Abstract
Chk2 is a critical regulator of the cellular DNA damage repair response. Activation of Chk2 in response to IR-induced damage is initiated by phosphorylation of the Chk2 SQ/TQ cluster domain at Ser19, Ser33, Ser35, and Thr68. This precedes autophosphorylation of Thr383/Thr387 in the T-loop region of the kinase domain an event that is a prerequisite for efficient kinase activity. We conducted an in-depth analysis of phosphorylation within the T-loop region (residues 366–406). We report four novel phosphorylation sites at Ser372, Thr378, Thr389, and Tyr390. Substitution mutation Y390F was defective for kinase function. The substitution mutation T378A ablated the IR induction of kinase activity. Interestingly, the substitution mutation T389A demonstrated a 6-fold increase in kinase activity when compared with wild-type Chk2. In addition, phosphorylation at Thr389 was a prerequisite to phosphorylation at Thr387 but not at Thr383. Quantitative mass spectrometry analysis revealed IR-induced phosphorylation and subcellular distribution of Chk2 phosphorylated species. We observed IR-induced increase in phosphorylation at Ser379, Thr389, and Thr383/Thr389. Phosphorylation at Tyr390 was dramatically reduced following IR. Exposure to IR was also associated with changes in the ratio of chromatin/nuclear localization. IR-induced increase in chromatin localization was associated with phosphorylation at Thr372, Thr379, Thr383, Thr389, Thr383/Thr387, and Thr383/Thr389. Chk2 hyper-phosphorylated species at Thr383/Thr387/Thr389 and Thr383/Thr387/Thr389/Tyr390 relocalized from almost exclusively chromatin to predominately nuclear expression, suggesting a role for phosphorylation in regulation of chromatin targeting and egress. The differential impact of T-loop phosphorylation on Chk2 ubiquitylation suggests a co-dependence of these modifications. The results demonstrate that a complex interdependent network of phosphorylation events within the T-loop exchange region regulates dimerization/autophosphorylation, kinase activation, and chromatin targeting/egress of Chk2.
Keywords: Cell Cycle, DNA Damage, DNA Repair, Mass Spectrometry (MS), Protein Phosphorylation, Signal Transduction
Introduction
The activation of checkpoint mechanisms is critical to maintenance of genomic integrity(1). In response to DNA damage activation of the damage checkpoints prevents cellular division so as to protect against permanent genetic change. The checkpoint effector kinase Chk2 performs an essential role in mediating upstream damage sensing signals to downstream regulators of cell cycle and apoptosis (2–4). The coordinated transfer of signal from damage sensing to cell response is dependant upon the temporal activation/deactivation of Chk2 kinase and is at least partially mediated through phosphorylation status.
Damage sensing, initial end processing, and repair involve the recruitment of cellular factors that in turn serve as signals to activate the checkpoint response. In the case of double strand breaks, the MRE11-RAD50-NBS1 complex processes double strand breaks to single strand DNA, which in turn recruits additional repair complexes. The presence of MRE11-RAD50-NBS1, single strand DNA, and a variety of other factors serves to initiate ATM/ATR/DNA-PK activation (5). Activated ATM/DNA-PK directly phosphorylates the N-terminal SQ/TQ domain of Chk2. The specific SQ/TQ residues involved are Ser19, Thr26, Ser28, Ser33/Ser35, Ser50, and Thr68 (6–8). Phosphorylation of the SQ/TQ regulatory domain serves to propagate the damage-induced signal and initiates the autophosphorylation of Thr383 and Thr387 within the T-loop region of the kinase domain. Autophosphorylation is essential for attaining conformation conducive to kinase activity (9, 10). The phosphorylation of Thr383/Thr387 is conducted in trans thus requiring a second Chk2 protein. The dimerization and oligomerization steps are also facilitated by the FHA protein-protein interaction domain (8, 10). Autophosphorylation subsequent to Chk2 kinase activation can also occur at Ser516 through a cis-acting mechanism. However, although Ser516 phosphorylation impacts downstream signaling, it is not a prerequisite to kinase activation (11).
Given the critical role of phosphorylation within the T-loop region of the Chk2 kinase domain, we targeted this region in a direct analysis of the global phosphorylation status. We utilized site-directed multiple reaction monitoring to identify and quantify the changes in phosphorylation in response to ionizing radiation. We identified three novel phosphorylation sites, including one tyrosine phosphorylation event at Tyr390, that are required for kinase function. Phosphorylation of one of these residues, Thr389, was a prerequisite for phosphorylation at Thr387. In response to ionizing radiation (IR), phosphorylation at individual sites within this region were associated with changes in chromatin versus soluble nuclear localization. Conversely, phosphorylation at multiple sites was associated with an increase in nuclear versus chromatin protein. We confirmed the dependence of ubiquitylation on phosphorylation at Ser379 and extended this finding for Thr383 as well. Interestingly, the kinase-inactive Y390F mutant retained ubiquitylation. Together, these results provide for a more complex model of Chk2 activation involving interdependent phosphorylation within the T-loop region that regulates kinase activity, early DNA damage-induced chromatin targeting, and subsequent chromatin egress.
EXPERIMENTAL PROCEDURES
Plasmid, Cell Culture, and Transfection
The S-Chk2 expression vector has been described (12). HEK293T17 cells (Invitrogen) were maintained in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Transient transfections were performed by standard calcium phosphate transfection method.
Subcellular Fractionation
Cells were washed twice with cold phosphate-buffered saline (PBS) and cytosolic (S2), soluble nuclear (S3), and chromatin-bound (P3) proteins were collected according to methods we have described previously (12).
Purification of Chk2 Protein from Mammalian Cells
S-Chk2 proteins were affinity-precipitated from 500–1000 μg of total protein sample either from whole protein lysates or from the three fractions (S2, S3, and P3) described above by incubation with 50–100 μl of S-proteinTM agarose (Novagen, Madison, WI). The pellet was washed three times with 1 ml of bind/wash buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100). The protein was eluted from the beads by re-suspension in an equal volume of Laemmli sample buffer with β-mercaptoethanol followed by boiling for 10 min. Eluates were electrophoresed in a NuPAGE® Novex 4–12% Bis-Tris2 Gel (Invitrogen) and visualized by Coomassie Blue staining with Bio-Safe Coomassie (Bio-Rad). Bands of interest were manually excised from the gel for further analysis.
LC-MS/MS Analysis
Multiple reaction monitoring (MRM) assays were developed using the MIDASTM workflow system and optimized for tryptic peptides derived from the activation loop of Chk2. In addition, MRM transition pairs capable of discriminating between individual phosphorylation sites in multiply phosphorylated peptides were empirically obtained from MS/MS scans derived from prior analysis of Chk2 using an LTQ mass spectrometer (12). The samples were analyzed by nano-LC-MRM/MS using a hybrid triple quadrupole/linear ion trap mass spectrometer 4000 (QTRAP® LC/MS/MS system, Applied Biosystems, Foster City, CA) coupled to a Tempo NanoLC system (Eksigent Technologies, Dublin, CA). The chromatography conditions were: Solvent A (0.1% formic acid, 0.005% heptafluorobutyric acid) and Solvent B (95% acetonitrile in 0.1% formic acid, 0.005% heptafluorobutyric acid). Tryptic digests (8-ml injections) were eluted at 500 nl/min with PicoFrit columns (75-mm inner diameter, 2-mm tip opening, New Objective, Woburn, MA) slurry-packed in house with 10 cm of reversed-phase, 5-mm 100 Angstrom Magic C18 resin (Michrom Bioresources, Auburn, CA) and with a gradient of 5–10% Solvent B in 3 min, 10–60% solvent B in 48 min, and 60–95% solvent B for 5 min before re-equilibration with 95% A for 7 min. Data acquisition was performed with an ion spray voltage of 2800 V, curtain gas of 20 p.s.i., nebulizer gas of 10 p.s.i., and an interface heater temperature of 125 °C. Collision energy, declustering potential, and collision cell exit potential were optimized for maximum transmission and sensitivity of each MRM transition. A dwell time of 10 ms was used for MRM analysis of the digests, and in all the experiments. Four MRM transitions per peptide were monitored and acquired at unit resolution both in the first and third quadrupole (Q1 and Q3) to maximize specificity. All LC-MRM/MS experiments were performed in triplicate. Two Chk-2 internal tryptic peptides of sequence 198VFVFFDLTVDDQSVYPK (IS1) 263EADPALNVETEIEILK (IS2) and that were empirically determined to have stable and reproducible ionization characteristics were used for normalization of all MRM pairs within a run. The internal peptide standards were evaluated over multiple runs to ensure that their relative area ratios (∼3.5:1) were consistent. Further analysis was only done in experiments with deviations of <12%.
Phosphopeptide quantitation analysis was performed automatically using MultiQuantTM software (Applied Biosystems). Internal standard peak areas were calculated, and the values were summed for each experiment. The highest value among the four runs (Chromatin/Nuclear/+IR/−IR) was used as a baseline and to develop a normalization factor for all values within an experiment. MS/MS spectra were searched against the SwissProt non-redundant data base (SwissProt 57.1) using an in house Mascot Daemon Server (Matrix Science). Search parameters included fixed carbamidomethylation of cysteines; trypsin digestion with one missed cleavage; variable modifications included oxidized methionines, phosphorylation of serine, threonine, and tyrosine residues; precursor mass tolerance of 50 ppm; product ion mass tolerance of 0.5 Da; and scoring settings for ESI Q Trap.
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) to introduce single amino acid changes by altering one nucleotide at specific sites in the S-Chk2 template. The primers containing the desired mutation were designed according to the manufacturer's protocol. The mutagenesis PCR reactions and subsequent bacterial transformations were performed according to the manufacturer's protocol. Plasmid DNA was isolated from colonies grown under ampicillin selection, and the constructs containing the desired mutation were identified by sequence analysis (Genewiz, South Plainfield, NJ). The plasmid DNA was amplified then isolated using the PureLink HiPure Plasmid Filter Maxiprep Kit (Invitrogen). Sequences of all mutants were verified by DNA sequencing. Expression was confirmed by transfection and Western analysis using anti-Chk2 monoclonal antibody (Upstate, Lake Placid, NY).
Western Analysis
Total protein concentrations were determined by Protein Assay Dye Reagent (Bio-Rad). Equal amount of protein lysate from different samples containing DTT and NuPAGE SDS Sample Buffer (Invitrogen) was heated at 70 °C for 10 min and then separated through a NuPAGE® Novex 4–12% Bis-Tris Gel (Invitrogen). The proteins were then transferred onto Immobilon-FL (Millipore, Billerica, MA) membrane using a Trans-blot SD semi-dry transfer cell (Bio-Rad) at 400 mA for 50 min. Following blocking for 1 h with Odyssey blocker (LI-COR Biosciences, Lincoln, NE), membranes were incubated in primary antibody mixture containing mouse monoclonal anti-Chk2 antibody diluted 1:4,000 (Upstate, Lake Placid, NY) and Chk2 phosphoryl-T383 antibody diluted 1:1,000 (Abcam, Cambridge, MA), Chk2 phosphoryl-T387 antibody diluted 1:500 (Cell Signaling, Danvers, MA), Chk2 phosphoryl-T68 antibody diluted 1:2000 (Rockland, Gilbertsville, PA), Chk2 phosphoryl-S516 antibody diluted 1:500 (Abcam, Cambridge, MA), Chk2 phosphoryl-S33/S35 antibody diluted 1:500 (Epitomics, Burlingame, CA) or rabbit anti-GFP antibody diluted 1:2,000 (Santa Cruz Biotechnology, Santa Cruz, CA) overnight. Membranes were washed 5 min four times with PBS-T prior to incubation with a mixture of goat anti-mouse and goat anti-rabbit secondary antibody, conjugated to IR 680Dye and IR 800Dye (LI-COR Biosciences, Lincoln, NE), respectively, diluted 1:20,000. Excess antibody was removed with three 5-min washes in PBS-T. Prepared membranes were scanned for fluorescence emission at 700 and 800 nm, using an Odyssey® infrared imaging system (LI-COR Biosciences). Densitometric analysis of fluorescent signals was performed using the Odyssey scanner software.
In Vitro Kinase Assays
Chk2 kinase assays were carried out using the K-LISA Checkpoint Activity Kit (Calbiochem, EMD Chemicals Inc., San Diego, CA). Expression and isolation of Chk2 was achieved as described above. The S-bead-bound Chk2 was washed three times with bind/wash buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Triton X-100) and once with the 1× kinase assay buffer from the kit. The kinase reaction was carried out in the presence of ATP, MgCl2, and biotinylated substrate peptide (KKKVSRSGLYRSPSMPENLNRPR), derived from Cdc25C. Reaction mixtures were incubated at 30 °C for 30 min, and the supernatant containing phosphorylated substrate peptide was transferred to Streptavidin-coated 96-well plates. The captured phosphorylated substrate was detected using the peptide specific phosphoserine detection antibody and HRP-conjugated secondary antibody. Activity was determined by the absorbance at dual wavelengths 450/570 nm. S-GFP isolated under the same conditions was used a negative control during the assays. The amount of Chk2 in the assay was determined by recovery of protein into Laemmli buffer and analysis by Western following SDS-PAGE separation. Standard deviation across triplicate reactions was used to generate error estimates.
In Vivo Ubiquitylation Assay
HEK293T17 cells were co-transfected with plasmids encoding S-Chk2 (WT and mutants) and HA-tagged ubiquitin using the CaCl2 precipitant method. At 48 h post-transfection, cells were washed twice with ice-cold PBS and lysed in M-PER mammalian protein extraction buffer (Pierce) supplemented with N-ethylmaleimide (5 mm), protease inhibitor mixture (Thermo, Rockford, IL), and phosphatase inhibitor mixture (Sigma). Ectopically expressed S-Chk2 was affinity precipitated as described above. The precipitates were subjected to SDS-PAGE separation and Western analysis. Rabbit anti-HA antibody (Zymed Laboratory, Carlsbad, CA, 1:2000 dilution in blocking buffer) and mouse anti-Chk2 antibody were used to detect ubiquitylated Chk2 and affinity-precipitated total Chk2.
RESULTS
Expression and Isolation of Human Chk2
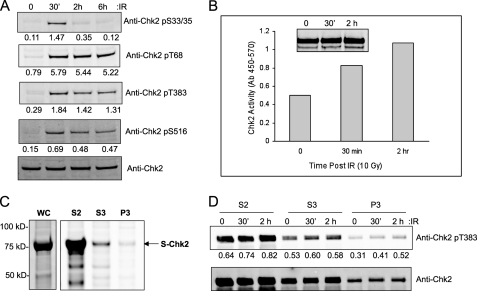
A comprehensive physical characterization of protein post-translational modification requires expression of sufficient amount of protein. We overexpressed Chk2 with an incorporated S tag for subsequent purification from the human cell line HEK293/T17. Because the cellular background can influence protein modification we examined endogenous Chk2 for IR-induced phosphorylation by immunoassay. In Fig. 1A, we show that this cell line displayed a normal response to IR in terms of the damage-response specific phosphorylation at Ser33/Ser35, Thr68, Thr383, and Ser516. We have previously demonstrated that this cell line displays intact downstream signaling by Chk2 in response to IR (13). These results suggest that our chosen cell model is appropriate to the proposed studies.
FIGURE 1.
Characterization of endogenous and exogenous Chk2 in HEK293T17. A, Western analysis of endogenous Chk2 protein phosphorylation following ionizing radiation (IR, 10 Gy) at indicated time points. The relative phosphorylation was determined by densitometry and normalized by total Chk2 protein. B, S-Chk2 was affinity-purified from HEK293T17 and subjected to in vitro kinase assay following exposure to IR. Shown is the relative kinase activity at the indicated time post exposure to 10 Gy IR. Total Chk2 was used as a normalization parameter (inset). C, S-Chk2 protein was purified from whole cell lysates and from the subcellular fractions, soluble cytoplasmic (S2), soluble nuclear (S3), and chromatin-bound protein (P3). The purified protein was then eluted, resolved by SDS-PAGE, and detected by Coomassie Blue staining. S-Chk2 protein is indicated. D, S-Chk2 purified from the subcellular fractions was also subjected to Western analysis for phosphorylation at Thr383. Chk2-expressing cells were subjected to 10 Gy IR and isolated at the indicated time points. The values from the analysis of the band intensity by densitometry are indicated. The relative densitometric analysis was normalized to total Chk2 protein as shown in the bottom panel.
It was into this background that we transiently expressed the Chk2 fusion protein and subsequently purified by affinity chromatography. The Chk2-expressing cells were subjected to IR prior to protein isolation and purification. The purified protein was normalized to total Chk2 and then assessed for kinase activity in vitro. As shown in Fig. 1B, the expressed and purified Chk2 protein displayed kinase activity, which was induced following exposure to IR. We then purified Chk2 protein from individual cytoplasmic, nuclear, and chromatin fractions, separated proteins by SDS-PAGE, and subjected to Coomassie staining for total protein (Fig. 1C). The relative amount of cytoplasmic Chk2 is in mass excess to protein in the nucleus. When we examined total purified transiently expressed Chk2 protein for IR-induced phosphorylation, we found that only residues Ser33/Ser35 displayed a measureable IR response (see Fig. 4). This is likely due to the overexpression and localization of the transfected Chk2 protein into the cytoplasm (Fig. 1C, S2 fraction). However, when we isolated Chk2 protein from the chromatin fraction we were able to observe additional IR-induced phosphorylation as we show for the T-loop residue Thr383 (Fig. 1D). Thus, the remainder of our experiments was conducted using Chk2 isolated from nuclear and chromatin fractions.
FIGURE 4.
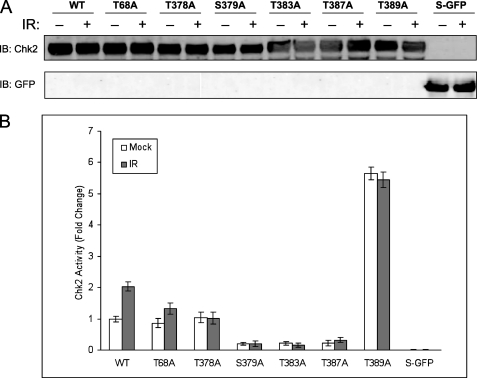
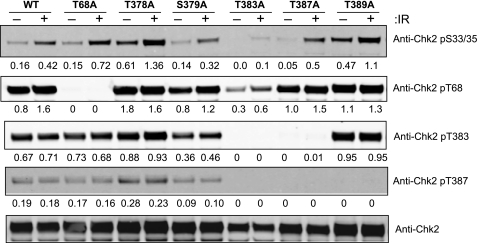
Effect of Chk2 substitution mutations on kinase activity. Cells were transfected with the appropriate plasmids to express wild-type S-Chk2 (WT) or the substitution mutants T68A, T378A, S379A, T383A, T387A, and T389A. Forty-six hours post-transfection, the cells were either mock or gamma-irradiated with 10 Gy IR. At 2 h post-IR, cells were harvested and whole cell lysates prepared. The Chk2 proteins were affinity-purified and subjected to in vitro kinase assay. A, Western analysis of Chk2 whole protein. B, kinase activity of all Chk2 proteins is shown. Mock and IR-treated values are indicated. The activity levels shown were normalized by the corresponding Chk2 total protein. Error bars indicate the standard error of the means from triplicate reactions. S-GFP protein was purified and evaluated as a negative control.
Identification of Novel Phosphorylation Sites within the T-loop Region of CHK2
MRM on a triple quadruple mass spectrometer allows for sensitive and specific quantitation of multiple post-translational modifications in proteins from highly complex samples (14–16). The essential requirement for the technology is the ability to detect a specific precursor ion (Q1 m/z) that is indicative of the targeted peptide, to isolate that ion for collision-induced fragmentation and, finally, to detect unique product ion (Q3 m/z), following fragmentation. We applied an MRM approach toward the discovery and quantitation of Chk2 T-loop phosphorylation species. We evaluated multiple putative phosphorylation sites in Chk2 by detecting signature ions unique for each specific modification event. Specifically we targeted eight sites in the activation T-loop of Chk2 protein (amino acid residues 365–406). This analysis is not possible using conventional antibody-based approaches due to the close proximity of multiple phosphorylation sites. MRM assays were developed using the MIDASTM workflow and optimized for tryptic peptides derived from the activation loop of Chk2. Two non-phosphorylated Chk-2 tryptic peptides were used for normalization of all MRM pairs across experimental runs. The sequence and the product ions selected for monitoring Chk2 peptides in the MRM assay are illustrated in supplemental Fig. 1 and listed in supplemental Table 1.
We isolated Chk2 protein from chromatin and nuclear fractions and conducted a survey for presence of phosphorylation in the target region. We were able to confirm the presence of known Chk2 phosphorylation at Ser379, Thr383, and Thr387. In addition, we identified four novel phosphorylation events at Ser372, Thr378, Thr389, and Tyr390. The modification at Tyr390 is the second report of a tyrosine phosphorylation of Chk2 (17). We could find no evidence of phosphorylation at Ser367.
Identification of IR-induced Chk2 Phosphorylation Species
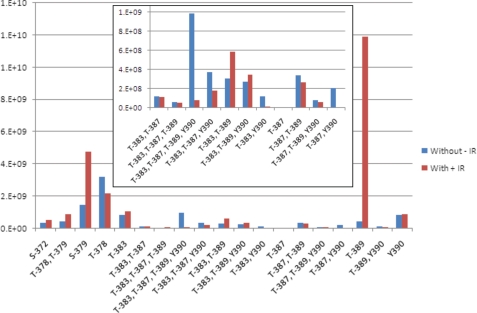
Quantitation of each Chk2 species in response to IR allowed for determination of IR-induced phosphorylation events. An example of the raw data is shown in supplemental Fig. 2, and the results for this analysis are summarized in Fig. 2. The most striking change response to IR was phosphorylation at Thr389. We observed a 26-fold increase in the monophosphorylated peptide. We also could confirm previous observations that Ser379 was IR-inducible. Monophosphorylation at Thr383 was only slightly increased following IR, but the double phosphorylation at Thr383/Thr389 nearly doubled following IR. Unique among all of the phosphorylated residues in this region, we saw no evidence of independent phosphorylation at Thr387 indicating that modification at this residue is a co-dependent process.
FIGURE 2.
Quantitative changes in phosphoryl-species in response to IR. The relative changes in the amounts of each observed peptide phosphoryl-species in response to ionizing radiation were calculated using quantitative multiple reaction monitoring mass spectrometry. The normalized ion counts for each species are indicated in the y axis graph. The target phosphoryl-species is indicated on the x axis. The value associated with each species before exposure to IR is indicated in blue (without −IR) and the values associated with the same species after exposure is indicated in red (with +IR). The inset contains an expansion of the y axis for those phosphoryl-species displaying lower values.
In some cases we observed a reduction in the presence of a phosphorylated species. We reasoned that this was the result of either IR-induced dephosphorylation or a phosphorylation event associated with inactive Chk2 (naïve to IR). Phosphorylation at Thr378 was not induced by exposure to IR but rather showed a damage-induced reduction in phosphorylated form. In addition, Chk2 phosphorylation species Thr383/Tyr390, Thr387/Tyr390, Thr389/Tyr390, and Thr383/Thr387/Tyr390 were also reduced following IR. Notable among the IR-induced reduction in phosphorylated species was the quadruple phosphorylated Chk2 species Thr383/Thr387/Thr389/Tyr390, which displayed a more than 12-fold decrease. In fact, most Chk2 proteins in which Tyr390 site was phosphorylated showed reduced levels following IR.
Quantitation of the Relative Distribution and Abundance of Chk2 Phosphorylation Species
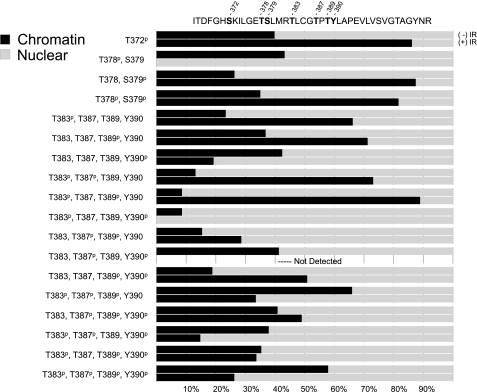
In this analysis we compared Chk2 species isolated from nuclear and chromatin cellular compartments. For each compartment we analyzed Chk2 before and after exposure to 5 Gy IR. We conducted a direct quantitation of each experimental condition and normalized to internal and external peptide controls. In this way we could achieve relative quantitation of the nuclear/chromatin distribution of each targeted Chk2 phosphorylation site. Our quantitation approach allowed for an assessment of IR-induced changes in the distribution of Chk2 species between chromatin and the nucleus (Fig. 3). We considered a 2-fold increase in the percent of a species in the chromatin as the criteria for chromatin targeting. Conversely we considered a 2-fold decrease in the percentage of a species in the chromatin as the criteria for chromatin egress. Designation as egress was contraindicated if the chromatin decrease was associated with a decrease in the percent of the same species in the nucleus. We observed a damage-induced chromatin targeting of Chk2 phosphorylated at Thr383 and Thr383/Thr387, consistent with these modifications signaling Chk2 activation and damage targeting. We observed a similar chromatin targeting of Chk2 phosphorylated at Ser372, Ser379, and Thr389. In addition, the double phosphorylation species Thr378/Ser379 and Thr383/Thr389 also displayed chromatin targeting following IR. Interestingly, only two species, the triple phosphorylation at Thr383/Thr387/Thr389 and the quadruple phosphorylation species at Thr383/Thr387/Thr389/Tyr390 displayed chromatin egress as defined in our study. This result would suggest that, although single and double phosphorylations in the T-loop signal recruitment to chromatin in response to damage, hyperphosphorylation of the same protein signals chromatin egress. In total these data support a model in which sequential phosphorylation of residues in the T-loop region regulate IR response.
FIGURE 3.
Changes in relative subcellular distribution of Chk2 phosphoryl-species in response to IR. The subcellular distribution of each Chk2 species was calculated as the relative amount identified in chromatin or nuclear cellular fractions. The percentage of total protein for each fraction is shown as a bar graph as indicated for each phosphoryl-species. The upper bar graph of each pair represents the relative distribution prior to exposure to IR. The lower bar graph of each pair represents the post-IR values. The targeted Chk2 region is shown at the top with separation of sequence to indicate tryptic sites. The amino acid residues for which phosphorylation was identified are indicated.
Novel S/T Phosphorylation Sites in the T-loop Region Involved in Kinase Activity
To determine whether the novel phosphorylation events were critical to activation of Chk2, we conducted site-directed mutational analysis. We engineered substitution of S/T > A to probe the functional significance of phosphorylation at specific T-loop region sites. The targeted substitution mutations T378A, S379A, T383A, T387A, and T389A were introduced into the S-Chk2 background. These T-loop region mutations, along with wild-type protein and the well characterized mutation at T68A, were transiently expressed in HEK293/T17 cells and subsequently isolated and purified. The purified protein was then assessed for kinase activity in vitro. The measured kinase activity from each study point was normalized to total purified protein as shown in Fig. 4A. As shown in Fig. 4B, wild-type protein displayed IR-inducible kinase activity. The T68A mutant retained kinase function but was reduced for response to IR. The substitution mutations S379A, T383A, and T387A showed dramatically reduced kinase activity. New to this study, we show that substitution at T378A retained wild-type basal kinase activity but failed to respond to IR. Interestingly, the substitution mutation of T389A resulted in a 6-fold increase in basal kinase activity when compared with wild-type protein. This result suggested that phosphorylation at Thr389 may represent a repressive phosphorylation event.
Phosphorylation at Thr389 Is a Prerequisite for Phosphorylation of Thr387
We next examined the dependence of specific Chk2 phosphorylation signaling events upon selected substitution mutations. Chk2 isolated from total cell lysates was examined for phosphorylation at Ser33/Ser35, Thr68, Thr383, and Thr387 (Fig. 5). As expected the substitution mutation at Thr68 was the only mutant that ablated reaction with anti-Chk2 pThr68. Although the overall intensity was altered, the strict dependence of Ser33/Ser35 phosphorylation upon DNA damage was unaffected by any of the tested mutants. Consistent with previous studies the phosphorylation of Thr383 and Thr387 were interdependent, because mutation at either site ablates recognition by relevant antibody to the other site. Interestingly, the T389A mutant protein displays phosphorylation at Thr383 but not at Thr387. Thus, phosphorylation at Thr389 may be a prerequisite for Thr387 modification. These results confirmed the interdependence of Thr383 and Thr387 and proposed a novel dependence of Thr387 phosphorylation upon Thr389.
FIGURE 5.
Effect of Chk2 substitution mutations on intramolecular phosphorylation. Cells were transfected with the appropriate plasmids and treated with IR, and whole cell lysates were prepared. Total protein was measured by BCA, and 40 μg was loaded for SDS-PAGE separation and subsequent Western analysis by the indicated antibodies. Densitometry was used to generate the relative expression values that were also corrected to the normalized value of total Chk2 protein. Phosphorylation in response to IR was determined and shown as indicated.
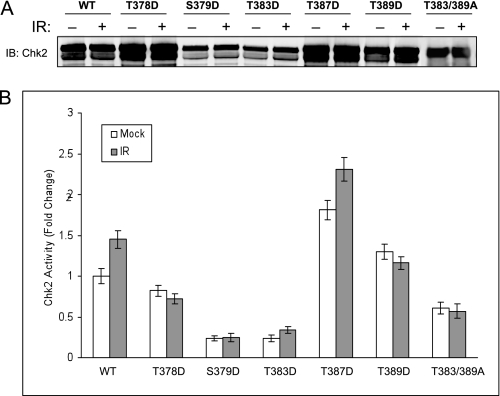
The use of so-called phosphorylation mimetics is problematic primarily due to the difficulty in interpretation of negative data. However, the use of mimetics in which protein function is restored can provide useful structural inferences. Therefore, we introduced aspartic acid substitutions for serine/threonine at T378D, S379D, T383D, T387D, and T389D (Fig. 6A). These mutations were generated to specifically rescue kinase activity of these mutations. Substitution mutations T378D, S379D, and T383D resulted in kinase activity indistinguishable from the relevant T/S > A mutation, and these results were not considered further. However, the substitution mutation T387D resulted in fully restored kinase function although less responsive to IR (Fig. 6B). These results confirm the dependence of Chk2 kinase function upon Thr387 structural changes consistent with phosphorylation and imply that the structural constraints placed upon Thr387 are more tolerant than those required for Thr383 phosphorylation. We also observed that T389D substitution mutation displayed functional kinase activity but at a level reduced to near wild-type Chk2 (Fig. 6B). We further reasoned that if phosphorylation at Thr387 is dependent upon prior phosphorylation of Thr389, then the T389A mutant must rely upon phosphorylation at Thr383 for kinase function. Therefore, the double mutation T383A/T389A should be kinase defective. As expected this double mutation was defective for kinase activity.
FIGURE 6.
Effect of Chk2 phosphoryl-mimetic substitution mutations on kinase activity. Cells were transfected with the appropriate plasmids to express wild-type S-Chk2 (WT) or the phosphoryl-mimetic substitution mutant of aspartic acid for serine/threonine (T378D, S379D, T383D, T387D, and T389D) and the double substitution mutant (T383A/T389A). Forty-six hours post-IR, the cells were either mock or gamma-irradiated with 10 Gy of IR. At 2-h post-IR, cells were harvested and lysed. Affinity-purified protein was subjected to in vitro kinase assay. A, Western analysis of isolated whole Chk2 protein. B, Chk2 kinase activity relative to wild-type protein was obtained. The activity measurements were normalized to corresponding isolated total Chk2 protein. Pre-and post-exposure to IR is indicated. Error bars indicate the standard error of the means from triplicate reactions.
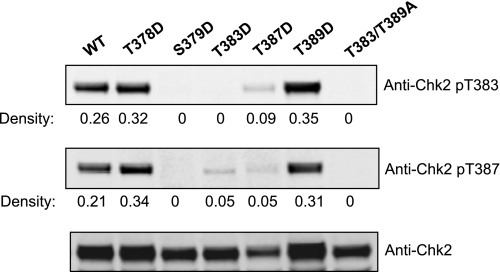
Interestingly, the T387D mutant restored antibody recognition of a phosphoryl-Thr387 species suggesting that the mimetic provided a structure consistent with phosphorylated Thr387 (Fig. 7). The T389D mimetic substitution also restored phosphorylation at Thr387 supporting the concept of interdependence of these two sites. These results underscore the difference in structural constraints placed upon Thr383 and Thr387 and further support a role for Thr389 phosphorylation in regulating modification at Thr387.
FIGURE 7.
Effect of phosphoryl-mimetic substitution mutations on Chk2 intramolecular phosphorylation. Cells expressing Chk2 (WT) and phosphoryl-mimetic mutants T378D, S379D, T383D, T387D, and T389D and the double substitution mutant T383A/T389A were harvested and protein-purified. Total protein was determined by BCA, and 40 μg of total protein was loaded onto SDS-PAGE for separation and Western analysis with the indicated antibody. Densitometry was used to generate the relative expression values that were also corrected to the normalized value of total Chk2 protein.
Phosphorylation of Chk2 at Tyr390 Is Required for Kinase Activity
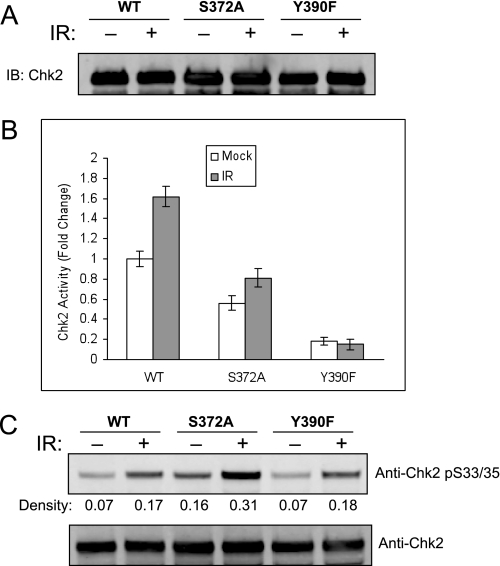
Our observation of phosphorylation at Tyr390 is the first report of specific modification of a tyrosine in Chk2 and suggests potential involvement of novel regulatory pathways. We had established that Tyr390 phosphorylation is not IR-induced, and here we examine the impact of Tyr390 modification on Chk2 function. The S372A protein was analyzed with Tyr390 and wild-type protein. Similar amounts of purified protein were expressed and isolated (Fig. 8A). The purified protein was assayed for kinase function as described. The S372A mutation retained near wild-type activity, although response to IR was impaired (Fig. 8B). However, the Y390F mutant was deficient for in vitro kinase activity. Each of the mutations displayed no effect upon early signal transduction as determined through specific phosphorylation at Ser33/Ser35 (Fig. 8C). These results demonstrate that Tyr390 phosphorylation is critical to Chk2 kinase function.
FIGURE 8.
Phosphorylation at Tyr390 is required for Chk2 kinase activity. Cells expressing S-Chk2 (WT) or S-Chk2 substitution mutants S372A and Y390F were exposed to either mock or 10 Gy of IR for 2 h and harvested. S-Chk2 protein was affinity-purified. A, the purified protein was normalized to total protein and loaded on SDS-PAGE for Western analysis with the indicated antibody. B, a portion of the same protein in A was subjected to in vitro kinase assay. The activity levels were normalized by the corresponding relative amount of whole protein. Error bars indicate the standard error of the means from triplicate reactions. C, a separate 40-μg portion of the purified protein was loaded on SDS-PAGE for Western analysis with the indicated antibody. Densitometry was used to generate the relative expression values that were also corrected to the normalized value of total Chk2 protein. Phosphorylation in response to IR was determined and shown as indicated.
Phosphorylation in the T-loop Region Has Variable Impact on Ubiquitylation
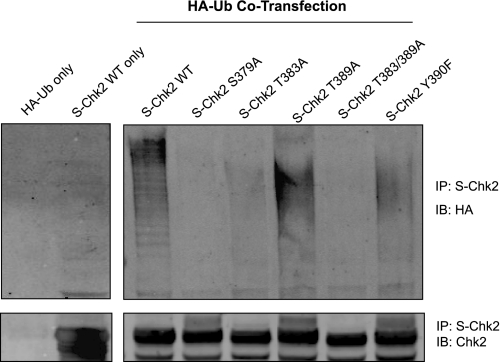
Recently, ubiquitylation of Chk2 has been shown to be essential to effective downstream repair-response function (18). In this same study it was reported that phosphorylation at Ser379 was a prerequisite to Chk2 ubiquitylation. In light of the relationship between phosphorylation and ubiquitylation of Chk2, we assessed the potential impact of phosphorylation of T-loop residues on Chk2 ubiquitylation. We show in Fig. 9 that wild-type protein was ubiquitylated efficiently as evidenced by presence of higher mass HA-ubiquitin-Chk2 species. As previously reported the S379A Chk2 mutation resulted in loss of Chk2 ubiquitylation. We also observed loss of Chk2 ubiquitylation in the kinase-deficient mutants T383A and T383A/T389A and preservation of Chk2 ubiquitylation in the hyperactive mutant T389A. Interestingly, the Y390F mutant retained Chk2 ubiquitylation and was the only kinase inactive mutation that was still ubiquitylated. These results suggest a complex relationship between phosphorylation of residues in the T-loop region and Chk2 ubiquitylation.
FIGURE 9.
Chk2 phosphorylation regulates ubiquitylation. Cells expressing S-Chk2 (WT) or the S-Chk2 substitution mutations (S379A, T383A, T389A, T383/389A, and Y390F) and HA-tagged ubiquitin (HA-Ub) were harvested, and the lysates were used for affinity purification (IP) of the Chk2 proteins. The proteins were separated on SDS-PAGE for Western analysis with the indicated antibody. Ubiquitylated proteins were visualized by immunoblotting (IB) with an antibody to HA tag. The total affinity-purified Chk2 protein was determined by IB with Chk2 antibody in the same filter. The experiment was conducted three times with comparable results.
DISCUSSION
Chk2 is a critical mediator of damage-dependent ATM/ATR signaling, and the T-loop exchange region within the kinase domain plays an essential role in Chk2 activation (2–4). DNA damage-induced activation of Chk2 initiates an intramolecular cascade of phosphorylation events. The initial signaling events are phosphorylation of residues in the SQ/TQ domain that appear to facilitate dimerization of two Chk2 molecules. The dimerization is required for trans-autophosphorylation of Thr383 and Thr387 within the activation T-loop exchange region, an event needed for full activation of kinase function. Chk2 thus activated in turn phosphorylates and activates a variety of target proteins involved in DNA repair and cell cycle regulation. Although specific phosphatases have been identified that dephosphorylate Chk2 at Thr68 (19–21), the independence of Thr383/Thr387 phosphorylation from Thr68 phosphorylation status suggests that other mechanisms exist to fine tune the kinase activation response (22).
The dimerization of Chk2 is mediated by direct interaction of the SQ/TQ domain of one molecule with the FHA domain of an adjacent molecule. This arrangement facilitates the trans-autophosphorylation of Thr383 on one molecule pair and Thr387 on the second molecule pair. The crystal structure of the Chk2 kinase domain in an activated state was recently solved (23) and revealed some physical detail of the T-loop exchange region. The crystal structure established that the T-loop consists of two α helices bordering a structured loop. Notably, residue Thr383 is within the “left-hand” helix (residues 377–386), whereas residue Thr387 lies within the more relaxed loop segment (residues 387–392). The positioning of Thr383 and Thr387 in structurally diverse regions suggests that the physical constraints upon Thr383 are different from those of Thr387 with the later being more flexible. Our study showed that T > D mimetic substitution of Thr387 could assume a structure recognized by a phospho-Thr387-specific Chk2 antibody, whereas T > D substitution at Thr383 failed to do so. In addition, the T387D mutant displayed kinase activity, whereas the T383D mutant was inactive. The entire loop segment, at least in the activated state, projects into a hydrophobic pouch formed by the second Chk2 molecule. Thus, our observation of a functional dependence upon phosphorylation at Ser389 and Tyr390 is consistent with the predicted steric impact of introduction of a phosphate group into a hydrophobic pocket.
The interdependence of Thr383 and Thr387 is assumed in fulfillment of an autophosphorylation model and has been demonstrated experimentally (6, 18). In our analysis we could confirm the dependence of phosphorylation at Thr387 on co-phosphorylation at Thr383. Interestingly, although we were not able to detect any instance of a single phosphorylation at Thr387, we did detect peptides with single phosphorylation at Thr383. This would suggest that phosphorylation at Thr383 precedes phosphorylation at Thr387. In fact our data showed evidence of singular phosphorylation events at all examined T-loop region sites (Thr372, Thr378, Ser379, Thr383, Thr389, and Tyr390) except for Thr387, suggesting a unique structural prerequisite for Thr387 phosphorylation.
Unique among all the residues examined in our study, Thr389 was the only phosphorylation event associated with a reduction in activity. Specifically, inhibition of phosphorylation at Thr389 via T > A substitution resulted in a dramatic increase in the kinase activity of Chk2. On the other hand introduction of a phosphomimetic substitution T > D at Thr389 reduced measureable kinase activity to levels equal to wild-type protein. Our findings also support a model in which phosphorylation of Thr387 is dependent upon phosphorylation at Thr389, because the T > A substitution at Thr389 abolished recognition of Chk2 by a phospho-Thr387-specific antibody. Conversely, the T > D mimetic substitution at Thr389 restored recognition of phospho-Thr387 Chk2. Thus, phosphorylation at Thr389 appears to be a prerequisite to phosphorylation at Thr387 but associated with reduced kinase function (albeit at wild-type protein levels).
One unique aspect to our study is the ability to acquire quantitative information on the relative abundance of phosphorylation at specific sites in response to exposure to IR. In addition, we could simultaneously obtain insight into the relative distribution of specific phosphorylation events between nuclear and chromatin compartments. This allows us to relate phosphorylation events to protein localization. We observed relative changes in abundance indicating IR-induced de novo phosphorylation and IR-induced loss of phosphorylation. Likewise we determined relative changes in the localization of Chk2 species consistent with chromatin targeting and chromatin egress. These results suggest a complex interrelationship between phosphorylation status of multiple T-loop sites and chk2 localization.
We could document a de novo increase in phosphorylation at Ser379, Thr389, and Thr383/Thr389 following exposure to IR. We also observed a modest de novo increase in single phosphorylation species at Thr383 following IR. There was a notable increase in Thr383 single phosphorylation species in the chromatin fraction following IR, but this was compensated for by a loss in the nuclear abundance of the same species. We failed to detect the existence of an independent phosphorylation at Thr387 under any conditions. These results suggest that phosphorylation at Thr387 is dependent upon prior phosphorylation. Our findings also suggest that Thr389 phosphorylation is an IR-induced de novo event and a prerequisite for Thr387 phosphorylation. It may be that the Thr383/Thr389 double phosphorylation species represents the initiating IR-dependent event that precedes phosphorylation at Thr387. Phosphorylation at Thr378, which had no impact upon kinase function, was dramatically reduced in the chromatin fraction following IR treatment. The tyrosine phosphorylation at Tyr390 was associated with IR-induced loss in Thr383/Tyr390, Thr387/Tyr390, and Thr389/Tyr390. We conclude that Thr378 and Tyr390 might be involved in an IR-induced dephosphorylation event or serve as potential signals for recovery of activated Chk2 into a naïve state. Further studies are required to evaluate these two hypotheses.
Chromatin targeting events post-IR were presumed to be phosphorylation changes associated with recruitment of Chk2 to sites of damage as a result of DNA damage response initiation. We observed phosphorylation at Thr372, Ser379, Thr383, and Thr389 to be associated with chromatin targeting after IR. We also observed double phosphorylation at Thr378/Ser379, Thr383/Thr387, and Thr383/Thr389 to be associated with chromatin targeting. In some cases the IR-induced increases in chromatin presence of these phosphorylated forms of Chk2 were reciprocal with the same fold decrease in the nucleus. This result is supportive of an IR-induced exchange and was noted for phosphorylation at Thr372, Thr383, and Thr383/Thr387. In other cases the increase in chromatin-localized Chk2 was associated with a de novo increase in specific phosphorylated forms, suggesting an IR-induced phosphorylation. This was observed for phosphorylation at Ser379, Thr389, Thr378/Ser379, and Thr383/Thr389. These results imply that de novo phosphorylation at these sites is a requisite to chromatin targeting.
Chromatin egress is an observed localization change in Chk2 associated with normal DNA damage response (24). It is speculated that egress may regulate downstream signaling, and the movement of Chk2 from sites of damage to the nucleoplasm is consistent with cellular target activation by Chk2. We defined chromatin egress as IR-induced reduction in the chromatin/nucleus ratio in the absence of reduced nuclear abundance of the same species. We observed chromatin egress of Chk2 to be associated with phosphorylation at Tyr390, Thr383/Thr387/Thr389, and Thr383/Thr387/Thr389/Tyr390. A reciprocal exchange was noted for Tyr390 and Thr383/Thr387/Thr389. Overall it seems that hyperphosphorylation of the 383–406 peptide was associated with chromatin egress. For some phosphorylation species, such as Thr387/Thr389, Thr383/Thr389/Tyr390, and Thr387/Thr389/Tyr390, the relative distribution and abundance did not change in response to IR. Thus, the functional significance of these events in IR-mediated signaling could not be established.
The critical role of ubiquitylation in regulating the molecular events that comprise cellular DNA damage response has recently come to light (25, 26). The observation that Chk2 function is dependent upon ubiquitylation suggests a possible role for this modification in regulating subcellular localization and/or protein turnover. We confirmed the requirement of phosphorylation at Ser379 and also showed that phosphorylation at Thr383 is required for Chk2 ubiquitylation. Because phosphorylation at Thr383 is required for kinase activation the dependence of ubiquitylation upon Thr383 phosphorylation may link this modification to kinase activity. Interestingly, we observed that the kinase inactive mutant Y390F was ubiquitylated. However, in contrast to either Ser379 or Thr383, phosphorylation at Tyr390 was almost always reduced in response to IR. Thus, ubiquitylation in response to IR can be maintained in the presence of altered kinase activity. The full implication of the relationship between phosphorylation, ubiquitylation, and chromatin targeting/egress needs further study.
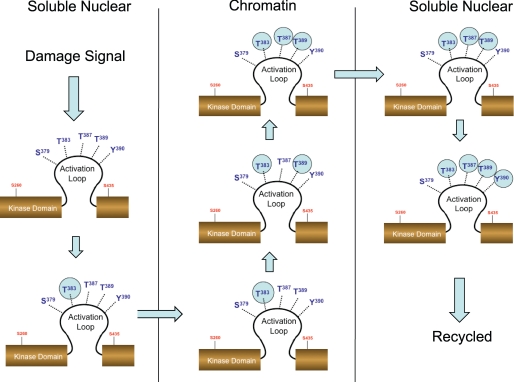
Our data suggests that IR induces de novo phosphorylation at Ser379, Thr389, and Thr383/Thr389. Of these events we suggest that the Thr383/Thr389 would be a prerequisite for phosphorylation at Thr387 and signal formation of oligomeric activated Chk2. The formation of the Chk2 species phosphorylated at Thr383/Thr387/Thr389 then signals chromatin egress of activated Chk2. The interplay between these three residues provides a mechanism for activation and egress of protein. Later events such as phosphorylation at Tyr390 may signal recycling of Chk2 from active to naïve state (Fig. 10). We confirm the interdependence of Thr383 and Thr387 and underscore the dependence of Thr387 phosphorylation on prior phosphorylation at both Thr383 and Thr389. These results support a model in which multiple interdependent phosphorylation events within the Chk2 T-loop region regulate chromatin targeting, kinase activation, chromatin egress, and recycling.
FIGURE 10.
Interdependent phosphorylation within the activation loop mediates damage-induced activation and subcellular redistribution of Chk2. Our model suggests that initial phosphorylation at Thr383 in response to IR results in localization of Chk2 to chromatin. Subsequent phosphorylation at Thr389 allows for autophosphorylation of Thr387. This triple phosphorylated form of Chk2 is capable of chromatin egress. Phosphorylation at Tyr390 may be a signal for recycling or degradation.
Supplementary Material
Acknowledgment
We thank Patrick Lundberg for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA76595 (to O. J. S.) from the NCI, U.S. Public Health Services. The authors declare they have no conflict of interest.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MRM
- multiple reaction monitoring.
REFERENCES
- 1.Lukas J., Lukas C., Bartek J. (2004) DNA Repair 3, 997–1007 [DOI] [PubMed] [Google Scholar]
- 2.Antoni L., Sodha N., Collins I., Garrett M. D. (2007) Nat. Rev. Cancer 7, 925–936 [DOI] [PubMed] [Google Scholar]
- 3.Bartek J., Falck J., Lukas J. (2001) Nat. Rev. Mol. Cell Biol. 2, 877–886 [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Poon R. Y. (2008) Front. Biosci. 13, 5016–5029 [DOI] [PubMed] [Google Scholar]
- 5.McGowan C. H., Russell P. (2004) Curr. Opin. Cell Biol. 16, 629–633 [DOI] [PubMed] [Google Scholar]
- 6.Buscemi G., Carlessi L., Zannini L., Lisanti S., Fontanella E., Canevari S., Delia D. (2006) Mol. Cell. Biol. 26, 7832–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuoka S., Nakagawa T., Masuda A., Haruki N., Elledge S. J., Takahashi T. (2001) Cancer Res. 61, 5362–5365 [PubMed] [Google Scholar]
- 8.Xu X., Tsvetkov L. M., Stern D. F. (2002) Mol. Cell. Biol. 22, 4419–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C. H., Chung J. H. (2001) J. Biol. Chem. 276, 30537–30541 [DOI] [PubMed] [Google Scholar]
- 10.Schwarz J. K., Lovly C. M., Piwnica-Worms H. (2003) Mol. Cancer Res. 1, 598–609 [PubMed] [Google Scholar]
- 11.Wu X., Chen J. (2003) J. Biol. Chem. 278, 36163–36168 [DOI] [PubMed] [Google Scholar]
- 12.Gupta S. K., Guo X., Durkin S. S., Fryrear K. F., Ward M. D., Semmes O. J. (2007) J. Biol. Chem. 282, 29431–29440 [DOI] [PubMed] [Google Scholar]
- 13.Haoudi A., Daniels R. C., Wong E., Kupfer G., Semmes O. J. (2003) J. Biol. Chem. 278, 37736–37744 [DOI] [PubMed] [Google Scholar]
- 14.Kondrat R. W., Cooks R. G., McLaughlin J. L. (1978) Science 199, 978–980 [DOI] [PubMed] [Google Scholar]
- 15.Unwin R. D., Griffiths J. R., Leverentz M. K., Grallert A., Hagan I. M., Whetton A. D. (2005) Mol. Cell Proteomics 4, 1134–1144 [DOI] [PubMed] [Google Scholar]
- 16.Unwin R. D., Griffiths J. R., Whetton A. D. (2009) Nat. Protoc. 4, 870–877 [DOI] [PubMed] [Google Scholar]
- 17.King J. B., Gross J., Lovly C. M., Rohrs H., Piwnica-Worms H., Townsend R. R. (2006) Anal. Chem. 78, 2171–2181 [DOI] [PubMed] [Google Scholar]
- 18.Lovly C. M., Yan L., Ryan C. E., Takada S., Piwnica-Worms H. (2008) Mol. Cell. Biol. 28, 5874–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto H., Onishi N., Kato N., Takekawa M., Xu X. Z., Kosugi A., Kondo T., Imamura M., Oishi I., Yoda A., Minami Y. (2006) Cell Death Differ. 13, 1170–1180 [DOI] [PubMed] [Google Scholar]
- 20.Liang X., Reed E., Yu J. J. (2006) Int. J. Mol. Med. 17, 703–708 [PubMed] [Google Scholar]
- 21.Oliva-Trastoy M., Berthonaud V., Chevalier A., Ducrot C., Marsolier-Kergoat M. C., Mann C., Leteurtre F. (2007) Oncogene 26, 1449–1458 [DOI] [PubMed] [Google Scholar]
- 22.Ahn J., Urist M., Prives C. (2004) DNA Repair 3, 1039–1047 [DOI] [PubMed] [Google Scholar]
- 23.Oliver A. W., Paul A., Boxall K. J., Barrie S. E., Aherne G. W., Garrett M. D., Mittnacht S., Pearl L. H. (2006) EMBO J. 25, 3179–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Stern D. F. (2005) J. Biol. Chem. 280, 37948–37956 [DOI] [PubMed] [Google Scholar]
- 25.Bennett R. A., Wilson D. M., 3rd, Wong D., Demple B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7166–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergink S., Jentsch S. (2009) Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.