Abstract
Eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) protect against cardiovascular disease by largely unknown mechanisms. We tested the hypothesis that EPA and DHA may compete with arachidonic acid (AA) for the conversion by cytochrome P450 (CYP) enzymes, resulting in the formation of alternative, physiologically active, metabolites. Renal and hepatic microsomes, as well as various CYP isoforms, displayed equal or elevated activities when metabolizing EPA or DHA instead of AA. CYP2C/2J isoforms converting AA to epoxyeicosatrienoic acids (EETs) preferentially epoxidized the ω-3 double bond and thereby produced 17,18-epoxyeicosatetraenoic (17,18-EEQ) and 19,20-epoxydocosapentaenoic acid (19,20-EDP) from EPA and DHA. We found that these ω-3 epoxides are highly active as antiarrhythmic agents, suppressing the Ca2+-induced increased rate of spontaneous beating of neonatal rat cardiomyocytes, at low nanomolar concentrations. CYP4A/4F isoforms ω-hydroxylating AA were less regioselective toward EPA and DHA, catalyzing predominantly ω- and ω minus 1 hydroxylation. Rats given dietary EPA/DHA supplementation exhibited substantial replacement of AA by EPA and DHA in membrane phospholipids in plasma, heart, kidney, liver, lung, and pancreas, with less pronounced changes in the brain. The changes in fatty acids were accompanied by concomitant changes in endogenous CYP metabolite profiles (e.g. altering the EET/EEQ/EDP ratio from 87:0:13 to 27:18:55 in the heart). These results demonstrate that CYP enzymes efficiently convert EPA and DHA to novel epoxy and hydroxy metabolites that could mediate some of the beneficial cardiovascular effects of dietary ω-3 fatty acids.
Keywords: Arachidonic Acid, Cytochrome P450, Eicosanoids Biosynthesis, Eicosapentaenoic Acid (EPA), Epoxygenase Pathway, Docosahexaenoic Acid, Hydroxylase
Introduction
Cytochrome P450 (CYP)3 enzymes initiate the so-called “third branch” of the arachidonic acid (AA; 20:4n-6) cascade (1). Members of the CYP2C and CYP2J subfamilies function as AA epoxygenases and produce isoform-specific sets of regio- and stereoisomeric epoxyeicosatrienoic acids (EETs) (2). In contrast, CYP4A and CYP4F isoforms are AA ω/(ω-1)-hydroxylases and produce 20-hydroxyeicosatetraenoic acid (20-HETE) as the main metabolite (3). EETs and 20-HETE serve as second messengers of various hormones and growth factors and play partially opposing roles in the regulation of vascular, renal, and cardiac function (4–9). Animal models and initial human studies revealed that alterations in CYP-dependent AA metabolism are associated with the development of hypertension and target organ damage (10–17).
Numerous studies demonstrated that diets rich in long-chain ω-3 polyunsaturated fatty acids (n-3 PUFA) protect against the development of cardiovascular disease (18–23). In particular, eicosapentaenoic acid (EPA, 20:5n-3) and docosahexaenoic acid (DHA, 22:6n-3), the major n-3 PUFAs contained in fish oil, have anti-inflammatory, anti-thrombotic, vasodilatory, hypolipidemic, and anti-arrhythmic properties and thus exert pleiotropic beneficial effects on cardiovascular function (18). The molecular mechanisms of n-3 PUFA action are only partially understood and include changes in membrane structures and gene expression, direct interactions with ion channels, and alterations in eicosanoid biosynthesis (24–29). EPA and DHA compete with AA for binding and conversion by cyclooxygenases and lipoxygenases and thus modulate the production and bioactivity of prostanoids and leukotrienes (30, 31). These interactions not only result in a partial replacement of the classical AA-derived eicosanoids by their less potent n-3 counterparts but also produce novel lipid mediators termed “resolvins” and “protectins” that have highly potent anti-inflammatory and pro-resolution properties (29). More recently, it became evident from several in vitro studies that n-3 PUFAs may also interfere with the CYP-branch of eicosanoid production (32–37). The principal CYP-dependent metabolites derived from EPA include five regioisomeric epoxyeicosatetraenoic acids (EEQs) and ω/(ω-1)-hydroxyeicosapentaenoic acids (20- and 19-HEPE), whereas DHA can be metabolized to six regioisomeric epoxydocosapentaenoic acids (EDPs) and ω/(ω-1)-hydroxydocosahexaenoic acids (22- and 21-HDoHE); see supplemental Fig. 1 (38, 39).
Considering the pivotal physiological and pathophysiological roles of the AA-derived EETs and 20-HETE, we hypothesize that their replacement by EPA- and DHA-derived epoxy and hydroxy metabolites may provide a novel mechanism of how dietary n-3 PUFAs influence cardiovascular function. However, to what extent such a replacement can indeed occur under in vivo conditions is unknown and may depend on both the relative availability of AA, EPA, and DHA and the substrate specificity of the individual CYP isoforms expressed in a given tissue. To address these questions, we have now analyzed the substrate specificity of major AA-metabolizing CYP isoforms from humans, rats, and mice; compared renal and hepatic microsomes for their capacity to metabolize AA, EPA, and DHA; and studied for the first time the effect of dietary EPA/DHA supplementation on the endogenous CYP-eicosanoid profile in different organs and tissues of rats.
EXPERIMENTAL PROCEDURES
Cloning and Expression of CYP Isoforms
CYP2C8, CYP2C9, CYP2C11, CYP2C23, Cyp4a12a, and Cyp4a12b were cloned and co-expressed with the human NADPH-CYP reductase (CPR) in a baculovirus/Sf9 insect cell system, and microsomes containing the recombinant CYP enzymes were prepared as described previously (33, 40). The CYP concentrations were estimated by means of CO difference spectra using a molar absorption coefficient of 91 mm−1 cm−1 (41). CPR activities were assayed in 50 mm Tris/HCl buffer (pH 7.5) containing 0.1 mm EDTA, 0.05 mm cytochrome c, 0.1 mm NADPH, and 2.2 mm KCN at 25 °C, using a molar absorption coefficient of 21 mm−1 cm−1 at 550 nm. For calculation of CPR concentration, we assumed that 4.5 units (μmol of cytochrome c/min) correspond to 1 nmol of CPR based on the specific activity of the purified 79-kDa enzyme (60 units/mg). The microsomes used for comparing the rates of AA, EPA, docosapentaenoic acid (DPA), and DHA metabolism contained CYP and CPR in a molar ratio of 1:0.8 (CYP2C8), 1:0.7 (CYP2C9), 1:0.7 (CYP2C11), 1:1.4 (CYP2C23), 1:1.4 (Cyp4a12a), and 1:1.2 (Cyp4a12b). The metabolite pattern produced by each isoform was analyzed with at least three independent microsomal preparations having similar CYP/CPR ratios. CYP2C19, CYP2J2, CYP2E1, CYP4A11, and CYP4F2 were purchased as supersomes from Gentest (BD Biosciences), which contained coexpressed human CPR and in the case of CYP4F2 additionally cytochrome b5.
Preparation of Hepatic and Renal Microsomes
Hepatic and renal microsomes from mice and rats were prepared as described previously (40, 42) using freshly dissected organs of 12–15-week-old male NMRI mice (Charles River Laboratories) and 7–8-week-old male Sprague-Dawley rats (Tierzucht, Schönwalde, Germany), respectively.
In Vitro Fatty Acid Metabolism
1-14C-Labeled AA, EPA, DPA (22:5n-3), and DHA with specific radioactivities of 53–55 mCi/mmol and radiochemical purities >99% were purchased from Hartmann Analytic GmbH (Braunschweig, Germany). Standard reactions with the recombinant CYP enzymes were performed in 100 μl of 100 mm potassium phosphate buffer (pH 7.2) containing 10 (CYP2C8, CYP2C9, CYP2C11, CYP2C23, CYP2E1, Cyp4a12a, and Cyp4a12b), 5 (CYP2C19), or 2 pmol of CYP (CYP4A11 and CYP4F2) and the respective substrate at a concentration of 10 μm. Before composing the reaction mixtures, Cyp4a12a, Cyp4a12b, and CYP4A11 were supplemented with cytochrome b5 in a 1:1 molar ratio by incubating the corresponding microsomes with highly purified cytochrome b5 (from Calbiochem) in a total volume of 15 μl for 10 min on ice. The recombinant enzymes were preincubated with the substrates for 10 min at 37 °C. Reactions were started with NADPH (final concentration of 1 mm) and terminated after 10 min with all CYP isoforms except CYP2C11 and CYP2C19 (after 5 min) and CYP2E1 (after 15 min) by adding 5 μl of 0.4 m citric acid. Control experiments included (i) omission of NADPH from the reaction mixtures and (ii) use of microsomes obtained after infecting the insect cells with an empty baculovirus. Both types of control reactions did not yield any detectable amounts of products (data not shown). Reaction products were extracted into ethyl acetate, evaporated under nitrogen, and resuspended in ethanol. The enzymatic activities of CYP2J2 were determined under the same conditions, using, however, 50 pmol of CYP, a reaction volume of 200 μl, and the substrates at a concentration of 40 μm. For the analysis of regioselectivities, the standard reactions were scaled up 4–10-fold. The substrate specificity of renal and hepatic microsomes was analyzed under the same conditions, using, however, a reaction time of 20 min and 800 μg of microsomal protein in 100 μl of 100 mm potassium phosphate buffer (pH 7.2) supplemented with 1 mm EDTA and 50 nm FAD/FMN.
HPLC Separation of Metabolites
Extracted reaction samples were first resolved by reversed phase high performance liquid chromatography (RP-HPLC; Shimadzu LC 10 Avp coupled to an online radioflow detector LB 509; Berthold), using a Nucleosil 100-5C18 HD column (250 × 4 mm; Macherey-Nagel) and a linear solvent gradient ranging from acetonitrile/water/acetic acid (50:50:0.1; v/v/v) to acetonitrile/acetic acid (100:0.1) over 40 min at a flow rate of 1 ml/min. This procedure separated the main product classes (dihydroxy, (ω-1)/ω-hydroxy, and epoxy metabolites (for representative examples, see supplemental Fig. 2). Hydroxylase activities were determined as the sum of the ω- and (ω-1)-hydroxy products formed/min and nmol of recombinant CYP or mg of microsomal protein. Total epoxygenase activities were calculated from the sum of epoxy and corresponding dihydroxy products.
The (ω-1)/ω-hydroxy metabolites of AA, EPA, DPA, and DHA eluting with retention times of about 16.1 (19-/20-HETE), 14.2 (19-/20-HEPE), 15.6 (21-/22-OH-DPA), and 14.8 min (21-/22-HDoHE) from RP-HPLC were collected and further resolved by subsequent normal phase (NP)-HPLC essentially as described previously (32, 43). NP-HPLC was done on a Nucleosil 100–5 column (250 × 4 mm; from Macherey-Nagel) using a linear gradient ranging from hexane/2-propanol/acetic acid (99:1:0.1, v/v/v) to hexane/2-propanol/acetic acid (98.3:1.7:0.1, v/v/v) over 40 min at a flow rate of 1 ml/min. Under these conditions, the (ω-1)-hydroxy products eluted first and were clearly separated from the ω-hydroxy metabolites: 19-/20-HETE at 20/28 min, 19-/20-HEPE at 19/26 min, 21-OH-/22-OH-DPA at 16/23 min, and 21-/22-DHoHE at 15/21 min (for representative examples, see supplemental Fig. 2).
The epoxy metabolites eluted from RP-HPLC as follows: AA-derived EETs, 14,15-EET (23.4 min), 11,12-EET (24.7 min), and 8,9-EET (25.3 min); EPA-derived EEQs, 17,18-EEQ (19.6 min) and other regioisomeric EEQs (21–23 min); DPA-derived epoxydocosatetraenoic acids, 19,20-epoxydocosatetraenoic acid (24.4 min) and other regioisomeric epoxydocosatetraenoic acids (25.6–27.5 min); DHA-derived EDPs, 19,20-EDP (23.0 min) and other regioisomeric EDPs (25.2–27 min). The regioisomeric ω-6 to ω-12 epoxyproducts, which eluted largely unresolved from RP-HPLC, were collected and further resolved by NP-HPLC on a Nucleosil 100-5 column (250 × 4 mm; from Macherey-Nagel) with an isocratic solvent system of hexane/isopropyl alcohol/acetic acid (100:0.3:0.1, v/v/v) over 40 min at a flow rate of 1.5 ml/min essentially as described previously (33, 43) (for representative examples, see supplemental Fig. 2). The EPA-derived epoxides eluted in the order 14,15-, 11,12-, and 8,9-EEQ at 14.5, 16.5, and 21.5 min; the DPA-derived epoxides in the order 16,17-, 13,14-, 10,11-, and 7,8-EDQ at 21.7, 23.2, 26.2, and 29.0 min; and the DHA-derived epoxides in the order 16,17-, 13,14-, 10,11-, and 7,8-EDP at 17.4, 19.5, 23.2, and 29.7 min. 5,6-EET, 5,6-EEQ, and 4,5-EDP were not detectable as products of the in vitro reactions, probably because of the intrinsic instability of these metabolites due to lactone formation.
Feeding Study with Rats
Male Sprague-Dawley rats received two types of diet (n = 6/group) starting at an age of 4 weeks. The standard chow (EF R/M; SNIFF Spezialitäten GmbH, Soest, Germany) was supplemented with either 5% (w/w) sunflower oil containing α-linolenic acid (ALA; 18:3n-3) as the only source of n-3 PUFAs (0.03% of total chow corresponding to 0.6% of total fatty acids) or additionally with 2.5% (w/w) OMACOR® oil consisting of pure EPA- and DHA-ethylesters (480 mg of EPA + 360 mg of DHA per ml; Pronova BioPharma (Sandefjord, Norway) and Solvay Arzneimittel GmbH (Hannover, Germany)). For the detailed composition of the diets used, see supplemental Table 1. The animals were allowed free access to the chow and drinking water ad libitum. After 3 weeks of feeding, the animals were sacrificed. Harvested organs, plasma samples, and pelleted and washed red blood cells were snap-frozen in liquid nitrogen and stored at −80 °C. For further analysis, organs were pulverized in liquid nitrogen using a Biopulverizer (Biospec Products).
Determination of Fatty Acid Profiles
Fatty acid compositions were analyzed according to the HS-Omega-3 Index® methodology as described previously (44). Fatty acid methyl esters were generated by acid transesterification and analyzed by gas chromatography using a GC2010 Gas Chromatograph (Shimadzu, Duisburg, Germany) equipped with a SP2560, 100-m column (Supelco, Bellefonte, PA) using hydrogen as carrier gas. Fatty acids were identified by comparison with a standard mixture of fatty acids characteristic of erythrocytes. The ω-3 index is given as EPA plus DHA expressed as a percentage of total identified fatty acids after response factor correction. The coefficient of variation for EPA plus DHA was 5%. Analyses were quality-controlled according to DIN ISO 15189.
Sample Preparation for the Analysis of the Endogenous CYP-eicosanoid Profiles
30-mg aliquots of homogenized tissue (or pelleted red blood cells) were mixed with 0.5 ml of distilled water, 0.5 ml of methanol, and 0.1 ml of internal standard containing 10 ng of 20-HETE-d6 and 10 ng of 14,15-EET-d8 in acetonitrile; the deuterated compounds were purchased from Cayman Chemicals and Biomol/Enzo Life Sciences Inc., respectively. The samples were then hydrolyzed after adding 300 μl of 10 m sodium hydroxide for 20 min at 60 °C. The solution was neutralized by adding 300 μl of 60% acetic acid and 2 ml of 1 m sodium acetate buffer, pH 6.0. The pH was controlled and if necessary adjusted to pH 6.0 by adding small volumes of either 0.1 m NaOH or 10% acetic acid. The hydrolyzed sample was cleared by centrifugation, and the metabolites were extracted from the supernatant as described below. Essentially the same protocol was used for plasma samples; however, we started with 200 μl of plasma and added only 100 μl of 10 m NaOH for alkaline hydrolysis. Subsequent solid-phase extraction was performed using Varian Bond Elut Certify II columns as described previously (45). The solid-phase extraction columns were preconditioned with 2 ml of methanol, followed by 2 ml of 0.1 m sodium acetate solution (pH 7) containing 5% methanol before application of the hydrolyzed samples. The columns were washed with 2 ml of methanol/water (1:1, v/v), and the CYP-metabolites and internal standards were eluted with 2 ml of hexane/ethyl acetate (75:25, v/v) containing 1% acetic acid. The eluate was evaporated to dryness on a heating block at 40 °C under a stream of nitrogen. Residues were then dissolved in 50 μl of acetonitrile and applied to LC-MS/MS as described below.
HPLC and MS Conditions for the Analysis of the CYP-eicosanoid Profile
20-μl samples were analyzed with an HPLC system of the Agilent 1200 series equipped with binary pump, autosampler, and column thermostat on a high resolution column (Zorbax Eclipse Plus-C18, 4.6 × 150 mm, 1.8 μm) using acetonitrile, 0.01 m ammonia acetate as the solvent system. Gradient elution was started with 30% acetonitrile. Acetonitrile was increased within 10 min to 90% and held for a further 10 min with a flow rate of 0.8 ml/min. The HPLC was coupled to the Agilent 6410 Triplequad mass spectrometer with electrospray ionization source. Analysis was performed with multiple reaction monitoring in negative mode, a gas temperature of 350 °C, a nitrogen stream of 12 liters/min, and a capillary voltage of 4000 V. Fragmentor voltage and collision energy were optimized for each individual metabolite using authentic standard compounds. The multiple reaction monitoring and MS conditions are shown in supplemental Table 2.
Authentic Standard Compounds
5,6-EET, 8,9-EET, 11,12-EET, 14,15-EET, 14,15-EEQ, 17,18-EEQ, 16,17-EDP, and 19,20-EDP as well as 20-HETE, EPA, DPA, and DHA were purchased from Cayman Chemicals. Other regioisomeric monoepoxides were synthesized by reacting the parental polyunsaturated fatty acid, supplemented with trace amounts of the corresponding 1-14C-labeled compound, with m-chloroperoxybenzoic acid as described for the chemical oxidation of AA to EETs (46). The reaction products were resolved and purified by RP- and subsequent NP-HPLC as described in detail above. The identity of the regioisomeric monoepoxides was confirmed by gas chromatography-mass spectrometry (GC-MS) after preparing the corresponding dihydroxy compounds and converting them into the trimethylsilylether-methylester derivatives as described previously (32, 33, 38). The GC-MS-electron impact spectra of these derivatives showed the characteristic fragments of α-cleavage products to -OSiMe3 groups at m/z 363, 233, and 131 (17,18-EEQ); 323, 273, and 171 (14,15-EEQ); 313, 283, and 211 (11,12-EEQ); 353, 251, and 243 (8,9-EEQ); 393, 291, and 203 (5,6-EEQ); 131, 233, and 391 (19,20-EDQ); 171, 273, and 351 (16,17-EDQ); 211, 311, and 313 (13,14-EDQ); 251, 271, and 353 (10,11-EDQ); 231, 291, and 393 (7,8-EDQ); 131, 389, and 491 (19,20-EDP); 171, 349, and 451 (16,17-EDP); 211, 309, and 411 (13,14-EDP); 251, 269, and 371 (10,11-EDP); 229, 291, and 331 (7,8-EDP); and 189, 291, and 331 (4,5-EDP).
Recombinant Cyp4a12a was used to produce the ω- and (ω-1)-hydroxy derivatives of AA, EPA, and DHA required as standard compounds for NP-HPLC and LC-MS/MS analysis. The standard reaction described above was scaled up to 4 ml, and the specific radioactivity of the substrates was adjusted to 2 mCi/mmol by adding appropriate amounts of the corresponding unlabeled polyunsaturated fatty acid. The reaction products were resolved by RP-HPLC and subsequent NP-HPLC as described above. The identity of the hydroxy metabolites was confirmed by MS based on their collision-induced dissociation spectra. 19- and 20-HETE showed the expected molecular ions at m/z 319, yielded common product ions at m/z 301 (loss of H2O) and 275 (loss of CO2), and characteristic fragments at m/z 231 (combined loss of CO2 and CH3CH2OH from 19-HETE) or 245 (combined loss of CO2 and CH2OH from 20-HETE). 19- and 20-HEPE showed analogous collision-induced dissociation spectra with peaks at m/z 317 (molecular ion), 299 (loss of H2O), and 277 (loss of CO2) and characteristic fragments at m/z 229 (loss of CO2 and CH3CH2OH from 19-HEPE) or 243 (loss of CO2 and CH2OH from 20-HEPE). The collision-induced dissociation spectra of 21- and 22-DHoHE (molecular ions at m/z 343) displayed common fragment ions at m/z 325 (loss of H2O) and 299 (loss of CO2) and characteristic fragments at m/z 255 (loss of CO2 and CH3CH2OH from 21-HDoHE) or 269 (loss of CO2 and CH2OH from 22-HDoHE).
Bioassay
Biological activities of CYP-dependent metabolites were determined using a bioassay essentially as described previously (47). Briefly, neonatal rat cardiomyocytes (NRCMs) were isolated from 1–3-day-old Wistar rats and cultured as monolayers on the bottom of Falcon flasks (12.5 cm) in 2.0 ml of Halle SM 20-I medium supplemented with 10% heat-inactivated fetal calf serum and 2 μm fluorodeoxyuridine. Spontaneously beating cell clusters occurred after 4–6 days (120–150 beats/min, monitored at 37 °C using an inverted microscope). The beating rates were determined for six individual clusters before and 5 min after the addition of the metabolites. Based upon the difference between the basal and compound-induced beating rate of the individual clusters, the chronotropic effects (changes in beats/min) were calculated and are given as mean ± S.E. values; n = 18–24 clusters originating from at least three independent NRCM cultures. All metabolites were prepared as 1000-fold stock solutions in ethanol and tested at a final concentration of 30 nm. EPA required a final concentration of 3.3 μm and 30 min of preincubation. The enantiomers of 17,18-EEQ and 19,20-EDP were prepared by chiral phase HPLC on a Chiralcel OB column (250 × 4.6 mm; Daicel) using hexane containing 0.3% isopropyl alcohol and 0.05% acetic acid as solvent (48). Authentic samples of (17R,18S)- and (17S,18R)-EEQ were produced using recombinant CYP1A1 and CYP102, respectively (34). It was assumed that the 19,20-EDP enantiomers elute in the same order as the 17,18-EEQ enantiomers; however, authentic standards were not available to verify their identities. 17,18-dihydroxyeicosatetraenoic acid was prepared by acetic acid-mediated hydrolysis of 17,18-EEQ, purified by reversed phase HPLC, and identified by GS-MS as described previously (32).
Statistical Analysis
All data derived from the animal experiments are presented as means ± S.E. and were analyzed by analysis of variance followed by the Tukey-Kramer multiple comparison test (GraphPad Software Inc.). p < 0.05 was considered statistically significant.
RESULTS
Metabolism of AA, EPA, DPA, and DHA by Recombinant CYP Isoforms
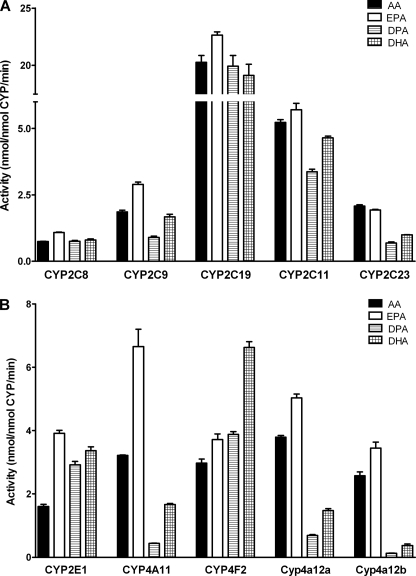
All CYP isoforms tested metabolized AA with reasonably high specific activities (Fig. 1) and thereby displayed their well known individual reaction specificities and regioselectivities (Tables 1 and 2). EPA was metabolized by the same CYP isoforms with at least equal or in part even significantly higher rates compared with AA. The relative conversion rates of EPA and AA ranged from about 1:1 with CYP2C23 and 1.2–1.4:1 with most of the isoforms to 2.1:1 with CYP4A11 and 2.4:1 with CYP2E1 (Fig. 1). Among all four PUFAs tested, EPA was the clearly preferred substrate of CYP4A11, but also most of the other CYP isoforms showed substrate specificities at least slightly in favor of EPA at the substrate concentration used (10 μm). DPA (22:5n-3) was utilized by some of the CYP isoforms with higher (CYP2E1 and CYP4F2) or similar rates (CYP2C8 and CYP2C19) compared with AA but represented the poorest substrate for most of the CYP isoforms tested (Fig. 1). In particular, CYP2C23, CYP4A11, Cyp4a12a, and Cyp4a12b showed largely reduced activities toward DPA that reached only 33, 14, 18, and 5% compared with AA. DHA was generally metabolized with higher rates than DPA, but the same enzymes that showed already weak activity toward DPA were also relatively inactive with DHA. In contrast, other CYP isoforms, such as CYP2C8, CYP2C19, and CYP2C11, metabolized DHA with rates almost as high as determined with AA (Fig. 1). Remarkably, DHA was the clearly preferred substrate of CYP4F2 that converted AA, EPA, DPA, and DHA with relative rates of 1:1.3:1.3:2.2. Moreover, also CYP2E1 showed about 2-fold higher activities toward DHA than AA. Compared with the other CYP isoforms, CYP2J2 showed rather weak activities that reached only 56 ± 4 pmol/nmol/min with AA but were about 17-fold higher with EPA (943 ± 17 pmol/nmol/min) and 4-fold higher with DHA (228 ± 8 pmol/nmol/min).
FIGURE 1.
Metabolisms of AA, EPA, DPA, and DHA by recombinant CYP isoforms functioning predominantly as epoxygenases (A) or hydroxylases (B). The data are mean values ± S.E. (error bars) from at least three determinations done in direct parallel with the four substrates. The specific activities were determined using the 1-14C-labeled substrates at a concentration of 10 μm and refer to total product formation (sum of epoxy and hydroxy metabolites) as analyzed by RP-HPLC; compare Tables 1 and 2 for the detailed regioisomeric composition of the metabolites.
TABLE 1.
Substrate-dependent regioselectivities of CYP isoforms in the epoxidation of long-chain PUFAs
| CYP | Substrate | Regioisomeric monoepoxidesa |
||||
|---|---|---|---|---|---|---|
| ω-3 | ω-6 | ω-9 | ω-12 | ω-15 | ||
| % | % | % | % | % | ||
| 2C8 | AA | 48 | 52 | NDb | ND | |
| EPA | 45 | 30 | 22 | 3 | ND | |
| DPA | 35 | 26 | 14 | 25 | ND | |
| DHA | 39 | 28 | 20 | 13 | ND | |
| 2C9 | AA | 52 | 30 | 18 | ND | |
| EPA | 7 | 45 | 28 | 20 | ND | |
| DPA | 0 | 16 | 0 | 73 | 10 | |
| DHA | 0 | 16 | 9 | 70 | 6 | |
| 2C19c | AA | 25 | 4 | 17 | ND | |
| EPA | 11 | 8 | ||||
| DPA | 13 | 20 | ||||
| DHA | 11 | 8 | 16 | 17 | ||
| 2C11 | AA | 39 | 42 | 19 | ND | |
| EPA | 33 | 27 | 20 | 20 | ND | |
| DPA | 24 | 12 | 8 | 18 | 39 | |
| DHA | 26 | 18 | 13 | 29 | 13 | |
| 2C23 | AA | 16 | 59 | 25 | ND | |
| EPA | 58 | 15 | 14 | 13 | ND | |
| DPA | 16 | 25 | 5 | 41 | 10 | |
| DHA | 27 | 24 | 5 | 44 | ND | |
| 2J2d | AA | 30 | 25 | 19 | ND | |
| EPA | 62 | 15 | 5 | 4 | ND | |
| DHA | 27 | 7 | 3 | 5 | 2 | |
a Data are mean values from at least three independent reactions and had S.D. values of ≤10% of the mean.
b ND, not detectable.
c CYP2C19 functioned predominantly as hydroxylase and produced with different regioselectivities the ω/(ω-1)-hydroxy products from AA (1:51), EPA (6:62), DPA (7:30), and DHA (4:43) as main products. The minor amounts of monoepoxides produced from EPA and DPA (8 and 20%, respectively) were not further resolved by NP-HPLC.
d ‖CYP2J2 metabolites were analyzed by LC-MS/MS. This enzyme functioned also as an ω/(ω-1)-hydroxylase and produced, in addition to the monoepoxides given in the table, XIX-/20-HETE (21 and 5% of total AA metabolites), 19-/20-HEPE (13.7 and 0.3% of total EPA metabolites) and 21-/22-HDoHE (50 and 7% of total DHA metabolites).
TABLE 2.
Substrate-dependent regioselectivities of CYP isoforms in the hydroxylation and ω-3 epoxidation of long-chain PUFAs
| CYP | Substrate | Regioisomeric productsa |
||
|---|---|---|---|---|
| ω | ω-1 | ω-3 | ||
| % | % | % | ||
| 2E1b | AA | 11 | 67 | |
| EPA | 5 | 44 | 37 | |
| DPA | 10 | 29 | 54 | |
| DHA | 5 | 42 | 47 | |
| 4A11 | AA | 78 | 22 | |
| EPA | 28 | 62 | 10 | |
| DPA | 54 | 45 | NDc | |
| DHA | 48 | 44 | 8 | |
| 4F2 | AA | 98 | 2 | |
| EPA | 80 | 16 | 4 | |
| DPA | 68 | 29 | 3 | |
| DHA | 81 | 18 | 1 | |
| 4a12a | AA | 86 | 14 | |
| EPA | 28 | 14 | 58 | |
| DPA | 46 | 17 | 37 | |
| DHA | 45 | 24 | 31 | |
| 4a12b | AA | 89 | 11 | |
| EPA | 24 | 14 | 72 | |
| DPA | 34 | 18 | 48 | |
| DHA | 25 | 31 | 44 | |
a Data are mean values from at least three independent reactions and had S.D. values of ≤10% of the mean.
b CYP2E1 additionally produced minor amounts of ω-6 to ω-12 epoxides from AA (22%) and EPA (6%) but not from DPA and DHA.
c ND, not detectable.
In addition to comparing single substrate turnovers, we also analyzed the relative rates of AA, EPA, and DHA metabolism, when these three substrates were simultaneously present in a 1:1:1 molar ratio (10 μm each). These experiments were performed with a typical AA ω-hydroxylase (CYP4F2), (ω-1)-hydroxylase (CYP2E1), and epoxygenase (CYP2J2). Confirming their distinct substrate preferences, the individual isoforms produced AA-, EPA-, and DHA-derived metabolites in ratios of 17:38:45 (CYP4F2), 19:58:23 (CYP2E1), and 15:63:22 (CYP2J2) (data in percentage of total metabolites) (supplemental Fig. 3).
All CYP isoforms tested displayed a substrate-dependent regioselectivity (Tables 1 and 2). Human CYP2C8 provided an example for an epoxygenase that converts AA with a rather strict regioselectivity. Among the four possible regioisomeric monoepoxides, CYP2C8 produced exclusively 14,15- and 11,12-EET (i.e. it attacked the ω-6 and ω-9 double bonds of AA). However, if the substrate contained an ω-3 double bond like EPA, CYP2C8 preferentially epoxidized this unique position and produced 17,18-EEQ as the main product. Concomitantly, CYP2C8 showed reduced ω-6 and ω-9 epoxygenase activities when metabolizing EPA instead of AA. A further change in regioselectivity occurred with DPA and DHA. These C22 n-3-PUFAs were epoxidized not only at the ω-3, -6, and -9 but significantly also at the ω-12 double bond. Similar profound changes in the regioselectivity were also observed with the other epoxygenases (Table 1). Notably, human CYP2C9 showed a rather broad regioselectivity with AA but became a predominant ω-12 epoxygenase with DHA. Rat CYP2C23 that epoxidized AA preferentially to 11,12-EET metabolized EPA to 17,18-EEQ and DHA to 7,8-EDP as main products. Human CYP2J2 showed a moderately expressed regioselectivity in favor of producing 14,15-EET as the main metabolite from AA. However, with EPA and DHA, the ω-3 double bond was by far the predominant epoxidation site. Moreover, CYP2J2 showed largely increased (ω-1)-hydroxylase activities when metabolizing DHA instead of AA (Table 1).
Also, typical AA hydroxylases showed different regioselectivities when metabolizing n-3 PUFAs as alternative substrates (Table 2). Human CYP2E1 preferentially functioned as (ω-1)-hydroxylase with AA but displayed in addition high ω-3 epoxygenase activities toward EPA, DPA, and DHA. Human CYP4A11 lost its rather strict regioselectivity for terminal hydroxylation as expressed with AA and showed markedly increased (ω-1)-hydroxylase activities and even weak ω-3 epoxygenase activities with the n-3 PUFAs. These effects were most pronounced with EPA that was metabolized by CYP4A11 to 20-HEPE, 19-HEPE, and 17,8-EEQ in a ratio of 28:62:10 compared with AA yielding 20-HETE and 19-HETE in a ratio of 78:22. Also human CYP4F2 that functioned almost exclusively as ω-hydroxylase with AA (20-HETE/19-HETE = 98:2) showed moderately increased (ω-1)-hydroxylase activities toward the n-3 PUFAs and metabolized DHA as its preferred substrate to 22-HDoHE, 21-HDoHE, and 19,20-EDP in a ratio of 81:18:1 (Table 2). The two mouse Cyp4a12 isoforms showed a clear regioselectivity in favor of producing 20-HETE from AA but featured amazingly high ω-3 epoxygenase activities toward EPA, DPA, and DHA (Table 2).
Metabolism of AA, EPA, DPA, and DHA by Renal and Hepatic Microsomes
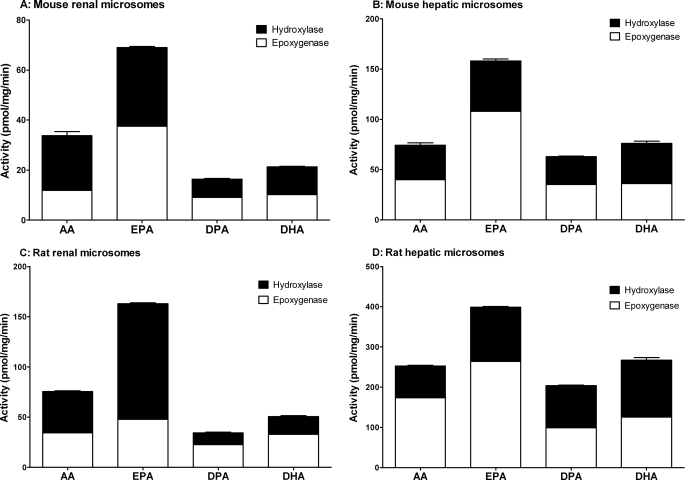
Renal and hepatic microsomes from mice and rats showed both hydroxylase and epoxygenase activities when metabolizing AA, EPA, DPA, and DHA (Fig. 2). EPA was converted with significantly higher rates than all other substrates tested. The microsomal activities were about 2.1-fold (mouse kidney), 1.4-fold (mouse liver), 2.2-fold (rat kidney), and 1.6-fold (rat liver) higher when determined with EPA instead of AA. In mouse renal and hepatic microsomes, this increase in total activities was associated with a significant shift of the relative hydroxylase/epoxygenase activities from 1:0.5 (with AA as substrate) to 1:1.2 (EPA) and from 1:1.2 to 1:2.2, respectively. The conversion rates of DPA reached only about 50% (renal microsomes) and 80% (hepatic microsomes) of that with AA. DHA was metabolized by the renal microsomes of both species with about 35% reduced activities but by the hepatic microsomes with rates almost identical to that of AA (Fig. 2).
FIGURE 2.
Metabolism of AA, EPA, DPA, and DHA by mouse and rat renal (A and C) and hepatic microsomes (B and D). The data are mean values ± S.E. (error bars) from at least three determinations done in direct parallel using the 1-14C-labeled substrates at a concentration of 10 μm. Hydroxylase activities were calculated from the generated (ω-1)/ω-hydroxy metabolites migrating largely unresolved in RP-HPLC and epoxygenase activities from the sum of regioisomeric epoxides and the corresponding diols.
Changes in Fatty Acid Composition upon Dietary EPA/DHA Supplementation
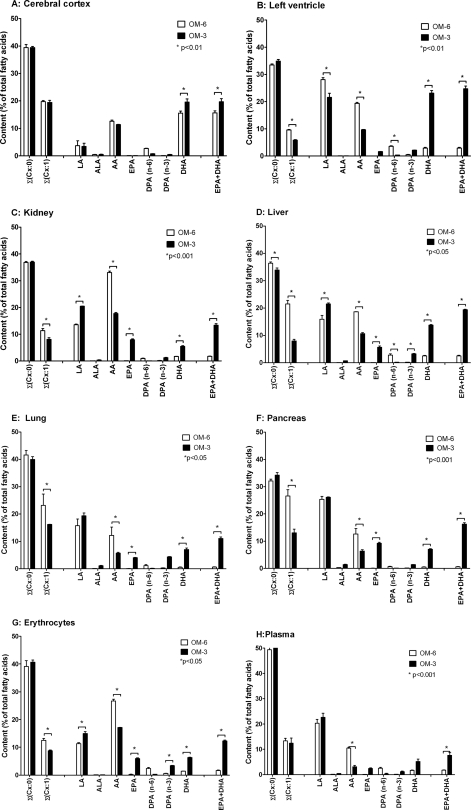
The organs and tissues of rats fed an ω-6 fatty acid-rich diet (OM-6 group) showed high linoleic acid (LA; 18:2n-6) but low ALA (18:3n-3) levels (Fig. 3), reflecting the composition of the sunflower oil used to prepare the experimental diet (see supplemental Table 1). AA was the predominant long-chain PUFA in most organs and tissues, and its content exceeded that of EPA + DHA 6–7-fold in the left ventricle, liver, and plasma and about 20-fold in the kidney, lung, pancreas, and red blood cells. The relative content of EPA was very low and ranged between 0.03% in the left ventricle and liver; 0.07–0.08% in the kidney, lung, pancreas, and plasma; and 0.2% of total fatty acids in red blood cells. The DHA levels were highest in the left ventricle (2.9%) and liver (2.5%), significantly lower in the kidney and plasma (1.7%) as well as red blood cells (1.4%), and very low in the lung and pancreas (0.5%). Compared with the other organs and tissues described above, the brain of OM-6-fed rats showed a unique fatty acid pattern characterized by high levels of both AA and DHA. In the cerebral cortex, AA represented 12.6% and DHA represented 15.6% of the total fatty acids (Fig. 3A).
FIGURE 3.
Effect of dietary EPA/DHA supplementation on the fatty acid composition of different organs and tissues in rat. Male Sprague-Dawley rats (n = 6/group) received for 3 weeks a diet either supplemented with 5% sunflower oil alone (ω-6 fatty acid-rich diet; OM-6 group) or additionally with 2.5% OMACOR® oil consisting of purified EPA- and DHA-ethylesters in a molar ratio of 1.4:1 (OM-3 group). The fatty acid profiles were determined for the harvested organs and tissues (A–H) of three animals per group, and the data represent the corresponding mean values ± S.E. (error bars). For the detailed composition of the diets used, see supplemental Tables 1 and 3 for the complete list of all individual fatty acids determined in the different organs and tissues.
Compared with OM-6-fed rats, dietary EPA/DHA supplementation resulted in a partial and tissue-specific replacement of AA by EPA and DHA (Fig. 3). The AA levels were reduced by about 50% in the left ventricle, kidney, liver, lung, and pancreas, 64% in red blood cells, and 70% in the plasma. Concomitantly, the percentage of EPA + DHA in total fatty acids increased dramatically from about 2.9 to 25% (left ventricle), 1.8 to 13% (kidney), 2.5 to 19% (liver), 0.6 to 11% (lung), 0.6 to 16% (pancreas), 1.6 to 12% (red blood cells), and 1.8 to 7.7% (plasma), comparing OM-6 and OM-3 fed rats. Some organs, like the kidney and pancreas, incorporated DHA and EPA almost in the same ratio as supplied via the diet (0.7:1), whereas others showed a relative enrichment of DHA over EPA: 1.05:1 in red blood cells, 1.2:1 in the lung, 2.1:1 in plasma, 2.4:1 in the liver, and even 14:1 in the left ventricle. Considering the whole extent of the tissue-specific changes in the fatty acid profiles, it is also of note that the partial replacement of AA by EPA/DHA was accompanied in the kidney, liver, lung, and red blood cells by a moderate increase of LA (Fig. 3). Moreover, a significant reduction of monounsaturated fatty acid levels (predominantly oleic acid, C18:1n-9) was obvious in most organs and tissues (Fig. 3 and supplemental Table 3). In contrast, the levels of saturated fatty acids remained largely unchanged comparing their tissue levels in OM-6 and OM-3 fed rats.
The brain showed a unique response to dietary EPA/DHA supplementation. The AA levels remained almost unchanged (12.6 versus 11.4%), and the DHA levels that were already high on the OM-6 diet (15.6) increased only moderately to 19.6% of total fatty acids. Moreover, the cerebral cortex contained only trace amounts of EPA (about 0.1%) independent of the diet (Fig. 3). The complete fatty acid patterns of the different organs and tissues after OM-6 and OM-3 feeding are given in supplemental Table 3.
Changes in the Endogenous CYP-eicosanoid Profile upon Dietary EPA/DHA-Supplementation
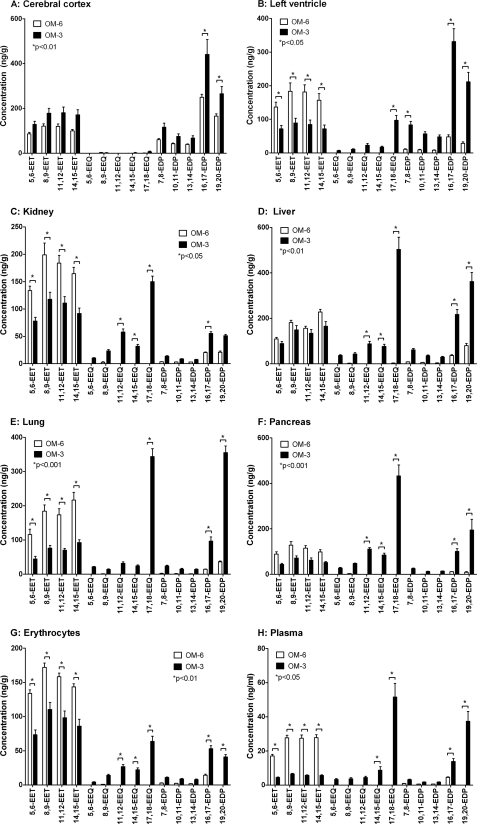
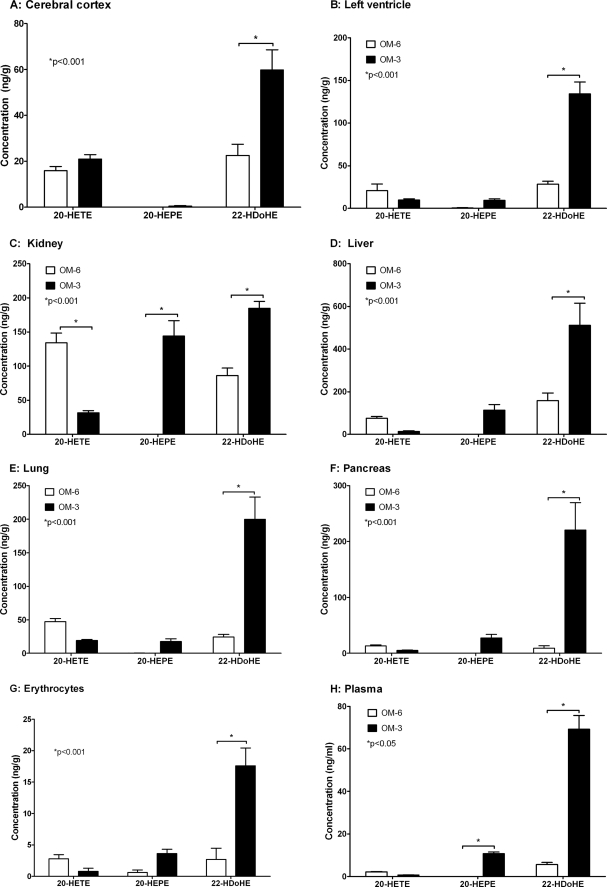
AA-derived CYP-eicosanoids, such as EETs and 20-HETE, were detectable in all organs and tissues analyzed from OM-6-fed rats (Figs. 4 and 5). The total EET levels ranged between nearly 100 ng/ml in the plasma, about 430 ng/g wet weight in the cerebral cortex and pancreas, 600 ng/g in red blood cells, and 660 to 680 ng/g in the left ventricle, kidney, liver, and lung (compare supplemental Fig. 4). 5,6-, 8,9-, 11,12-, and 14,15-EET contributed to the total EETs with relative percentages of 20:28:28:23 (cerebral cortex), 21:28:27:24 (left ventricle), 20:29:27:24 (kidney), 16:27:23:34 (liver), 17:27:25:31 (lung), 20:30:27:23 (pancreas), 22:28:25:23 (red blood cells), and 16:28:28:28 (plasma). The EPA-derived monoepoxides (EEQs) were undetectable in most organs and tissues and reached levels not higher than 2–5 ng/g wet weight in the kidney and liver (Fig. 4). In contrast, the DHA-derived monoepoxides, among them primarily 16,17- and 19,20-EDPs, occurred already in significant amounts in the OM-6-fed rats (Fig. 4). However, compared with EETs, the total EDP levels were severalfold lower in the left ventricle (6.3-fold), kidney (12.6-fold), liver (4.8-fold), lung (12.1-fold), pancreas (16.0-fold), red blood cells (26.4-fold), and plasma (16.7-fold). It is noteworthy that high EDP levels occurred in different regions of the brain isolated from the OM-6-fed rats. The EET/EDP ratios were about 0.8:1 in the cerebral cortex and diencephalon and 0.7:1 in the cerebellum (Fig. 4 and supplemental Fig. 5).
FIGURE 4.
Effect of dietary EPA/DHA supplementation on the profile of AA-, EPA-, and DHA-derived monoepoxides in different organs and tissues of rat. The animals received either an ω-6 fatty acid-rich (OM-6 group) or an EPA/DHA-supplemented diet (OM-3 group) as described in the legend to Fig. 3. AA-derived EETs, EPA-derived EEQs and DHA-derived EDPs were determined by LC-MS/MS in the harvested organs and tissues (A–H) of six animals per group, and the data represent the corresponding mean values ± S.E. (error bars) for the individual metabolites. For a summarized depiction of the changes in total EET, EEQ, and EDP levels, see supplemental Fig. 4.
FIGURE 5.
Effect of dietary EPA/DHA supplementation on the profile of AA-, EPA-, and DHA-derived ω-hydroxy metabolites in different organs and tissues of rat. The animals were fed an ω-6 fatty acid-rich diet (OM-6 group) or an EPA/DHA-supplemented diet (OM-3 group) as described in the legend to Fig. 3. AA-derived 20-HETE, EPA-derived 20-HEPE and DHA-derived 22-HDoHE were determined by LC-MS/MS in the harvested organs and tissues (A–H) of six animals per group, and the data represent the corresponding mean values ± S.E. (error bars) for the individual metabolites.
Among the ω-hydroxy metabolites determined in OM-6-fed rats (Fig. 5), the AA-derived 20-HETE occurred at the highest levels in the kidney (about 134 ng/g wet weight), followed by the liver (76 ng/g), lung (47 ng/g), cerebral cortex (16 ng/g), pancreas (13 ng/g), left ventricle (9 ng/g), red blood cells (2.8 ng/g), and plasma (2.3 ng/ml). The EPA-derived 20-HEPE was not detectable in any of the organs and tissues analyzed (Fig. 5). In contrast, the DHA-derived 22-HDoHE was present in amounts that partially reached or even exceeded the 20-HETE levels in a given tissue (Fig. 5). The relative contribution of 20-HETE and 22-HDoHE to the total ω-hydroxy-metabolites (20-HETE + 22-HDoHE set to 100%) was 41:59 (cerebral cortex), 27:73 (left ventricle), 61:39 (kidney), 32:68 (liver), 66:44 (lung), 59:41 (pancreas), 51:49 (red blood cells), and 29:71 in the plasma.
Compared with the endogenous CYP-eicosanoid patterns described above for OM-6-fed rats, dietary EPA/DHA supplementation induced a profound and tissue-specific shift from AA- to the EPA- and DHA-derived metabolites (Figs. 4 and 5). The EET levels were reduced by 52% (left ventricle), 42% (kidney), 21% (liver), 59% (lung), 47% (pancreas), 40% (red blood cells), and 78% (plasma). Concomitantly, a large increase of EEQs and EDPs occurred that was associated with a tissue-specific increase of the total monoepoxide levels: left ventricle (1.6-fold), kidney (1.1-fold), liver (2.4-fold), lung (1.6-fold), pancreas (2.8-fold), red blood cells (unchanged), and plasma (1.4-fold); compare supplemental Fig. 4. EEQs, the EPA-derived monoepoxides that were almost undetectable after OM-6 feeding, became highly abundant upon dietary EPA/DHA supplementation (Fig. 4). In general, the different regioisomers were present in the order 17,18-EEQ ≫ 11,12-EEQ ≥ 14,15-EEQ > 8,9-EEQ ≥ 5,6-EEQ. In some organs, such as the kidney, liver, and pancreas, 17,18-EEQ was even the predominant metabolite among all regioisomeric monoepoxides derived from AA, EPA, and DHA (Fig. 4). The DHA-derived EDPs increased from their low but significant levels in OM-6-fed rats severalfold in the left ventricle (about 7.0-fold), kidney (2.5-fold), liver (5.0-fold), lung (8.9-fold), pancreas (14-fold), red blood cells (5.3-fold), and plasma (9.7-fold); compare supplemental Fig. 4. The regioisomeric composition varied between the different organs and tissues. In general, however, 19,20-EDP and 16,17-EDP were clearly predominant, followed by 13,14-, 7,8-, and 10,11-EDP (Fig. 4 and supplemental Fig. 5).
Again, the brain provided an exception. In contrast to all other tissues analyzed, the EET levels were not decreased upon EPA/DHA supplementation, as shown in Fig. 4 for the cerebral cortex and in supplemental Fig. 5 also for the cerebellum and diencephalon. Moreover, EEQs remained largely undetectable except for 17,18-EEQ, which reached low but clearly measurable levels of 5–7 ng/g. Total EDPs, among them primarily 16,17- and 19,20-EDP, were moderately but significantly increased by a factor of 1.7 compared with OM-6-fed rats.
EPA/DHA supplementation also had a marked effect on the profile of the ω-hydroxy metabolites (Fig. 5). The AA-derived 20-HETE was strongly reduced in all organs and tissues except the cerebral cortex. The remaining 20-HETE levels were 53% (left ventricle), 66% (kidney), 81% (liver), 60% (lung and pancreas), and 70% (red blood cells and plasma) lower compared with those in OM-6-fed rats. The EPA-derived 20-HEPE, which was almost undetectable in OM-6-fed rats, became a prominent metabolite in the kidney, liver, lung, pancreas, and plasma but occurred only in minor amounts in the cerebral cortex (0.45 ng/g). The DHA-derived 22-HDoHE increased from its already significant amounts in OM-6-fed rats in the cerebral cortex (2.7-fold), left ventricle (5.2-fold), kidney (2.1-fold), liver (3.2-fold), lung (8.3-fold), pancreas (24.3-fold), and plasma (21.1-fold).
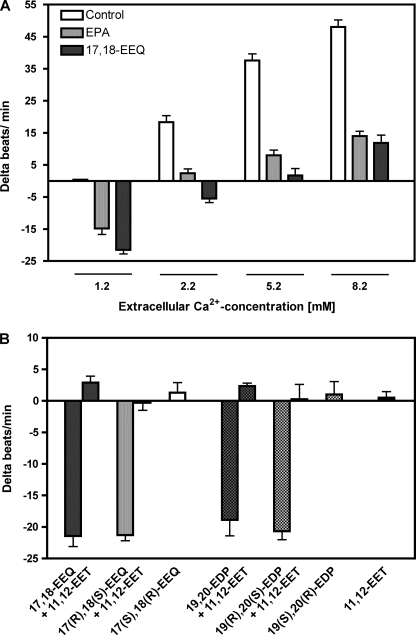
Effects of EPA and 17,18-EEQ on the Contractility of Neonatal Rat Cardiomyocytes (NRCMs)
Cultured NRCMs responded to increases in extracellular Ca2+ concentrations with largely increased beating rates (Fig. 6A, white bars). This response was strongly repressed after preincubating the cells for 30 min with 3.3 μm EPA (Fig. 6A, gray bars). 17,18-EEQ mimicked the effect of its parental fatty acid (Fig. 6A, black bars). However, 17,18-EEQ was almost immediately effective and inhibited the Ca2+ response of cardiomyocytes already at a concentration of 30 nm. As shown in Fig. 6B, the effect of 17,18-EEQ was stereoselective. Both the racemate and the R,S-enantiomer of 17,18-EEQ clearly reduced the spontaneous beating rate of NRCMs. In contrast, (17S,18R)-EEQ was inactive when tested at the same concentration (30 nm). Moreover, 17,18-dihydroxyeicosatetraenoic acid, the hydrolysis product of 17,18-EEQ, showed no activity (data not shown). Similar to 17,18-EEQ, 19,20-EDP also reduced the contractility of NRCMs in a stereoselective manner (Fig. 6B). The AA-derived metabolite 11,12-EET (30 nm) strongly antagonized the effects of both 17,18-EEQ and 19,20-EDP; Fig. 6B.
FIGURE 6.
Effects of EPA and selected epoxy metabolites on spontaneously beating NRCMs. A, incubation of cultured NRCMs with EPA (3.3 μm, 30 min) or 17,18-EEQ (30 nm, 5 min) reduced the beating rate under basal conditions (1.2 mm Ca2+) and attenuated the response to increased extracellular calcium ion concentrations. The vehicle (0.1% ethanol) had no effect on the basal and Ca2+-induced beating rates. B, 17,18-EEQ and 19,20-EDP exerted negative chronotropic effects that were abolished by adding 11,12-EET. Only the R,S- and not the S,R-enantiomers of 17,18-EEQ and 19,20-EDP were active. All metabolites were tested at a final concentration of 30 nm. Data are mean values ± S.E. (error bars) from n = 18–24 cell clusters originating from at least three independent NRCM cultures.
DISCUSSION
The present study shows that EPA and DHA are efficient alternative substrates of AA-metabolizing CYP enzymes in vitro. Furthermore, we are the first to demonstrate that dietary EPA/DHA supplementation causes a profound and tissue-specific shift of the CYP-eicosanoid profile under in vivo conditions. On the one hand, these findings may come as no surprise, considering the fact that many CYP enzymes are well known to have rather broad substrate specificities. On the other hand, this very feature makes CYP-dependent signaling pathways highly susceptible to changes in dietary intake and subject to other environmental factors. Because CYP-eicosanoids are involved in the regulation of vascular, renal, and cardiac function, this state of affairs may have important implications for the development of cardiovascular and other chronic diseases. Moreover, the fact that AA-metabolizing CYP isoforms accept EPA and DHA as highly efficient alternative substrates appears unique compared with the cyclooxygenase- and lipoxygenase-dependent branches of the AA cascade. In those cascade branches, EPA and DHA are generally considered as relatively poor substrates (25). Searching for the molecular mechanisms mediating the beneficial effects of n-3 PUFAs, it is also remarkable that the CYP isoforms preferentially attacked the ω-3 double bond, which results in a predominant accumulation of 17,18-EEQ and 19,20-EDP. These metabolites are novel products in most organs and tissues upon dietary EPA/DHA supplementation.
Most of the recombinant CYP enzymes tested displayed almost equal or in part even higher catalytic activities when metabolizing EPA or DHA instead of AA. Considering the results of earlier studies, we would conclude that the capacity to utilize EPA and DHA as alternative substrates is shared by virtually all of the AA-metabolizing CYP isoforms that belong to the subfamilies 1A (34, 35, 49), 2C (33, 35), 2E (35), 2J (35), 2U (50), 4A (32, 36, 40), and 4F (36, 37). The ω-3 double bond that distinguishes EPA and DHA from their n-6 PUFA counterparts was a major site of epoxidation, as catalyzed by CYP2J2 and most of the CYP2C isoforms. Among the CYP hydroxylases, CYP4A and somewhat less so the CYP4F isoforms showed largely increased (ω-1)-hydroxylase activities when utilizing EPA and DHA instead of AA as substrates. Mouse Cyp4a12a, human CYP4A11, and, as reported previously, also rat CYP4A1 (32) even displayed significant epoxygenase activities when an ω-3 double bond in the fatty acid substrate was available. High ω-3 epoxygenase activities toward EPA and DHA were also observed with human CYP1A1 (34, 35) and CYP2E1. These enzymes hydroxylated AA predominantly at the (ω-1)-position. Similar results were accrued with human CYP4F8 and CYP4F12 that metabolized AA by (ω-2)/(ω-3)-hydroxylation (51). Thus, CYP enzymes generally respond to the altered double-bond structure and chain length of their fatty acid substrates with remarkable changes in the regioselectivity. Presumably, they behave similarly in terms of the stereoselectivity of their product formation. This state of affairs may have important physiological implications because the biological activities of CYP-eicosanoids are dependent on the regio- and stereoisomeric position of their functional epoxy or hydroxy groups.
The rats received two types of dietary fatty acid supplementation. We gave sunflower oil alone or sunflower oil combined with OMACOR® oil to partially mimic the nutritional behaviors prevalent in current Western countries and in populations traditionally living on fish and other seafood. The selected diets correspond to the possible extremes of ingesting a plant-derived food that is characterized by LA (18:2n-6) ≫ ALA (18:3n-3), the complete absence of long-chain n-6 and n-3 PUFAs, and a relatively low content of saturated fatty acids or a food containing the same plant-derived fatty acids and additionally high amounts of preformed EPA and DHA. Other standard rodent chows are often prepared with soybean oil containing higher amounts of ALA (e.g. sniff R-Z) or even with a fishmeal additive (e.g. NIH-31 or -07) that provides some EPA and DHA. Our OM-6 diet resulted in a 10–20-fold excess of AA over EPA/DHA in the membrane phospholipids of most organs except the brain, which maintained an about 1:1.2 ratio of AA to DHA. After dietary EPA/DHA supplementation, the AA levels were generally reduced by 40–50% and partially replaced by EPA and DHA. The relative enrichment of DHA over EPA in several organs indicated systemic and local interconversion of these long-chain n-3 PUFAs. EPA is converted to circulating DHA primarily in the liver. In the heart, a combination of selective uptake and degradation mechanisms may be responsible for the relative enrichment of DHA (52). Preferential DHA uptake and low turnover are believed to allow the adult brain to preserve its unique fatty acid composition much longer than other organs upon dietary changes in the n-6/n-3 PUFA ratio (53). The exceptionally low levels of EPA may be attributed to the high β-oxidation rates of EPA after its uptake into the brain (54).
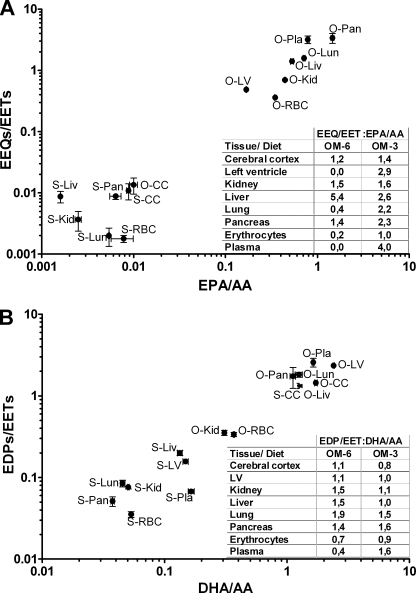
In vivo, EETs and 20-HETE are de novo synthesized in response to extracellular signals that activate phospholipases A2 and thus provide free AA to the CYP enzymes. However, once produced, EETs and 20-HETE are partially re-esterified into the sn-2-position of phospholipids, which generates a membrane pool of preformed CYP-eicosanoids that is also accessible to phospholipases A2 (7, 10, 55–57). Our results indicate that the same principles of synthesis, storage, and release may also apply to the epoxy and hydroxy derivatives of EPA and DHA. The diet-induced partial replacement of AA by EPA and DHA was accompanied by a marked shift of the endogenous CYP-eicosanoid profile. EETs were largely replaced by EEQs and EDPs. 20-HEPE and 22-HDoHE substituted for 20-HETE. The extent of the metabolite exchange in a given tissue was obviously determined by first, the extent of the replacement of AA for EPA and DHA and second, the relative efficiencies by which the different precursor fatty acids were metabolized to their epoxy and hydroxy derivatives. Our data indicate that the conversion of EPA to EEQs proceeded with higher efficiencies compared with that of AA to EETs and of DHA to EDPs in most tissues (Fig. 7). Our data also suggest an efficient competition between EPA and DHA. The formation of EDPs was largely repressed in organs containing almost equal amounts of DHA and EPA (kidney and pancreas) and increased less than proportionally with higher DHA/EPA ratios, as exemplified by the liver and heart. These findings are in agreement with the in vitro studies showing that most of the CYP-epoxygenases accept DHA and AA as equally efficient substrates but exhibit significantly higher specific activities when utilizing EPA instead of AA or DHA. Also, the tissue-specific exchange of 20-HETE for 20-HEPE and 22-HDoHE may be partially explained by the relative availability of the three precursor fatty acids and the substrate specificity of the CYP-hydroxylases. However, the formation of 22-HDoHE in organs, such as the kidney, liver, and heart, appears much higher than expected from the in vitro microsomal hydroxylase activities. The formation levels are also largely overproportional, considering the hydroxy metabolite/precursor fatty acid ratios in these organs. Actually, the formation of endogenous CYP-eicosanoid pools is a multienzyme process whose selectivity is not only determined by the CYP enzymes (55, 58). Several key questions must be answered to understand the specific accumulation of certain metabolites in a given tissue. The list begins with defining extracellular signal-induced activation of phospholipases that may either indiscriminately provide free AA, EPA, and DHA to the CYP enzymes or show distinct fatty acid and phospholipid selectivities, as increasingly recognized in the brain (59). Also, not yet elucidated are the final steps, namely ATP-dependent acyl-CoA formation and esterification into different phospholipid classes that may be substrate-selective in terms of the acyl-backbone and regio- and stereoisomeric position of the functional oxygen group, as shown in detail for the incorporation of regioisomeric EETs in the rat liver (58).
FIGURE 7.
Relative efficiencies of precursor fatty acid to epoxy metabolite conversions. A shows the ratio of total EEQ and EET levels versus the ratio of EPA and AA in the given organs and tissues. B shows the ratio of EDP and EET levels versus the ratio of DHA and AA as the corresponding precursor fatty acid. The calculated data (insets) show that the relative conversions of EPA to EEQs (A) and of DHA to EDPs (B) deviate from that of AA to EETs in a diet- and tissue-specific manner. S, sunflower oil (OM-6) diet; O, OMACOR® oil (OM-3) diet; CC, cerebral cortex; LV, left ventricle of the heart; Liv, liver; Kid, kidney; Lun, lung; Pan, pancreas; RBC, red blood cells; Pla, plasma. Error bars, S.E.
Compared with the CYP-expressing tissues, the relative efficiencies of EPA to EEQ and DHA to EDP conversion were much lower in red blood cells (Fig. 7) that produce EETs by a CYP-independent, presumably peroxidation-initiated mechanism (60). Nonetheless, red blood cells represented the major reservoir of all circulating n-6 and n-3 CYP-epoxyeicosanoids. Because red blood cells release EETs upon deformation, hypoxia, and ATP stimulation (60, 61), they may also distribute significant amounts of the EPA- and DHA-derived metabolites to various tissues during passage of the microvasculature. Lipoproteins circulating in the blood plasma potentially represent another systemic source of CYP-eicosanoids that may become accessible to other tissues after lipoprotein lipase-mediated release (62, 63). Also, in this compartment, dietary EPA/DHA supplementation resulted in a significant replacement of EETs and 20-HETE by EEQs/EDPs and 20-HEPE/22-HDoHE.
Studies on the physiological effects of CYP-dependent EPA and DHA metabolites are only beginning. Mimicking the effects of EETs in the vasculature (4), EEQs and EDPs are powerful activators of calcium-activated potassium (BK) channels and mediate the dilation of coronary and mesenteric microvessels (32, 64–66). In rat cerebral artery vascular smooth muscle cells, BK channel activation was highly regio- and stereoselective, and the effect of (17R,18S)-EEQ largely exceeded that of (11R,12S)-EET, the most powerful AA metabolite (32). 17,18-EEQ also hyperpolarizes and relaxes human pulmonary artery and bronchial smooth muscle cells (67). DHA-derived epoxides were up to 1000-fold more potent than EETs in activating BK channels in rat coronary arterioles (66). Thus, an exchange of EETs for EEQs and EDPs could explain the observation that n-3 PUFAs improve vascular function by increasing the response to vasodilator hormones. Recently, (19R)-HETE was shown to act as an antagonist of 20-HETE (68), raising the possibility that the enhanced formation of (ω-1)-hydroxylase products from EPA and DHA may interfere with vasoconstrictor signaling.
In the heart, EETs reduce infarction injury and improve functional recovery after ischemia/reperfusion (8, 69), whereas 20-HETE plays an opposite detrimental role under the same pathological conditions (70, 71). Mitochondrial and sarcolemmal KATP channels known to play a key role in cardiac protection are activated by EETs but obviously blocked by 20-HETE. The capacity of EETs to activate KATP channels in cardiomyocytes is largely exceeded by their EPA- and DHA-derived counterparts (72). Protection against arrhythmia and sudden cardiac death is probably the most prominent benefit from dietary EPA/DHA supplementation (23, 73). We have shown previously that the high mortality and arrhythmia inducibility can be strongly reduced in a rat model of sudden cardiac death by adding 2.5% OMACOR® oil to the normal chow (22). In the present study, we used the same EPA/DHA supplement and demonstrate that 17,18-EEQ and 19,20-EDP become the predominant cardiac epoxy metabolites under these conditions. Moreover, we found that (17R,18S)-EEQ mimics the effects of EPA on the contractility of neonatal cardiomyocytes. Neonatal cardiomyocytes have been introduced by other authors as an in vitro model to analyze the anti-arrhythmic mechanisms elicited by EPA and DHA (23, 74). In agreement with our results, EPA (2–10 μm) was reported to reduce the spontaneous beating rate of these cells and to protect them against Ca2+ overload (74). Our results demonstrate that (17R,18S)-EEQ exerts the same effects as EPA but is active already in the low nanomolar range. Thus, we hypothesize that (17R,18S)-EEQ and presumably also (19R,20S)-EDP may play an important role in mediating the anti-arrhythmic effect of n-3 PUFAs. Of note, CYP2J2, the major AA epoxygenase in the human heart (75), shows largely increased activities toward EPA and DHA and a clear preference for epoxidizing the ω-3 double bond. Thus, similar to our rat model, the human heart can be expected to accumulate 17,18-EEQ and 19,20-EDP upon dietary ω-3 fatty acid supplementation.
A recent study revealed that 17,18-EEQ is also a potent regulator of human lung inflammation, and it remains to be seen whether this EPA metabolite may provide protection against critical inflammatory bronchial disorders (76). In the brain, EETs are involved in neurovascular signaling (77) and neuroprotection (78). Surprisingly, however, almost nothing has been known about the potential role of EDPs that are highly abundant in this organ with its uniquely high DHA/AA ratio.
More work will be required to establish the role of CYP-dependent EPA/DHA metabolites in the cardiovascular system; however, the reward could be substantial. After appropriate dietary n-3 PUFA supplementation, EEQs/EDPs and 20-HEPE/22-HDoHE have the capacity to take over the roles of EETs and 20-HETE as second messengers. On the one hand, they may thus enhance or dampen various signaling pathways regulating vascular, renal, and cardiac function. On the other hand, some of the unique metabolites, such as 17,18-EEQ or 19,20-EDP, may also exhibit novel biological activities not gradually shared by the AA-derived metabolites but essential to mediate specific effects attributed to EPA and DHA, such as protection against cardiac arrhythmias. There is increasing evidence for the existence of CYP-eicosanoid receptors (79). Future studies will extend the hypothesis that distinct receptors may specifically interact with selected sets of epoxy and hydroxy metabolites derived from n-6 and n-3 PUFAs.
Supplementary Material
Acknowledgments
We thank Christel Andrée, Jutta Meisel, and Ramona Zummach for excellent technical assistance.
This work was supported in part by grants SCHU822/5 and FOR1054 from the Deutsche Forschungsgemeinschaft (to D. N. M. and W.-H. S.). The animal studies were supported by Solvay Arzneimittel GmbH.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Tables 1–3.
- CYP
- cytochrome P450
- AA
- arachidonic acid
- EET
- epoxyeicosatrienoic acid
- 20-HETE
- 20-hydroxyeicosatetraenoic acid
- EPA
- eicosapentaenoic acid
- DHA
- docosahexaenoic acid
- EEQ
- epoxyeicosatetraenoic acid
- EDP
- epoxydocosapentaenoic acid
- HEPE
- hydroxyeicosapentaenoic acid
- HDoHE
- hydroxydocosahexaenoic acid
- PUFA
- polyunsaturated fatty acid
- DPA
- docosapentaenoic acid (22:5n-3)
- BK
- calcium-activated potassium channel
- CPR
- CYP reductase
- RP
- reversed phase
- NP
- normal phase
- ALA
- α-linolenic acid
- NRCM
- neonatal rat cardiomyocyte.
REFERENCES
- 1.Capdevila J. H., Falck J. R. (2002) Prostaglandins Other Lipid Mediat. 68, 325–344 [DOI] [PubMed] [Google Scholar]
- 2.Zeldin D. C. (2001) J. Biol. Chem. 276, 36059–36062 [DOI] [PubMed] [Google Scholar]
- 3.Kroetz D. L., Xu F. (2005) Annu. Rev. Pharmacol. Toxicol. 45, 413–438 [DOI] [PubMed] [Google Scholar]
- 4.Campbell W. B., Falck J. R. (2007) Hypertension 49, 590–596 [DOI] [PubMed] [Google Scholar]
- 5.Fleming I. (2008) Trends Cardiovasc. Med. 18, 20–25 [DOI] [PubMed] [Google Scholar]
- 6.Miyata N., Roman R. J. (2005) J Smooth Muscle Res. 41, 175–193 [DOI] [PubMed] [Google Scholar]
- 7.McGiff J. C., Quilley J. (1999) Am. J. Physiol. 277, R607–R623 [DOI] [PubMed] [Google Scholar]
- 8.Seubert J. M., Zeldin D. C., Nithipatikom K., Gross G. J. (2007) Prostaglandins Other Lipid Mediat. 82, 50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spector A. A. (2009) J. Lipid Res. 50, (suppl.) S52–S56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roman R. J. (2002) Physiol. Rev. 82, 131–185 [DOI] [PubMed] [Google Scholar]
- 11.Capdevila J. H., Falck J. R., Imig J. D. (2007) Kidney Int. 72, 683–689 [DOI] [PubMed] [Google Scholar]
- 12.Imig J. D., Hammock B. D. (2009) Nat. Rev. Drug Discov. 8, 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laffer C. L., Laniado-Schwartzman M., Wang M. H., Nasjletti A., Elijovich F. (2003) Circulation 107, 574–578 [DOI] [PubMed] [Google Scholar]
- 14.Gainer J. V., Bellamine A., Dawson E. P., Womble K. E., Grant S. W., Wang Y., Cupples L. A., Guo C. Y., Demissie S., O'Donnell C. J., Brown N. J., Waterman M. R., Capdevila J. H. (2005) Circulation 111, 63–69 [DOI] [PubMed] [Google Scholar]
- 15.Spiecker M., Darius H., Hankeln T., Soufi M., Sattler A. M., Schaefer J. R., Node K., Börgel J., Mügge A., Lindpaintner K., Huesing A., Maisch B., Zeldin D. C., Liao J. K. (2004) Circulation 110, 2132–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward N. C., Puddey I. B., Hodgson J. M., Beilin L. J., Croft K. D. (2005) Free Radic. Biol. Med. 38, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 17.Minuz P., Jiang H., Fava C., Turolo L., Tacconelli S., Ricci M., Patrignani P., Morganti A., Lechi A., McGiff J. C. (2008) Hypertension 51, 1379–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kris-Etherton P. M., Harris W. S., Appel L. J. (2002) Circulation 106, 2747–2757 [DOI] [PubMed] [Google Scholar]
- 19.Mozaffarian D. (2008) Am. J. Clin. Nutr. 87, 1991S–1996S [DOI] [PubMed] [Google Scholar]
- 20.Simopoulos A. P. (2008) Exp. Biol. Med. 233, 674–688 [DOI] [PubMed] [Google Scholar]
- 21.Lavie C. J., Milani R. V., Mehra M. R., Ventura H. O. (2009) J. Am. Coll. Cardiol. 54, 585–594 [DOI] [PubMed] [Google Scholar]
- 22.Fischer R., Dechend R., Qadri F., Markovic M., Feldt S., Herse F., Park J. K., Gapelyuk A., Schwarz I., Zacharzowsky U. B., Plehm R., Safak E., Heuser A., Schirdewan A., Luft F. C., Schunck W. H., Muller D. N. (2008) Hypertension 51, 540–546 [DOI] [PubMed] [Google Scholar]
- 23.Leaf A., Kang J. X., Xiao Y. F., Billman G. E. (2003) Circulation 107, 2646–2652 [DOI] [PubMed] [Google Scholar]
- 24.Chapkin R. S., McMurray D. N., Davidson L. A., Patil B. S., Fan Y. Y., Lupton J. R. (2008) Br. J. Nutr. 100, 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jump D. B. (2002) J. Biol. Chem. 277, 8755–8758 [DOI] [PubMed] [Google Scholar]
- 26.Deckelbaum R. J., Worgall T. S., Seo T. (2006) Am. J. Clin. Nutr. 83, 1520S–1525S [DOI] [PubMed] [Google Scholar]
- 27.Xiao Y. F., Sigg D. C., Leaf A. (2005) J. Membr. Biol. 206, 141–154 [DOI] [PubMed] [Google Scholar]
- 28.Calder P. C. (2006) Am. J. Clin. Nutr. 83, 1505S–1519S [DOI] [PubMed] [Google Scholar]
- 29.Serhan C. N., Chiang N., Van Dyke T. E. (2008) Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz G., Ecker J. (2008) Prog. Lipid Res. 47, 147–155 [DOI] [PubMed] [Google Scholar]
- 31.Wada M., DeLong C. J., Hong Y. H., Rieke C. J., Song I., Sidhu R. S., Yuan C., Warnock M., Schmaier A. H., Yokoyama C., Smyth E. M., Wilson S. J., FitzGerald G. A., Garavito R. M., Sui de X., Regan J. W., Smith W. L. (2007) J. Biol. Chem. 282, 22254–22266 [DOI] [PubMed] [Google Scholar]
- 32.Lauterbach B., Barbosa-Sicard E., Wang M. H., Honeck H., Kärgel E., Theuer J., Schwartzman M. L., Haller H., Luft F. C., Gollasch M., Schunck W. H. (2002) Hypertension 39, 609–613 [DOI] [PubMed] [Google Scholar]
- 33.Barbosa-Sicard E., Markovic M., Honeck H., Christ B., Muller D. N., Schunck W. H. (2005) Biochem. Biophys. Res. Commun. 329, 1275–1281 [DOI] [PubMed] [Google Scholar]
- 34.Schwarz D., Kisselev P., Ericksen S. S., Szklarz G. D., Chernogolov A., Honeck H., Schunck W. H., Roots I. (2004) Biochem. Pharmacol. 67, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 35.Fer M., Dréano Y., Lucas D., Corcos L., Salaün J. P., Berthou F., Amet Y. (2008) Arch. Biochem. Biophys. 471, 116–125 [DOI] [PubMed] [Google Scholar]
- 36.Fer M., Corcos L., Dréano Y., Plée-Gautier E., Salaün J. P., Berthou F., Amet Y. (2008) J. Lipid Res. 49, 2379–2389 [DOI] [PubMed] [Google Scholar]
- 37.Harmon S. D., Fang X., Kaduce T. L., Hu S., Raj Gopal V., Falck J. R., Spector A. A. (2006) Prostaglandins Leukot. Essent. Fatty Acids 75, 169–177 [DOI] [PubMed] [Google Scholar]
- 38.Van Rollins M., Frade P. D., Carretero O. A. (1988) Biochim. Biophys. Acta 966, 133–149 [DOI] [PubMed] [Google Scholar]
- 39.VanRollins M. (1990) Lipids 25, 481–490 [DOI] [PubMed] [Google Scholar]
- 40.Muller D. N., Schmidt C., Barbosa-Sicard E., Wellner M., Gross V., Hercule H., Markovic M., Honeck H., Luft F. C., Schunck W. H. (2007) Biochem. J. 403, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Omura T., Sato R. (1964) J. Biol. Chem. 239, 2370–2378 [PubMed] [Google Scholar]
- 42.Kaergel E., Muller D. N., Honeck H., Theuer J., Shagdarsuren E., Mullally A., Luft F. C., Schunck W. H. (2002) Hypertension 40, 273–279 [DOI] [PubMed] [Google Scholar]
- 43.Capdevila J. H., Falck J. R., Dishman E., Karara A. (1990) Methods Enzymol. 187, 385–394 [DOI] [PubMed] [Google Scholar]
- 44.Harris W. S., Von Schacky C. (2004) Prev. Med. 39, 212–220 [DOI] [PubMed] [Google Scholar]
- 45.Rivera J., Ward N., Hodgson J., Puddey I. B., Falck J. R., Croft K. D. (2004) Clin. Chem. 50, 224–226 [DOI] [PubMed] [Google Scholar]
- 46.Falck J. R., Yadagiri P., Capdevila J. (1990) Methods Enzymol. 187, 357–364 [DOI] [PubMed] [Google Scholar]
- 47.Wallukat G., Muñoz Saravia S. G., Haberland A., Bartel S., Araujo R., Valda G., Duchen D., Diaz Ramirez I., Borges A. C., Schimke I. (2010) J. Am. Coll. Cardiol. 55, 463–468 [DOI] [PubMed] [Google Scholar]
- 48.Kulas J., Schmidt C., Rothe M., Schunck W. H., Menzel R. (2008) Arch. Biochem. Biophys. 472, 65–75 [DOI] [PubMed] [Google Scholar]
- 49.Schwarz D., Kisselev P., Chernogolov A., Schunck W. H., Roots I. (2005) Biochem. Biophys. Res. Commun. 336, 779–783 [DOI] [PubMed] [Google Scholar]
- 50.Karlgren M., Backlund M., Johansson I., Oscarson M., Ingelman-Sundberg M. (2004) Biochem. Biophys. Res. Commun. 315, 679–685 [DOI] [PubMed] [Google Scholar]
- 51.Stark K., Wongsud B., Burman R., Oliw E. H. (2005) Arch. Biochem. Biophys. 441, 174–181 [DOI] [PubMed] [Google Scholar]
- 52.Gao F., Kiesewetter D., Chang L., Ma K., Bell J. M., Rapoport S. I., Igarashi M. (2009) J. Lipid Res. 50, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Kim H. Y. (2007) J. Biol. Chem. 282, 18661–18665 [DOI] [PubMed] [Google Scholar]
- 54.Chen C. T., Liu Z., Ouellet M., Calon F., Bazinet R. P. (2009) Prostaglandins Leukot. Essent. Fatty Acids 80, 157–163 [DOI] [PubMed] [Google Scholar]
- 55.Capdevila J. H., Falck J. R., Harris R. C. (2000) J. Lipid Res. 41, 163–181 [PubMed] [Google Scholar]
- 56.Kaduce T. L., Fang X., Harmon S. D., Oltman C. L., Dellsperger K. C., Teesch L. M., Gopal V. R., Falck J. R., Campbell W. B., Weintraub N. L., Spector A. A. (2004) J. Biol. Chem. 279, 2648–2656 [DOI] [PubMed] [Google Scholar]
- 57.Carroll M. A., Balazy M., Huang D. D., Rybalova S., Falck J. R., McGiff J. C. (1997) Kidney Int. 51, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 58.Karara A., Dishman E., Falck J. R., Capdevila J. H. (1991) J. Biol. Chem. 266, 7561–7569 [PubMed] [Google Scholar]
- 59.Rapoport S. I. (2008) Prostaglandins Leukot. Essent. Fatty Acids 79, 153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H. (2007) Prostaglandins Other Lipid Mediat. 82, 4–10 [DOI] [PubMed] [Google Scholar]
- 61.Jiang H., Zhu A. G., Mamczur M., Falck J. R., Lerea K. M., McGiff J. C. (2007) Br. J. Pharmacol. 151, 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karara A., Wei S., Spady D., Swift L., Capdevila J. H., Falck J. R. (1992) Biochem. Biophys. Res. Commun. 182, 1320–1325 [DOI] [PubMed] [Google Scholar]
- 63.Shearer G. C., Newman J. W. (2009) Curr. Atheroscler. Rep. 11, 403–410 [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y., Oltman C. L., Lu T., Lee H. C., Dellsperger K. C., VanRollins M. (2001) Am. J. Physiol. Heart Circ. Physiol. 280, H2430–H2440 [DOI] [PubMed] [Google Scholar]
- 65.Hercule H. C., Salanova B., Essin K., Honeck H., Falck J. R., Sausbier M., Ruth P., Schunck W. H., Luft F. C., Gollasch M. (2007) Exp. Physiol. 92, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 66.Ye D., Zhang D., Oltman C., Dellsperger K., Lee H. C., VanRollins M. (2002) J. Pharmacol. Exp. Ther. 303, 768–776 [DOI] [PubMed] [Google Scholar]
- 67.Morin C., Sirois M., Echave V., Rizcallah E., Rousseau E. (2009) Am. J. Physiol. Lung Cell Mol. Physiol. 296, L130–L139 [DOI] [PubMed] [Google Scholar]
- 68.Cheng J., Ou J. S., Singh H., Falck J. R., Narsimhaswamy D., Pritchard K. A., Jr., Schwartzman M. L. (2008) Am. J. Physiol. Heart Circ Physiol. 294, H1018–H1026 [DOI] [PubMed] [Google Scholar]
- 69.Seubert J., Yang B., Bradbury J. A., Graves J., Degraff L. M., Gabel S., Gooch R., Foley J., Newman J., Mao L., Rockman H. A., Hammock B. D., Murphy E., Zeldin D. C. (2004) Circ. Res. 95, 506–514 [DOI] [PubMed] [Google Scholar]
- 70.Gross E. R., Nithipatikom K., Hsu A. K., Peart J. N., Falck J. R., Campbell W. B., Gross G. J. (2004) J. Mol. Cell Cardiol. 37, 1245–1249 [DOI] [PubMed] [Google Scholar]
- 71.Nithipatikom K., Gross E. R., Endsley M. P., Moore J. M., Isbell M. A., Falck J. R., Campbell W. B., Gross G. J. (2004) Circ. Res. 95, e65–71 [DOI] [PubMed] [Google Scholar]
- 72.Lu T., VanRollins M., Lee H. C. (2002) Mol. Pharmacol. 62, 1076–1083 [DOI] [PubMed] [Google Scholar]
- 73.Marchioli R., Barzi F., Bomba E., Chieffo C., Di Gregorio D., Di Mascio R., Franzosi M. G., Geraci E., Levantesi G., Maggioni A. P., Mantini L., Marfisi R. M., Mastrogiuseppe G., Mininni N., Nicolosi G. L., Santini M., Schweiger C., Tavazzi L., Tognoni G., Tucci C., Valagussa F. (2002) Circulation 105, 1897–1903 [DOI] [PubMed] [Google Scholar]
- 74.Kang J. X., Leaf A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9886–9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu S., Moomaw C. R., Tomer K. B., Falck J. R., Zeldin D. C. (1996) J. Biol. Chem. 271, 3460–3468 [DOI] [PubMed] [Google Scholar]
- 76.Morin C., Sirois M., Echave V., Albadine R., Rousseau E. (2009) Am. J. Respir. Cell Mol. Biol. [DOI] [PubMed] [Google Scholar]
- 77.Harder D. R., Alkayed N. J., Lange A. R., Gebremedhin D., Roman R. J. (1998) Stroke 29, 229–234 [DOI] [PubMed] [Google Scholar]
- 78.Iliff J. J., Alkayed N. J. (2009) Future Neurol. 4, 179–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang W., Tuniki V. R., Anjaiah S., Falck J. R., Hillard C. J., Campbell W. B. (2008) J. Pharmacol. Exp. Ther. 324, 1019–1027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.