Summary
With the ready availability of spatial databases and geographical information system software, statisticians are increasingly encountering multivariate modelling settings featuring associations of more than one type: spatial associations between data locations and associations between the variables within the locations. Although flexible modelling of multivariate point-referenced data has recently been addressed by using a linear model of co-regionalization, existing methods for multivariate areal data typically suffer from unnecessary restrictions on the covariance structure or undesirable dependence on the conditioning order of the variables. We propose a class of Bayesian hierarchical models for multivariate areal data that avoids these restrictions, permitting flexible and order-free modelling of correlations both between variables and across areal units. Our framework encompasses a rich class of multivariate conditionally autoregressive models that are computationally feasible via modern Markov chain Monte Carlo methods. We illustrate the strengths of our approach over existing models by using simulation studies and also offer a real data application involving annual lung, larynx and oesophageal cancer death-rates in Minnesota counties between 1990 and 2000.
Keywords: Lattice data, Linear model of co-regionalization, Markov chain Monte Carlo methods, Multivariate conditionally autoregressive model, Spatial statistics
1. Introduction
The last decade has seen an explosion of interest in disease mapping, with recent developments in advanced spatial statistics and increasing availability of computerized geographic information system technology. For example, the databases from the National Center for Health Statistics or from the ‘Surveillance, epidemiology, and end results’ (SEER) programme of the National Cancer Institute, which are publicly available to anyone with a Web browser, provide an enormous supply of georeferenced data.
Disease mapping is an epidemiological technique that is used to describe the geographic variation of disease and to generate aetiological hypotheses about the possible causes for apparent differences in risk. Disease maps are used to highlight geographic areas with high and low incidence or mortality rates of a specific disease, and the variability of such rates over a spatial domain. They can also be used to detect spatial clusters which may be due to common environmental, demographical or cultural effects that are shared by neighbouring regions. However, mapping of crude incidence or mortality rates can be misleading when the population sizes for some of the units are small, resulting in large variability in the estimated rates, and making it difficult to distinguish chance variability from genuine differences. The correct geographic allocation of health care resources would be greatly enhanced by the development of statistical models that allow a more accurate depiction of ‘true’ rates of disease and their relation to explanatory variables (e.g. covariates).
For reasons of confidentiality or practicality, disease incidence or mortality data are often reported as counts or rates at a regional level (county, census tract, zip code, etc.). Conditionally autoregressive (CAR) models have been widely used for disease mapping with such data. They allow the borrowing of strength across regions by using not only the data from a given region but also the data from neighbouring regions. When we have multivariate areal (lattice) data (say, counts of p ≥ 2 diseases over the same regions), an obvious first choice would be to use p separate univariate CAR models. But correlation across diseases may occur if they share the same set of (spatially distributed) risk factors, or are linked by aetiology, a common risk factor or an affected organ. Moreover, the presence of one disease might encourage or inhibit the presence of another over a region. A multivariate areal model can permit modelling of dependence between those diseases while maintaining spatial dependence between regions. Identifying similar patterns in geographical variation of related diseases in a multivariate way may provide more convincing evidence for any real clustering in the underlying risk than would be available from the analysis of any single disease separately.
Several multivariate areal models have been proposed to date, any of which could be applied to multiple-disease mapping. The primary underlying challenge of multivariate areal modelling is to formulate valid probability models that account for association between different variables (e.g. diseases) within areal units along with the spatial association between areal units. Although they provide valuable theoretical insight into joint modelling of areal data, current methods often fall short of offering a template that is at once versatile and practical. Mardia (1988) described the theoretical background for multivariate Gaussian Markov random-field specifications, extending the pioneering work of Besag (1974), but used separable models that force identical spatial smoothing for all variables. The ‘twofold CAR’ model of Kim et al. (2001) offers richer spatial covariance structures for counts of two different diseases over each areal unit, but its extension to the case of more than two variables is unclear. Knorr-Held and Best (2001) developed a latent variable ‘shared component’ model for bivariate disease mapping; here extension to more than two diseases is possible (Held et al., 2005) but can be awkward. Sain and Cressie (2002) discussed a multiobjective version of the CAR model that allows for flexible modelling of the spatial dependence structure, the cross-correlations in particular, but may become computationally prohibitive. Carlin and Banerjee (2003) and Gelfand and Vounatsou (2003) developed essentially equivalent multivariate CAR (MCAR) models for hierarchical modelling to include non-separable models, but they left room for further generality in the covariance structures.
Adapting the multivariate point level data approach of Royle and Berliner (1999), Jin et al. (2005) proposed a generalized MCAR (GMCAR) model for areal data that formulates the joint distribution for a multivariate Markov random field by specifying simpler conditional and marginal models. These models are computationally efficient and allow sufficient flexibility in the specifications of the spatial covariance structure. Indeed, many of the above models arise as special cases of the GMCAR model. However, an inherent problem with these methods is that their conditional specification imposes a potentially arbitrary order on the variables being modelled, as they lead to different marginal distributions depending on the conditioning sequence. This problem is somewhat ameliorated in certain (e.g. medical and environmental) contexts where a natural order is reasonable, but in many disease mapping contexts this is not so. Although Jin et al. (2005) suggested using model comparison techniques to decide on the proper order of modelling, since all possible permutations of the variables would need to be considered this seems feasible only with relatively few variables. In any case, the principle of choosing between conditioning sequences by using model comparison metrics is perhaps not uncontroversial.
In this paper we develop an order-free framework for multivariate areal modelling that allows versatile spatial structures yet is computationally feasible for many variables. Our approach is based on a linear model of co-regionalization (LMC) that has recently been proposed for multivariate point-referenced data (Wackernagel, 2003; Schmidt and Gelfand, 2003; Gelfand et al., 2004). Essentially, the idea is to develop richer spatial association models by using linear transformations of much simpler spatial distributions. In this paper, we apply the LMC approach to the analysis of multivariate areal data, with an eye towards developing models for multipledisease mapping. In the process, we arrive at a very versatile framework, which encompasses a rich class of MCAR models (including most of the existing models) as special cases. In particular, we consider modelling the annual mortality rates from lung, larynx and oesophageal cancer between 1990 and 2000 in Minnesota counties, a setting in which association would be expected both within and across the areal units.
The format of our paper is as follows. In Section 2, we briefly review the spatial modelling of a single disease and multiple diseases. In Section 3, we introduce new LMC-based ways of multivariate spatial modelling. Bayesian computing issues are discussed in Section 4, whereas Section 5 evaluates our approach in terms of the average mean-square error (AMSE) and the deviance information criterion DIC via simulation. Section 6 then illustrates our approach in the aforementioned multiple-cancer data mapping setting. Finally, Section 7 summarizes our findings and suggests avenues for future research in this area.
2. Spatial modelling for disease mapping
Disease incidence or mortality data are often reported as counts or rates at a regional level (county, census tract, zip code, etc.) and are called areal (or lattice) data. Markov random-field models for lattice data are based on the Markov property, where the conditional distribution of a site's response given the responses of all the other sites depends only on the observations in the neighbourhood of this site. In this paper we define the neighbourhood by area adjacency, although other definitions are sometimes used (e.g. regions with centroids within a given fixed distance).
2.1. Spatial modelling of a single disease
Let Yi be the observed number of cases of a certain disease in region i, i = 1, … n, and Ei be the expected number of cases in this same region. Here the Yi are thought of as random variables, whereas the Ei are thought of as fixed and known (and are often simply taken as proportional to the number of people who are at risk in the region). For rare diseases, a Poisson model approximation to a binomial sampling distribution for disease counts is often used. Thus, a commonly used likelihood when mapping a single disease is
| (1) |
where . The xi are explanatory, region level spatial covariates, having parameter coefficients β. The parameter μi represents the log-relative-risk, estimates of which are often based on the departures of observed from expected counts. We place a form of Gaussian Markov random-field model, which is commonly referred to as the CAR prior, on the random effects ϕ = (ϕ1, …, ϕn)′, i.e.
| (2) |
where Nn denotes the n-dimensional normal distribution, D is an n × n diagonal matrix with diagonal elements mi that denote the number of neighbours of area i and W is the adjacency matrix of the map (i.e. Wii = 0, and Wii′ = 1 if i′ is adjacent to i and Wii′ = 0 otherwise). In the joint distribution (2), τ− is the spatial dispersion parameter, and α is the spatial autocorrelation parameter. The CAR prior corresponds to the following conditional distribution of ϕi:
| (3) |
where i ~ j denotes that region j is a neighbour (which is typically defined in terms of spatial adjacency) of region i. The CAR structure (2) reduces to the well-known intrinsic CAR (ICAR) model (Besag et al., 1991) if α = 1, or an independence model if α = 0. The ICAR model induces ‘local’ smoothing by borrowing strength from the neighbours, whereas the independence model assumes independence of spatial rates and induces ‘global’ smoothing. The smoothing parameter α in the CAR prior (2) controls the strength of spatial dependence between regions, though it has long been appreciated that a fairly large α may be required to deliver significant spatial correlation; see Wall (2004) for recent discussion and exemplification.
A similar approach proposes a Gaussian convolution prior for the modelling of the random effects ϕ. The random effects ϕ are assumed to be the sum of the two independent components with one having a Gaussian independence prior and the other a Gaussian ICAR prior (Besag et al., 1991). With such a convolution prior, we may capture both the relative contributions of regionwide heterogeneity and local clustering. Although this method has limitations (see for example Banerjee et al. (2004), pages 163–165), since the convolution process priors are among the most widely used we implement this model for our Minnesota cancer data analysis in Section 6.
2.2. Spatial modelling of multiple diseases
Now let Yij be the observed number of cases of disease j in region i, i = 1, …, n, j = 1, …, p, and let Eij be the expected number of cases for the same disease in this same region. As in Section 2.1, the Yij are thought of as random variables, whereas the Eij are thought of as fixed and known. For the first level of the hierarchical model, conditionally on the random effects ϕij, we assume that the Yij are independent of each other such that
| (4) |
where the xij are explanatory, region level spatial covariates for disease j having (possibly region-specific) parameter coefficients βj.
Carlin and Banerjee (2003) and Gelfand and Vounatsou (2003) generalized the univariate CAR model (2) to a joint model for the random effects ϕij under a separability assumption, which permits modelling of correlation between the p diseases while maintaining spatial dependence across space. Separability assumes that the association structure separates into a non-spatial and a spatial component. More precisely, the joint distribution of ϕ is assumed to be
| (5) |
where , Λ is a p × p positive definite matrix that is interpreted as the non-spatial precision (inverse dispersion) matrix between cancers and ‘⊗’ denotes the Kronecker product. We denote the distribution in expression (5) by MCAR(α, Λ). This distribution can be further generalized by allowing different smoothing parameters for each disease, i.e.
| (6) |
where , j = 1, … p. We denote the distribution in expression (6) by MCAR (α1, …, αp, Λ). Note that the off-diagonal block matrices (the Ris) in the precision matrix in expression (6) are completely determined by the diagonal blocks. Thus, the spatial precision matrices for each disease induce the cross-covariance structure in expression (6).
Recently, Jin et al. (2005) developed a more flexible GMCAR model for the random effects ϕ. For example, in the bivariate case (p = 2), they specified the conditional distribution ϕ1|ϕ2 as N[(η0I + η1W)ϕ2, {τ1(D − α1W)}−1], and the marginal distribution of ϕ2 as N[0, {ϕ2(D − α2W)}−], both of which are univariate CAR as in model (2). This formulation yields the models of Kim et al. (2001) as a special case and recognizes explicit smoothing parameters (η0 and η1) for the cross-covariances, unlike the MCAR models in expression (6) where the cross-covariances are not smoothed explicitly.
Kim et al. (2001) and Jin et al. (2005) demonstrated that explicit smoothing of the crosscovariances yield better model fits to areally referenced bivariate data. However, to model the random effects ϕ with the GMCAR model, we need to specify the order of conditioning, since different orders of conditioning will result in different marginal distributions for ϕ1 and ϕ2 and, hence, different joint distributions for ϕ. As mentioned in Section 1, in disease mapping contexts a natural conditioning order is often not evident—which is a problem that is exacerbated when we have more than two diseases. What we seek, therefore, are models that avoid this dependence on conditional ordering, yet are computationally feasible with sufficiently rich spatial structures.
3. Order-free multivariate conditionally autoregressive distributions
Our primary methodological objective is to formulate MCAR distributions that allow explicit smoothing of cross-covariances while not being hampered by conditional ordering. The most natural model here would parameterize the cross-covariances themselves as D − γijW, instead of using the Rjs as in expression (6). Unfortunately, except in the separable model with only one smoothing parameter α, constructing such dispersion structures is not trivial and leads to issues of identifiability on the γs (see, for example, Gelfand and Vounatsou (2003)). Kim et al. (2001) resolved these identifiability issues in the bivariate setting by using diagonal dominance but recognized the difficulty in extending this to the multivariate setting. We address this problem by using an LMC. The LMC is a well-established tool that is used in multivariate geostatistics (Chilés and Delfiner, 1999; Wackernagel, 2003; Banerjee et al., 2004) to incorporate different spatial ranges for each variable. However, to date this technique has not been employed in areal modelling, which has instead traditionally relied on conditional specifications.
It is worth pointing out that our use of the LMC here is somewhat broader than usually encountered in geostatistics. In geostatistics we typically transform independent latent effects, which suffices in meeting the primary goal of introducing a different spatial range for each variable. This is akin to introducing different smoothing parameters for each variable and indeed, as we show below in Section 3.2, independent latent effects lead to the MCAR(α1, …, αp; Λ) model in expression (6). However, to smooth the cross-covariances explicitly with identifiable parameters, we shall relax the independence of latent effects. Still, in our ensuing parameterization, we can derive conditions that yield valid joint distributions. To be precise, let be an np × 1 vector, where each ϕj = (ϕ1j, …, ϕnj)′ is n × 1 representing the spatial effects corresponding to disease j. We can write ϕ = (A ⊗ In×n)u, where is n p × 1 with each uj being an n × 1 areal process. Indeed, a proper distribution for u ensures a proper distribution for ϕ subject only to the non-singularity of A. The flexibility of this approach is apparent: we obtain different multivariate lattice models with rich spatial covariance structures by making different assumptions about the p spatial processes uj.
3.1. Case 1: independent and identical latent processes
First, we shall assume that the random spatial processes uj, j = 1, …, p, are independent and identical. Since each spatial process uj is a univariate process over areal units, we might adopt a CAR structure (2) for each of them, i.e.
| (7) |
Since the uj are independent of each other, the joint distribution of is u ~ Nnp {0, Ip×p ⊗ (D − αW)−1}. Recall that, since ϕ = (A ⊗ In × n) u, the joint distribution of ϕ is
| (8) |
defining Σ = AA′. We denote the distribution in expression (8) by MCAR(α, Σ). Note that the joint distribution of expression (8) is identifiable up to Σ = AA′ and is independent of the choice of A. Thus, without loss of generality, we can specify the matrix A as the upper triangular Cholesky decomposition of Σ. The MCAR(α, Σ) distribution that is given in expression (8) is exactly the same as the MCAR(α, Λ) structure in expression (5), as in Carlin and Banerjee (2003) and Gelfand and Vounatsou (2003) with Σ corresponding to Λ−1. Since ϕ = (A ⊗ In × n) u, a valid joint distribution of ϕ requires valid joint distributions of the uj, which happens if and only if 1/ξmin < α < 1/ξmax, where ξmin and ξmax are the minimum and maximum eigenvalues of D−1/2 WD−1/2. If α=1 in CAR structure (7), which is an ICAR model, the joint distribution of ϕ in expression (8) becomes the multivariate ICAR model (Gelfand and Vounatsou, 2003).
Currently, the WinBUGS package (http://www.mrc-bsu.cam.ac.uk/bugs/welcome.shtml) can fit the MCAR(α = 1, Σ) distribution (using its mv.car distribution), but not the MCAR(α, Σ) distribution. However, through the LMC approach we still can fit the MCAR(α, Σ) distribution in WinBUGS by writing ϕ = (A ⊗ (In×n)u and assigning proper CAR priors (via the car.proper distribution) for each uj j = 1, …, p, with a common smoothing parameter α. Regarding the prior on A, note that, since AA′ = Σ and A is the Cholesky decomposition of Σ, there is a one-to-one relationship between the elements of Σ and A. In Section 4, we argue that assigning a prior to Σ is computationally preferable.
3.2. Case 2: independent but not identical latent processes
In case 1, we assumed that the random spatial processes uj, j = 1,…, p, were independent and identical. However, it will often be preferable to have p different spatial processes. In this subsection, we shall continue to assume that the uj are independent, but we relax their being identically distributed. Adopting the CAR structure (2), the distribution of uj is assumed to be
| (9) |
where αj is the smoothing parameter for the jth spatial process. Since the ujs are independent of each other and ϕ = (A ⊗ In×n/u, the joint distribution of ϕ is
| (10) |
where Σ = AA′ and Γ is an n p × n p block diagonal matrix with n × n diagonal entries Γj = D − αjW, j = 1,…, p. We denote the distribution in expression (10) by MCARα1,…, αp, Σ).
In this case, from the joint distribution in expression (10) it can be seen that different joint distributions of ϕ having different covariance matrices emerge under different linear transformation matrices A. To ensure that A is identifiable, we could again specify it to be the upper triangular Cholesky decomposition of Σ, although this might not be the best choice computationally. Through the LMC approach in this case, the distribution in expression (10) is similar to the MCAR(α1,…, αp, Λ) structure (6), which was developed in Carlin and Banerjee (2003) and Gelfand and Vounatsou (2003). All of these have the same number of parameters, and there is no unique joint distribution for ϕ with the MCAR(α1,…, αp, Λ) structure, since there is not a unique Rj-matrix such that RjR′j = RjPP′j = D − αjW (P being an andarbitrary orthogonal matrix). Carlin and Banerjee (2003) took Rj as the Cholesky decomposition of D − αjW, whereas Gelfand and Vounatsou (2003) instead recommended a spectral decomposition.
Again, a valid joint distribution in expression (10) requires p valid distributions for uj, i.e. 1=ξmin < αj < 1=ξmax, j = 1,…, p. Through the LMC approach, we can also fit the data with the MCAR.α1,…, αp, Σ/ prior distribution (10) on ϕ in WinBUGS as in Section 3.1 by writing ϕ = (A ⊗ In×n)u and assigning proper CAR priors (via the car.proper distribution) with a distinct smoothing parameter αj for each uj, j = 1,…, p. As mentioned in Section 3.1, we assign a prior to AA′ = Σ (e.g. an inverse Wishart prior) and determine A from the one-to-one relationship between the elements of Σ and A; Section 4 provides details.
3.3. Case 3: dependent and not identical latent processes
Finally, in this case we shall assume that the random spatial processes uj = (u1j,…, unj)′, j= 1,…, p, are neither independent nor identically distributed. We now assume that uij and ui,l≠j are independent given uk≠,j and uk≠i,l≠j, where l, j = 1,…, p and i, k = 1,…, n implying that latent effects for different diseases in the same region are conditionally independent given those for diseases in the neighbouring regions. On the basis of the Markov property and similarly to the conditional distribution that is given by expression (3) in the univariate case, we specify the ijth conditional distribution as Gaussian with mean
and conditional variance var(uij|uk≠i,j, ui,l≠j, uk≠i,l≠j) ∝ 1/mi, where bjj denotes the spatial autocorrelation for the random spatial process uj whereas bjl (l≠j, l, j=1,…, p) denotes the cross-spatial correlation between the random spatial process uj and ul. Putting these conditional distributions together reveals the joint distribution of to be
| (11) |
where I is a p × p identity matrix and B is a p × p symmetric matrix with elements bjl, j, l = 1,…, p. As long as the dispersion matrix in expression (11) is positive definite, which boils down to (Ip × p⊗D−B⊗W) being positive definite, expression (11) is itself a valid model. To assess non-singularity, note that
Denoting the eigenvalues for D−1/2 WD−1/2 as ξi, i = 1,…, n, and the eigenvalues for B as ζj, j = 1,…, p, we find (see, for example, Harville (1997), theorem 21.11.1) the eigenvalues for B⊗(D−1/2WD−1/2) as ξi × ζj, i = 1,…, n, j = 1,…, p. Hence, the conditions for Ip×p⊗D−B⊗W being positive definite become ξiζj < 1, i.e. 1/ξmin < ζj < 1/ξmax, i = 1,…,n, j = 1,…, p, where ξmin and ξmax are the minimum and maximum eigenvalues of D−1/2WD−1/2. Thus, 1/ξmin < ζj < 1, j = 1,…, p, ensures the positive definiteness of the matrix Ip × p⊗D−B⊗W and, hence, the validity of the distribution of u given in expression (11). In fact, ξmax = 1 and ξmin < 0 (see, for example, Banerjee et al. (2004)), which makes this formulation easier to work with in practice (e.g. in choosing priors; see Section 4) than the alternative constraint 1/ζmin < ξj < 1/ζmax.
Model (11) introduces smoothing parameters in the cross-covariance structure through the matrix B but unlike the MCAR models in Sections 3.1 and 3.2 does not have the Σ-matrix to capture non-spatial variances. To remedy this, we model ϕ = (A⊗In×n) u so that the joint distribution random effects ϕ is
| (12) |
Since ϕ = (A⊗In×n)u, it is immediate that the validity of model (11) ensures a valid joint distribution for expression (12). We denote distribution (12) by MCAR(B,Σ), where Σ=AA′. Again, A identifies with the upper triangular Cholesky square root of Σ. With Σ=I we recover expression (11), which we henceforth denote as MCAR(B, I).
To see the generality of expression (12), we find that the joint distribution of ϕ reduces to the MCAR(α1,…,αp,Σ) distribution (10) if bjl=0 and bjj=αj, or the MCAR(α,Σ) distribution (8) if bjl=0 and bjj=α, in both cases for j, l=1,…,p. Also note that the distribution in expression (12) is invariant to orthogonal transformations (up to a reparameterization of B) in the following sense: let T = AP with P being a p×p orthogonal matrix such that TT′=APP′ A′Σ. Then the covariance matrix in expression (12) can be expressed as
where C=P′BP. Without loss of generality, then, we can choose the matrix A as the upper triangular Cholesky decomposition of Σ.
To understand the features of the MCAR(B,Σ) distribution (12), we illustrate in the bivariate case (p=2). Define
and
where
and
Note that the γs are not identifiable from the matrix Λ and our reparameterization in terms of B must be used to conduct posterior inference on B and Λ (see Section 4), from which the cross-covariances may be recovered. The above expression does allow the MCAR(B,Σ) distribution (12) to be rewritten as
| (13) |
which is precisely the general dispersion structure that we set out to achieve.
To see how the parameters in expression (13) affect smoothing, we obtain the conditional means
and
and the conditional variances and , i, k = 1, … n. (The conditional moments for special cases such as the separable model (5) arise by simply setting γ1 = γ2 = γ12 = α.) We note that −Λ12/Λ11 and −Λ12/Λ22 appear as regression parameters, regressing one component of ϕ at a given site on the other. The spatial smoothing is incorporated by an autoregressive term for each site in the neighbour set which is corrected at each site for the regression on the other component. The smoothing parameters are different for each spatial association and cross-spatial association. For neighbouring regions i and k, expression (13) yields the partial correlations corr(ϕi1, ϕk1|ϕ1\(ik),ϕ2)=γ1/√(mimk), corr(ϕi2, ϕk2|ϕ2\(ik),ϕ1) = γ2/√(mimk), corr(ϕi1,ϕi2|ϕ1\i,ϕ2\i)=−Λ12/√(Λ11Λ22), and corr(ϕi1, ϕk2|ϕ1\i, ϕ2\\k) = corr(ϕk1,ϕi2|ϕ2\i)= γ12Λ12/√(mimk×Λ11Λ22), where ϕl = (ϕ1l,…,ϕnl)′ and ϕl\i=(ϕ1l,…ϕi−1,ϕi+1,l,…,ϕnl)′, for l = 1, 2 and i, k = 1,…, n. With the MCAR(α, Λ) distribution from expression (5) or the MCAR(α,Σ) distribution from expression (8), we have corr(ϕi1, ϕk1|ϕ1\(ik), ϕ2)=corr(ϕi2, ϕk2|ϕ2\(ik),ϕ1)=α/√(mimk), and corr(ϕi1,ϕk2|ϕ1\i,ϕ2\k)=−corr(ϕi1, ϕk1|ϕ1\(ik),ϕ2) corr(ϕi1 ϕk|ϕ1\(ik,|ϕ2) corr(ϕi, ϕi2|ϕ1\i, ϕ2\i)= α Λ12/√(mimkΛ11Λ22), in both cases for i, k = 1, …, n. Thus case 3 offers the most flexible model for the conditional correlation structure.
4. Bayesian computation
Our proposed MCAR(B, Σ) model is straightforwardly implemented in a Bayesian frame-work by using Markov chain Monte Carlo (MCMC) methods. As in Section 3.3, we write ϕ = (A ⊗ In×n)u, where u = (u1, u2)′ and uj = (u1j,…, unj/′. The joint posterior distribution is
| (14) |
where Y1 = (Y11, …, Yn1)′ and Y2 = (Y12, …, Yn2)′ and L(Y, Y2|u, σ2, A) is the data likelihood.
The second term on the right-hand side of expression (14) is p(u|B) = Nn p {0, (Ip × p ⊗ D − B ⊗ W)−1}. As mentioned in Section 3.3, propriety of this distribution requires the eigenvalues ζj of B to satisfy 1/ξmin < ζj < 1 (j = 1,…, p). When p is large, it is difficult to determine the intervals over the elements of B that result in 1/ξmin < ζj < 1, and thus designing priors for B that guarantee this condition is awkward. In principle, we might impose the constraint numerically by assigning a flat prior or a normal prior with a large variance for the elements of B, and then simply check whether the eigenvalues of the corresponding B-matrix are in that range during a random-walk Metropolis–Hastings update. If the resulting eigenvalues are out of range, the values are thrown out since they correspond to prior probability 0; otherwise we perform the standard Metropolis–Hastings comparison step. In our experience, however, this does not work well, especially when p is large.
Instead, here we outline a different strategy to update the matrix B. Our approach is to represent B by using the spectral decomposition, which we write as B = PΔP′, where P is the corresponding orthogonal matrix of eigenvectors and Δ is a diagonal matrix of ordered eigen-values, ζ1,…, ζp. We parameterize the p × p orthogonal matrix P in terms of the p(p − 1)/2 Givens angles θij for i = 1,…, p − 1 and j = i + 1,…, p (Daniels and Kass, 1999). The matrix P is written as the product of p(p− 1)/2 matrices, each associated with a Givens angle. Specifically, P = G12G13 … G1p … Gp−1)p where i and j are distinct and Gij is the p × p identity matrix with the ith and jth diagonal elements replaced by cos(θij), and the (i, j)th and (j, i)th elements replaced by ± sin(θij). Since the Givens angles θij are unique with a domain (−π/2, π/2) and the eigenvalues ζj of B are in the range (1/ξmin, 1), we then put a U(−π/2, π/2) prior on the θij and a U(1/ξmin, 1) prior on the ζj. To update θijs or ζjs by using random-walk Metropolis–Hastings steps with Gaussian proposals, we need to transform them to have support equal to the whole real line. A straightforward solution here is to use
a transformation having Jacobian . In practice, the ζj must be bounded from 1 (say, by insisting that 1=ξmin < ζj < 0:999, j = 1,…, p) to maintain identifiability and hence computational stability. In fact, with our approach it is also easy to calculate the determinant of the precision matrix, i.e. , where ξi are the eigenvalues of D−1/2 WD−1/2, which can be calculated before any MCMC iteration. For the special case of the MCAR(α1, …, αp, Σ) models, we could assign each αi ~ U(0, 1), which would be sufficient to ensure a valid model (e.g. Carlin and Banerjee (2002)). However, we also investigated with more informative priors on the αis such as the beta(2, 18) prior that centres the smoothing parameters closer to 1 and leads to greater smoothing.
With respect to the prior distribution p(A) on the right-hand side of expression (14), we can put independent priors on the individual elements of A, such as inverse gamma for the square of the diagonal elements of A and normal for the off-diagonal elements. In practice, we cannot assign non-informative priors here, since then MCMC convergence is poor. In our experience it is easier to assign a vague (i.e. weakly informative) prior on Σ than to put such priors on the elements of A in terms of letting the data drive the inference and obtaining good convergence. Since Σ is a positive definite covariance matrix, the inverse Wishart prior distribution renders itself as a natural choice, i.e. Σ−1 ~ Wishart{ν, (νR)−1} (see for example Carlin and Louis (2000), page 328). Hence, we instead place a prior directly on Σ and then use the one-to-one relationship between the elements of Σ and A. Then the prior distribution p(A) becomes
where |∂Σ/aij| is the Jacobian. For example, when p = 2, the Jacobian is . Rather than updating Σ as a block by using a Wishart proposal, updating the elements aij of A offers better control. These are updated via a random-walk Metropolis step, using log-normal proposals for the diagonal elements and normal proposals for the off-diagonal elements. With regard to choosing ν and R in the Wishart{ν, (νR)−1 prior, since E(Σ−1) = R−1, if there is no information about the prior mean structure of Σ, a diagonal matrix R can be chosen, with the scale of the diagonal elements being judged by using ordinary least squares estimates based on independent models for each response variable. Although this leads to a data-dependent prior, it typically lets the data drive the results, leading to robust posterior inference. In this study we adopt ν = 2 (i.e. the smallest value for which this Wishart prior is proper) and R = diag(0.1, 0.1). Finally, for the remaining terms on the right-hand side of expression (14), flat priors are chosen for β1 and β2, whereas σ2 is assigned a vague inverse gamma prior, i.e. an IG(1, 0.01) prior parameterized so that E(σ2) = b/(a − 1). In this study, β and σ2 have closed form full conditionals and so can be directly updated by using Gibbs sampling.
5. Simulation study
To evaluate our new approach for modelling multivariate areal data, we begin with some simulation studies. The studies use the spatial lay-out of the 87 counties in the state of Minnesota, which is a fairly typical areal arrangement and the one that is used by our Section 6 data set. We generated count data from a Poisson distribution, as is typical in disease mapping settings:
| (15) |
where Yij is the observed number of cases of cancer type j (one of two types) in region i and log(μij) = βj + ϕij with the βjs being fixed as β1 = −0.05 and β2 = − 0.01. This set-up mimics oesophageal (relatively low incidence) and lung (higher incidence) Minnesota cancer incidence data and uses the actual internally standardized expected mortalities Eij from our SEER data.
To assess the relative performance of our proposed model, we designed five simulation studies. In study 1, we generate ϕ1 = (ϕ11,…, ϕ1n)′ and ϕ2 = (ϕ21,…, ϕ2n)′ from the MCAR(B, Σ) distribution (12), where D = diag(mi) and the adjacency matrix W are based on the Minnesota county map. We specify the distribution in expression (12) by setting
which yields a non-spatial correlation of 0.32, and
These choices yielded ϕijs ranging between (−1.5, 1.5) with correlations between the spatial effects ranging from 0.08 to 0.73. In study 2, we generate ϕ from the MCAR(B, I) model (11) retaining the same B as in study 1, whereas, in study 3, we generate them from the MCAR(α, Σ) model (8) with the same A as in study 1. In study 4, we generate the ϕ from the GMCAR(α1, α2, η0, η1, τ1, τ2) models under the conditioning order ϕ1|ϕ2. We choose the true parameter values in this GMCAR distribution to be α1 = 0.1, α2 = 0.8, η0 = 0.4, η1 = 0:3, τ1 = 10 and τ2 = 10. Finally, study 5 examines the effect of model misspecification by using a geostatistical model (instead of an MCAR model) as the true random-effect distribution. Here ϕ1 and ϕ2 arise from a Gaussian process with exponential covariance functions exp(−0.01dij) and exp(−0.05dij) respectively, where dij is the distance between the centroids of counties i and j, and is calculated using the rdist.earth() function in R using the fields package. In this model we take the same values of a11, a12, a21 and a22 as in study 2.
The data that are analysed in this paper and the programs that were used to analyse them can be obtained from http://www.blackwellpublishing.com/rss
5.1. Simulation results: mean-squared error
To evaluate the performance of our proposed model, we simulated N = 1000 data sets and fitted several multivariate models to each. In each study of Table 1, model 1 is the MCAR(B, Σ) model that is specified by expression (12), whereas model 2 is the MCAR(B, I) model in expression (11). Model 3 is the MCAR(α, Σ) model from Section 3.2 or, equivalently, the MCAR(α, Λ) model in expression (5). Models 4 and 5 are the order-specific GMCAR(α1, α2, η0, η1, τ1, τ2) models (Jin et al., 2005) with the conditioning order ϕ1|ϕ2 and the reverse conditioning order ϕ2|ϕ1 respectively. Finally, model 6 is a bivariate independent and identically distributed (IID) model.
Table 1.
AMSE, associated Monte Carlo standard errors se and percentage change in AMSE (Δ) relative to the true model in each simulation study with Poisson responses†
| Study | Model | AMSE1 |
AMSE2 |
Overall AMSE |
|||
|---|---|---|---|---|---|---|---|
| Mean (× 10−2) (se (× 10−5)) |
Δ (%) | Mean (× 10−2) (se (× 10−5)) |
Δ (%) | Mean (× 10−2) (se (× 10−5)) |
Δ (%) | ||
| 1 | 1, MCAR(B, Σ) | 7.51 (8.51) | — | 1.34 (0.49) | — | 4.43 (8.54) | — |
| 2, MCAR(B, I) | 8.36 (8.53) | 11.32 | 2.17 (1.09) | 61.94 | 5.26 (4.30) | 18.74 | |
| 3, MCAR(α, Σ) | 7.90 (8.48) | 5.19 | 1.54 (1.15) | 14.92 | 4.72 (4.28) | 6.54 | |
| 4, GMCAR | 8.16 (8.55) | 8.65 | 1.71 (1.19) | 27.61 | 4.93 (4.32) | 11.29 | |
| 5, GMCAR (reverse order) | 7.75 (8.51) | 3.19 | 1.39 (0.69) | 3.73 | 4.57 (4.27) | 3.16 | |
| 6, bivariate IID | 9.92 (8.52) | 32.09 | 2.91 (1.18) | 117.16 | 6.42 (4.30) | 44.92 | |
| 2 | 1, MCAR(B, Σ) | 7.90 (8.41) | −0.13 | 2.15 (1.33) | 3.86 | 5.03 (4.26) | 0.80 |
| 2, MCAR(B, I) | 7.91 (8.39) | — | 2.07 (1.19) | — | 4.99 (4.24) | — | |
| 3, MCAR(α, Σ) | 8.07 (8.41) | 2.02 | 2.58 (1.31) | 24.64 | 5.33 (4.25) | 6.81 | |
| 4, GMCAR | 8.19 (9.51) | 3.54 | 2.82 (1.45) | 36.23 | 5.51 (4.81) | 10.42 | |
| 5, GMCAR (reverse order) | 7.96 (8.51) | 0.63 | 2.39 (1.34) | 15.46 | 5.18 (4.31) | 3.81 | |
| 6, bivariate IID | 10.53 (10.39) | 33.12 | 3.70 (2.29) | 78.74 | 7.11 (5.32) | 42.48 | |
| 3 | 1, MCAR(B, Σ) | 8.07 (8.27) | 0.25 | 1.98 (2.25) | −2.94 | 5.03 (4.18) | −0.40 |
| 2, MCAR(B, I) | 8.75 (8.29) | 8.70 | 2.61 (1.19) | 27.94 | 5.68 (4.19) | 12.48 | |
| 3, MCAR(α, Σ) | 8.05 (8.31) | — | 2.04 (0.87) | — | 5.05 (4.18) | — | |
| 4, GMCAR | 8.31 (8.57) | 3.23 | 2.26 (1.24) | 10.78 | 5.29 (4.33) | 4.75 | |
| 5, GMCAR (reverse order) | 8.28 (9.48) | 2.86 | 2.17 (1.30) | 6.37 | 5.23 (4.78) | 3.56 | |
| 6, bivariate IID | 9.57 (10.34) | 18.89 | 3.38 (2.11) | 65.68 | 6.48 (5.28) | 28.32 | |
| 4 | 1, MCAR(B, Σ) | 7.80 (8.02) | 0.91 | 1.68 (2.45) | 7.01 | 4.74 (4.19) | 1.93 |
| 2, MCAR(B, I) | 8.23 (8.12) | 6.47 | 2.22 (3.39) | 41.40 | 5.22 (4.40) | 12.26 | |
| 3, MCAR(α, Σ) | 7.95 (8.20) | 2.85 | 1.94 (2.57) | 23.57 | 4.95 (4.30) | 6.45 | |
| 4, GMCAR | 7.73 (7.98) | — | 1.57 (2.42) | — | 4.65 (4.17) | — | |
| 5, GMCAR (reverse order) | 8.08 (9.38) | 4.53 | 2.01 (3.31) | 28.02 | 5.04 (4.97) | 8.38 | |
| 6, bivariate IID | 9.68 (9.31) | 25.23 | 3.38 (3.31) | 115.29 | 6.53 (4.94) | 40.43 | |
| 5 | 1, MCAR(B, Σ) | 9.26 (8.72) | — | 2.07 (2.63) | — | 5.66 (4.55) | — |
| 2, MCAR(B, I) | 10.03 (8.82) | 8.32 | 2.65 (3.03) | 28.02 | 6.34 (4.66) | 12.01 | |
| 3, MCAR(α, Σ) | 9.98 (8.70) | 7.77 | 2.44 (2.93) | 17.87 | 6.21 (4.59) | 9.72 | |
| 4, GMCAR (reverse order) | 10.28 (10.03) | 11.01 | 3.95 (3.11) | 90.82 | 7.12 (5.25) | 25.79 | |
| 5, GMCAR | 10.19 (9.99) | 1.40 | 3.67 (3.02) | 9.66 | 6.93 (5.22) | 3.00 | |
| 6, bivariate IID | 14.20 (9.51) | 53.35 | 3.98 (3.52) | 92.27 | 9.09 (5.07) | 60.61 | |
Except for study 5, the dash indicates that the model is the true model for this study.
For each data set and model in each study, we first ran a few initially overdispersed parallel MCMC chains, and monitored them by using measurements of sample autocorrelations within the chains, cross-correlations between parameters and plots of sample traces. From these, we decided to use 20000 iterations for the preconvergence burn-in period, and then a further 20000 iterations as our ‘production’ run for posterior summarization. Unfortunately, the complexity of model (12) precluded us from using the WinBUGS package, so we instead relied on our own programs written in C and executed in R (http://www.r-project.org) using the .C function. Random-number generation and posterior summarization were also implemented in R.
To evaluate the relative performance of the models, we compare their AMSE. Since the true ϕij-values are known in the simulation, the AMSE for each disease can be estimated as
with associated Monte Carlo standard error estimate
where is the posterior mean estimate for disease j at county i based on the rth data set. In our case we have N = 1000, n = 87 and j = 1, 2.
Table 1 gives the estimated AMSEj-values and their associated Monte Carlo standard errors for each disease and each model in each simulation study. The estimated overall AMSE values are calculated by aggregating over the two diseases. Here we also calculate the percentage change in estimated AMSE for each model compared with the true model in each study, i.e. for models k = 1,…,5; negative values would indicate superiority over the true model.
From Table 1, the performance of the MCAR(B, Σ) model appears quite impressive. Not surprisingly, in study 1 where it is the true model it excels over the other MCAR and GMCAR models. In study 2 its performance is highly comparable with the true model MCAR(B, I). In fact, we find a marginally lower AMSE score for ϕ1 (−0.13%) and a virtually identical overall AMSE (0.80%). The story is similar for study 3, where the MCAR(α, Σ) model is the true model, and now we find a marginally lower overall AMSE score (−0.40%). In study 4, where the GMCAR model is the true model, it performs a little worse (the overall AMSE score exceeds that of the true model by 1.93%) but is still appreciably better than the other competing models. Finally, in study 5 where all the models are misspecified, the MCAR(B, Σ) model remains the best, with considerably lower overall AMSE scores and only a hint of a competition from the GMCAR (reverse order) model.
The performance of the MCAR(B, I) model is disappointing. The only study where it performs competitively is in study 2, where it is in fact the true model. In all the remaining studies its overall AMSE scores beat only those of the bivariate IID model and are usually much higher than those of the GMCAR models (especially that having the better ordering) and the MCAR(α, Σ) model. That this occurs despite the smoothing of the cross-covariances through the matrix B is probably a reflection of the model's inadequacy in capturing the scaling of the spatial effects that are offered by Σ. The MCAR(α, Σ) model, unlike the MCAR(B, I) model, incorporates a single smoothing parameter but captures the within-site association between the two diseases through Σ. The MCAR(α, Σ) model's overall AMSE performance is typically seen to lie between those of the MCAR(B, Σ) and the better GMCAR model.
The GMCAR model also presents some interesting features. Recall that its parameterization is quite rich and it does allow spatially adaptive smoothing of the variances and cross-covariances. However, its primary drawback lies in its sensitivity to the order of conditioning, and this is apparent in Table 1. Except for study 4, where ϕ1|ϕ2 is the true model, ϕ2|ϕ1 appears to be offering much better estimation. In fact, its performance is seen to be only marginally inferior to the MCAR(B, Σ) model and its overall AMSE score never exceeds 3.81% of that of the true model. It typically beats the MCAR(α, Σ) model, except in study 3 where the latter is the true model and has an overall AMSE score lesser by 3.56%. In study 5, where all the models are misspecified, the better GMCAR model's AMSE score exceeds that of the MCAR(B, Σ) model by only 3% and is considerably better than all the remaining models.
In summary, although the MCAR(B, Σ) model consistently produces the lowest AMSEs-cores in Table 1, the absolute differences in AMSE among most of the models are rather small, of the order of 10−2 in our scale. The associated Monte Carlo standard errors of these AMSEs are of the order of 10−5, making these differences highly reliable computationally. However, in disease mapping exercises we would not expect to see practical differences between the models. Rather, the main message here is that, despite the rich parameterization of the MCAR(B, Σ) model, it does not appear to be suffering from excessive overfitting (i.e. few negative Δs with respect to the true model) while offering very robust estimation compared with its competitors.
5.2. Simulation results. deviance information criterion
To investigate the predictive performance of our methods, we now turn to the deviance information criterion DIC (Spiegelhalter et al., 2002) as our model choice criterion. This criterion is based on the posterior distribution of the deviance statistic, D(θ) =−2 log {f(y|θ) 2 log {h(y)}, where f(y|θ) is the likelihood function for the observed data vector y given the parameter vector θ on which we focus, and h(y) is some standardizing function of the data alone (which thus has no influence on model selection and which we set to 0). DIC is defined analogously to the Akaike information criterion as the posterior expected deviance plus the ‘effective’ number of parameters, i.e. DIC = D̄ + pD. Spiegelhalter et al. (2002) showed that pD is reasonably defined as Eθ|y[D] − D(Eθ|y[θ]) = D̄ − D(θ̄), i.e. the expected deviance minus the deviance evaluated at the expectations. Since small values of D̄ indicate good fit and small values of pD indicate a parsimonious model, small values of the sum (DIC) indicate preferred models. Note that a model with more parameters can be ‘more parsimonious’ using pD, since this criterion measures effective model size (i.e. the dimension of the posterior space spanned by the parameters after accounting for shrinkage of the random effects towards their grand mean). DIC is scale free (because D̄ is), and so no particular score has any intrinsic meaning; only the ordering of DIC-scores across models is meaningful. The use of pD and DIC is not without controversy; in particular, Celeux et al. (2006) highlighted several missing data settings where negativepD is a real risk. However, we did not experience this problem with any of our multivariate data models, and so we adopt this approach here owing to its flexibility and ease of use.
To calculate DIC in our MCMC setting, we need to calculate only the average deviance D̄, and the deviance of the posterior mean, D(θ̄). Here D(θ) is the same for the models that we wish to compare since they differ only in their random-effect distributions p(ϕ|B, Σ), which we do not consider to be part of the likelihood. Specifically, setting 2 log {h(y)} = 0 in D(θ), we have D(θ) ≡ D(β, ϕ) = −2 log{L(Y1, Y2|β, ϕ)}, where L is the likelihood the Poisson for model (15).
Table 2 gives the simulated results for D̄, pD and DIC for the same six models and five studies as were considered in Table 1. In Table 2 we redefine Δ as the absolute (not relative) change when compared with the true model in each case. Thus positive Δ-values indicate poorer fit, larger effective model size and poorer overall model selection scores for D̄, pD and DIC respectively.
Table 2.
Simulated D̄, pD and DIC-scores and associated standard deviations, along with absolute change (Δ) relative to the true model in each simulation study†
| Study | Model | D̄ |
pD |
DIC |
|||
|---|---|---|---|---|---|---|---|
| Mean (sd) | Δ | Mean (sd) | Δ | Mean (sd) | Δ | ||
| 1 | 1, MCAR(B, Σ) | 1004.3 (20.6) | — | 71.0 (3.6) | — | 1075.3 (20.8) | — |
| 2, MCAR(B, I) | 1071.9 (21.1) | 67.5 | 59.0 (3.8) | −12.0 | 1130.8 (21.5) | 55.6 | |
| 3, MCAR(α, Σ) | 1039.9 (20.4) | 35.6 | 64.1 (3.5) | −6.8 | 1105.0 (20.6) | 28.9 | |
| 4, GMCAR | 1052.8 (20.1) | 48.5 | 57.9 (3.9) | −13.0 | 1110.7 (20.7) | 35.5 | |
| 5, GMCAR (reverse order) | 1026.9 (21.6) | 32.6 | 58.2 (3.4) | −12.7 | 1085.2 (22.0) | 19.9 | |
| 6, bivariate IID | 1112.8 (23.8) | 108.4 | 55.0 (3.8) | −15.9 | 1167.8 (24.0) | 92.5 | |
| 2 | 1, MCAR(B, Σ) | 1004.9 (20.6) | −22.6 | 68.9 (3.7) | 13.8 | 1073.8 (20.9) | −8.8 |
| 2, MCAR(B, I) | 1027.5 (21.5) | — | 55.1 (3.5) | — | 1082.6 (21.8) | — | |
| 3, MCAR(α, Σ) | 1043.0 (19.8) | 15.5 | 63.9 (3.6) | 8.8 | 1106.9 (20.2) | 24.3 | |
| 4, GMCAR | 1071.1 (21.4) | 43.6 | 65.0 (3.8) | 9.8 | 1136.1 (21.7) | 53.5 | |
| 5, GMCAR (reverse order) | 1022.3 (21.9) | −5.2 | 63.0 (3.8) | 7.9 | 1085.3 (22.3) | 2.7 | |
| 6, bivariate IID | 1106.2 (24.7) | 78.7 | 51.0 (3.8) | −4.1 | 1157.3 (25.0) | 74.7 | |
| 3 | 1, MCAR(B, Σ) | 1000.5 (20.5) | −39.5 | 70.0 (3.9) | 9.9 | 1070.5 (20.7) | −29.6 |
| 2, MCAR(B, I) | 1092.0 (20.6) | 51.0 | 53.0 (3.7) | −7.1 | 1145.0 (21.1) | 44.9 | |
| 3, MCAR(α, Σ) | 1040.0 (20.6) | — | 60.1 (3.6) | — | 1100.1 (20.9) | — | |
| 4, GMCAR | 1078.9 (21.1) | 38.9 | 54.0 (3.8) | −6.1 | 1132.9 (21.4) | 32.8 | |
| 5, GMCAR (reverse order) | 1064.6 (22.9) | 24.5 | 52.1 (3.5) | −8.0 | 1116.7 (23.3) | 16.6 | |
| 6, bivariate IID | 1134.9 (24.5) | 94.9 | 51.1 (3.9) | −9.0 | 1186.0 (24.7) | 85.9 | |
| 4 | 1, MCAR(B, Σ) | 987.2 (20.3) | −7.3 | 75.0 (3.6) | 11.0 | 1062.2 (20.6) | 3.7 |
| 2, MCAR(B, I) | 1028.6 (21.5) | 34.1 | 73.9 (3.7) | 9.9 | 1102.5 (21.7) | 44.1 | |
| 3, MCAR(α, Σ) | 1021.9 (20.3) | 27.4 | 77.8 (3.5) | 13.8 | 1099.7 (20.6) | 41.2 | |
| 4, GMCAR | 994.5 (21.6) | — | 64.0 (3.9) | — | 1058.5 (21.7) | — | |
| 5, GMCAR (reverse order) | 1020.5 (22.0) | 26.0 | 66.1 (3.6) | 2.1 | 1086.5 (22.4) | 28.0 | |
| 6, bivariate IID | 1095.7 (24.9) | 101.2 | 60.0 (3.8) | −4.0 | 1155.8 (25.2) | 97.3 | |
| 5 | 1, MCAR(B, Σ) | 1152.7 (30.0) | — | 84.9 (6.5) | — | 1237.6 (30.6) | — |
| 2, MCAR(B, I) | 1227.7 (30.5) | 75.0 | 83.9 (6.8) | −1.0 | 1311.6 (31.3) | 74.0 | |
| 3, MCAR(α, Σ) | 1191.4 (38.6) | 38.7 | 87.9 (6.5) | 3.0 | 1279.3 (39.0) | 41.7 | |
| 4, GMCAR | 1205.4 (37.6) | 52.7 | 73.9 (7.8) | −11.0 | 1279.4 (38.7) | 41.8 | |
| 5, GMCAR (reverse order) | 1220.2 (38.4) | 67.5 | 75.9 (7.5) | −9.0 | 1296.1 (39.0) | 58.6 | |
| 6, bivariate IID | 1395.3 (44.8) | 242.6 | 59.5 (7.8) | −25.4 | 1454.8 (45.1) | 217.2 | |
Except for study 5, the dash indicates that the model is the true model for this study.
Overall, the results are similar to those which were seen for the mean-squared error, with generally strong support for the MCAR(B, Σ) model. The large improvements in fit that are brought about by this model are typically not offset by the relatively modest increases in effective model size, leading to significant improvements in overall DIC-score (the ‘sd’ measures in Table 2 are simply sample standard deviations of the MCMC draws themselves, not Monte Carlo standard errors as in Table 1). Not only does it excel in study 1, but it actually outperforms the true models in terms of actual DIC in studies 2 and 3. In fact, in terms of D̄ it outperforms all the true models in studies 2–4 but marginally loses out to the true GMCAR model in terms of overall DIC owing to a substantial increase in the effective number of parameters. The full MCAR(B, Σ) model also dominates the other models in study 5 even though it, like the rest of the models, is not designed to capture the true geostatistical nature of the simulated data. Although the differences in the average DIC-scores in Table 2 are not very large relative to the standard deviations, the ranking of the models is consistently preserved. For instance, the percentage of replicate data sets for which the MCAR(B, Σ) model had the lowest DIC-score was roughly 99% in study 1, 57% in study 2, 86% in study 3, 47% in study 4 (the true GMCAR model had lower DIC in the remaining 53% of the replications) and a full 100% in study 5.
The behaviour of the remainder of the models is also quite similar with results seen for the AMSE and probably with similar reasonings to those given there. For instance, we again note the typically poorer performance of the MCAR(B, I) model relative to the other spatial models; the GMCAR model with the better ordering is the closest competitor to the MCAR(B, Σ) model whereas the MCAR(α, Σ) model performs somewhere between the two GMCAR models in studies 1 and 2, performs better than either of them in study 3 (the true model) and study 5 (marginally over the better GMCAR model) and worse in study 4.
6. Data example: Minnesota cancer data
We now turn from the Gaussian likelihood with p = 2 to a non-Gaussian likelihood with p = 3 in a disease mapping context, and we illustrate our methods with a data set that was extracted from the public use SEER mortality database (http.//seer.cancer.gov). SEER county level mortality databases (National Cancer Institute, 2003) provide the numbers of deaths and corresponding numbers of person-years at risk in quinquennial age brackets for each county in a particular state and each cancer site. Our data consist of the numbers of deaths due to cancers of the lung, larynx and oesophagus in the years from 1990 to 2000 at the county level in Minnesota. The larynx and oesophagus are sites of the upper aerodigestive tract, so they are closely related anatomically. Epidemiological evidence shows a strong and consistent relationship between exposure to alcohol and tobacco and the risk of cancer at these two sites (Baron et al., 1993). Meanwhile, lung cancer is the leading cause of cancer death for both men and women. An estimated 160440 Americans will have died in 2004 from lung cancer, accounting for 28% of all cancer deaths. It has long been established that tobacco, and particularly cigarette smoking, is the major cause of lung cancer. More than 87% of lung cancers are smoking related (http.//www.lungcancer.org).
These cancers are sufficiently rare relative to the population in each county that the Poisson spatial regression model (4) in Section 2.2 is appropriate. To calculate the expected counts Eij, we must take each county's age distribution (over the 18 age groups) into account. To do so, we calculate the expected age-adjusted number of deaths due to cancer j in county i as , i = 1,…,87, j = 1, 2, 3, k = 1,…, 18, where
is the age-specific death-rate due to cancer j for age group k over all Minnesota counties, is the number of deaths in age group k of county i due to cancer j and is the total population at risk in county i and age group k, which we assume to be the same for each type of cancer.
6.1. Model comparison
To compare models, we might set aside the values for some of the cancers in some of the counties, impute them by using the posterior predictive distribution based on the remaining data and then compare the resulting total mean-squared prediction error across models. However, several practical difficulties arise, including the number and precise choice of validation counties, and which cancer counts (one or all three) to delete in each. The dependence structure that is imposed by the county lattice and the problem of spatial edge effects also suggest that this would not offer a general model checking method in our context. As such, we again compare models by using the DIC-criterion.
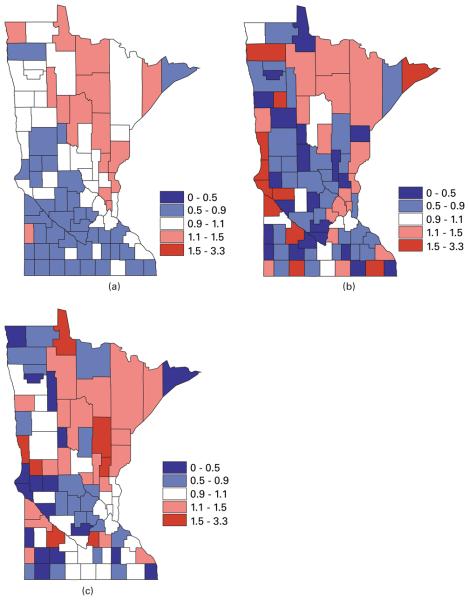
The county level maps of the raw age-adjusted standardized mortality ratios (i.e. SMRijYij/Eij) that are shown in Fig. 1 exhibit evidence of correlation both across space and among the cancers, motivating use of our proposed multivariate lattice model. Using the likelihood in expression (4), we model the random effects ϕij by using our proposed MCAR(B, Σ) model (12). In what follows we compare it with other MCAR models, including the MCAR(α, Σ) and MCAR(1, Σ) models from Section 3.1, a ‘three separate CAR structure’ model ignoring correlation between cancers and a trivariate IID model ignoring correlations of any kind. We also compare one of the MCAR(α1, α2, α3, Σ) models that is given in expression (10) of Section 3.2 by choosing the matrix A as the upper triangular Cholesky decomposition of Σ. However, we do not consider the order-specific GMCAR model (Jin et al., 2005) since, with no natural causal order for these three cancers, it is difficult to choose between the six possible conditioning orders.
Fig. 1.
Maps of raw standardized mortality ratios SMR for (a) lung, (b) larynx and (c) oesophageal cancer in the years from 1990 to 2000 in Minnesota
As in the previous section, the deviance D(θ) is the same for the models that we wish to compare since they differ only in their random-effect distributions p(ϕ|B, Σ). Specifically, we now have D(θ) ≡ D(β, ϕ) = −2 log{L(Y1, Y2, Y3|β, ϕ)}, where L is the likelihood for the Poisson model (4). We choose the same prior distributions for each parameter as in Section 5 and again set the deviance standardizing function h(y) = 0. Since p = 3 in this example, we choose the inverse Wishart distribution with ν = 3 and R = diag(0.1, 0.1, 0.1) for Σ. For a fair comparison of DICs, we retain the same ‘focus’ parameters and likelihood across the models. We used 20000 preconvergence burn-in iterations followed by a further 20000 production iterations for posterior summarization.
In what follows, models 1–6 are multivariate lattice models with different assumptions about the smoothing parameters. Model 1 is the full model MCAR(B, Σ) (with a 3 × 3 matrix B whose elements are the six smoothing parameters) whereas model 2 is the MCAR(B, I) model. Model 2 is the MCAR(α1, α2, α3, Σ) model (10) with a different smoothing parameter for each cancer. Model 3 assumes a common smoothing parameter α and model 4 fits the three separate univariate CAR model, whereas model 6 is the trivariate IID model. Fit measures D̄, effective numbers of parameters pD and DIC-scores for each model are seen in Table 3. We find that the MCAR(B, Σ) model has the smallest D̄ and DIC-values for this data set. The MCAR(B, I) model again disappoints, excelling over the non-spatial model and the separate CAR models only (very marginally over the latter). The MCAR(α, Σ) and MCAR(α1, α2, α3, Σ) models perform slightly worse than the MCAR(B, Σ) model, suggesting the need for different spatial autocorrelation and cross-spatial-correlation parameters for this data set. Note that the effective numbers of parameters pD in model 3 is a little larger than in model 1, even though the latter has three extra parameters. Finally, the MCAR models do better than the separate CAR model or the IID trivariate model, suggesting that it is worth taking account of the correlations both across counties and among cancers. Model 6 exhibits a large pD-score, suggesting that it does not seem to allow sufficient smoothing of the random effects. This is what we might have expected, since the spatial correlations are missed by this model.
Table 3.
Model comparison using DIC-statistics, Minnesota cancer data analysis
| Model | D̄ | pD | DIC |
|---|---|---|---|
| 1, MCAR(B, Σ) | 138.8 | 82.5 | 221.3 |
| 2, MCAR(B, I) | 147.6 | 81.4 | 229.0 |
| 3, MCAR(α1, α2, α3, Σ) | 139.6 | 86.4 | 226.0 |
| 4, MCAR(α,Σ) | 143.4 | 81.9 | 225.3 |
| 5, separate CAR | 147.6 | 82.8 | 230.4 |
| 6, trivariate IID | 146.8 | 91.3 | 238.1 |
| 7, MCAR(B, Σ) + trivariate IID | 129.6 | 137.6 | 267.2 |
| 8, MCAR(B, I) + trivariate IID | 139.5 | 155.2 | 294.7 |
| 9, MCAR(α1, α2, α3, Σ) + trivariate IID | 137.4 | 155.0 | 292.4 |
| 10, MCAR(α, Σ) + trivariate IID | 138.2 | 151.0 | 289.2 |
| 11, separate CAR + trivariate IID | 139.2 | 162.8 | 302.0 |
Models 7-11 are the convolution prior models corresponding to models 1–5 that are formed by adding IID random effects (following N(0, τ2)) to the ϕijs. Here the distinctions between the models are somewhat more pronounced owing to the added variability in the models that is caused by the IID effects. The relative performances of the models remain the same with the MCAR(B, Σ) plus IID model emerging as best. Interestingly, none of the convolution models perform better than their purely spatial counterparts, as the improvements in D̄ in the former are insignificant compared with the increase in the effective dimensions. This is indicative of the dominance of the spatial effects over the IID random effects.
6.2. Results from the model selected
In this subsection, we begin by summarizing our results from the MCAR(B, Σ) model, or model 1 in Table 3. Table 4 provides posterior means and associated standard deviations for the parameters β, Σ and bij in this model, where bij is the element of the symmetric matrix B. Instead of reporting Σ12, Σ13 and Σ23, we provide the mean and associated 95% credible intervals for the correlation parameters ρ12, ρ13 and ρ23, which are calculated as ρij = Σij/√(ΣiiΣjj). Table 4 reveals several positive correlations between cancers; in particular, between lung and oesophagus cancer (ρ13). This might explain why the DIC-scores for models 1–4 are smaller than that under the separate CAR model.
Table 4.
Posterior summaries of parameters in the MCAR(B, Σ) model for the Minnesota cancer data
| Parameters | Results for the following cancers: |
||
|---|---|---|---|
| Lung |
Larynx |
Oesophagus |
|
| Median (2.5%, 97.5%) | Median (2.5%, 97.5%) | Median (2.5%, 97.5%) | |
| β1, β2, β3 | −0.093 (−0.179,−0.006) | −0.128 (−0.316, 0.027) | −0.080 (−0.194, 0.025) |
| Σ11, Σ22, Σ33 | 0.048 (0.030, 0.073) | 0.173 (0.054, 0.395) | 0.107 (0.044, 0.212) |
| ρ12, ρ13 | 0.277 (−0.112, 0.643) | 0.378 (−0.022, 0.716) | |
| ρ 23 | 0.337 (−0.311, 0.776) | ||
| b11, b22, b33 | 0.442 (−0.302, 0.921) | 0.036 (−0.830, 0.857) | 0.312 (−0.526, 0.901) |
| b12, b13 | 0.323 (−0.156, 0.842) | 0.389 (−0.028, 0.837) | |
| b 23 | 0.006 (−0.519, 0.513) | ||
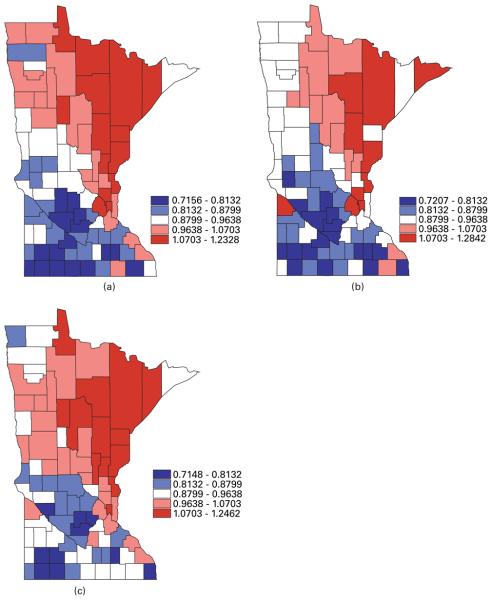
Turning to geographical summaries, Fig. 2 maps the posterior means of the fitted standard mortality ratios SMR of lung, larynx and oesophageal cancer from our MCAR(B, Σ) model. The correlation between the cancers is apparent, with higher fitted ratios extending from the Twin Cities metro area to the north and north-east (an area where cigarette smoking may be more common). In Fig. 1, the range of the raw SMRs is seen to be from 0 to 3.3, whereas, in Fig. 2, the range of the fitted SMRs is from 0.7 to 1.3, owing to spatial shrinkage in the random effects.
Fig. 2.
Maps of posterior means of the fitted standard mortality ratios SMR for (a) lung, (b) larynx and (c) oesophageal cancer in the years from 1990 to 2000 in Minnesota from the MCAR(B, Σ) model
7. Summarizing remarks and future research
In this paper we have applied the notion of the linear model of co-regionalization to the analysis of multivariate areal data and proposed the MCAR(B, Σ) model for mapping multiple diseases. As with the existing MCAR(α, Λ) and multivariate IID models in the literature, the MCAR(B, Σ) model is order free, i.e. independent of the ordering of the variables in the hierarchical model. But the MCAR(B, Σ) model is much more flexible for modelling spatial correlations in multivariate areal data. Our simulations and data example demonstrate the improved performance of the MCAR(B, Σ) model over the existing alternatives as measured by the AMSE or DIC, as well as its efficient implementation by using MCMC algorithms. Our approach is also readily extended to higher dimensional (p> 2) settings; the computational burden does not increase much with dimension p since it does not involve large matrix calculations.
Though our emphasis has been on mapping multiple diseases, our proposed MCAR model may also be useful for mapping a single disease. For example, consider a spatially varying-coefficients model (Assunção, 2003) extending model (1) in Section 2.1 to
where the xi are explanatory, region level spatial covariates having parameter coefficients β and zi1 and zi2 are a subset of xi having spatially varying coefficients ςi1 and ςi2 respectively. We might suspect that ςi1, ςi2 or ϕi corresponding to counties in geographic proximity to each other might also be similar in magnitude. This in turn means that it might be worth fitting a multivariate areal model. We could take our proposed MCAR(B, Σ) model for , where ς1 = (ς11,…, ςn1)′, ς2 = (ς12,…, ςn2)′ ans ϕ = (ϕ1, …, ϕn)′.
In our current work, we have only considered mapping the geographic pattern of multiple diseases at a single point in time. However, disease data are often reported over a series of time periods. For example, the SEER database currently provides cancer mortality information for the years from 1969 to 2001. In such cases, we may be interested in temporal effects as well as spatial effects. This motivates an extension of the proposed MCAR model to multivariate spatiotemporal data. Knorr-Held (2000) proposed a general framework for spatiotemporal modelling of a single disease by using ICAR models, which could perhaps be extended to our multivariate setting. Alternatively, we may work with a multivariate Gaussian AR(1) time series of spatial processes in the setting of dynamic models (Gelfand et al., 2005). This would extend model (4) in Section 2.2 to
where the xijt are explanatory, region level spatial covariates for disease j at time period t having parameter coefficients βjt. Let ϕj, t+1 = Hjϕj, t + εjt, where Hj is an n × n matrix, ϕjt = (ϕ1jt,…, ϕnjt)′ and εjt = (ε1jt,…, εnjt)′, j = 1,…, p. Then we can assume that Hj = H = θ0I or Hj = H = θ0I + θ1W, where W is an adjacency matrix and I is an n × n identity matrix. The random effects follow a multivariate areal model, such as our proposed MCAR(Bt, Σt) model, or perhaps the MCAR(B, Σ) model if the parameters of the MCAR model do not change over time.
Finally, here we have studied methods for mapping rare disease rates, with the rarity required to ensure the validity of the Poisson approximation to a binomial sampling distribution. However, it is easy to adapt the statistical methods that we have presented here to analyse non-rare diseases by replacing the Poisson likelihood with a binomial distribution for the data (MacNab, 2003). Also, although in this paper we considered only mapping disease rates, we also can apply our MCAR(B, Σ) model methodologies when modelling random effects in a hazard function (Carlin and Banerjee, 2003).
Acknowledgements
The work of all three authors was supported in part by National Institutes of Health grants 2-R01-ES07750 and 1-R01-CA95955, and that of the second and third authors was supported in part by National Institutes of Health grant 1-R01-CA112444.
References
- Assunção RM. Space-varying coefficient models for small area data. Environmetrics. 2003;14:453–473. [Google Scholar]
- Banerjee S, Carlin BP, Gelfand AE. Hierarchical Modeling and Analysis for Spatial Data. Chapman and Hall–CRC; Boca Raton: 2004. [Google Scholar]
- Baron AE, Franceschi S, Barra S, Talamini R, La Vecchia C. Comparison of the joint effect of alcohol and smoking on the risk of cancer across sites in the upper aerodigestive tract. Cancer Epidem. Biomark. Prevn. 1993;2:519–523. [PubMed] [Google Scholar]
- Besag J. Spatial interaction and the statistical analysis of lattice systems (with discussion) J. R. Statist. Soc. B. 1974;36:192–236. [Google Scholar]
- Besag J, York JC, Mollié A. Bayesian image restoration, with two applications in spatial statistics (with discussion) Ann. Inst. Statist. Math. 1991;43:1–59. [Google Scholar]
- Carlin BP, Banerjee S. Hierarchical multivariate CAR models for spatio-temporally correlated survival data (with discussion) In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics 7. Oxford University Press; Oxford: 2003. pp. 45–63. [Google Scholar]
- Carlin BP, Louis TA. Bayes and Empirical Bayes Methods for Data Analysis. 2nd edn. Chapman and Hall–CRC; Boca Raton: 2000. [Google Scholar]
- Celeux G, Forbes F, Robert CP, Titterington DM. Deviance information criteria for missing data models (with discussion) Bayes. Anal. 2006;1:651–706. [Google Scholar]
- Chilés JP, Delfiner P. Geostatistics: Modelling Spatial Uncertainty. Wiley; New York: 1999. [Google Scholar]
- Daniels MJ, Kass RE. Nonconjugate Bayesian estimation of covariance matrices and its use in hierarchical models. J. Am. Statist. Ass. 1999;94:1254–1263. [Google Scholar]
- Gelfand AE, Banerjee S, Gamerman D. Spatial process modelling for univariate and multivariate dynamic spatial data. Environmetrics. 2005;16:1–15. [Google Scholar]
- Gelfand AE, Schmidt AM, Banerjee S, Sirmans CF. Nonstationary multivariate process modelling through spatially varying coregionalization (with discussion) Test. 2004;13:263–312. [Google Scholar]
- Gelfand AE, Vounatsou P. Proper multivariate conditional autoregressive models for spatial data analysis. Biostatistics. 2003;4:11–25. doi: 10.1093/biostatistics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Harville DD. Matrix Algebra from a Statistician's Perspective. Springer; New York: 1997. [Google Scholar]
- Held L, Natário I, Fenton SE, Rue H, Becker N. Towards joint disease mapping. Statist. Meth. Med. Res. 2005;14:61–82. doi: 10.1191/0962280205sm389oa. [DOI] [PubMed] [Google Scholar]
- Jin X, Carlin BP, Banerjee S. Generalized hierarchical multivariate CAR models for areal data. Biometrics. 2005;61:950–961. doi: 10.1111/j.1541-0420.2005.00359.x. [DOI] [PubMed] [Google Scholar]
- Kim H, Sun D, Tsutakawa RK. A bivariate Bayes method for improving the estimates of mortality rates with a twofold conditional autoregressive model. J. Am. Statist. Ass. 2001;96:1506–1521. [Google Scholar]
- Knorr-Held L. Bayesian modelling of inseparable space-time variation in disease risk. Statist. Med. 2000;19:2555–2567. doi: 10.1002/1097-0258(20000915/30)19:17/18<2555::aid-sim587>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Knorr-Held L, Best NG. A shared component model for detecting joint and selective clustering of two diseases. J. R. Statist. Soc. A. 2001;164:73–85. [Google Scholar]
- MacNab YC. Hierarchical Bayesian spatial modelling of small-area rates of non-rare disease. Statist. Med. 2003;22:1761–1773. doi: 10.1002/sim.1463. [DOI] [PubMed] [Google Scholar]
- Mardia KV. Multi-dimensional multivariate Gaussian Markov random fields with application to image processing. J. Multiv. Anal. 1988;24:265–284. [Google Scholar]
- National Cancer Institute . Mortality—All COD, Public-use with County, Total U.S. for Expanded Races (1990-2000) <18 Age Groups>. National Cancer Institute; Bethesda: 2003. [Google Scholar]
- Royle JA, Berliner LM. A hierarchical approach to multivariate spatial modeling and prediction. J. Agric. Biol. Environ. Statist. 1999;4:29–56. [Google Scholar]
- Sain SR, Cressie N. Multivariate lattice models for spatial environmental data. Proc. Statist. Environ. Sect. Am. Statist. Ass. 2002:2820–2825. [Google Scholar]
- Schmidt AM, Gelfand AE. A Bayesian coregionalization approach for multivariate pollutant data. J. Geophys. Res. Atmos. 2003;108 article 8783. [Google Scholar]
- Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion) J. R. Statist. Soc. 2002;64:583–639. [Google Scholar]
- Wackernagel H. Multivariate Geostatistics: an Introduction with Applications. 3rd edn. Springer; New York: 2003. [Google Scholar]
- Wall MM. A close look at the spatial structure implied by the CAR and SAR models. J. Statist. Planng Inf. 2004;121:311–324. [Google Scholar]