Abstract
The rapid emergence of drug-resistant variants of human immunodeficiency virus, type 1 (HIV-1), has limited the efficacy of anti-acquired immune deficiency syndrome (AIDS) treatments, and new lead compounds that target novel binding sites are needed. We have determined the 3.15 Å resolution crystal structure of HIV-1 reverse transcriptase (RT) complexed with dihydroxy benzoyl naphthyl hydrazone (DHBNH), an HIV-1 RT RNase H (RNH) inhibitor (RNHI). DHBNH is effective against a variety of drug-resistant HIV-1 RT mutants. While DHBNH has little effect on most aspects of RT-catalyzed DNA synthesis, at relatively high concentrations it does inhibit the initiation of RNA-primed DNA synthesis. Although primarily an RNHI, DHBNH binds >50 Å away from the RNH active site, at a novel site near both the polymerase active site and the non-nucleoside RT inhibitor (NNRTI) binding pocket. When DHBNH binds, both Tyr181 and Tyr188 remain in the conformations seen in unliganded HIV-1 RT. DHBNH interacts with conserved residues (Asp186, Trp229) and has substantial interactions with the backbones of several less well-conserved residues. On the basis of this structure, we designed substituted DHBNH derivatives that interact with the NNRTI-binding pocket. These compounds inhibit both the polymerase and RNH activities of RT.
Human immunodeficiency virus, type 1 (HIV-1), reverse transcriptase (RT) is essential for HIV replication. RT converts the single-stranded viral genomic RNA into a linear double-stranded DNA that can be integrated into the host chromosomes (reviewed in ref 1). The enzyme has two activities, (i) a DNA polymerase that can use either RNA or DNA as a template and (ii) an RNase H (RNH) that selectively degrades the RNA strand of an RNA–DNA heteroduplex. The RNH activity of RT is required for virus replication; cellular RNH cannot substitute for the retroviral enzyme (2). The RNH activity degrades the genomic RNA during first-strand (“minus-strand”) DNA synthesis, which allows the newly synthesized DNA to be used as a template for second-strand (“plus-strand”) DNA synthesis.
HIV-1 RT is a heterodimer consisting of 66 kDa (p66) and 51 kDa (p51) subunits. The two polypeptide chains have 440 N-terminal amino acid residues in common. These comprise four polymerase subdomains: the thumb, palm, fingers, and connection (3, 4). The C-terminus of p66 contains an additional 120 amino acid residues that form the bulk of the RNH domain. Despite having identical N-terminal sequences, the arrangement of the subdomains in the two subunits differs dramatically. The p66 subunit contains a large cleft formed by the fingers, palm, and thumb subdo-mains that can accommodate double-stranded nucleic acid template–primers (3–6). Although the p51 subunit contains the same four subdomains, it does not form a nucleic acid binding cleft.
Because of its pivotal role in the HIV life cycle, HIV RT is a primary target for antiretroviral agents. All RT inhibitors currently approved for the treatment of acquired immune deficiency syndrome (AIDS) inhibit the polymerase activity of HIV-1 RT; there are no anti-AIDS drugs that specifically inhibit RNH. There are two major classes of anti-RT drugs: nucleoside/nucleotide RT inhibitors (both called NRTIs for simplicity), and non-nucleoside RT inhibitors (NNRTIs). NRTIs block reverse transcription because they lack a hydroxyl group at the 3′-position of the ribose ring and, when incorporated into viral DNA by RT, act as chain terminators. The NNRTIs, in contrast to NRTIs, bind in a hydrophobic pocket ∼10 Å from the polymerase active site (Figure 1) and act noncompetitively. Binding an NNRTI does not prevent the binding of the nucleic acid or nucleoside triphosphate substrates to RT; rather, the NNRTI blocks the chemical step of the polymerization reaction (7, 8). Crystallographic studies (9, 10) have shown that the binding of an NNRTI causes conformational changes near the polymerase active site of HIV-1 RT, including a displacement of the β12-β13-β14 sheet that contains the polymerase primer grip (9–12), which is important for properly positioning the nucleic acid relative to the polymerase active site. Binding an NNRTI can also influence the geometry at the polymerase catalytic site (13–15). Many NNRTIs do not affect RNH activity; however, certain NNRTIs, rather than inhibit RNH activity, have been reported to increase the number of RNH cleavages and the rate of RNH activity under certain conditions (16–18).
Figure 1.
HIV-1 RT bound with DHBNH. Although DHBNH primarily inhibits the RNH activity, it binds >50 Å away from the RNH subdomain, at a site that partially overlaps the NNRTI-binding pocket. The subdomains of the p66 subunit are color-coded (fingers in blue, palm in red, thumb in green, connection in yellow, and RNH in gold). Upper left inset: a close-up of the DHBNH binding site. The inhibitor is shown in magenta. The position that would be occupied by an NNRTI is shown in gray (the NNRTI pocket is not occupied in the current structure). Upper right inset: chemical structure of DHBNH.
ⓦ See a full 3D interactive version of this figure on the ACS Chemical Biology web page. Access to the full structure is available with the molecular visualization tool FirstGlance in Jmol (http://molvis.sdsc.edu/fgij/index.htm).
The early successes of highly active antiretroviral therapy are now threatened by the emergence of drug-resistant viral variants, which arise from the rapid and error-prone replication of the virus (reviewed in ref 19). Because the virus can be suppressed but not eradicated in patients, drug treatments are life-long. This makes the toxicity of many of the existing drugs a significant problem in AIDS therapy. It is important to develop new inhibitors of HIV-1 that will target novel binding sites and inhibit essential viral functions that are not affected by existing drugs. Cross-resistance between such new inhibitors and existing drugs is unlikely. One such target is the RNH activity of HIV-1 RT.
Several classes of HIV-1 RNH inhibitors (RNHIs) have been reported, some of which have sub-micromolar activity. In contrast, most of the effective anti-AIDS drugs have IC50 values in the nanomolar or sub-nanomolar range, so the potency of the current RNHIs needs to be substantially improved. One of the most potent classes of RNHIs is the N-acyl hydrazone (NAH) analogues that are derivatives of N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone (BBNH) (20, 21). NAH compounds have been shown to inhibit either the polymerase or the RNH activity of RT and, in some cases, both (20, 21).
The development of effective RNHIs has been hampered by the lack of detailed knowledge of how the current lead compounds interact with HIV-1 RT. To better understand the mechanisms of the RNH inhibition and to help design improved inhibitors, we solved the crystal structure of HIV-1 RT in complex with the novel NAH analogue (E)-3,4-dihydroxy-N′-((2-methoxynaphthalen-1-yl)methylene)benzohydrazide (DHBNH) at 3.15 Å resolution.
RESULTS AND DISCUSSION
Inhibitory Activity of DHBNH
DHBNH inhibited the RNH activity of HIV-1 RT with an IC50 of 0.5 µM (Table 1); this inhibition was noncompetitive with respect to the hybrid duplex nucleic acid substrate (data not shown). In contrast, DHBNH was unable to inhibit the RNA-dependent DNA polymerase (RDDP) activity of HIV-1 RT in standard processive RDDP assays using poly(rA)– oli-go(dT) as template–primer. DHBNH was also ∼40-fold less potent at inhibiting a catalytically active isolated HIV-1 RT-RNH domain (22, 23). This suggests that the binding pocket for interaction of DHBNH with RT may be outside the RNH domain of the enzyme.
TABLE 1.
Some inhibitory properties of DHBNH
| Parameter | IC50 (µM)a |
|---|---|
| Inhibition of RT-RNH (intact enzyme) |
0.5 ± 0.2 (noncompetitive) |
| Inhibition of RT RDDP activity | >25 |
| Inhibition of p15-EC RNH fragment |
18.5 ± 3.4 |
Values are the means ± standard deviation (SD) determined from at least five separate experiments, each carried out in duplicate.
Overall Protein Conformation
The RT/DHBNH complex crystallizes with an overall RT conformation similar to that observed in RT/NNRTI complexes. In this conformation, the cleft between the fingers and the thumb of p66 is wider than that in HIV-1 RTs complexed with either DNA–DNA (4, 5, 15) or RNA–DNA (6) template–primers. In the RT/DHBNH structure, the inhibitor does not bind in the vicinity of the RNH active site; instead, it binds to a novel site >50 Å away, between the NNRTI-binding pocket and the polymerase active site (Figure 1 and Figure 2). The binding site, located in the palm of p66, is formed by the β12–β13 loop (including the polymerase primer grip, residues 229–231), Val108 of the β6 strand, and the β10 strand (including residues 186–188). The inhibitor binds within 3.5 Å of the catalytic YMDD sequence in the β9–β10 turn (p66 residues 183–186). DHBNH is oriented with its benzoyl ring partially entering the NNRTI pocket and the naphthyl ring system near the polymerase active site and the polymerase primer grip (Figure 2).
Figure 2.
Electron density map of the bound inhibitor. A simulated-annealing Fo − Fc omit map is shown at the 3σ contour level, generated with DHBNH omitted from the phase calculation. The binding site of DHBNH is adjacent to the polymerase active site (green), the polymerase primer grip (cyan), and the NNRTI-binding pocket (gray). The naphthyl ring appears to be positioned so that most of its contacts are made with Leu228. The carbonyl oxygen of Leu228 forms a hydrogen bond with the nitrogen of the DHBNH hydrazone directly adjacent to the benzoyl group. The DHBNH benzoyl ring sits snugly between Trp229 and Tyr188. The electron density appears to favor placement of a hydroxyl at both meta positions, suggesting that the benzoyl ring may adopt two alternative conformations.
ⓦ See a full 3D interactive version of this figure on the ACS Chemical Biology web page. Access to the full structure is available with the molecular visualization tool FirstGlance in Jmol (http://molvis.sdsc.edu/fgij/index.htm).
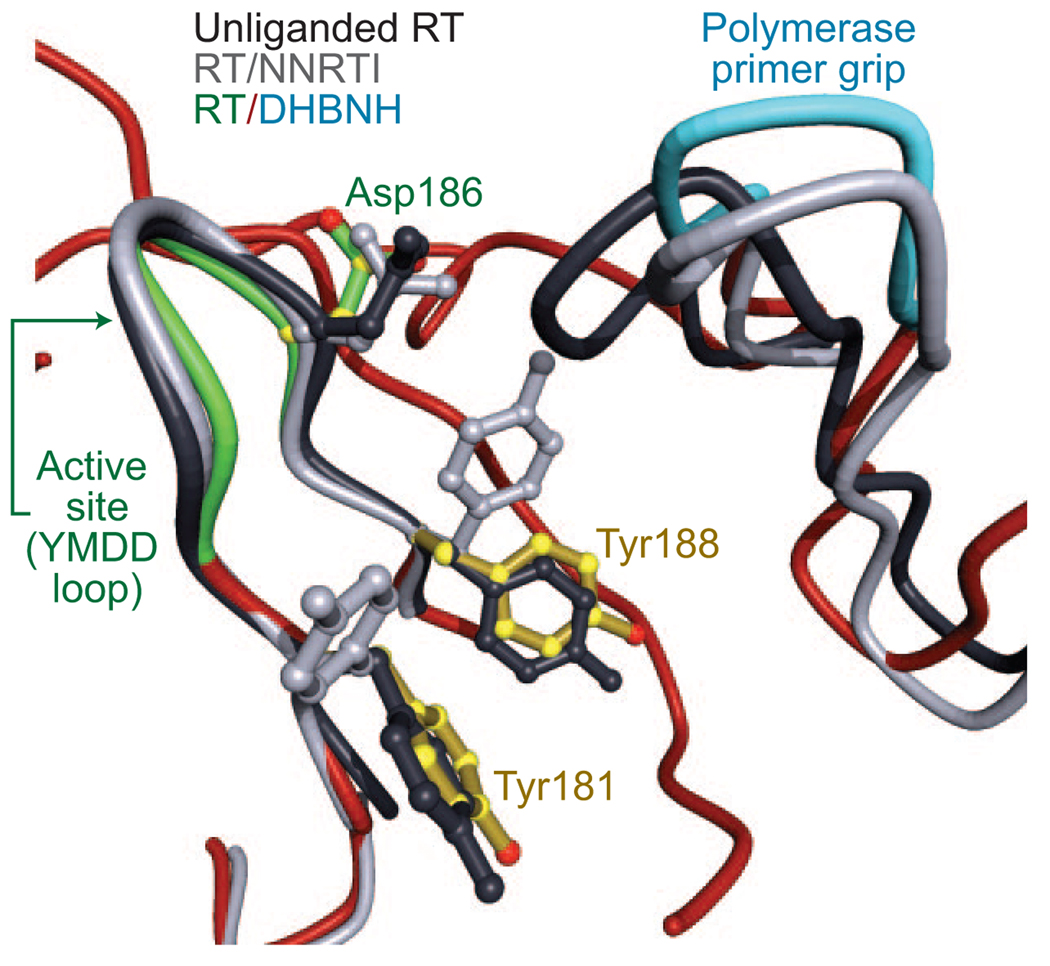
In structures of unliganded RT and RT complexed with nucleic acid, the side chains of Tyr181, Tyr188, and Trp229 fill the NNRTI pocket. Indeed, in the available structures, the pocket does not exist in the absence of an NNRTI. In the RT/DH-BNH structure, the side chain of Trp229 is displaced from the pocket, as it is in RT/NNRTI structures; in the RT/DH-BNH structure, the Trp229 side chain is positioned to interact with the benzoyl moiety of DHBNH. In structures of RT/NNRTI complexes, the side chains of Tyr181 and Tyr188 normally point toward the polymerase active site, helping to form the hydrophobic pocket in which the NNRTI binds (3, 9, 10, 24, 25). By contrast, in the RT/DHBNH structure, the side chains of both tyrosines point away from the active site and are in positions similar to those in unliganded RT (9, 10) and RT complexed with nucleic acids (4–6, 15) (Figure 3). Although the side chains of Tyr181 and Tyr188 are in positions similar to those of unliganded RT and fill some of the NNRTI-binding pocket, an unoccupied cavity is present in the part of the pocket adjacent to the DHBNH hydroxyl on the 4-position of the benzoyl ring.
Figure 3.
Inhibitor binding sites and conformational changes in the polymerase site. Shown are the superimposed structures of unliganded RT (10) (black), RT/TMC125-R165335 (28) (an RT/NNRTI complex, gray), and RT/DHBNH (in color). For clarity, the inhibitors are omitted from the diagram. Superposition is based on p66 residues 107–112 and 178–215. The positions and conformations of the polymerase primer grip differ in each structure. The primer grip of unliganded RT repositions to fill a major part of the NNRTI pocket. In the RT/DHBNH structure, although no NNRTI is present, the primer grip lifts up and away from the NNRTI pocket, leaving a cavity that is only partially filled by Tyr181 and Tyr188. The side-chain conformations of Tyr181 and Tyr188 are similar in unliganded RT and RT/DHBNH, whereas in RT/NNRTI complexes these tyrosine side chains typically rotate to form part of the NNRTI pocket. The active site YMDD loop assumes a similar conformation in RT/DHBNH and RT/NNRTI complexes.
ⓦ See a full 3D interactive version of this figure on the ACS Chemical Biology web page. Access to the full structure is available with the molecular visualization tool FirstGlance in Jmol (http://molvis.sdsc.edu/fgij/index.htm).
As in RT/NNRTI structures, the polymerase primer grip of the RT/DHBNH structure is substantially displaced from its position in unliganded RT, but its position in RT/DHBNH is significantly different from that seen in many RT/NNRTI complexes. When the NNRTI-binding pocket of the RT/DHBNH structure is superposed on a typical RT/NNRTI structure (the superposition is based on p66 residues 107–112 and 178–215), the position of the catalytic YMDD β9–β10 turn is similar in both structures, whereas the position of the polymerase primer grip in the RT/DHBNH structure is further from the active site by >1.4 Å compared with typical RT/NNRTI structures (Figure 3).
Protein–Ligand Interactions
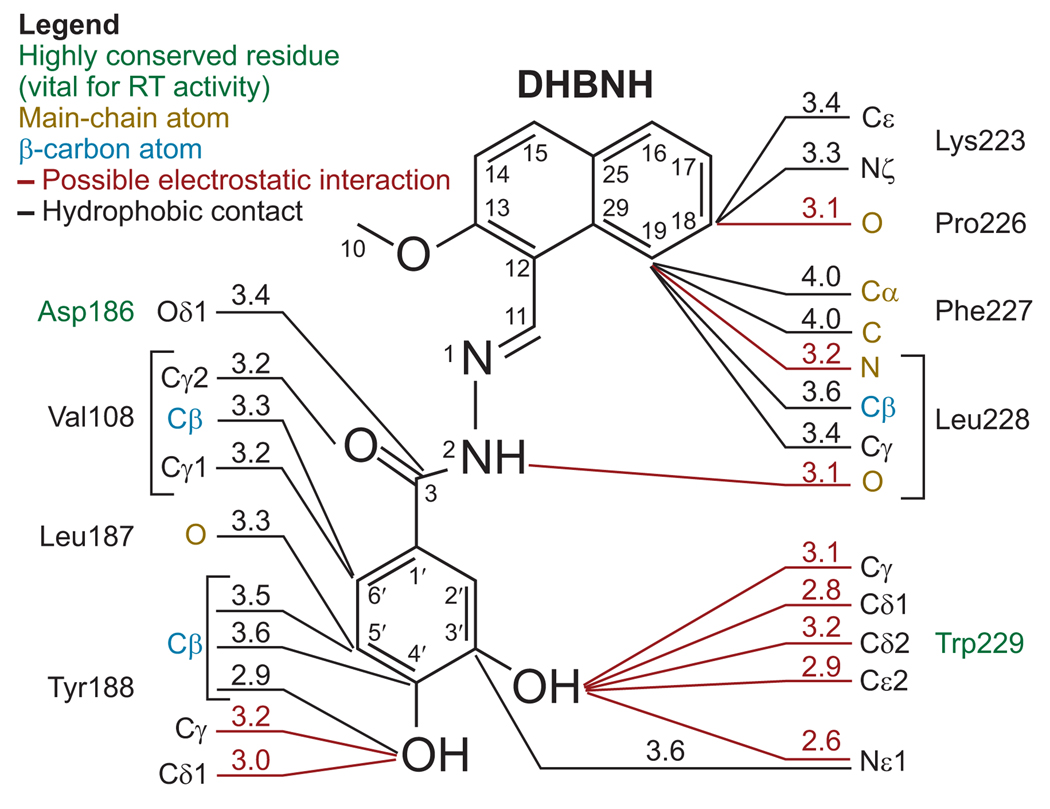
DHBNH appears to have specific interactions with several amino acid residues, including the highly conserved residues Trp229 and Asp186. DHBNH interacts with other residues, including Val108, Leu187, Tyr188, Lys223, Phe227, and Leu228 (Figure 4, Supplementary Table 1). Contacts with most of these residues involve interactions with main-chain or Cβ atoms or both, suggesting that inhibitors that bind to the same site as DHBNH may be effective against viruses that carry most of the common mutations that give rise to NNRTI resistance. As expected, DHBNH retains full inhibitory potency against the RNH activities of HIV-1 RT mutants that are resistant to a variety NNRTIs and NRTIs, including Tyr188Leu (Table 2).
Figure 4.
Possible protein–inhibitor contacts. Selected RT/DHBNH interactions are shown with contact distances (≤4.0 Å). Inhibitor atom numbering is indicated. All residues in contact with the inhibitor are from the p66 subunit of RT. Highly conserved residues that are vital for RT enzymatic activity are shown in green. Hydrogen bonds and other potential electrostatic interactions are designated by red lines, and hydrophobic interactions are depicted in black. The naphthyl ring has a large number of hydrophobic contacts with Leu228. Electrostatic interactions play a significant role in protein interactions with the rest of the inhibitor. Many RT-inhibitor contacts involve main-chain and β-carbon protein atoms. See Supplementary Table 1 for a comprehensive list of interactions.
TABLE 2.
Inhibitory activity of DHBNH against drug-resistant HIV-1 RT variants
| Virus/enzyme | IC50 (µM) a |
|
|---|---|---|
| DHBNH | Efavirenz (EFV) | |
| Inhibition of RT-RNH | ||
| Y181C RT | 0.65 ± 0.1 | b |
| Y188L RT | 1.2 ± 0.4 | b |
| V106A+Y181C RT | 0.85 ± 0.25 | b |
| L100I+K103N RT | 0.7 ± 0.15 | b |
| D67N+K70R+T215F+K219Q RT | 0.5 ± 0.1 | b |
| Antiviral activity | ||
| Wild type | 5.5 ± 1.7 | 0.002 ± 0.0005 |
| NVP-resistant (Y181C) | 8.2 ± 2.5 | 0.032 ± 0.002 |
| UC781-resistant | 6.7 ± 1.4 | 0.2 ± 0.01 |
| (V106A+Y181C) | ||
| EFV-resistant (L100I+K103N) | 7.7 ± 3.5 | 7.9 ± 0.3 |
| AZT-resistant | 5.6 ± 1.5 | 0.003 ± 0.001 |
| (D67N+K70R+T215F+K219Q) | ||
| Cytotoxicity (CC50) | ||
| MT-2 cells | >100 | c |
| Peripheral blood mononuclear cells | >100 | c |
Values are the means ± SD determined from at least three separate experiments.
No inhibition.
Not determined.
The naphthyl ring of DHBNH is solvent-exposed. Most of the stabilizing contacts are with both the side chain and main chain of Leu228, including a possible hydrogen bond with the main-chain nitrogen (Figure 4, Supplementary Table 1). The main-chain carbonyl oxygen of Pro226 also appears to form a hydrogen bond with the naphthyl ring. It is possible that the side chain of Lys223 interacts with the naphthyl ring directly or indirectly (i.e., through a water molecule), but the electron density is ambiguous. The position of the side chain of Lys223 appears to be stabilized by interactions with Glu224 and Pro226. Loop modeling using the program Prime suggests an alternative conformation for Lys223 in which a salt bridge is formed with Asp110. This lower-energy conformation also restricts the binding pocket and may provide additional stability to the bound DHBNH. 3Fo – 2Fc difference maps of the naphthyl region suggest the possibility that there may be alternative binding modes for the naphthyl ring. The modest resolution produces a large electron density envelope around DHBNH, leading to some uncertainty about the geometry and position of the central (hydrazone) region of the inhibitor. In a crystal structure of the related NAH, BBNH, the free inhibitor was co-planar, except for substituents on the benzoyl ring (M. A. Parniak and G. I. Dmitrienko, unpublished data). Quantum mechanical calculations were performed to generate torsional energy profiles for rotation around each of the DHBNH core angles. Several structures from the Cambridge Structural Database (26) were identified that contained the same core motifs as DHBNH, and these structures were consistent with the energy minima determined for the core torsional angles. These energy profiles were used as guides to model DHBNH into the observed electron density. In the structure, the main-chain carbonyl of Leu228 is 3.1 Å away from the hydrazone nitrogen adjacent to the benzoyl group (Figure 4), which appears to be oriented appropriately to form a hydrogen bond.
Structure–Activity Relationship (SAR) Analysis
The benzoyl ring of DHBNH fits into a tunnel formed by Val108, Tyr188, Phe227, Leu228, and Trp229 that leads directly into the NNRTI-binding pocket. The hydroxyl at one of the meta positions on the benzoyl ring is <2.9 Å away from the indole ring of Trp229. The electron density for this portion of DHBNH suggests that there may be partial occupancy for a hydroxyl at the second meta position. This would imply that when bound to HIV-1 RT, the benzoyl ring of DHBNH may exist in two conformations. The second conformation would require a small adjustment in the position of the inhibitor, because there are close contacts with Trp229 and Tyr188, and this adjustment may account for the relatively broad electron density envelope for DHBNH.
The structure predicts that DHBNH derivatives with bulky substitutions at the para position of the benzoyl ring would access part of the NNRTI-binding pocket, which could cause increased inhibition of the RT polymerase activity while retaining the ability to inhibit RNH (Figure 2). To test this possibility, we prepared a series of NAHs with increasingly bulky substituents at the para position of the benzoyl ring (“A”-ring, Table 3). As predicted, the ability of NAH to inhibit RT DNA polymerase activity is substantially enhanced when the size of the para substituent is increased. In contrast, these same substitutions do not significantly affect the ability of the compounds to inhibit the RNH activity of HIV-1 RT.
TABLE 3.
Effect of “A”-ring size on inhibitory potency of NAH
 | |||
|---|---|---|---|
| “A”-ring | Connolly molecular area (Å2) |
IC50(µM)RT polymerase |
IC50(µM)RT RNH |
| 100.165 | >50 | 15 | |
| 118.819 | >50 | 15 | |
| 126.407 | >50 | 12 | |
 |
146.252 | >50 | 2 |
 |
163.147 | 2 | 3 |
| 169.97 | 0.5 | 5 | |
 |
180.448 | 0.4 | 7 |
| 201.066 | 1 | 7.5 | |
A recently published structure for RT in a complex with the NNRTI CP-94,707 (27) suggests the idea that there is an opportunity to develop DHBNH derivatives that have NNRTI-like activity. The RT/CP-94,707 structure is similar to the RT/DHBNH structure in that the side chains of Tyr181 and Tyr188 are in the conformation seen in unliganded RT, and the overall conformations of the polymerase active site and NNRTI-binding pocket are very similar to those of RT/DHBNH (Figure 5). However, the binding of CP-94,707 differs from that of DHBNH in that CP-94,707 binds well inside the NNRTI-binding pocket with its benzo-thiazolidinone ring between Trp229 and Tyr188. The benzo-thiazolidinone ring of CP-94,707 appears to play a role similar to the benzoyl ring of DHBNH, which could account for the similarity in the overall conformations of the two structures. When the polymerase active sites of the two structures are superimposed (Figure 5), only the benzo-thiazolidinone ring of CP-94,707 overlaps the benzoyl ring of DHBNH. This superposition suggests that it may be possible to develop inhibitors that contact both the DHBNH and NNRTI binding sites and that such compounds would inhibit both the RNH and polymerase activities of HIV-1 RT.
Figure 5.
Comparison of binding modes for DHBNH and CP-94,707. The superimposed binding sites of RT complexed with DHBNH and CP-94,707 (27) are shown. In both complexes, Tyr181 and Tyr188 assume conformations similar to those in unliganded RT. However, CP-94,707 binds to a site that is distinct from the DHBNH binding site. CP-94,707 binds to a site closer to the typical NNRTI binding pocket than DHBNH.
ⓦ See a full 3D interactive version of this figure on the ACS Chemical Biology web page. Access to the full structure is available with the molecular visualization tool FirstGlance in Jmol (http://molvis.sdsc.edu/fgij/index.htm).
A “dual inhibitor” could have both disadvantages and advantages. A DHBNH-like inhibitor with substituents that form contacts in the NNRTI pocket may be subject to cross-resistance from mutations that cause resistance to NNRTIs. A properly designed inhibitor, however, could avoid this pitfall. For example, flexibility can be built into key positions on the NNRTI-like substituent so that the drug could assume multiple conformations in response to mutations in the NNRTI pocket. Flexibility of the compound is believed to contribute to the resilience of certain diarylpyrimidine-series NNRTIs, which show considerable promise in clinical trials against many NNRTI-resistant mutant strains (28–31). In addition, the DHBNH-like binding contacts could help to compensate for the loss of protein–inhibitor interactions that occur when residues in the NNRTI-binding pocket are mutated. An inhibitor with both DHBNH-like and NNRTI-like contacts could be highly specific for HIV-1 RT. Ongoing studies should help us determine the benefit of substitutions at the para position of the benzoyl ring of DHBNH. It should be noted that DHBNH derivatives without substitutions at this position are effective against a variety of NNRTI-resistant HIV-1 mutants, including variants with multiple mutations in the NNRTI-binding pocket (Table 2 and ref 20).
Mechanism of Action
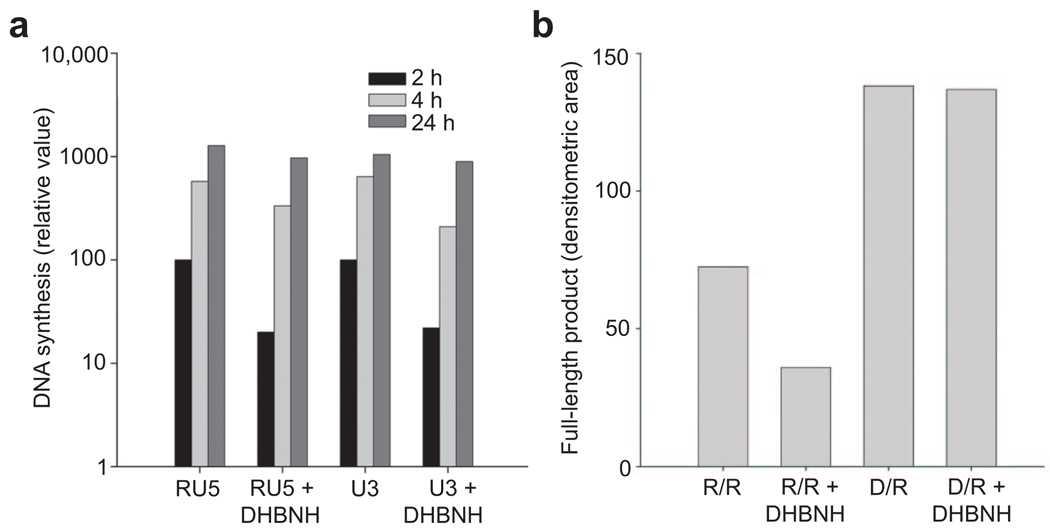
DHBNH is a sub-micromolar inhibitor of the RNH activity of HIV-1 RT and a very weak inhibitor of RT polymerase activity as measured in standard RT RDDP assays using a DNA primer (IC50 > 25 µM). The superposition of the RT/DHBNH coordinates with structures of RT complexed with either RNA–DNA (6) or DNA– DNA (15) suggests that DHBNH would not prevent the binding of the template–primer or the dNTP substrates. This is consistent with the weak inhibitory activity of DHBNH against RT-catalyzed DNA synthesis. DHBNH does, however, inhibit to some extent the initiation of reverse transcription during HIV replication in vivo (Figure 6, panel a) as well as RNA-primed RT-catalyzed DNA polymerization in vitro (Figure 6, panel b), although at concentrations substantially higher than those needed to inhibit RNH activity. Nonetheless, real-time polymerase chain reaction (PCR) analysis suggests that inhibition of the initiation of reverse transcription might make a significant contribution to the antiviral activity of DHBNH. The basis for the inhibition of the initiation of viral DNA synthesis is presently unclear, and additional studies are needed.
Figure 6.
DHBNH inhibits RNA-primed initiation of reverse transcription. a) Real-time PCR analysis of intracellular reverse transcription. The amounts of RU5 and U3 DNA (see ref 6) were measured using real-time PCR to monitor the initiation of reverse transcription and minus-strand DNA transfer, respectively. The y-axis shows the relative amounts of RU5 and U3 DNA 2, 4, and 24 h after infection in the presence or absence of DHBNH. b) DHBNH partially inhibits RNA-primed but not DNA-primed RDDP activity in vitro. Reactions were carried out with a 42-nucleotide RNA template annealed to a 21-nucleotide RNA primer (R/R) or to the same sequence DNA primer (D/R) as described in Methods. The figure shows the amount of full-length 21-nucleotide extended DNA polymerization product formed by RT in 10 min. The final concentration of DHBNH in reactions containing the inhibitor was 10 µM. Only starting primer and full-length polymerization products were seen in these assays. The lack of intermediate products suggests that DHBNH was primarily affecting the initiation of reverse transcription and not subsequent elongation.
The similarity in conformation between RT/DHBNH and RT/CP-94,707 (27) suggests that these two compounds may have a similar mode of action. It is possible, for example, that CP-94,707 might also inhibit RNH activity. CP-94,707 was originally identified as an inhibitor of initiation of transfer-RNA (tRNA) primed DNA synthesis (27), supporting the idea that DHBNH may effectively inhibit initiation of tRNA-primed synthesis of minus-strand DNA.
If DHBNH interferes with correct positioning of a DNA–RNA substrate (e.g., by affecting the position of the polymerase primer grip), then this effect might be even more pronounced with an RNA–RNA substrate. The initiation of HIV RT-catalyzed DNA synthesis with an RNA–RNA primer–template is intrinsically less efficient than that from a DNA–RNA primer–template even in the absence of inhibitor (Figure 6, panel b). Because the RT polymerase primer grip interacts with the 3′-terminus of the primer, even a slight distortion of the conformation at the primer grip could affect the correct positioning of the primer 3′-terminal nucleotide, and the consequences could be more pronounced when the 3′-terminus is a ribonucleotide (possibly due to altered interaction with the 2′-hydroxyl of the ribonucleotide). Furthermore, duplex RNA is intrinsically more rigid than DNA–RNA and is expected to be more refractory to forming the ∼40° bend between A- and B-forms of the template–primer as seen in complexes of RT with duplex nucleic acid (6, 15). Both of these factors may contribute to the substantially reduced levels of DNA synthesized in assays in which an RNA–RNA substrate is provided to RT in the presence of DHBNH.
Our current structural data for DHBNH are consistent with a mechanism of inhibition that involves the binding of DHBNH proximal to the polymerase active site (Figure 7). We believe this causes structural changes in the polymerase primer grip that may alter the trajectory of the template–primer between the polymerase and RNH domains, so that the sugar–phosphate backbone of the RNA template would not be properly positioned at the RNH active site and could not be cleaved. Changes in specificity and efficiency of RNA cleavage caused by changes in the polymerase domain of RT have been reported previously. For example, mutations in the polymerase primer grip and thumb of p66 have been shown to dramatically alter the specificity of RNH cleavage of the RNA template strand (32–37). Furthermore, crystallographic studies suggest that the polymerase primer grip plays a role in positioning the RNA template relative to the RNH active site (6, 15). Moreover, there is excellent agreement between the structural data, the model, and the SAR that predicts which of the substituted compounds will interact with the NNRTI-binding pocket.
Figure 7.
Possible mechanisms for inhibition by DHBNH. Two alternative modes of RNHI activity are represented schematically. 1) As seen in the current RT/DHBNH crystal structure, the inhibitor binds adjacent to the polymerase active site and induces a repositioning of the polymerase primer grip (cyan). This may redirect the trajectory of the substrate so that the RNA strand is not close enough to the RNH active site for cleavage to occur. 2) The inhibitor may bind at a second site close to the RNH domain to inhibit its activity. These two alternative modes of action would not necessarily be exclusive.
We cannot exclude the possibility that DHBNH also binds at a second site near the RNH active site. We previously suggested, based on kinetic analysis of the inhibition, that NAHs may inhibit RT polymerase and RNH activities by binding to two different sites on the enzyme (20, 21). A second DHBNH binding site is supported by the observation that DHBNH is equally effective at inhibiting RT RNH activity in the presence or the absence of 20 µM of the NNRTI nevirapine (data not shown), a concentration that would saturate the NNRTI-binding site. When nevirapine is bound to RT, the side chain of Tyr188 rotates toward the polymerase active site (3, 9, 10, 24, 25) into a position that would block the binding of the benzoyl ring of DHBNH observed in the current structure. This hypothetical second binding site could be formed in part by the RNA–DNA substrate and would not be present in the crystals we prepared, because nucleic acid was not cocrystallized with the protein. We are trying to obtain a crystal structure of DHBNH bound to the RT–nucleic acid complex.
Conclusions
We have refined a 3.15 Å resolution crystal structure of HIV-1 RT complexed with DHBNH, an NAH-class RNHI. The crystal structure shows that DHBNH binds to a novel site adjacent to the polymerase active site and the NNRTI-binding pocket, >50 Å away from the RNH active site. Binding of DHBNH directly affects the position of the polymerase primer grip, as well as the thumb, which is located adjacent to the primer grip. This finding is consistent with the possibility that the inhibitor perturbs the trajectory of the template–primer so that RNH cannot cleave the RNA strand of an RNA–DNA duplex. Preliminary SAR studies show that DHBNH derivatives with substituents on the benzoyl ring can interact with residues in the NNRTI-binding pocket to inhibit the polymerase activity, as predicted from the RT/DHBNH structure. The DHBNH binding site provides opportunities to develop new inhibitors that can inhibit the polymerase activity, the RNH activity, or both. More importantly, the prevalence of DHBNH interactions with main-chain and Cβ atoms suggests that inhibitors developed based on chemical modifications of DHBNH have the potential to be effective against a wide range of drug-resistant mutant strains of RT.
METHODS
2-Methoxy-1-naphthaldehyde was obtained from Sigma-Aldrich (St. Louis, MO). 3,4-Dihydroxybenzhydrazide and other acid hydrazides were obtained from TransWorld Chemical (Rockville, MD). [3H]-TTP and the homopolymeric template–primer poly(rA)– oligo(dT)12–18 were products of Amersham Bio-sciences. The oligonucleotides 5′-GAU CUG AGC CUG GGA GCU-fluorescein-3′ and 5′-dabcyl-AGC TCC CAG GCT CAG ATC-3′ were synthesized and provided as an annealed RNA–DNA duplex by TriLink Biotechnologies (San Diego, CA). Trilink Biotechnologies also provided all other oligonucleotides used in these studies.
Synthesis of NAHs
DHBNH and other NAHs were synthesized by condensation of the aromatic aldehyde with the corresponding acid hydrazide, essentially as described previously (38). As an example, DHBNH was prepared by the dropwise addition of 3,4-dihydroxybenzyhydrazide (1.1 mM in 10 mL of ethanol) to a solution of 2-methoxy-1-naphthaldehyde (1 mM in 4% acetic acid in ethanol) with stirring while heating in a boiling water bath. Heating and stirring were continued for 20 min following completion of addition of the ethanolic acid hydrazide solution, and then the mixture was allowed to cool to room temperature. The precipitate was collected by filtration, washed with cold ethanol and diethyl ether, and dried. Elemental and mass spectral analyses were consistent with the expected structure (for summary of DHBNH characterization data, see Supplementary Figures 1–3).
Protein Preparation and Purification
HIV-1 RT was prepared as described previously (39). The RT/DHBNH crystallization complex was prepared by mixing 7.4 µL of 20 mM inhibitor in dimethyl sulfoxide (DMSO) with 2.5 µL of 20% β-octyl glucopyranoside. Of this solution, 7.5 µLwas combined with 65.0 µL of 40 mg mL−1 RT and 57.5 µL of additional RT buffer (10 mM trishydroxymethylaminoethane (Tris), pH 8.0, 75 mM NaCl) on ice. A catalytically active chimeric isolated HIV-1 RNH domain protein containing an α-helical substrate-binding loop derived from Escherichia coli RNHI (22, 40) (termed p15-EC, a kind gift from C. Shaw-Reid, Merck Research Laboratories, West Point, PA) was expressed and purified as described previously (40).
Assay of RT RDDP Activity
HIV-1 RT RDDP activity was generally determined by a fixed time assay as previously described (20). Briefly, reaction mixtures (100 µL total volume) contained 50 mM Tris-HCl (pH 7.8, 37 °C), 60 mM KCl, 10 mM MgCl2, 10–25 ngof p51/p66 RT, 0.5 units of template–primer, and 5 µM [3H]-TTP substrate. Stock solutions of DHBNH and other NAHs were prepared in DMSO. Aliquots of these DMSO solutions containing the inhibitor were added such that the final DMSO concentration was <2%. Reaction assays were incubated at 37 °C for 10–20 min and then quenched with 500 µL of cold 20 mM sodium pyrophosphate in 10% trichloroacetic acid (TCA). After 30 min on ice, the samples were filtered on Whatman 934-AH glass fiber filters and washed with 10% TCA and ethanol, and the radioactivity determined by liquid scintillation spectrometry.
RNA-primed and DNA-primed RT RDDP activity was evaluated using a 42-nucleotide RNA template of the sequence 5′-GGAAAAUCUCUACGAGUGGCGCCCGAACAGGGACCUGACCAG-3′ annealed to the complementary 21-nucleotide RNA primer 5′-CUGGUCAGGUCCCUGUUCGGG-3′ or to the complementary 21-nucleotide DNA primer 5′-CTGGTCAGGTCCCTGTTCGGG-3′. Reaction mixtures (15 µL final volume) contained 50 nM template–primer, 250 nM p66/p51 RT, and 25 µM of each dNTP ([α-32P]-dCTP was included as tracer) in 50 mM Tris, pH 8.0, containing 60 mM KCl and 5 mM MgCl2, in the presence or the absence of 10 µM DHBNH. Briefly, RT and template–primer were incubated at 37 °C for 5 min, and then the reactions were started by the addition of the dNTPs and MgCl2 to the final concentrations indicated. After a 10 min incubation at 37 °C, reactions were quenched by the addition of an equal volume of sequencing gel loading buffer (98% deionized formamide, 10 mM EDTA, and 1 mg mL−1 each of bromophenol blue and xylene cyanol) followed by heating at 95 °C for 5 min. Reaction products were resolved by denaturing gel electrophoresis, and the amount of full-length 42-nucleotide polymerization product was quantified by phosphorimaging.
Assay of RT RNH Activity
RT RNH activity was measured using a FRET-based microplate fluorescence assay that we have recently described (41). Briefly, 50 µL of a 0.5 µM solution of RNA–DNA hybrid duplex in 50 mM Tris, pH 8.0, containing 60 mM KCl, was added to individual wells of a 96-well microplate. Reactions were initiated by the addition of 50 µL of 5 nM HIV-1 RT in 50 mM Tris, pH 8.0, containing 60 mM KCl and 10 mM MgCl2 and allowed to proceed at 37 °C for 30 min. Reactions were quenched by the addition of 50 µL of 0.5 M EDTA, pH 8.0. Fluorescence intensity was assessed using an excitation wavelength of 490 nm and an emission wavelength of 528 nm, with cutoff filter set to 515 nm.To assess the effect of inhibitors, 1 µL of a DMSO inhibitor solution was added to the microplate well prior to the addition of substrate and RT solutions in order to ensure adequate mixing and suspension of the hydrazone in the aqueous reaction medium.
Cell Culture and Antiviral Assays
Antiviral assays were carried out by infection of MT-2 lymphoblastoid cells in the presence of varying concentrations of DHBNH essentially as previously described (20). Briefly, MT-2 cells (4 × 105 cells mL−1) were incubated overnight in the absence or in the presence of varying concentrations of DHBNH and were then infected with HIV-1 (NL4-3, moi of 0.01). The extent of HIV-1 replication was evaluated 5 d post-infection by microscopic evaluation of HIV-induced cytopathic effect and by analysis of HIV-1 p24 antigen levels in cell-free culture supernatants. Cytotoxicity was evaluated in MT-2 cells and in peripheral blood mononuclear cells using (2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide, XTT in a commercially available kit (Roche Diagnostics, Indianapolis IN) according to manufacturer’s directions. Cells were incubated with varying concentrations of DHBNH (0–200 µM) for 5 d prior to evaluation of XTT reactivity.
Real Time PCR Analysis of Intracellular Reverse Transcription Products
293 and HOS cells were maintained in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 5% fetal bovine serum, 5% newborn calf serum, and penicillin (50 units mL−1) plus streptomycin (50 µg mL−1; Quality Biological). VSV-g pseudotyped HIV vectors that undergo a single cycle of replication were generated by cotransfecting 293 cells with 5 µg of pNLNgoMIVR-E-.HSA and 3 µg of pHCMV-g using the calcium phosphate method. The 293 cells were washed with 10 mL of phosphate-buffered saline (PBS) 8, 24, and 32 h after transfection. The 48-h supernatants were harvested, clarified by centrifugation, and filtered through a 0.45 µm filter. The supernatants were treated with 100 units of RNase-free DNase I (Roche) for 30 min at room temperature and were then concentrated to 2 mL using the 300,000 MWCO Vivaspin 20 mL concentrators. The viruses were diluted to a total volume of 13 mL in complete media, and then 2 mL of diluted virus was used to infect 2 × 105 HOS cells. Stocks (10 mM) of DHBNH were prepared in DMSO and diluted to a final concentration of 10 µM in the treated groups, which were incubated for 4 h with DHBNH prior to viral infection. HOS cells were washed with 2 mL of PBS 2 h after infection, and then fresh medium was added. Total DNA was isolated 2, 4, and 24 h after infection using the EZ-1 DNA Tissue protocol (Qiagen). Real-time PCR reactions were used to quantitate the amounts of DNA in the infected cells as previously described (42). Virions containing the D110E polymerase active site mutation were also generated and used to demonstrate that the plasmid DNA used in the transfection did not significantly contribute to the amount of viral DNA measured.
Crystallization and Data Collection
RT/DHBNH was crystallized by vapor diffusion in microseeded hanging drops containing equal volumes of protein (above) and reservoir solution (50 mM imidazole, pH 6.4, 100 mM ammonium sulfate, 15 mM magnesium sulfate, 5% glucose, 11.5% poly(ethylene glycol) (PEG) 8000) at 4.0 °C. RT/DHBNH crystals were transferred to a stabilization solution containing mother liquor and 15% PEG 8000. This was replaced stepwise at 5–10 min intervals with stabilization solutions containing increasing concentrations of sucrose in 5% increments until the sucrose concentration was 25%. The crystals were subsequently flash-cooled and stored in liquid N2. X-ray data were collected at 100 K at the National Synchrotron Light Source at Brookhaven National Laboratories, Beamline X25. The data were processed using HKL-DENZO-SCALEPACK (43). Crystallographic statistics are shown in Supplementary Table 2).
Computational Methods
Density functional theory calculations were carried out with the Gaussian 98 software package (44) using the B3LYP functional model (45, 46) and the 6–31G* basis set. Each of the four torsional angles in the DHBNH core was scanned in 15° increments followed by geometry optimization. Loop modeling of p66 residues Gln222 through Leu228 was performed using Prime (Schrödinger, LLC) with the AGBNP solvation model (47, 48).
Structure Determination and Refinement
Phases were determined by molecular replacement with the program AMoRe (49) using the HIV-1 RT/R100943 structure (PDB accession number 1S6P) as an initial search model. Stepwise model building and torsional simulated annealing refinement protocols were conducted using the O graphics package (50) and CNS (51) with a bulk solvent correction. Ligand geometry was optimized using the IMPACT software package (Schrödinger, LLC). Restrained minimization of the entire complex, in which hydrogen atoms were added to the structure, side-chain hydroxyl groups were reoriented, and potential steric clashes were alleviated, was performed using Schrödinger’s FirstDiscovery protein preparation facility (Schrödinger, LLC). Results from the energetic calculations performed in FirstDiscovery and Prime (above) were iteratively incorporated into the crystallographic refinement (unpublished procedure). Water molecules were added manually in the final stages of refinement and were only built in where they could be justified by hydrogen bonds and Fo − Fc electron density at or above the 2.5σ contour level.
Acknowledgments
This research was supported by National Institutes of Health (NIH) Grants AI 27690 (MERIT Award to E.A.) and P01GM066671 (to E.A. and M.A.P.), GM64375 (to R.M.L.), and F32 AI 060300 (NIH NRSA fellowship to D.M.H.). We are grateful to synchrotron staff members at Brookhaven National Light Source and other members of our laboratories for their assistance and helpful discussions. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-98CH10886. This publication has been funded in part with federal funds from the National Cancer Institute, NIH, under contract no. NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, as well as the National Institute of General Medical Sciences.
Footnotes
Accession Codes: The atomic coordinates for the refined structure of RT/DHBNH have been deposited in the Protein Data Bank (PDB accession number 2I5J).
Supporting Information Available: This material is free of charge via the Internet.
REFERENCES
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.Tisdale M, Schulze T, Larder BA, Moelling K. Mutations within the RNase H domain of HIV-1 RT abolish virus infectivity. J. Gen. Virol. 1991;72:59–66. doi: 10.1099/0022-1317-72-1-59. [DOI] [PubMed] [Google Scholar]
- 3.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 4.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr, Ju X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 6.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rittinger K, Divita G, Goody RS. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodgers DW, Camblin SJ, Harris BA, Ray S, Culp JS, Hellmig B, Woolf DJ, Debouck C, Harrison SC. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U.S.A. 1995;92(1):1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsiou Y, Ding J, Das K, Clark AD, Jr, Hughes SH, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: Implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 11.Tantillo C, Ding J, Jacobo-Molina A, Nanni RG, Boyer PL, Hughes SH, Pauwels R, Andries K, Janssen PAJ, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase: implications for mechanisms of drug inhibition and resistance. J. Mol. Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 12.Das K, Ding J, Hsiou Y, Clark AD, Jr, Moereels H, Koymans L, Andries K, Pauwels R, Janssen PAJ, Boyer PL, Clark P, Smith RH, Jr, Smith MBK, Michejda CJ, Hughes SH, Arnold E. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J. Mol. Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 13.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart DI. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat. Struct. Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 14.Ding J, Das K, Hsiou Y, Zhang W, Arnold E. Structural studies of HIV-1 reverse transcriptase and implications for drug design. In: Veerapandian P, editor. Structure-based Drug Design. New York: Marcel Dekker, Inc; 1997. pp. 41–82. [Google Scholar]
- 15.Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jr, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- 16.Palaniappan C, Fay PJ, Bambara RA. Nevirapine alters the cleavage specificity of ribonuclease H of human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 1995;270:4861–4869. doi: 10.1074/jbc.270.9.4861. [DOI] [PubMed] [Google Scholar]
- 17.Shaw-Reid CA, Feuston B, Munshi V, Getty K, Krueger J, Hazuda DJ, Parniak MA, Miller MD, Lewis D. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry. 2005;44:1595–1606. doi: 10.1021/bi0486740. [DOI] [PubMed] [Google Scholar]
- 18.Gopalakrishnan V, Benkovic S. Effect of a thiobenzimidazolone derivative on DNA strand transfer catalyzed by HIV-1 reverse transcriptase. J. Biol. Chem. 1994;269:4110–4115. [PubMed] [Google Scholar]
- 19.Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Borkow G, Fletcher RS, Barnard J, Arion D, Motakis D, Dmitrienko GI, Parniak MA. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry. 1997;36:3179–3185. doi: 10.1021/bi9624696. [DOI] [PubMed] [Google Scholar]
- 21.Sluis-Cremer N, Arion D, Parniak MA. Destabilization of the HIV-1 reverse transcriptase dimer upon interaction with N-acyl hydrazone inhibitors. Mol. Pharmacol. 2002;62:398–405. doi: 10.1124/mol.62.2.398. [DOI] [PubMed] [Google Scholar]
- 22.Keck JL, Marqusee S. Substitution of a highly basic helix/loop sequence into the RNase H domain of human immunodeficiency virus reverse transcriptase restores its Mn(2+)-dependent RNase H activity. Proc. Natl. Acad. Sci. US.A. 1995;92:2740–2744. doi: 10.1073/pnas.92.7.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Dan-zeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 24.Ding J, Das K, Moereels H, Koymans L, Andries K, Janssen PAJ, Hughes SH, Arnold E. Structure of HIV-1 RT/ TIBO R 86183 complex reveals similarity in the binding of diverse non-nucleoside inhibitors. Nat. Struct. Biol. 1995;2:407–415. doi: 10.1038/nsb0595-407. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 26.Allen FH. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 27.Pata JD, Stirtan WG, Goldstein SW, Steitz TA. Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant reverse transcriptases resistant to other non-nucleoside inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10548–10553. doi: 10.1073/pnas.0404151101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das K, Clark AD, Jr, Lewi PJ, Heeres J, de Jonge MR, Koymans LMH, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, Corte BD, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, Béthune M-Pd, Boyer PL, Clark P, Hughes SH, Janssen PAJ, Arnold E. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 29.Das K, Lewi PJ, Hughes SH, Arnold E. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog. Biophys. Mol. Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Guillemont J, Pasquier E, Palandjian P, Vernier D, Gaurrand S, Lewi PJ, Heeres J, de Jonge MR, Koymans LMH, Daeyaert FFD, Vinkers MH, Arnold E, Das K, Pauwels R, Andries K, de Béthune M-P, Bettens E, Hertogs K, Wigerinck P, Timmerman P, Janssen PAJ. Synthesis of novel diarylpyrimi-dine analogues and their antiviral activity against human immunodeficiency virus type 1. J. Med. Chem. 2005;48:2072–2079. doi: 10.1021/jm040838n. [DOI] [PubMed] [Google Scholar]
- 31.Janssen PAJ, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, Kukla M, Ludovici D, Andries K, de Béthune M-P, Pauwels R, Das K, Clark AD, Jr, Frenkel YV, Hughes SH, Medaer B, De Knaep F, Bohets H, De Clerck F, Lampo A, Williams P, Stoffels P. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoetenyl]-2-6-dimethylphenyl]amino]-2-pyrimidinyl]amino]-benzonitrile (R278474-rilpivirine) J. Med. Chem. 2005;48:1901–1909. doi: 10.1021/jm040840e. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh M, Jacques PS, Rodgers DW, Ottman M, Darlix J-L, Le Grice SFJ. Alterations to the primer grip of P66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry. 1996;35:8553–8562. doi: 10.1021/bi952773j. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh M, Williams J, Powell MD, Levin JG, Le Grice SF. Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry. 1997;36:5758–5768. doi: 10.1021/bi963045e. [DOI] [PubMed] [Google Scholar]
- 34.Palaniappan C, Wisniewski M, Jacques PS, Le Grice SF, Fay PJ, Bambara RA. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 35.Powell MD, Ghosh M, Jacques PS, Howard KJ, Le Grice SF, Levin JG. Alanine-scanning mutations in the “primer grip” of P66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J. Biol. Chem. 1997;272:13262–13269. doi: 10.1074/jbc.272.20.13262. [DOI] [PubMed] [Google Scholar]
- 36.Gao HQ, Boyer PL, Arnold E, Hughes SH. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 37.Powell MD, Beard WA, Bebenek K, Howard KJ, Le Grice SF, Darden TA, Kunkel TA, Wilson SH, Levin JG. Residues in the alphaH and alphaI helices of the HIV-1 reverse transcriptase thumb subdomain required for the specificity of RNase H-catalyzed removal of the polypurine tract primer. J. Biol. Chem. 1999;274:19885–19893. doi: 10.1074/jbc.274.28.19885. [DOI] [PubMed] [Google Scholar]
- 38.Edward JT, Gauthier M, Chubb FL, Ponka P. Synthesis of new acylhydrazones as iron-chelating compounds. J. Chem. Eng. Data. 1988;33:538–540. [Google Scholar]
- 39.Clark AD, Jr, Jacobo-Molina A, Clark P, Hughes SH, Arnold E. Crystallization of human immunodeficiency virus type 1 reverse transcriptase with and without nucleic acid substrates, inhibitors and an antibody Fab fragment. Methods Enzymol. 1995;262:171–185. doi: 10.1016/0076-6879(95)62017-6. [DOI] [PubMed] [Google Scholar]
- 40.Carroll SS, Sardana V, Yang Z, Jacobs AR, Mizenko C, Hall D, Hill L, Zugay-Murphy J, Kuo LC. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: Implications for viral replication and in vitro assays. Biochemistry. 2000;39:8243–8249. doi: 10.1021/bi991992s. [DOI] [PubMed] [Google Scholar]
- 41.Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Julias JG, Ferris AL, Boyer PL, Hughes SH. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 2001;75:6537–6546. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. DENZO and SCALEPACK, in. In: Rossmann MG, Arnold E, editors. Crystallography of Biological Macromolecules. Boston: Kluwer Academic Publishers; 2001. pp. 26–235. [Google Scholar]
- 44.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Jr, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson BG, Chen W, Wong MW, Andres JL, Head-Gordon M, Replogle ES, Pople JA. Gaussian 98. Pittsburgh, PA: Gaussian, Inc; 1998. revision A.9. [Google Scholar]
- 45.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 46.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B: Condens. Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 47.Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA. A hierarchical approach to allatom loop prediction. Proteins: Struct., Funct., Bioinf. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 48.Gallicchio E, Levy RM. AGBNP: an analytic implicit solvent model suitable for molecular dynamics simulations and high-resolution modeling. J. Comput. Chem. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
- 49.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- 50.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved experimental procedures for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 51.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]