Abstract
Investigations into the use of baboons as organ donors for human transplant recipients, a procedure called xenotransplantation, have raised the specter of transmitting baboon viruses to humans and possibly establishing new human infectious diseases. Retrospective analysis of tissues from two human transplant recipients with end-stage hepatic disease who died 70 and 27 days after the transplantation of baboon livers revealed the presence of two simian retroviruses of baboon origin, simian foamy virus (SFV) and baboon endogenous virus (BaEV), in multiple tissue compartments. The presence of baboon mitochondrial DNA was also detected in these same tissues, suggesting that xenogeneic “passenger leukocytes” harboring latent or active viral infections had migrated from the xenografts to distant sites within the human recipients. The persistence of SFV and BaEV in human recipients throughout the posttransplant period underscores the potential infectious risks associated with xenotransplantation.
Baboons harbor several exogenous retroviruses that exist as persistent, life-long infections.1 Of these, simian T cell lymphotropic virus (STLV) induces lymphomas and leukemias, while others such as simian foamy virus (SFV) cause little or no apparent disease in baboons.2,3 SFV is infectious for humans, having been transmitted from nonhuman primates to humans at primate facilities in the United States and elsewhere.4 Baboons also carry a replication-competent endogenous retrovirus (BaEV) that is found at high copy number and distributed throughout the baboon genome and infects human cells in vitro.5 It is conceivable that simian retroviruses may represent possible infectious disease risks in human transplant recipients. Previous studies have shown the existence of chimerism established in both allogeneic and xenogeneic transplant recipients in which lymphocytes from donor organs were found to circulate in the recipient.6,7 Although no infectious diseases have been apparent that could have been attributed to the transfer of a baboon virus to humans, there is the potential for migrating baboon cells to disseminate simian viruses to the human recipients.1 This study was undertaken to determine if human xenotransplant recipients had evidence of simian retroviruses of possible public health concern.
The first baboon-to-human liver transplant was attempted in June 1992 to an HIV-infected patient and the second was in January 1993; patient 2 was a 62-year-old male who also received donor bone marrow intravenously and survived for 27 days.7,8 Both patients had hepatitis B virus (HBV)-associated cirrhosis and were HBV carriers. Patient 1 was a 35-year-old male who survived 70 days. Neither transplanted liver functioned normally. In addition, both patients developed renal failure and multiple posttransplant infectious complications. Both patients received an immunosuppressive regimen of FK-506, prednisone, and cyclophosphamide. The two adult male baboons were screened against a panel of simian and human viruses and were negative for STLV, SIV, and simian retro-virus (SRV).
Since evidence of antiviral antibody responses generally corresponds with active infection, antibodies against SFV in donor baboons were assessed by Western blotting. Immunoblot strips were prepared with gel-purified SFV viral proteins. The established criterion used for SFV infection has been serologic reactivity to the two gag-related proteins p74 and p70, which are highly immunogenic and have been shown to correlate directly with SFV infection.9 As seen in Fig. 1A, anti-p74/p70 antibodies were detected in samples from both donor baboons prior to transplantation whereas the human patients were nonreactive at several time points during the posttransplantation period; however, a faint p74 band was visible at day 22 (Fig. 1B). The failure of patient 2 to generate a significant antiviral antibody response may have been due to the insufficient time between transplantation and death. For patient 1, HIV infection may have had a role in suppressing antiviral responses. In addition, the immunosuppressive regimens used to treat both recipients inhibit B and T cell responsiveness.7,8
FIG. 1.
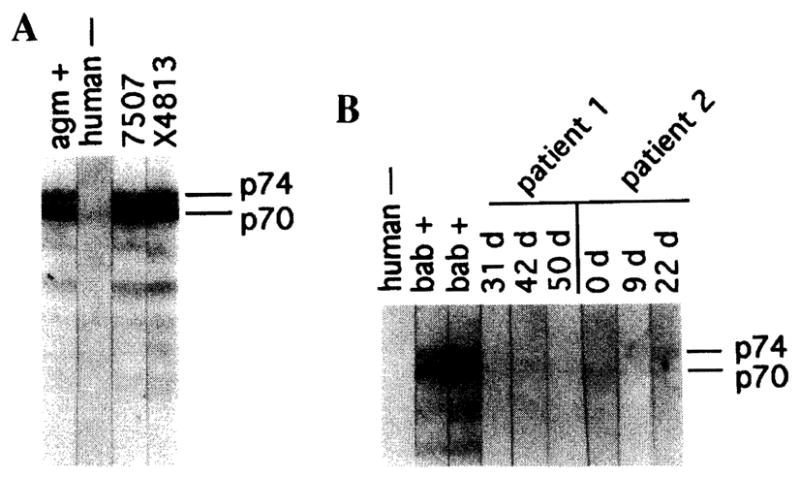
Immunoblot analysis of serum samples from donor baboons and human transplant recipients. Serum samples from the baboon donors (A) or human transplant recipients (B) were diluted 1:40 and reacted with SFV immunoblot strips. Positive and negative controls were used at a 1:80 dilution. Immunoblotting was performed as previously described with SFV-Bab1 as the antigen.3 SFV-Bab1 used for making Western blot reagents was isolated from peripheral blood lymphocytes of a baboon that developed lymphoma and has been previously described.3 Virus was grown in primary human foreskin fibroblasts (HFSM) and harvested during the cytopathic phase of infection. HSFM cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, penicillin (50 U/ml), and streptomycin (50 μg/ml).
To assess directly the presence of SFV in tissues of these patients, DNA extracted from tissue specimens were subjected to polymerase chain reaction (PCR) amplification using a set of SFV primers specific to the R-U5 region of the long terminal repeat (LTR). This region is highly conserved and has been used to amplify SIV from a variety of African and Asian nonhuman primates.3,9 The amplified products were further analyzed by Southern blotting using a 32P-labeled LTR probe to increase sensitivity and as a means for confirming the identity of the amplified products. As seen in Fig. 2, an identifiable LTR band of approximately 329 bp was visible in multiple tissue samples from both human recipients. In patient 2, SFV DNA was detected in the liver graft on day 24 but not day 12. The liver sample taken from patient 1 on day 16 was also negative, yet SFV was detected in both lymph node and kidney on day 70. It is possible that the levels of SFV in the liver of patient 2 were initially below the detection limits of this assay, while the later positive signal resulted from an increase in the number of active or latently infected cells. Because the sizes of the baboon livers were small in comparison with the recipients and were found to regenerate within 3 weeks to the appropriate liver mass for those recipients, it is conceivable that this proliferative phase also allowed for replication of viruses or cell types that may have harbored SFV. This would account for the delayed presence in patient 2 of SFV DNA in the graft. To ensure the integrity of the DNA preparations in those samples that were SFV negative, we successfully amplified the β-globin gene from each sample (not shown). We also repeated PCR analysis of the tissue samples from both patients, and the only difference was a negative signal in the DNA sample taken from the small intestine.
FIG. 2.
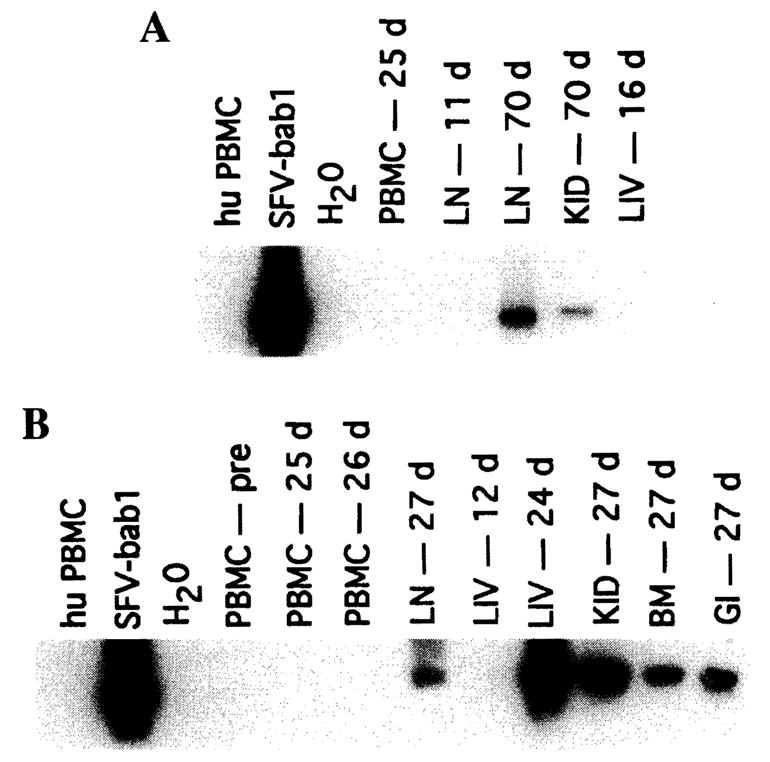
Detection of SFV DNA in tissues from human liver transplant patients. PCR amplification of SFV DNA using LTR primers was performed with DNA from (A) patient 1 and (B) patient 2. Designations are as follows: PBMC, peripheral blood mononuclear cells; LN, lymph node; LIV, liver, KID, kidney; BM, bone marrow; and GI, small intestine. PBMC DNA samples from an SFV-seronegative human and an SFV+ baboon were used as negative and positive controls, respectively. A total of 100 ng of total cellular DNA was added to 200 μM dNTPs, 50 mM KCl, 10 mM Tris, 2.5 mM MgCl2, bovine serum albumin (BSA, 5 mg/ml), 30 pmol of primer, and 1 U of Taq polymerase (Promega, Madison, WI). Amplification was performed using the following primers: PBF1 (5′-CACTACTCGCTGCGTCGAGAGGTGT-3′) and PBF2 (5′-GGAATTTTGTATATTGATTATCC-3.′). A 329-bp fragment within the SFV LTR (nt 786-1115, based on the HFV sequence) was amplified. Conditions for LTR primers were as follows: 1 cycle at 94°C for 1 min, 52°C for 45 sec and 72°C for 1 min; 1 cycle at 94°C for 1 min, 50°C for 45 sec, and 72°C for 1 min; 1 cycle at 94°C for 1 min, 48°C for 45 sec, and 72°C for 1 min; 1 cycle at 94°C for 1 min, 46°C for 45 sec, and 72°C for 1 min; 25 cycles at 94°C for 1 min, 45°C for 45 sec, and 72°C for 1 min; followed by extension at 72°C for 7 min. Amplified fragments were separated on agarose gels, blotted onto Nytran Plus membranes, and then hybridized with 32P-labeled SFV-Bab1 LTR probe.
The life cycle of SFV includes integration of proviral DNA into the host genome. Our ability to amplify SFV may reflect either active infection of human tissues or simply the migration of SFV-infected baboon cells into those tissue compartments. Primers to the mitochondrial cytochrome oxidase subunit II (COII) gene were used to detect baboon-specific genes in the xenograft and patient samples.10 In addition, we attempted to amplify BaEV DNA. While endogenous viruses do not typically replicate in their natural host, infection in the transplant recipients may also result from either active infection or circulating baboon cells. A summary of these results is presented in Table 1. Mitochondrial DNA and BaEV were detected in every sample in which SFV was seen. In addition, baboon mitochondrial DNA was also detected in liver from both patients at early time points and in the lymph node (day 11) from patient 1 in the absence of detectable SFV. Baboon cells contain both BaEV and mitochondrial DNA at high copy number, while SFV infection is generally more limited in cell type and copy number. Consequently, our inability to amplify SFV DNA in some tissue samples that have evidence of baboon DNA may have resulted from these differences and possibly differences in sensitivity of the assays.
Table 1.
Detection of Baboon-Related DNA in Human Liver Transplant Patients
| Sample | SFV (LTR)a | BaEVb | Baboon mitochondrial DNAc |
|---|---|---|---|
| Baboon PBMCs (positive control) | +++/+++ | ++ | +++ |
| Human PBMCs (negative control) | −/− | – | – |
| Patient 1 | |||
| PBMCs (day 25) | −/− | – | + |
| LN (day 11) | −/− | + | + |
| LN (day 70) | +/+ | ++ | +++ |
| Liver (day 16) | −/− | ++ | +++ |
| Kidney (day 70) | +/+ | + | +++ |
| Patient 2 | |||
| PBMCs (pretransplantation) | −/− | – | – |
| PBMCs (day 25) | −/− | – | – |
| LN (day 27) | +/+ | + | +++ |
| Liver (day 12) | −/− | ++ | +++ |
| Liver (day 27) | +++/+++ | ++ | +++ |
| Kidney (day 27) | +/+ | + | + |
| Bone marrow (day 27) | +/− | ++ | ++ |
Simian foamy virus LTR DNA was amplified on two different occasions with separate samples that had been stored in another laboratory. Signal intensity was scored from (+) weak to (+ + +) strong.
A 389-bp BaEV proviral fragment was amplified by PCR using pol primers BPOLF4 and BPOLR5. Amplification conditions included 40 cycles, with 1 min of denaturation (94°C), annealing (50°C), and extension (72°C). Detection of the PCR product was made by Southern blot hybridization with the 32P-labeled BaEV internal probe BPOLP3. The sequences of the primers and probe are as follows and are based on the GenBank accession number M16550: BPOLF4 (5′-AGAAAACCAACCAAGCCCACACG-ATT-3′), BPOLR5 (5′-CAGCTTTGTTCTTGATTICCTTTCCTTCTGAG-3′), and BPOLP3 (5′-GGGCACAATCACTACCTCCTGGCA-3′).
Baboon mitochondrial DNA was detected by amplifying a 312-bp fragment from the mitochondrial cytochrome oxidase subunit II (COII) gene, using baboon-specific primers as previously described10: BCOIIFI (5′-CACTAACATCACGGACGCCCAA-3′) and BCOIIR1 (5′-GCCTGGTCGTG-TGGCTGTAAATAC-3′). Products were electrophoresed on 1.5% agarose gels followed by Southern blotting using 32P-labeled probe BCOIIP3 (5′-TCTAAATCCAGGTGACCTTC-3′).
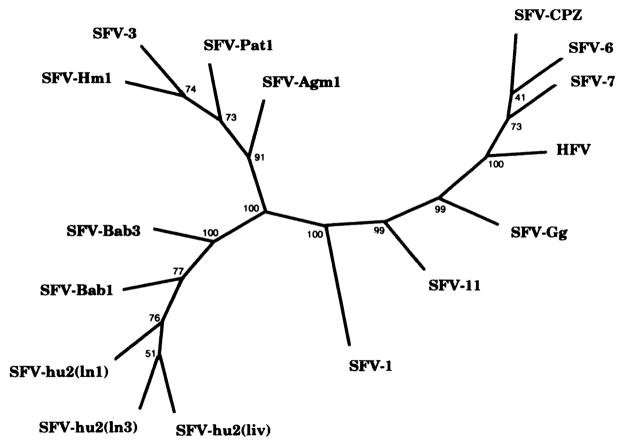
Early reports suggested widespread foamy virus infection in humans; however, more recent studies have shown that the human foamy virus (HFV) is not endemic in human populations.11 Our previous studies have shown that baboons harbor host-specific viruses, based on the genetic relatedness of both SFV pol and LTR sequences.3 To determine if the foamy virus DNA detected in these patients is of baboon origin and not due to prior infection with an HFV, PCR-amplified FV DNA from patient 2 on day 27 was sequenced. Viral DNA from both liver and lymph node of patient 2 was found to be most closely related toSFV-Bab viruses (Fig. 3). The phylogenetic tree shows that the SFV sequences (SFV-hu2) clustered with the SFV-Bab group of viruses and away from other simian and human SFV-like sequences. To rule out possible contamination of samples, PCR amplification and DNA sequencing were repeated with the liver sample stored in another laboratory and thus unlikely to be contaminated with SFV. Greater than 99% homology between liver samples was noted (not shown).
FIG. 3.
Unrooted consensus tree of SFV-hu2 LTR DNA from human transplant recipient. The designations for foamy viruses are as follows: SFV-hu2(ln1), -hu2(ln3), -hu2(liv) (human recipient), SFV-1 (macaque), SFV-3, -Agm1 (African green monkey), SFV-Bab1, -Bab3 (baboon), SFV-Gg (gorilla), SFV-6, -7, and CPZ (chimpanzee), SFV-11 (orangutan), SFV-Pat (patas), and SFV-Hm1 (Hamlyn’s guenon). LTR-amplified products were cloned with a TA cloning kit (Invitrogen, San Diego, CA) and sequenced as previously described.3 Two clones from each of the human samples were randomly selected and purified using a S.N.A.P. miniprep kit (Invitrogen). An Applied Biosysterns (Foster City, CA) automated sequencer model 373A, version 1.2.0, was used to derive the DNA sequences, which were then aligned using the GCG package, version 7.0, and manipulated by hand. Phylogenies were done using 100 bootstrap replicates in generating unrooted consensus trees with PHYLIP version 3.5p. Distances were calculated with Kimura’s two-parameter method (GCG Package version 7.0). Nucleotide sequences have been submitted to GenBank and are available under accession numbers AFO62309, AFO62310, and AFO62311.
These findings demonstrate the potential for both exogenous and endogenous viruses to reside in human recipients of animal organs for a significant period after transplantation. It is possible that these circulating xenogeneic cells could also act as conduits for new human infections. Although attempts to isolate SFV from these patients was unsuccessful, it remains to be determined if SFV is expressed in these recipients (M.G. Michaels, unpublished data, 1997). Studies of SFV-infected animal workers would suggest that SFV might replicate in humans yet, to date, there has been no evidence of retrovirus-related diseases. It must be emphasized that treatment of transplant recipients with immunosuppressive agents could enhance the risks from xenogeneic infections. Since retroviruses commonly exist as persistent latent infections, with an incidence of disease that varies because of both host and viral factors, the possibility that baboon foamy viruses might cause disease in humans remains a consideration in discussing future animal sources for xenotransplantation. Theoretically, other yet to be characterized viruses carried by baboons might also be transmitted to human recipients. In addition to baboons, pigs are also being considered as donors for human transplantation, and studies raise the possibility that porcine viruses may be of public health significance to humans. 12 Further studies are warranted that closely define the range of infectious agents present in donor species, and the potential for these agents to be transmitted to recipients and thus the population at large.
Acknowledgments
This study was funded in part from NIH Grant NIAID RO1 AI28273. We also thank Richard Heberling for generously providing human foreskin fibroblast cultures.
References
- 1.Allan JS. Xenotransplantation at a crossroads: Prevention versus progress. Nature Med. 1996;2:18–21. doi: 10.1038/nm0196-18. [DOI] [PubMed] [Google Scholar]
- 2.Mone J, Whitehead EM, Leland MM, Hubbard G, Allan JS. Simian T-cell leukemia virus type I infection in captive baboons. AIDS Res Hum Retroviruses. 1992;8:1653–1661. doi: 10.1089/aid.1992.8.1653. [DOI] [PubMed] [Google Scholar]
- 3.Broussard SR, Comuzzie AG, Leighton KL, Leland MM, Whitehead EM, Allan JS. Characterization of new simian foamy viruses (SFV) from African nonhuman primates. Virology. 1997;237:349–359. doi: 10.1006/viro.1997.8797. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. Nonhuman primate spumavirus infections among persons with occupational exposure–United States. MMWR. 1997;46:129–131. [PubMed] [Google Scholar]
- 5.van der Kuyl AC, Dekker JT, Goudsmit J. Full-length proviruses of baboon endogenous virus (BaEV) and dispersed BaEV reverse transcriptase retroelements in the genome of baboon species. I Virol. 1995;69:5917–5924. doi: 10.1128/jvi.69.9.5917-5924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starzl TE, Demetris AJ, Murase N, Trucco M, Thomson AW, Rao AS. The lost chord: Microchimerism. Immunol Today. 1996;17:577–584. doi: 10.1016/s0167-5699(96)10070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starzl TE, Valdivia LA, Murase N, Demetris AJ, Fontes P, Rao AS, Manez R, Marino I, Todo S, Thomson AW, Fung JJ. The biologic basis of and strategies for clinical xenotransplantation. Immunol Rev. 1994;141:213–244. doi: 10.1111/j.1600-065x.1994.tb00879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starzl TE, Fung J, Tzakis A, Todo S, Demetris AJ, Marino IR, Doyle H, Zeevi A, Warty V, Michaels M, et al. Baboon-to-human liver transplantation. Lancet. 1993;341:65–71. doi: 10.1016/0140-6736(93)92553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieniasz PD, Rethwilm A, Pitman R, Daniel MD, Chrystie I, McClure MO. A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology. 1995;207:217–228. doi: 10.1006/viro.1995.1068. [DOI] [PubMed] [Google Scholar]
- 10.Heneine W, Switzer WM. Highly sensitive and specific polymerase chain reaction assays for detection of baboon and pig cells following xenotransplantation in humans. Transplantation. 1996;62:1360–1362. doi: 10.1097/00007890-199611150-00033. [DOI] [PubMed] [Google Scholar]
- 11.Schweizer M, Turek R, Hahn H, Schliephake A, Netzer KO, Eder G, Reinhardt M, Rethwilm A, Neumann-Haefelin D. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: Appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res Hum Retroviruses. 1995;11:161–170. doi: 10.1089/aid.1995.11.161. [DOI] [PubMed] [Google Scholar]
- 12.Patience C, Takeuchi Y, Weiss RA. Infection of human cells by an endogenous retrovirus of pigs. Nature Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]