic and resolving Cys residues, unlike MsrB1. One exception is the X. campestris MsrB that evolved a new resolving Cys, Cys31 (36) and requires structural rearrangements. Together with the structure of MsrB1, the X. campestris structure shows flexibility in the use of resolving Cys by MsrBs.
The difference in proximal and distal disulfide/selenenylsulfide bond formation between most bacterial proteins and mammalian MsrB1 is also evident from the analysis of their secondary structure. The presence of the α-helical structures in the N-terminal region of bacterial MsrBs suggests that large structural alterations are unlikely in these enzymes due to the rigidity imposed by α-helices. Instead, the absence of any secondary structure in the N terminus of Sec-containing MsrB1 does not impose any rigidity constraints. Thus, the formation of the selenenylsulfide (disulfide) bridge between Cys4 and Sec/Cys95 separated by 17.5–43.5 Å in the reduced form of the protein is possible as also confirmed by the tentative oxidized structure (supplemental Fig. S2). The presence of flexible Gly8-Gly9 and Pro18-Gly19 hinge motifs further supports the proposed catalytic mechanism.
Comparative structural analysis of mouse MsrB1 and bacterial MsrBs indicates structural differences between selenoprotein and non-selenoprotein MsrBs that result in the different mechanisms of the redox reaction catalyzed by MsrBs. Such peculiarities of catalytic reactions between Sec- and Cys-containing proteins are the result of evolutionary changes of catalytic Cys to Sec accompanied by other adaptations such as distinct resolving Cys and flexible N terminus.
Among MsrBs, 60% have a conserved resolving Cys in their sequence. The remaining 40%, including mammalian MsrB2 and MsrB3, lack this resolving Cys. Recent studies have shown that the sulfenic acid intermediate in these MsrBs can be directly reduced by thioredoxin (11, 12). The structures of MsrBs determined so far, including our own MsrB1 structure, show common and distinct features of the catalytic mechanism due to distinct function of the resolving Cys.
CONCLUSION
We determined the first selenoprotein MsrB1 structure using high resolution NMR. The structure consists of two antiparallel β-sheets. Both N- and C-terminal tails are poorly structured and easily mobile. Location of the zinc ion, coordinated by four Cys residues, is consistent with a structural role of this metal ion in MsrB1 molecule, as previously proposed. The catalytic site of MsrB1 is composed of Sec/Cys95 (catalytic residue), Cys4 (resolving residue) and Trp43, His80, Phe82, Asp83, Arg93, and Phe97, which assist in the reduction of the oxidized methionine. The hydrophobic interactions between the substrate and the active site aromatic residues play an important role in the MsrB1 function. Comparison of structural and functional properties of mammalian MsrB1 and bacterial MsrBs allowed us to pinpoint some crucial differences between these MsrB classes. From a structural point of view, secondary structures in the N-terminal region of prokaryotic MsrBs distinguish their structures from those of mammalian Sec-containing MsrB1s. Indeed, we show that the evolution of a short and unstructured N-terminal region is coupled with (i) the evolution of Sec as the catalytic residue, and (ii) a new resolving Cys in position 4. We suggest that the N terminus plays a crucial role in MsrB1: its unstructured nature and hydrophobic compatibility with the active site allow it to move toward the active site in a specific response to substrate binding. The enzyme completes the second phase of the catalytic cycle by modulating the access of the resolving Cys to the active site.
Supplementary Material
Acknowledgments
We thank Dr. Rebecca Del Conte for energy minimization with AMBER and Geun-Hee Kwak (Yeungnam University, Korea) for the pH-dependence activity experiment.
This work was supported, in whole or in part, by National Institutes of Health Grant AG021518 (to V. N. G.). This work was also supported by the Faculty of Natural Sciences and Technology at the Norwegian University of Science and Technology (A. D.) and National Research Foundation of Korea Grant 2010-0001240 (to H.-Y. K.). Access to the 800-MHz NMR spectrometer at the NMR center at the University of Florence was within the EUNMR project Contract RII3-026145.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (code 2KV1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MetSO
- methionine sulfoxide
- HSQC
- heteronuclear single quantum coherence
- MetSO2
- methionine sulfone
- Msr
- methionine sulfoxide reductase
- PDB
- Protein Data Bank
- Sec
- selenocysteine.
REFERENCES
- 1.Weissbach H., Etienne F., Hoshi T., Heinemann S. H., Lowther W. T., Matthews B., St. John G., Nathan C., Brot N. (2002) Arch. Biochem. Biophys. 397, 172–178 [DOI] [PubMed] [Google Scholar]
- 2.Kim H. Y., Gladyshev V. N. (2007) Biochem. J. 407, 321–329 [DOI] [PubMed] [Google Scholar]
- 3.Lin Z., Johnson L. C., Weissbach H., Brot N., Lively M. O., Lowther W. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9597–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee B. C., Le D. T., Gladyshev V. N. (2008) J. Biol. Chem. 283, 28361–28369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S. R., Bar-Noy S., Kwon J., Levine R. L., Stadtman T. C., Rhee S. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stadtman T. C. (1996) Annu. Rev. Biochem. 65, 83–100 [DOI] [PubMed] [Google Scholar]
- 7.Kim H. Y., Gladyshev V. N. (2004) Mol. Biol. Cell 15, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowther W. T., Weissbach H., Etienne F., Brot N., Matthews B. W. (2002) Nat. Struct. Biol. 9, 348–352 [DOI] [PubMed] [Google Scholar]
- 9.Kumar R. A., Koc A., Cerny R. L., Gladyshev V. N. (2002) J. Biol. Chem. 277, 37527–37535 [DOI] [PubMed] [Google Scholar]
- 10.Olry A., Boschi-Muller S., Branlant G. (2004) Biochemistry 43, 11616–11622 [DOI] [PubMed] [Google Scholar]
- 11.Kim H. Y., Kim J. R. (2008) Biochem. Biophys. Res. Commun. 371, 490–494 [DOI] [PubMed] [Google Scholar]
- 12.Lee T. H., Kim H. Y. (2008) Arch. Biochem. Biophys. 478, 175–180 [DOI] [PubMed] [Google Scholar]
- 13.Kim H. Y., Gladyshev V. N. (2005) PLoS Biol. 3, e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y. K., Shin Y. J., Lee W. H., Kim H. Y., Hwang K. Y. (2009) Mol. Microbiol. 72, 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sal L. S., Aachmann F. L., Kim H. Y., Gladyshev V. N., Dikiy A. (2007) Biomol. NMR Assign. 1, 131–133 [DOI] [PubMed] [Google Scholar]
- 16.Keller R. L. J. (2004) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment, CANTINA Verlag, Zürich [Google Scholar]
- 17.Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 18.Kay L. E., Torchia D. A., Bax A. (1989) Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 19.Güntert P., Braun W., Wüthrich K. (1991) J. Mol. Biol. 217, 517–530 [DOI] [PubMed] [Google Scholar]
- 20.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 21.Chantalat L., Leroy D., Filhol O., Nueda A., Benitez M. J., Chambaz E. M., Cochet C., Dideberg O. (1999) EMBO J. 18, 2930–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colonna-Cesari F., Perahia D., Karplus M., Eklund H., Brädén C. I., Tapia O. (1986) J. Biol. Chem. 261, 15273–15280 [PubMed] [Google Scholar]
- 23.Mechulam Y., Schmitt E., Maveyraud L., Zelwer C., Nureki O., Yokoyama S., Konno M., Blanquet S. (1999) J. Mol. Biol. 294, 1287–1297 [DOI] [PubMed] [Google Scholar]
- 24.Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kryukov G. V., Kumar R. A., Koc A., Sun Z., Gladyshev V. N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boschi-Muller S., Olry A., Antoine M., Branlant G. (2005) Biochim. Biophys. Acta 1703, 231–238 [DOI] [PubMed] [Google Scholar]
- 27.Kornhaber G. J., Snyder D., Moseley H. N., Montelione G. T. (2006) J. Biomol. NMR 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 28.Kauffmann B., Aubry A., Favier F. (2005) Biochim. Biophys. Acta 1703, 249–260 [DOI] [PubMed] [Google Scholar]
- 29.Cheighton T. E. (1996) Protein: Structure and Molecular Properties, 2nd Ed., W. H. Freeman and Company, New York [Google Scholar]
- 30.Neiers F., Sonkaria S., Olry A., Boschi-Muller S., Branlant G. (2007) J. Biol. Chem. 282, 32397–32405 [DOI] [PubMed] [Google Scholar]
- 31.Olry A., Boschi-Muller S., Yu H., Burnel D., Branlant G. (2005) Protein Sci. 14, 2828–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deleted in proof
- 33.Li H., Robertson A. D., Jensen J. H. (2005) Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 34.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano W. L. (2002) The PyMOL User's Manual, DeLano Scientific, San Carlos, CA [Google Scholar]
- 36.Ranaivoson F. M., Neiers F., Kauffmann B., Boschi-Muller S., Branlant G., Favier F. (2009) J. Mol. Biol. 394, 83–93 [DOI] [PubMed] [Google Scholar]
- 37.Neiers F., Kriznik A., Boschi-Muller S., Branlant G. (2004) J. Biol. Chem. 279, 42462–42468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.