Abstract
A large set of chemokines is highly up-regulated in human chondrocytes in response to IL-1β (Sandell, L. J., Xing, X., Franz, C., Davies, S., Chang, L. W., and Patra, D. (2008) Osteoarthr. Cartil. 16, 1560–1571). To investigate the mechanism of transcriptional regulation, deletion constructs of selected chemokine gene promoters, the human CCL3 (MIP-1α) and CCL4 (MIP-1β), were transfected into human chondrocytes with or without IL-1β. The results show that an IL-1β-responsive element is located between bp −300 and −140 of the CCL3 promoter and between bp −222 and −100 of the CCL4 promoter. Because both of these elements contain CCAAT/enhancer-binding protein β (C/EBPβ) motifs, the function of C/EBPβ was examined. IL-1β stimulated the expression of C/EBPβ, and the direct binding of C/EBPβ to the C/EBPβ motif was confirmed by EMSA and ChIP analyses. The −300 bp CCL3 promoter and −222 bp CCL4 promoter were strongly up-regulated by co-transfection with the C/EBPβ expression vector. Mutation of the C/EBPβ motif and reduction of C/EBPβ expression by siRNA decreased the up-regulation. Additionally, another cytokine-related transcription factor, NF-κB, was also shown to be involved in the up-regulation of chemokines in response to IL-1β, and the binding site was identified. The regulation of C/EBPβ and NF-κB was confirmed by the inhibition by C/EBPβ and NF-κB and by transfection with C/EBPβ and NF-κB expression vectors in the presence or absence of IL-1β. Taken together, our results suggest that C/EBPβ and NF-κB are both involved in the IL-1β-responsive up-regulation of chemokine genes in human chondrocytes. Time course experiments indicated that C/EBPβ gradually and steadily induces chemokine up-regulation, whereas NF-κB activity was highest at the early stage of chemokine up-regulation.
Keywords: C/EBP Transcription Factor, Cellular Regulation, Chemokines, Interleukin, NF-kappaB, Human Chondrocytes
Introduction
Chemokines are associated with several diseases, including cardiovascular diseases, neuroinflammation, cancer, and HIV-associated diseases (1). As mediators of cell recruitment, chemokines are known to be important in inflammatory diseases, including rheumatoid arthritis, osteoarthritis (OA),2 inflammatory bowel disease, multiple sclerosis and transplant rejection (1, 2). Chemokines are a specific class of cytokines that classically mediate chemoattraction (chemotaxis) between cells. However, the production and function of chemokines in organs and tissues have recently been recognized, and knock-out of the chemokine receptors CXCR4 and CXCR2 suggests a role in development and cell senescence (3, 4), and the chemokine CCL3 is a potent osteoclastogenic factor (5). There exist over 50 chemokine ligands and 20 G protein-coupled receptors (1). Chemokines have similar protein structures, being 8–10 kDa, with two major subclasses having conserved cysteine residues either adjacent (CC) or separated by one amino acid (CXC) (2).
IL-1β is an important cytokine in rheumatoid and osteoarthritic joint diseases. Generally, IL-1β is viewed as a catabolic factor for cartilage, inducing enzymes that degrade the extracellular matrix and reducing synthesis of the primary cartilage components type II collagen (COL2A1) and aggrecan (6–8), although it can also induce BMP-2 (bone morphogenetic protein 2) potentially to initiate a repair response (9). In joint diseases, IL-1β is synthesized by synovial cells (10) and cartilage chondrocytes (6, 11); therefore, its effect on chondrocytes is highly relevant to the fate of cartilage.
We have shown by microarray analysis, that a large set of chemokine genes is up-regulated by the proinflammatory cytokine IL-1β in adult normal cartilage and from patients with OA (6). It can be expected that this increase in a wide range of chemokines will have a significant impact on the cells of cartilage and other related joint tissues and should be considered in the pathophysiology of OA. Recently, we demonstrated that the adipokine, resistin, present in injured joints (12), also increased chemokine genes at both the transcriptional and post-transcriptional levels (13) in temporal patterns similar to the IL-1β patterns (6). The post-transcriptional regulation of chemokines was further investigated by us in chondrocytes with resistin (13) and by others in fibroblasts (14).
In the present study, we focused on the transcriptional regulation of chemokine genes. As we reported (6), IL-1β increased expression of chemokines CCL3, CCL4, CCL20, CCL3L1, CXCL1, CXCL2, CXCL3, CXCL6, and CXCL8 (IL-8) from 25- to 75-fold in human articular chondrocytes. A computational analysis of these co-regulated genes identified NF-κB, C/EBPβ, and MEF-3 (myocyte enhancer binding factor 3), as candidate transcriptional regulators. NF-κB has been shown to regulate a specific subset of chemokines (15–18); however, Amos et al. (19) recently demonstrated that inhibition of NF-κB activity did not inhibit all inflammatory mediators; therefore, there are probably other transcriptional mechanisms involved. C/EBP has been shown to regulate a set of chemokines (4, 20, 21). We have previously shown that C/EBPβ is associated with IL-1β-induced and tumor necrosis factor-α (TNF-α)-induced down-regulation of matrix genes in chondrocytes and the repression of cartilage gene expression in non-cartilaginous tissues (22–25). Here, we investigated the roles of C/EBPβ and NF-κB in the up-regulation of two chemokine genes, CCL3 (MIP-1α; macrophage inflammatory protein 1α) and CCL4 (MIP-1β), in human chondrocytes in response to IL-1β.
EXPERIMENTAL PROCEDURES
Materials
The materials used in this work were purchased as follows. Dulbecco's modified Eagle's medium (DMEM) and Ham's F-12 medium were from Mediatech Inc. (Herndon, VA). Fetal bovine serum, pfx polymerase, SuperScript® II reverse transcriptase, restriction enzymes, Alexa fluor® 488 goat anti-mouse IgG, and Alexa fluor® 594 goat anti-rabbit IgG were from Invitrogen; penicillin/streptomycin solution, ascorbic acid, actinomycin D, Tween 20, Triton X-100, and the CelLyticTMNu-Clear extraction kit were from Sigma; 16% paraformaldehyde was from Electron Microscopy Science (Hatfield, PA); recombinant human IL-1β was from R&D Systems, Inc. (Minneapolis, MN); the RNeasy Mini Kit, QIAshredder, and DNase I were from Qiagen, Inc. (Valencia, CA); [γ-32P]dATP was from PerkinElmer Life Sciences; FuGENE® 6 Transfection Reagent, X-tremeGENE siRNA Transfection Reagent, Quick Spin Sephadex G-50 and G-25 columns, Pronase, and Collagenase P were from Roche Applied Science; pGL3-basic vector, reporter lysis buffer, luciferase assay reagent, and β-galactosidase were from Promega (Madison, WI); QuikChange® II site-directed mutagenesis kit was from Stratagene (La Jolla, CA); the Ready Gel Tris-HCl precast gels and nonfat dry milk were from Bio-Rad; anti-C/EBPβ, SREBP1, anti-c-Rel, normal rabbit IgG, and actin antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); SuperSignal® West Pico chemiluminescent substrate was from Thermo Fisher Scientific Inc. (Waltham, MA); SYBR Green PCR Master Mix was from Applied Biosystems (Foster City, CA); cell-permeable NEMO binding domain (NBD) synthetic peptides (IKK-NBD peptide and IKK-NBD control peptide) were from BIOMOL (Plymouth Meeting, PA); SB203580 was from Calbiochem; and EZ-ChIPTM, Immobilon-P, and Immobilon-FL transfer membrane were from Millipore (Bedford, MA).
Plasmid Constructs
A series of CCL3 and CCL4 promoter 5′-deletion constructs were made by PCR and subcloned into pGL3-basic vector using the pGL2-CCL3(−1972/+75) and pGL3-CCL4(−1281/+12) as described previously (26, 27). The CCL3 and CCL4 promoter constructs, C/EBPβ and IKK2 (IκB kinase 2) expression vector, and pNF-κB luciferase reporter were provided by the following. The human pGL2-CCL3(−1972/+75) was from Dr. G. David Roodman (University of Pittsburgh) (26); human pGL3-CCL4(−1281/+12) was obtained from Dr. Sheau-Farn Yeh (National Yang-Ming University, Taipei) (27); human IKK2 in the pCDNA3 vector and pNF-κB luciferase reporter were from Dr. Yousef Abu-Amer (Washington University) (28); full-length human C/EBP in the pCDNA3 vector was from Dr. Erika Crouch (Washington University) (29). The empty expression vectors were made by excision of cDNAs from the corresponding C/EBP expression vectors (22). To facilitate subcloning of the amplified fragments, the antisense primer contained a HindIII restriction site adaptor and the sense primer contained an XhoI or SmaI site. The PCR fragments and the luciferase expression vector pGL3-basic vector were digested with XhoI or SmaI and HindIII before ligation. All constructs were confirmed by DNA sequence analysis using a GL2 primer and RV3 primer.
Cell Culture
Human primary chondrocytes were obtained from articular cartilage obtained at the time of total joint replacement or from above the knee amputation, with approval of the Washington University Human Studies Review Board and permission of the patient (Institutional Review Board 05-0279). Chondrocytes were isolated following previously published procedures (6) and plated at a density of 2.5 × 105 cells/cm2 in DMEM/F-12 medium plus 10% fetal bovine serum (FBS), 50 μg/ml ascorbate, and antibiotics (50 units/ml penicillin and 50 μg/ml streptomycin). Cells were allowed to rest for 24 h, and IL-1β was added at the concentrations and times indicated. IL-1β was reconstituted in sterile phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin. The T/C-28a2 human chondrocyte cell line was also used (a gift from Dr. Mary B. Goldring, Cornell University) and cultured like human articular chondrocytes. For RNA or nuclear protein extraction, the T/C-28 human chondrocyte cell line was plated in 3 × 104 cells/cm2 densities and cultured overnight. Human IL-1β was then added to the medium at the concentrations indicated.
Transient Transfection and Luciferase Assay
DNA transfections of T/C-28a2 cells were performed using FuGENE 6TM transfection reagent or X-tremeGENE siRNA transfection reagent. 2 × 105 of T/C-28a2 cells were cultured in a 6-well plate overnight. The transfection mixture containing 3–9 μl of transfection reagent (6:1 ratio of transfection reagent (μl) to DNA (μg)), 500 ng of various promoter constructs, and 100 or 200 ng of pCMV-β-gal was then added, and the cells were cultured for 8 or 24 h with or without IL-1β as indicated. For the co-transfection assay, 500 ng of C/EBPβ expression vectors or empty vector and a 162 pm concentration of siRNAs were added to the 100-μl transfection mixture as indicated. The sequences of siRNAs used were described previously (21). Due to low translation efficiency of C/EBPβ, we used higher amounts of plasmid (500 ng) for transfection, as shown previously (22, 24). FBS was added to transfection medium 4 h later to a final concentration of 10%. After 24 h of incubation, cells were replaced with fresh complete medium and incubated for an additional 8 or 24 h with or without IL-1β. The cells were then harvested with Reporter Lysis BufferTM, and the lysate was analyzed for luciferase activity using Promega Luciferase Assay ReagentTM. The β-galactosidase activities were also measured to normalize variations in transfection efficiency. Each transfection experiment was performed in duplicate or triplicate and repeated at least three times.
Preparation of Nuclear Extracts
Nuclear extracts from T/C-28a2 cells were isolated using Nu-Clear Extraction KitTM according to the manufacturer's instructions. Protein concentration of nuclear extract was determined using the Bio-Rad protein assay kit.
Electrophoretic Mobility Shift Assay (EMSA)
Fragment A (between −300 and −141 bp relative to the human CCL3 translation start site) and Fragment B (between −222 and −101 bp relative to the human CCL4 translation start site) were amplified by PCR. All oligonucleotides were synthesized by Invitrogen, and complementary oligonucleotide was annealed to make double-stranded oligonucleotide (30). The Fragment A, Fragment B, and various double-stranded oligonucleotides were end-labeled using T4 polynucleotide kinase and [γ-32P]dATP. Bandshifts were performed by incubating 5 μg of nuclear extracts in the mobility shift buffer (10 mm Hepes-KOH, pH 7.9, 100 mm NaCl, 1 mm EDTA, 10% glycerol, and 1 mm DTT) and 2 μg of poly(dI-dC) with the DNA probe on ice for 30 min. For the competition studies, the cold DNA fragments were added at a 100-fold molar excess compared with the probe and incubated for 30 min on ice before adding the DNA probe. For the antibody interference experiments, the nuclear extracts and 2 μl of antibody were preincubated in the buffer for 1 h at 4 °C. DNA protein complexes were resolved on a 5% polyacrylamide gel at 100 V for about 90 min. The gels were dried and autoradiographed.
Western Blotting
Fifteen μg of nuclear extracts were denatured in SDS sample buffer containing 0.1 m dithiothreitol at 100 °C for 3 min and separated on a 12% Bio-Rad Ready Gel in Tris/glycine/SDS buffer. The gels were then transferred to Immobilon-P transfer membrane in Tris/glycine buffer, pH 8.3, containing 20% methanol. The membranes were saturated in 5% nonfat dry milk in PBS at 4 °C overnight and reacted with anti-C/EBPβ antibody diluted to 1:1000 or actin antibody diluted to 1:500 in PBS containing 0.05% Tween 20. The bound antibodies were recognized by IgG antibodies coupled to horseradish peroxidase, and the secondary antibodies were detected by autoradiograph using SuperSignal chemiluminescent substrate from Thermo Scientific. For Western blotting using the Odyssey imaging system, 10 μg of nuclear extracts were used, and the gels were transferred and scanned according to the protocol recommended by the manufacturer (LI-COR).
RNA Isolation and Real Time Quantitative PCR
Total RNA was isolated from T/C-28 cells and human primary articular chondrocytes with an RNeasy minikit with DNase I treatment, following the protocol recommended by the manufacturer (Qiagen). Total RNA (1 μg) was reverse-transcribed with a SuperScriptTM II reverse transcriptase to synthesize cDNA. The cDNA was then used for real-time quantitative PCR. Real-time quantitative PCR was performed in a total volume of 20 μl of reaction mixture containing 10 μl of SYBR Green PCR Master Mix, 2.5 μl of cDNA, and 200 nm primers using a 7300 real-time PCR system (Applied Biosystems) and done in triplicate. Primers used for quantitative PCR were optimized for each gene, and the dissociation curve was determined by the real-time PCR System. The parameters of primer design included a primer size of 18–21 bp, a product size of 80–150 bp, a primer annealing temperature of 59–61 °C, and a primer GC content of 45–55%. Results were normalized to glyceraldehyde-3- phosphate dehydrogenase (GAPDH). The primer sequences are as follows: human GAPDH, 5′-ACCCAGAAGACTGTGGATGG-3′ (sense) and 5′-GAGGCAGGGATGATGTTCTG-3′ (antisense); human C/EBPβ, 5′-CTCGCAGGTCAAGAGCAAGG-3′ (sense) and 5′-TCGTCGCTGTGCTTGTCC-3′ (antisense) (22); human CCL3, 5′-GCAACCAGTTCTCTGCATCA-3′ (sense) and 5′-TGGCTGCTCGTCTCAAAGTA-3′ (antisense); human CCL4, 5′-GCTTTTCTTACACTGCGAGGA-3′ (sense) and 5′-CCAGGATTCACTGGGATCAG-3′ (antisense); human NF-κB1 (p50), 5′-CCTGGATGACTCTTGGGAAA-3′ (sense) and 5′-TCAGCCAGCTGTTTCATGTC-3′ (antisense); human NF-κB2 (p52), 5′-GAACAGCCTTGCATCTAGCC-3′ (sense) and 5′-TTTTCAGCATGGATGTCAGC-3′ (antisense); human RelA (p65), 5′-TCTGCTTCCAGGTGACAGTG-3′ (sense) and 5′-GCCAGAGTTTCGGTTCACTC-3′ (antisense); human c-Rel, 5′-CGAACCCAATTTATGACAACCG-3′ (sense) and 5′-TTTTGTTTCTTTGCTTTATTGCCG-3′ (antisense) (31); human RelB, 5′-CTGCTTCCAGGCCTCATATC-3′ (sense) and 5′-CGCAGCTCTGATGTGTTTGT-3′ (antisense); human IκBα (inhibitor of κB), 5′-GATCCGCCAGGTGAAGGG-3′ (sense) and 5′-GCAATTTCTGGCTGGTTGG-3′ (antisense) (32). The cycle threshold (Ct) values for GAPDH and those of genes of interest were measured for each sample, and the relative transcript levels were calculated as χ = 2−ΔΔCt, where ΔΔCt = Δtreatment − ΔC and Δtreatment = Cttreatment − CtGAPDH; ΔC = Ctcontrol − CtGAPDH.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitations (IPs) were carried out using the EZ ChIPTM assay kit following the protocol recommended by the manufacturer. One 15-cm dish of T/C-28 cells (4 × 106) in 15-cm culture dishes was used for one IP reaction. Cross-linking of DNA-proteins was induced by the addition of formaldehyde (1% final concentration) directly to the culture medium for 10 min at 37 °C. Cells were lysed, and DNA in the supernatant was sheared by sonication. A 10-μl aliquot of the sonicated chromatin sample (“input”) was removed for PCR analysis, and the remainder was used for IPs. To each IP was added either 5 μg of C/EBPβ antibody or normal rabbit IgG, and reactions were incubated overnight with constant mixing at 4 °C. Protein G beads were collected by centrifugation, and protein-DNA complexes were eluted from the beads followed by a cross-link reversal step by the addition of 5 m NaCl (8 μl) to the eluted sample (200 μl) overnight at 65 °C. DNA from each IP reaction (2 μl) was used for PCR with primer pairs to amplify the 140-bp region of interest in CCL3 (−300/−140) and the 110 bp in CCL4 (−222/−100): CCL3, 5′-TCATGCACAGACCAGTTCTTATGA-3′ (sense primer) and 5′-CTCTAACTCTCAGCTCTC-AACTCA-3′ (antisense primer); CCL4, 5′-CTGTACCACTTCCCTTTTCTTCTC-3′ (sense primer) and 5′-CTGAAGCTAGCTGAGTGAGGAGTT-3′ (antisense primer). Input or immunoprecipitated DNA was amplified by PCR (94 °C, 20 s; 59 °C, 30 s; 72 °C, 2 min) for 32 cycles (CCL3) or 34 cycles (CCL4).
Immunofluorescence
5 × 104 human articular chondrocytes were cultured in each well of 8-well chamber slides (from Lab Tek). Cells were allowed to rest for 24 h, and IL-1β was then added at the times indicated. Cells were incubated in 4% paraformaldehyde in PBS for 10 min, 0.2% Triton X-100 in PBS for 5 min, and 10% normal goat serum in PBS for 2 h at room temperature. Cells were reacted with rabbit anti-C/EBPβ and mouse anti-c-Rel antibodies diluted to 1:400 in 2% normal goat serum in PBS for overnight at 4 °C. The secondary antibodies, Alexa fluor 488 dye-labeled goat anti-mouse IgG diluted to 1:250 and Alexa fluor 594 dye-labeled goat anti-rabbit IgG diluted to 1:400 in 2% normal goat serum in PBS, were then added to the cells for 1 h at room temperature. Immunoreactivity was detected by fluorescence microscopy.
RESULTS
IL-1β Stimulates the Expression of Chemokine Genes CCL3 and CCL4 in Normal Human Articular Chondrocytes and the Chondrocyte-derived Cell Line, T/C-28
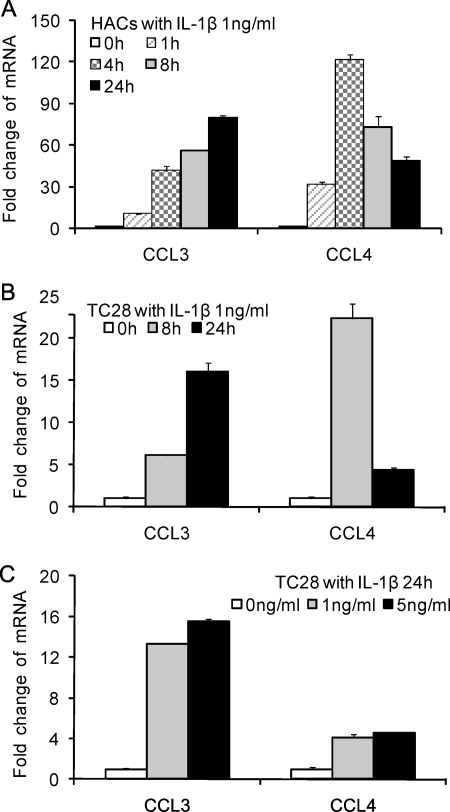
We have reported that a large set of chemokines that are up-regulated by the IL-1β in adult normal cartilage and from patients with OA (6). Here, we showed that two representative genes, CCL3 and CCL4, were increased in human articular chondrocytes and in the cell line T/C-28 with the same kinetics in the 24-h time period, although the expression level of CCL3 and CCL4 in human primary chondrocytes was higher than in the T/C-28 chondrocyte cell line (Fig. 1, A–C). CCL3 demonstrated a continuous increase to 24 h to an approximately 80-fold increase in primary cells. CCL4 was up-regulated by 4 h but then decreased during the remaining time period to about a 45-fold increase at 24 h. A dose-response test of IL-1β showed that 1 ng/ml is adequate for significant up-regulation (Fig. 1C). The primary chondrocytes and T/C-28 chondrocyte cell line showed a similar pattern of induction (Fig. 1, A and B).
FIGURE 1.
IL-1β stimulates the expression of CCL3 and CCL4 in normal human articular chondrocytes (HACs) and T/C-28 (TC28) chondrocyte cells. A, human articular chondrocytes were treated with 1 ng/ml IL-1β for various times as indicated. B, T/C-28 cells were treated with 1 ng/ml IL-1β for various times as indicated. C, T/C-28 cells were treated for various concentrations for 24 h. The relative expression levels were examined by the quantitative real time PCR method. Each bar represents the mean ± S.D. (error bars) from three experiments.
An IL-1β-responsive Element Is Located between −300 and −140 bp of the CCL3 Promoter and between −222 and −100 bp of the CCL4 Promoter
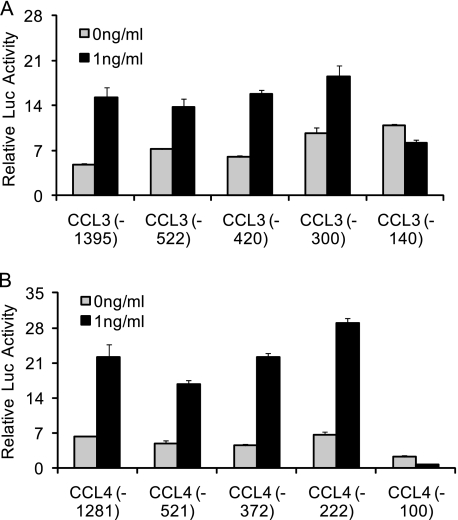
To identify the IL-1β-responsive element within CCL3 and CCL4 promoters, five 5′-deletion constructs were transiently transfected into T/C-28 cells and incubated in the absence or presence of IL-1β (Fig. 2, A and B). Although there are many exceptions, most transcriptional regulation occurs within the first 1000 nucleotides of the promoter (33). For the CCL3 promoter constructs, the −1395, −522, −420, and −300 bp CCL3 promoter constructs were up-regulated by the IL-1β treatment; however, the response was lost in the −140 bp CCL3 constructs (Fig. 2A). For CCL4 promoter constructs, the expression of the −1281, −521, −372, and −222 bp CCL4 promoter constructs were up-regulated by the IL-1β treatment, whereas the response was lost in the −100 bp CCL4 construct (Fig. 2B). These data suggest that in these constructs, the IL-1β-responsive element is located between −300 and −140 bp of the CCL3 promoter and between −222 and −100 bp of the CCL4 promoter.
FIGURE 2.
IL-1β-responsive element is located between −300 and −140 bp of the CCL3 promoter and between −222 and −100 bp of the CCL4 promoter. 5′-Deletion constructs were transiently transfected into T/C-28a2 cells and incubated for 24 h and then for a further 24 h (CCL3) (A) or 8 h (CCL4) (B) in the absence or presence of IL-1β (1 ng/ml) with fresh complete medium. Luciferase activities were measured and expressed relative to the activity of promoterless pGL3b (set as 1). Each bar represents the mean ± S.D. of at least three independent experiments.
IL-1β Stimulates the Expression of C/EBPβ in Human Articular Chondrocytes and T/C-28 Cells
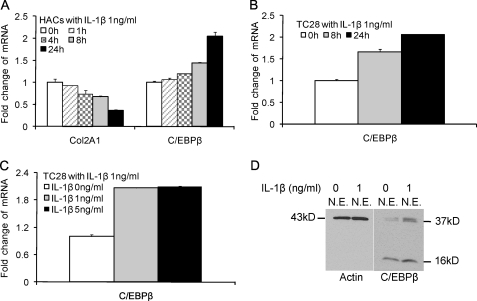
Our computational data base of regulatory motifs revealed that there were potential binding sites for C/EBPβ between −300 and −140 bp of the CCL3 promoter (2 sites) and between −222 and −100 bp of the CCL4 promoter (1 site); therefore, we examined whether IL-1β stimulates the expression of C/EBPβ in primary chondrocytes and T/C-28 cells. The expression level of C/EBPβ mRNA was stimulated by IL-1β over 24 h (Fig. 3, A–C). Type II collagen mRNA decreased to about 60% within 24 h in human chondrocytes as expected (Fig. 3A) as we have shown previously for rat chondrosarcoma cells (22) and human articular chondrocytes (6). Western blots for C/EBPβ revealed that in the absence of IL-1β treatment, C/EBPβ protein was present in the nuclei. When cells were stimulated with IL-1β, C/EBPβ protein expression was increased about 3-fold (Fig. 3D). Multiple C/EBPβ isoforms with stimulatory or inhibitory activity can be translated from a single mRNA by use of alternative translation initiation sites within the same open reading frame (34, 35). The IL-1β treatment stimulated the expression of two of C/EBPβ forms, 38- and 36-kDa liver-enriched transcriptional activator proteins (LAP), and 16-kDa liver-enriched inhibitory protein (LIP) (23, 36); however, the up-regulation of protein expression of C/EBPβ LAP was highest.
FIGURE 3.
IL-1β stimulates the expression of C/EBPβ in normal human articular chondrocytes (HACs) and T/C-28 chondrocyte cells. The relative expression levels were examined in normal chondrocytes from human articular cartilage and T/C-28 cells treated with IL-1β (1 ng/ml) using the quantitative real-time PCR method. A and B, cells were treated with 1 ng/ml IL-1β for various times as indicated in human articular chondrocytes (A) and T/C-28 cells (B). C, T/C-28 cells were treated with various concentrations of IL-1β for 24 h. D, C/EBPβ proteins were examined by Western blot of nuclear extracts (N.E.) from T/C-28 cells treated or without IL-1β (1 ng/ml). IL-1β increased all of the isoforms of C/EBPβ, LAP (38 and 36 kDa), and LIP (16 kDa), and both isoforms of C/EBPβ-LAP were more significantly increased than LIP. Each bar represents the mean ± S.D. from 3–5 experiments.
C/EBPβ Binds to the IL-1β-responsive Element of the CCL3 and CCL4 Promoters
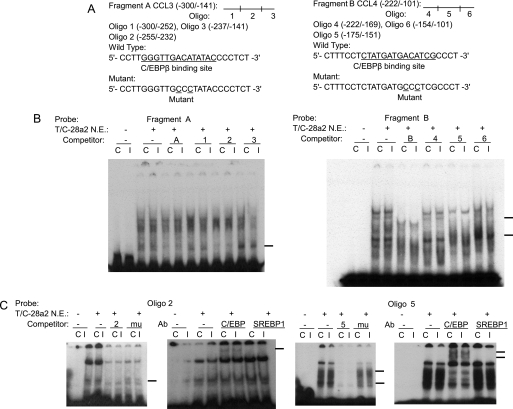
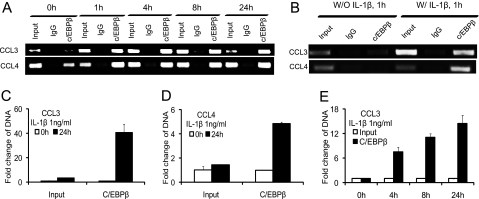
To determine whether C/EBPβ functions within the IL-1β-responsive element, EMSA was carried out using nuclear extract from the T/C-28 cells. Fragment A, containing the sequence between −300 and −141 bp of the CCL3, and Fragment B, containing the sequence between −222 and −101 bp of the CCL4, were used as probes, and various cold oligonucleotides were used as competitors (Fig. 4A). Competitors 2 and 5, which have a potential binding site for C/EBPβ at −251/−238 bp of CCL3 (site S3 in Fig. 9) and −168/−155 bp of CCL4 (site S14 in Fig. 9), competed for the binding of nuclear proteins and the probe (Fig. 4B). As expected, the mutant oligonucleotides 2 and 5, in which the C/EBPβ-binding sites were altered, did not compete for the binding. Supershift analysis confirmed that C/EBPβ bound to the sequence of oligonucleotide 2, in the CCL3 gene and oligonucleotide 5 in the CCL4 gene (Fig. 4C). Site 4 in CCL3 did not bind to the Fragment A (supplemental Fig. 1).
FIGURE 4.
C/EBPβ binds the sequence between −251 and −238 bp of the CCL3 promoter and between −168 and −155 bp of the CCL4 promoter. A, a diagram showing DNA fragments used for EMSA. For CCL3, Fragment A from −300 to −141 bp and the relative locations of oligonucleotides 1, 2, and 3 are shown. For CCL4, Fragment B from −222 to −101 bp and the relative locations of oligonucleotides 4, 5, and 6 are shown. Oligonucleotide 2 from −255 to −232 bp and oligonucleotide 5 from −175 to −151 bp contain the C/EBPβ motif (underlined). Mu, mutant oligonucleotides 2 and 5 containing two base pair mutations (AA to CC) is underlined within the C/EBPβ motif. B, EMSA for T/C-28a2 nuclear extracts using Fragment A or B as a probe and various cold competitors at a 100-fold molar excess. Oligonucleotides 2 and 5 competed with the binding of nuclear proteins to the probe. C, EMSA and supershift analysis for T/C-28a2 nuclear extracts using oligonucleotides 2 and 5 as the probe. Mutant oligonucleotides 2 and 5 did not compete with the binding of nuclear proteins to the probe. Antibodies against C/EBPβ or SREBP1 were preincubated with the nuclear extracts. The lines show the shift band of the complex of C/EBPβ antibody (Ab) and specific retarded bands (C, control; I, IL-1β treatment).
FIGURE 9.
Transcriptional regulation in the highly expression of CCL3 and CCL4. A, the candidate C/EBPβ and NF-κB binding sites in human CCL3 (−1395) and CCL4 (−1281) constructs. B, C/EBPβ and IKK2 stimulate the expression of CCL3 (−1395) and CCL4 (−1281) promoter in T/C-28 human chondrocytes. The CCL3 (−1395) and CCL4 (−1281) promoter constructs were co-transfected with C/EBPβ and IKK2 expression plasmid into T/C-28a2 cells with or without IL-1β (1 ng/ml) for 24 h (CCL3) or 8 h (CCL4). Relative luciferase activity indicates the -fold expression relative to the activity of the construct co-transfected with empty vector (set as 1) in the presence of IL-1β after each co-transfection compared with their activities in the absence of IL-1β. Each bar represents the mean ± S.D. (error bars) of three independent experiments.
To further confirm that C/EBPβ binds to the specific CCL3 and CCL4 promoter regions, we performed ChIP assays using anti-C/EBPβ antibody. Endogenous binding of C/EBPβ was stimulated by treatment with IL-1β up to 24 h (Fig. 5A). Treatment with IL-1β enhanced the binding of C/EBPβ to the CCL3 promoter region from −300 to −140 bp and the CCL4 promoter region from −222 to −100 bp (Fig. 5, A and B), quantified by real-time PCR (Fig. 5, C and D). As a control, the ChIP analysis with and without IL-1β at 1 h is shown in Fig. 5B. IL-1β significantly increased binding of C/EBPβ to its cognate sites. IL-1β stimulation gradually increased the binding activity of C/EBPβ over time (Fig. 5E).
FIGURE 5.
C/EBPβ binding activity to the CCL3(−300/−141) and CCL4(−222/−101) promoter regions is increased in the presence of IL-1β. A chromatin immunoprecipitation assay was performed using anti-C/EBPβ antibodies in T/C-28 cells treated with IL-1β for various times as indicated. Treatment with IL-1β enhanced the binding of C/EBPβ to the promoter. A, T/C-28 cells were treated with 1 ng/ml IL-1β for 0, 1, 4, 8, and 24 h. B, T/C-28 cells were treated with or without IL-1β (1 ng/ml) for 1 h. C and D, T/C-28 cells were treated with 1 ng/ml IL-1β for 0 and 24 h, and the relative expression levels of input and C/EBPβ antibody DNAs of CCL3 (C) and CCL4 (D) were examined by the quantitative real time PCR method. The value was normalized to GAPDH, and the expression levels of input and C/EBPβ at 0 h were set as 1. E, T/C-28 cells were treated with 1 ng/ml IL-1β for 0, 4, 8, and 24 h, and the relative expression levels of CCL3 of input and C/EBPβ antibody DNAs were examined by the quantitative real-time PCR method. The value was normalized to GAPDH and input, and the expression levels of C/EBPβ at 0 h were set as 1. Error bars, S.D.
C/EBPβ Functions as an Activator for the CCL3 and CCL4 Promoter Activities in Response to IL-1β Treatment
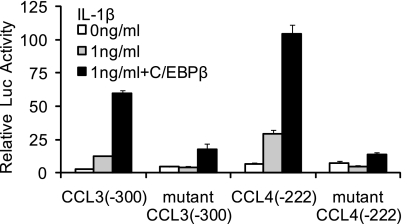
To confirm the function of C/EBPβ-responsive sites on the CCL3 and CCL4 genes, the C/EBPβ-binding sites were mutated in the −300 bp CCL3 construct and −222 bp CCL4 construct using mutant oligonucleotides 2 and 5, respectively. The constructs were transfected into T/C-28 cells and incubated in the absence or presence of IL-1β for an additional 8 or 24 h. The promoter activity of the wild type −300 bp CCL3 construct was about 9-fold stronger than that of mutant −300 bp CCL3 construct, and the −222 bp CCL4 construct was about 6.5-fold stronger than the mutant −222 bp construct after transient transfection into T/C-28 cells, confirming that C/EBPβ is acting as an activator (Fig. 6). The mutation greatly decreased the response to IL-1β in both gene constructs (Fig. 6).
FIGURE 6.
C/EBPβ functions as an activator for the CCL3 and CCL4 promoter activities, and mutation of the C/EBP-binding site down-regulates the promoter activity of the CCL3(−300/−1) and CCL4(−222/−1) constructs. Site-directed mutagenesis was performed within the C/EBP-binding site of the CCL3(−300/−1) and CCL4(−222/−1) constructs using the mutant oligonucleotide 2 and 5 sequence. The mutant and wild type constructs were transiently transfected into T/C-28a2 cells and incubated with or without IL-1β. Luciferase activities were measured and expressed relative to the activity of promoterless pGL3b (set as 1). Further, the wild type and mutant of CCL3(−300/−1) and CCL4(−222/−1) were co-transfected with C/EBPβ expression plasmid into T/C-28a2 cells with IL-1β (1 ng/ml). Relative luciferase activity indicates the -fold expression relative to the activity of promoterless pGL3b, which co-transfected with empty vector (set as 1) in the presence of IL-1β (1 ng/ml). Each bar represents the mean ± S.D. (error bars) of at least three independent experiments.
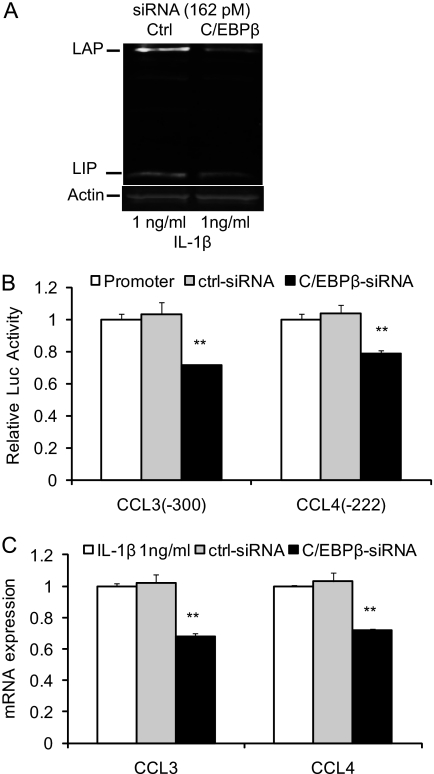
In order to demonstrate the requirement for C/EBPβ, we examined the induction of CCL3 and CCL4 after reducing the C/EBPβ expression by siRNA. Knockdown of C/EBPβ expression was confirmed by Western blot analysis (Fig. 7A). The knockdown of C/EBPβ suppressed the IL-1β-induced CCL3 and CCL4 promoter activities (Fig. 7B) as well as CCL3 and CCL4 mRNA expression (Fig. 7C). Taken together, these results indicate that the C/EBPβ sites at −251/−238 bp of CCL3 and −168/−155 bp of CCL4 promoters are required for the functional response to IL-1β and suggest that C/EBPβ is a key factor in this response.
FIGURE 7.
Silencing of C/EBPβ suppresses the IL-1β-induced CCL3 and CCL4 transcriptional activation. A, Odyssey Western blot of C/EBPβ proteins in T/C-28 cells transfected with control (Ctrl) or C/EBPβ siRNAs (162 pm), and stimulated with 1 ng/ml IL-1β for 8 h. B, T/C-28 cells were transfected with CCL3 and CCL4 promoters and, where indicated, cotransfected with control (ctrl) or C/EBPβ siRNAs (162 pm). Twenty-four h after transfection, T/C-28 cells were stimulated with 1 ng/ml IL-1β for 24 h (CCL3) or 8 h (CCL4). Relative luciferase activity indicates the -fold expression relative to the activity of promoter with IL-1β (1 ng/ml) only (set as 1). C, T/C-28 cells transfected with control or C/EBPβ siRNAs were stimulated for 8 h with IL-1β (1 ng/ml). The relative expression levels were examined by the quantitative real time PCR method. The p value of C/EBPβ siRNA was compared with IL-1β alone based on Student's t test (**, p < 0.01). Each bar represents the mean ± S.D. (error bars) of at least three independent experiments.
NF-κB Is Involved in the Up-regulation of Chemokine Genes by Human Chondrocytes in Response to IL-1β
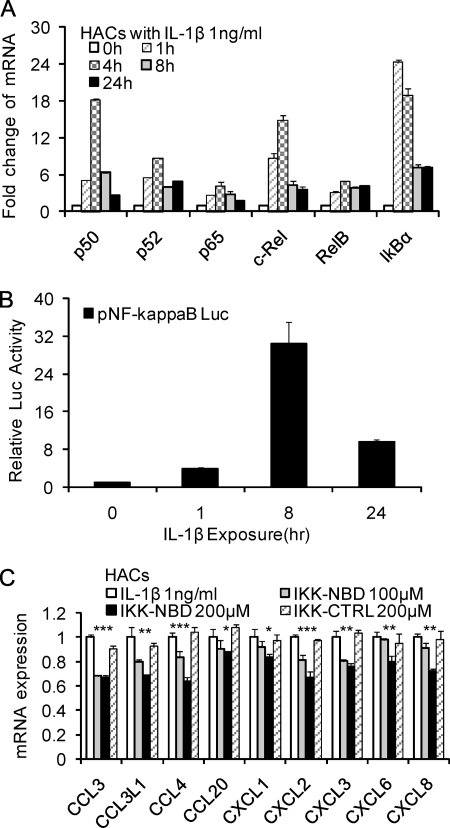
Because the transcription complex NF-κB has been shown to be involved in the regulation of multiple cytokine and chemokine genes, we sought to determine the relative roles of C/EBPβ and NF-κB. The expression of NF-κB in normal human articular chondrocytes was examined. The levels of NF-κB1 (p50), NF-κB2 (p52), RelA (p65), c-Rel, RelB, and IκBα mRNAs were investigated in IL-1β-treated chondrocytes at various times. NF-κB isoforms were rapidly increased, but they decreased after 4 h (Fig. 8A). When compared with the concentration of C/EBPβ in cells, we show that the NF-κB isoforms are lower. In human chondrocytes, ΔC of C/EBPβ averages 3.98 and thus is lower than ΔC of NF-κB1 (6.09), NF-κB2 (6.81), RelA (5.11), c-Rel (9.16), and RelB (5.92). However, these studies also provided insight into the likely NF-κB subunits used in chondrocytes in response to IL-1β. p50 and c-Rel showed the highest relative increase.
FIGURE 8.
Involvement of NF-κB in the up-regulation of chemokines in human chondrocytes in response to IL-1β. A, human articular chondrocytes were treated with 1 ng/ml IL-1β for various times as indicated. B, effect of IL-1β (1 ng/ml) on the expression of pNF-κB Luc reporter in T/C-28 human chondrocytes. Relative luciferase activity indicates the -fold expression relative to the activity of zero time (set as 1) in the presence of IL-1β (1 ng/ml). C, human articular chondrocytes were pretreated with vehicle (DMSO), IKK-NBD peptide (100 and 200 μm), or IKK-NBD control peptide (200 μm) for 1 h and then exposed to IL-1β (1 ng/ml) for 4 h. The p value of IKK-NBD (200 μm) was compared with IL-1β alone based on Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Each bar represents the mean ± S.D. (error bars) of three independent experiments.
Due to the complicated regulatory mechanism of NF-κB, we also looked directly at NF-κB function by using pNF-κB luciferase reporter. The activity of pNF-κB luciferase reporter was up-regulated at 1 h, peaked at 8 h, and was reduced by 24 h (Fig. 8B).
To further evaluate the potential role of NF-κB in primary human articular chondrocyte IL-1β-induced chemokine activation, IKK-NBD, a specific NF-κB inhibitor, was added to the culture before the addition of IL-1β. Following 4 h of IL-1β treatment, the mRNA from these cells showed a modest but dose-dependent suppression of chemokine activity in the medium of cultures treated with IKK-NBD compared with non-inhibited cultures, whereas control IKK-NBD had no significant effect (Fig. 8C). The modest suppression can be attributed to the use short time periods and the use of primary chondrocytes in the present study as opposed to previous experiments, where only cell lines were used (13). Therefore, these results indicated that NF-κB is involved in the expression of chemokines, particularly at 4–8 h of exposure.
C/EBPβ and NF-κB Enhance the Expression of CCL3 and CCL4 in Human Chondrocytes in Response to IL-1β
The previous results indicated that both C/EBPβ and NF-κB contributed to the transcriptional up-regulation of CCL3 and CCL4. In order to determine the relative roles of C/EBPβ and NF-κB in transcription of CCL3 and CCL4, we determined the location of binding sites on the genes and performed transfection studies with CCL3 (−1395) constructs (26), CCL4 (−1281) constructs (27), C/EBPβ expression vector (29), and IKK2 expression vector (28). The CCL3 (−1395) and CCL4 (−1281) constructs contain several high probability candidate C/EBPβ and NF-κB binding sites as assessed by our computational data base (6, 37, 38) (Fig. 9A), although only a limited number of sites are present in the IL-1β response domain. Based on the reported NF-κB binding studies in chemokines (20), the IL-1β-responsive elements had c-Rel binding sites at −210 to −206 bp of the CCL3 promoter (site S5) and −174 to −169 bp of the CCL4 promoter (site S13).
To investigate the C/EBPβ and NF-κB function on CCL3 and CCL4 in detail, C/EBPβ and IKK2 (IKKβ) expression vectors were co-transfected with the −1395 bp CCL3 and −1281 bp CCL4 promoter constructs. IKK-2 is a protein subunit of IκB kinase complex (39, 40). Activated IKK-2 phosphorylates IκBα, which binds NF-κB and inhibits its function (40, 41). Phosphorylated IκBα undergoes proteasomal degradation, thus permitting nuclear translocation of NF-κB subunits, where they activate various genes involved in inflammation and other immune responses (40, 42).
The promoter activities of the −1395 bp CCL3 and −1281 bp CCL4 constructs were up-regulated in a dose-dependent manner with or without IL-1β stimulation after being transfected with C/EBPβ and IKK2, suggesting that C/EBPβ and IKK2 function as activators (Fig. 9B). Experiments were terminated at the peak of mRNA expression (i.e. 24 h for CCL3 and 8 h for CCL4). Consequently, CCL3 appears more responsive to C/EBPβ, whereas CCL4 is responsive to C/EBPβ and NF-κB.
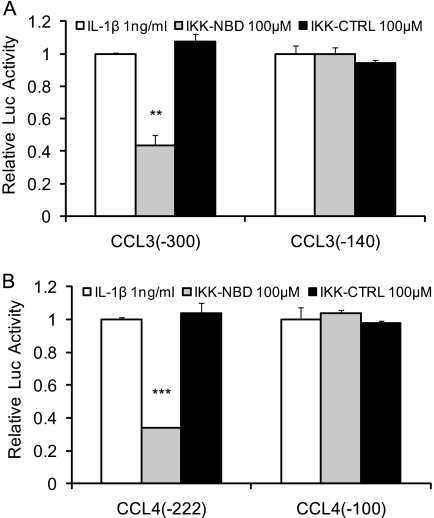
The involvement of NF-κB in the IL-1β-responsive elements was further confirmed by inhibiting its activity. The promoter activities were significantly decreased in the CCL3 (−300) and CCL4 (−222) by the IKK-NBD following 4 h of IL-1β treatment, whereas the control IKK-NBD had no significant effect, and there was no effect on the shorter promoter constructs (Fig. 10, A and B).
FIGURE 10.
Involvement of NF-κB in IL-1β-responsive elements of chemokines in human chondrocytes. The CCL3 (−300) (A), CCL3 (−140) (A), CCL4 (−222) (B), and CCL4 (−100) (B) promoter constructs were co-transfected into T/C-28a2 cells. Twenty-four h after transfection, T/C-28 cells were pretreated with vehicle (DMSO), IKK-NBD peptide (100 μm), or IKK-NBD control peptide (100 μm) for 1 h and then exposed to IL-1β (1 ng/ml) for 4 h. The p value of IKK-NBD (100 μm) was compared with IL-1β alone based on Student's t test (**, p < 0.01; ***, p < 0.001). Each bar represents the mean ± S.D. (error bars) of three independent experiments.
The effects of inhibition of C/EBPβ and NF-κB were tested by inhibition of each transcription factor separately and together. SB303580, an inhibitor of C/EBPβ via inhibition of p38MAPK (21, 43), and IKK-NBD reduced the expression of CCL3 and CCL4 with IL-1β stimulation (Fig. 11, A and B). IKK-NBD and SB303580 added together further decreased mRNA expression of CCL3 and CCL4 (Fig. 11A). Last, to confirm that both transcription factors are functional in transcription, promoter activity was determined after inhibition. The result showed that the promoter activities of CCL3 and CCL4 were significantly decreased by the inhibition of both factors (Fig. 11B).
FIGURE 11.
IKK-NBD and SB303580 (SB) co-enhance the inhibition of chemokines by human chondrocytes with IL-1β treatment. A, T/C-28 cells were pretreated with vehicle (DMSO), IKK-NBD peptide (100 μm), SB303580 (100 μm), or IKK-NBD control peptide (100 μm) for 1 h and then exposed to IL-1β (1 ng/ml) for 4 h. After IL-1β treatment, total RNA was isolated, and real-time quantitative PCR was performed. B, the CCL3 (−1395) and CCL4 (−1281) promoter constructs were transfected into T/C-28a2 cells and incubated for 24 h and then were pretreated with vehicle (DMSO), IKK-NBD peptide (100 μm), SB303580 (100 μm), and IKK-NBD control peptide (100 μm) for 1 h and exposed to IL-1β (1 ng/ml) for 4 h. Relative luciferase activity indicates the -fold expression relative to the activity of the construct co-transfected with empty vector (set as 1) in the presence of IL-1β (1 ng/ml). The p value of IKK-NBD (100 μm) or SB303580 (100 μm) was compared with IL-1β alone based on Student's t test (**, p < 0.01; ***, p < 0.001). Each bar represents the mean ± S.D. (error bars) of at least three independent experiments.
Immunohistochemistry of C/EBPβ and NF-κB
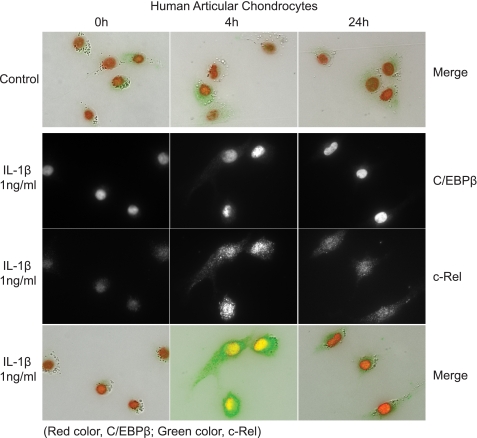
Our biochemical studies provided evidence that the regulation of CCL3 and CCL4 is coordinated in time with the NF-κB response early and transient and that the C/EBPβ response is early and sustained. To support this conclusion, we traced the location of these transcription factors over time by immunohistochemistry (Fig. 12). Fig. 12 shows double immunohistochemistry with antibodies to C/EBPβ and c-Rel after stimulation with IL-1β. At zero time, c-Rel was localized in the cytoplasm, and then it was localized in the nucleus at 4 h and was greatly reduced by 24 h. C/EBPβ was present at the beginning and was increased at 4 and 24 h.
FIGURE 12.
Subcellular localization of C/EBPβ and c-Rel in response to IL-1β. The top panel shows a merged image of immunohistochemistry for C/EBPβ and the NF-κB subunit c-Rel; C/EBPβ is present in the nucleus of cells even without IL-1β exposure, whereas c-Rel is located diffusely throughout the cell. With the addition of IL-1β, C/EBPβ is increased in the nucleus, and c-Rel is increased and translocates to the nucleus. At 24 h, C/EBPβ remains high in the nucleus, and c-Rel is significantly reduced.
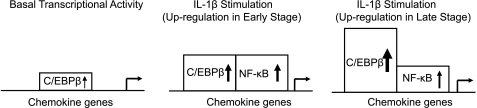
Our studies strongly suggest that both C/EBPβ and NF-κB are important for expression and response to IL-1β in human chondrocytes (Fig. 13). The transcription factors, C/EBPβ and NF-κB, are both involved in the up-regulation of chemokine genes in human chondrocytes in a time-ordered manner; NF-κB provides an initial up-regulation, and C/EBPβ sustains increased gene expression. Because C/EBPβ is present in untreated chondrocytes and can stimulate chemokine gene expression in the absence of IL-1β (13), we predict that C/EBPβ participates in basal regulation of these chemokine genes.
FIGURE 13.
A transcriptional model for the up-regulation of chemokine genes in human chondrocytes in response to IL-1β. Upon IL-1β stimulation, C/EBPβ protein and DNA binding are increased up to the 24 h tested. NF-κB showed little function without IL-1β but increased up to 8 h, after which the function was greatly reduced. The distribution of the transcription factors (see Fig. 12) is consistent with the functional parameters.
DISCUSSION
In this study, we demonstrate a critical role for C/EBPβ in the increased expression of chemokine genes in response to IL-1β in human chondrocytes. We studied in detail the regulation of two representative chemokines that are known to be increased in osteoarthritis and many other inflammatory conditions, CCL3 and CCL4. We also show the cooperative regulation of these genes by both C/EBPβ and NF-κB that is both time- and function-dependent. We conclude from these data that C/EBPβ is involved in constitutive regulation of chemokine genes and is also responsive to IL-1β. From this work on chemokine gene stimulation and our previous work on extracellular matrix gene suppression by C/EBPβ, we hypothesize that C/EBPβ is one of the most important regulators of gene activity in chondrocytes in response to IL-1β. In fact, C/EBPβ may be one of the pivotal regulators of chondrocyte function in that we have shown that C/EBPβ is responsible for the suppression of cartilage matrix genes in non-cartilaginous tissues (25), and Kawaguchi and co-workers (44) have shown that C/EBPβ is critical for chondrocyte hypertrophy, where it increases type X collagen and MMP-13 (matrix metalloproteinase 13), suppresses type II collagen synthesis, and activates p57kip2. This group also showed that removal of C/EBPβ protects against osteoarthritis.
In addition to C/EBPβ, we demonstrate a complementary role for the classic proinflammatory mediator, NF-κB. Like C/EBPβ, NF-κB activity is increased by IL-1β; however, in contrast to C/EBPβ, the increase in NF-κB activity is transient. Fig. 13 demonstrates the timing and functional activity of each transcription factor. Considering the time course of expression of CCL3 and the gradual increase of C/EBPβ binding to DNA, we suggest that C/EBPβ is involved in the up-regulation of chemokine genes over time, especially for the regulation of chemokine genes like CCL3, which are induced more slowly and where the mRNA was gradually and steadily increased, not reaching peak expression even in the 24-h observation period. In contrast, NF-κB appears to be involved in the up-regulation of chemokines in the early stage, with involvement gradually decreasing in the late stage. In CCL4, which was up-regulated by 4 h but then quickly decreased during the remaining time period, NF-κB is probably more important than C/EBPβ. In addition, the level of gene expression is regulated at both transcriptional and post-transcriptional levels in eukaryotic cells, including fibroblasts and chondrocytes (14, 45, 46). As Baltimore and colleagues have reported (14), the expression of genes like CCL4 is also highly regulated by mRNA stability.
We showed that the IL-1β-responsive elements of CCL3 and CCL4 promoters have C/EBPβ binding sites, and co-transfection with C/EBPβ could enhance the promoter activity. Within the IL-1β-responsive elements of these genes were also NF-κB-binding sites. Of note is that the increase in gene expression due to C/EBPβ was greater than that induced by the same concentration of IKK2. Because the NF-κB inhibitor, IKK-NBD, inhibited only about 30% of the CCL3 and CCL4 mRNA expression, C/EBPβ could be the more important regulator of the expression of chemokine genes in this cell type.
Co-regulation by NF-κB and C/EBPβ was recently shown in several genes in different cells (4, 21, 47, 48). Teti and colleagues (21) reported that Pseudomonas aeruginosa induced IL-8 (CXCL8) promoter expression and protein production in conjunctival epithelial cells by activating RelA and C/EBPβ and by promoting the cooperative binding of these transcription factors to the IL-8 promoter that in turn activates transcription. They also showed that C/EBPβ regulated the IL-8 basal transcriptional activity but not NF-κB, which is consistent with our data on the IL-1β effect in chondrocytes. Gil and colleagues (4) reported that IL-8, CXCL1, and CXCL5, which are CXCR2-binding chemokines, are up-regulated during replicative and oncogene-induced senescence. They demonstrated a function of NF-κB and C/EBPβ in controlling the secretory program associated with cell senescence, although they did not investigate the mechanism.
Studies have shown that the RelA (p65) subunit of NF-κB is involved in regulation of some lipopolysaccharide (LPS)-stimulated chemokines in human monocytic cells (17, 18). However, we show in chondrocytes that, of the NF-κB subunits, c-Rel mRNA was increased more than p65 with IL-1β stimulation, and the computational analysis in chemokine genes CCL3 and CCL4 showed that c-Rel binding sites were predicted in the IL-1β-responsive elements. Ohashi and colleagues (49) recently reported that c-Rel was an important transcription factor in the regulation of the induction of proinflammatory cytokines; therefore, c-Rel would be an interesting target for further investigation.
Additional transcription factors have been shown to regulate CCL3 and CCL4. In multiple myeloma, AML-1A (acute myeloid leukemia 1A) and AML-1B (also known as Runx1 (Runt-related transcription factor 1)) are principle regulators of CCL3 (26), although C/EBPβ was later found to be important (50). In our experiments, Runx1 transfection did not increase gene expression.3 C/EBPβ is thought to be responsible for regulating growth, proliferation, and antiapoptotic responses by regulating the expression of other key transcription factors. In an early publication, Grove and Plumb (20) demonstrated that C/EBP, NF-κB, and cEts family members were important in the LPS-induced stimulation of CCL3.
Levels of the proinflammatory cytokine IL-1β are increased in synovial fluid in joint diseases, such as OA and rheumatoid arthritis, and can induce cartilage damage and bone resorption. Chemokines function in the recruitment of neutrophils, monocytes, immature dendritic cells, B cells, and activated T cells (51). However, chemokines also have functions at the individual tissue level as it has been reported that the CXC family of chemokines are important in the regulation of angiogenesis, and CCL2, CCL3, and CCR2 stimulate osteoclastogenesis (52–54). In summary, the data presented here show that chondrocytes react in a cell-specific manner to IL-1β, utilizing C/EBPβ and NF-κB in a combinatorial regulation of chemokine gene expression. The activity of C/EBPβ is augmented by a transient increase in activity of NF-κB, and both transcription factors act independently on the chemokine genes, CCL3 and CCL4. These studies provide the foundation for the control of chemokine gene expression in chronic joint disease.
Supplementary Material
Acknowledgments
We thank Drs. John C. Clohisy, Robert L. Barrack, Douglas McDonald, Ryan Nunley, and Rick W. Wright and Head Nurse Keith Foreman for normal cartilage and cartilage from patients with osteoarthritis. We also thank Drs. G. David Roodman and Sheau-Farn Yeh for the generous gifts of CCL3 and CCL4 plasmids. We also thank Drs. Xiaoyun Xing, Chikashi Kobayashi, Yousef Abu-Amer, and Audrey McAlinden at the Washington University School of Medicine for valuable assistance.
This work was supported, in whole or in part, by National Institutes of Health, NIAMS, Grants R01 AR050847 and R01 AR036994, R01 65554 and ARRA Supplement, and Center for Musculoskeletal Research P30 AR057235.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Z. Zhang and L. J. Sandell, unpublished results.
- OA
- osteoarthritis
- C/EBPβ
- CCAAT/enhancer-binding protein
- IKK-NBD
- cell-permeable NEMO binding domain
- LAP
- liver-enriched transcriptional activator proteins
- LIP
- liver-enriched inhibitory protein
- IP
- immunoprecipitation.
REFERENCES
- 1.Gerard C., Rollins B. J. (2001) Nat. Immunol. 2, 108–115 [DOI] [PubMed] [Google Scholar]
- 2.Haringman J. J., Ludikhuize J., Tak P. P. (2004) Ann. Rheum. Dis. 63, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Q., Jones D., Borghesani P. R., Segal R. A., Nagasawa T., Kishimoto T., Bronson R. T., Springer T. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9448–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acosta J. C., O'Loghlen A., Banito A., Guijarro M. V., Augert A., Raguz S., Fumagalli M., Da Costa M., Brown C., Popov N., Takatsu Y., Melamed J., d'Adda di Fagagna F., Bernard D., Hernando E., Gil J. (2008) Cell 133, 1006–1018 [DOI] [PubMed] [Google Scholar]
- 5.Han J. H., Choi S. J., Kurihara N., Koide M., Oba Y., Roodman G. D. (2001) Blood 97, 3349–3353 [DOI] [PubMed] [Google Scholar]
- 6.Sandell L. J., Xing X., Franz C., Davies S., Chang L. W., Patra D. (2008) Osteoarthr. Cartil. 16, 1560–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamasaki K., Nakasa T., Miyaki S., Ishikawa M., Deie M., Adachi N., Yasunaga Y., Asahara H., Ochi M. (2009) Arthritis Rheum. 60, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyaki S., Nakasa T., Otsuki S., Grogan S. P., Higashiyama R., Inoue A., Kato Y., Sato T., Lotz M. K., Asahara H. (2009) Arthritis Rheum. 60, 2723–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukui N., Zhu Y., Maloney W. J., Clohisy J., Sandell L. J. (2003) J. Bone Joint Surg. Am. 85, Suppl. 3, 59–66 [DOI] [PubMed] [Google Scholar]
- 10.Martel-Pelletier J., Alaaeddine N., Pelletier J. P. (1999) Front. Biosci. 4, D694–D703 [DOI] [PubMed] [Google Scholar]
- 11.Tiku K., Thakker-Varia S., Ramachandrula A., Tiku M. L. (1992) Cell. Immunol. 140, 1–20 [DOI] [PubMed] [Google Scholar]
- 12.Lee J. H., Ort T., Ma K., Picha K., Carton J., Marsters P. A., Lohmander L. S., Baribaud F., Song X. Y., Blake S. (2009) Osteoarthr. Cartil. 17, 613–620 [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z., Xing X., Hensley G., Chang L. W., Liao W., Abu-Amer Y., Sandell L. J. (2010) Arthritis Rheum. 162, 1193–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao S., Baltimore D. (2009) Nat. Immunol. 10, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulai J. I., Chen H., Im H. J., Kumar S., Hanning C., Hegde P. S., Loeser R. F. (2005) J. Immunol. 174, 5781–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezzonico R., Imbert V., Chicheportiche R., Dayer J. M. (2001) Blood 97, 2932–2940 [DOI] [PubMed] [Google Scholar]
- 17.Lakshmanan U., Porter A. G. (2007) J. Immunol. 179, 8480–8490 [DOI] [PubMed] [Google Scholar]
- 18.Lim C. A., Yao F., Wong J. J., George J., Xu H., Chiu K. P., Sung W. K., Lipovich L., Vega V. B., Chen J., Shahab A., Zhao X. D., Hibberd M., Wei C. L., Lim B., Ng H. H., Ruan Y., Chin K. C. (2007) Mol. Cell 27, 622–635 [DOI] [PubMed] [Google Scholar]
- 19.Amos N., Lauder S., Evans A., Feldmann M., Bondeson J. (2006) Rheumatology 45, 1201–1209 [DOI] [PubMed] [Google Scholar]
- 20.Grove M., Plumb M. (1993) Mol. Cell. Biol. 13, 5276–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venza I., Cucinotta M., Visalli M., De Grazia G., Oliva S., Teti D. (2009) J. Biol. Chem. 284, 4191–4199 [DOI] [PubMed] [Google Scholar]
- 22.Okazaki K., Li J., Yu H., Fukui N., Sandell L. J. (2002) J. Biol. Chem. 277, 31526–31533 [DOI] [PubMed] [Google Scholar]
- 23.Imamura T., Imamura C., Iwamoto Y., Sandell L. J. (2005) J. Biol. Chem. 280, 16625–16634 [DOI] [PubMed] [Google Scholar]
- 24.Imamura T., Imamura C., McAlinden A., Davies S. R., Iwamoto Y., Sandell L. J. (2008) Arthritis Rheum. 58, 1366–1376 [DOI] [PubMed] [Google Scholar]
- 25.Okazaki K., Yu H., Davies S. R., Imamura T., Sandell L. J. (2006) J. Cell. Biochem. 97, 857–868 [DOI] [PubMed] [Google Scholar]
- 26.Choi S. J., Oba T., Callander N. S., Jelinek D. F., Roodman G. D. (2003) Blood 101, 3778–3783 [DOI] [PubMed] [Google Scholar]
- 27.Tseng P. C., Hsu H. C., Janmanchi D., Lin C. H., Kuo Y. H., Chou C. K., Yeh S. F. (2008) Biochem. Pharmacol. 76, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 28.Otero J. E., Dai S., Alhawagri M. A., Darwech I., Abu-Amer Y. (2010) J. Bone Miner. Res. 25, 1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y., Crouch E. (2002) J. Biol. Chem. 277, 19530–19537 [DOI] [PubMed] [Google Scholar]
- 30.McAlinden A., Liang L., Mukudai Y., Imamura T., Sandell L. J. (2007) J. Biol. Chem. 282, 24444–24454 [DOI] [PubMed] [Google Scholar]
- 31.Lu K. T., Sinquett F. L., Dryer R. L., Song C., Covey L. R. (2006) Blood 108, 3769–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anest V., Cogswell P. C., Baldwin A. S., Jr. (2004) J. Biol. Chem. 279, 31183–31189 [DOI] [PubMed] [Google Scholar]
- 33.Kristiansson E., Thorsen M., Tamás M. J., Nerman O. (2009) Mol. Biol. Evol. 26, 1299–1307 [DOI] [PubMed] [Google Scholar]
- 34.Descombes P., Schibler U. (1991) Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 35.Poli V. (1998) J. Biol. Chem. 273, 29279–29282 [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y., Saunders M. A., Yeh H., Deng W. G., Wu K. K. (2002) J. Biol. Chem. 277, 6923–6928 [DOI] [PubMed] [Google Scholar]
- 37.Chang L. W., Nagarajan R., Magee J. A., Milbrandt J., Stormo G. D. (2006) Genome Res. 16, 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies S. R., Chang L. W., Patra D., Xing X., Posey K., Hecht J., Stormo G. D., Sandell L. J. (2007) Genome Res. 17, 1438–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zandi E., Rothwarf D. M., Delhase M., Hayakawa M., Karin M. (1997) Cell 91, 243–252 [DOI] [PubMed] [Google Scholar]
- 40.Häcker H., Karin M. (2006) Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 41.DiDonato J. A., Hayakawa M., Rothwarf D. M., Zandi E., Karin M. (1997) Nature 388, 548–554 [DOI] [PubMed] [Google Scholar]
- 42.Rothwarf D. M., Karin M. (1999) Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 43.Basak C., Pathak S. K., Bhattacharyya A., Mandal D., Pathak S., Kundu M. (2005) J. Biol. Chem. 280, 4279–4288 [DOI] [PubMed] [Google Scholar]
- 44.Hirata M., Kugimiya F., Fukai A., Ohba S., Kawamura N., Ogasawara T., Kawasaki Y., Saito T., Yano F., Ikeda T., Nakamura K., Chung U. I., Kawaguchi H. (2009) PLoS One 4, e4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukui N., Ikeda Y., Ohnuki T., Hikita A., Tanaka S., Yamane S., Suzuki R., Sandell L. J., Ochi T. (2006) J. Biol. Chem. 281, 27229–27241 [DOI] [PubMed] [Google Scholar]
- 46.Tebo J. M., Datta S., Kishore R., Kolosov M., Major J. A., Ohmori Y., Hamilton T. A. (2000) J. Biol. Chem. 275, 12987–12993 [DOI] [PubMed] [Google Scholar]
- 47.El-Asmar B., Giner X. C., Tremblay J. J. (2009) J. Mol. Endocrinol. 42, 131–138 [DOI] [PubMed] [Google Scholar]
- 48.Sow F. B., Alvarez G. R., Gross R. P., Satoskar A. R., Schlesinger L. S., Zwilling B. S., Lafuse W. P. (2009) J. Leukocyte Biol. 86, 1247–1258 [DOI] [PubMed] [Google Scholar]
- 49.Lu Y. C., Kim I., Lye E., Shen F., Suzuki N., Suzuki S., Gerondakis S., Akira S., Gaffen S. L., Yeh W. C., Ohashi P. S. (2009) J. Immunol. 182, 7212–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal R., Janz M., Galson D. L., Gries M., Li S., Jöhrens K., Anagnostopoulos I., Dörken B., Mapara M. Y., Borghesi L., Kardava L., Roodman G. D., Milcarek C., Lentzsch S. (2009) Blood 114, 3890–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borzì R. M., Mazzetti I., Cattini L., Uguccioni M., Baggiolini M., Facchini A. (2000) Arthritis Rheum. 43, 1734–1741 [DOI] [PubMed] [Google Scholar]
- 52.Binder N. B., Niederreiter B., Hoffmann O., Stange R., Pap T., Stulnig T. M., Mack M., Erben R. G., Smolen J. S., Redlich K. (2009) Nat. Med. 15, 417–424 [DOI] [PubMed] [Google Scholar]
- 53.Miyamoto K., Ninomiya K., Sonoda K. H., Miyauchi Y., Hoshi H., Iwasaki R., Miyamoto H., Yoshida S., Sato Y., Morioka H., Chiba K., Egashira K., Suda T., Toyama Y., Miyamoto T. (2009) Biochem. Biophys. Res. Commun. 383, 373–377 [DOI] [PubMed] [Google Scholar]
- 54.Rudolph E. H., Woods J. M. (2005) Curr. Pharm. Des. 11, 613–631 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.