Abstract
The emergence of geographical information systems and related softwares nowadays enables medical databases to incorporate the geographical information on patients, allowing studies in spatial associations. Public health administrators and researchers are often interested in detecting variation in survival patterns by region or county in order to understand the possible factors that contribute towards such spatial discrepancies. These issues have led statisticians to develop survival models that account for spatial clustering and variation. Additionally, with rapid developments in medical and health sciences, researchers increasingly encounter data sets where a substantial portion of patients are cured. Models accounting for cure in the population assist in the prognosis of potentially terminal diseases. This article proposes a Bayesian modelling framework that models spatial associations for areally referenced survival data using a general class of cure models proposed by Cooner et al. The special models we outline are alternatives to the traditional proportional hazards models and can be fitted using standard Bayesian software such as WinBUGS.
1 Introduction
With the advent of geographical information systems, medical and health databases now often include geographical information on patients (e.g., county of residence) that allow the researchers to investigate spatial associations in health phenomena. Spatial disease mapping, an area that has attracted much attention over the last several years,1–3 concerns the development of statistical and cartographic methods that map disease rates accounting for different types of extraneous variation. Lawson and Williams,3 in particular, discuss several aspects of modelling and software implementations.
In recent times, there has been growing interest in capturing spatial trends in the survival patterns of patients suffering from potentially terminal diseases. Health care administrators and public health researchers are often interested in detecting variation in survival patterns by counties for determining the possible factors that contribute towards such variability. These issues have led statisticians to develop survival models that account for spatial clustering and variation. Banerjee et al.4 investigated the spatially correlated frailties in traditional parametric survival models adopting a hierarchical Bayesian approach. Instead, Li and Ryan5 address this problem from a classical perspective.
Generally, the spatial survival modelling mentioned above is treated in a proportional hazards framework. With rapid developments in medical and health sciences, scientists and health professionals encounter more data sets where the patients are expected to be cured. Formulation and estimation of models that account for cure are important for understanding prognosis in potentially terminal diseases. Traditional parametric survival models such as Weibull or Gamma6 do not account for cure, assuming instead that individuals who do not experience the event are censored. Although subtle, in these contexts one distinguishes between the concepts of censoring and cure: censoring refers to a subject who does not fail within the time window of the experiment, while cure refers to one who will never fail. Indeed the latter is an abstraction as we never ‘observe’ a cure (due to a finite monitoring time). Still estimating the probability of such an outcome, especially in various cancer-relapse settings, can help expose unknown health issues concerning that population.
Statistical models address this conceptually challenging problem by parametrizing the probability of a cure, called the cure fraction. One of the earliest such models was a class of mixture models by Berkson and Gage7 that generated subsequent investigations by Farewell,8,9 Goldman,10 and Ewell and Ibrahim11 among others. Yakovlev et al.12 and Chen et al.13 offered an alternative approach to formulating cure models14,15 that assume a latent biological process in generating the observed failure (say, cancer relapse). Recently, Cooner et al.16 proposed a very general class of cure models assuming (possibly several) latent factors or latent risks (e.g., metastasis-competent tumours that lead to cancer relapse) corresponding to each patient. For an individual to be at risk of failure, one must be exposed to at least one of these latent factors. If not, the individual is not at risk and is considered cured. Failure is observed when some (perhaps one or all) of these latent factors become activated.
In the modelling of spatial association using random effects and, more generally, regressors in cure models, the modeller faces some conceptually interesting alternatives. Certain data sets may be better modelled by assuming that the cure fractions are spatially associated. Alternatively, one may model regressors and spatial effects in the distribution of the latent factors. Richer models incorporating such random effects in both the cure fraction and the latent distributions can also be conceptualized. However, identifiability and estimability of such models raise concerns that have hitherto gone unaddressed with most of the spatial-survival analysis literature focussing upon proper survival models. Recently, Banerjee and Carlin17 demonstrated data analysis from a spatially referenced interval-censored smoking-cessation study in southern Minnesota using a binary cure model interpreted as one latent factor. However, their models preclude studying spatial effects in the cure fraction, as such effects are not estimable.
This article explores spatial cure rate models by extending the modelling framework of Cooner et al.16 We propose a methodological framework that enables versatile and flexible modelling of spatial associations for survival data in which a prominent proportion of subjects might be cured. We adopt a Bayesian hierarchical framework that allows very rich modelling structures estimated using Markov Chain Monte Carlo (MCMC) methods.18 It is important to point out that we limit our discussion to areal settings where the geographical referencing for each subject is by the county they live in, rather than by the coordinates of each subject’s residence (point-referencing). Areal summaries are frequently found in public health data due to data privacy issues.
With information at the county level, spatial modelling will proceed from the neighbourhood information of the counties. More precisely, we consider a vector of univariate latent random variables ϕ = (ϕ1,…,ϕn) in n areal locations (such as counties, states) that follow a spatial distribution. Unlike the independent random effects that specify ϕ ~ N(0, τ2I), a spatial distribution will model ϕ ~ N(0, Σ−1), where Σ−1 is a choice of the spatial precision matrix. Typically, these effects are modelled after adjusting for covariates or risk information and hence are zero-centered. Specifying Σ−1 based on the underlying neighbourhood configurations follow from the theory of Markov Random Fields (see e.g., the works of Rue and Held19 for details). We work with univariate and multivariate conditionally autoregressive (CAR) models that accrue computational benefits, while incorporating spatial effects. In particular, we investigate a Generalized multivariate CAR (GMCAR) distribution, recently investigated by Jin et al.20 that encompasses very rich multivariate spatial associations.
The remainder of this paper evolves as follows. In Sections 2 and 3 we outline the cure rate and spatial modelling framework that we work with. Section 4 discusses the Bayesian implementation algorithms. Section 5 provides the analysis of a breast cancer survival data set obtained from the National Cancer Institute’s Surveillance, Epidemiology and End Results (SEER) database (http://www.seer.cancer.gov/). Finally, Section 6 summarizes and indicates future areas for research.
2 Cure rate modelling under latent activation schemes
Cure models based upon latent activation schemes16 involves failure times at two different levels: an observed failure time T corresponding to the observed time when an individual fails, as well as the latent event times, Yk, k = 1,…,N, the activation times for the N latent factors that generate the observed failure at time T. Note that if N = 0 then the individual is not exposed to any of the latent factors and is considered immune from failure; thus the individual is cured and T = ∞. For a given are assumed to be independently and identically distributed (i.i.d.) with a survival distribution P(Y > t) = S(t) that does not depend upon N. We call this the latent survival function and denote the corresponding distribution function by F(t) = 1 − S(t).
To derive the distribution of T, we assume that r out of N latent factors need to be activated for the subject to fail, so T = Y(r), r = 1,…, N where Y(1) < … < Y(N) are the ordered Yk’s. Cooner et al.16 investigate the different modelling choices for r. While in general r itself can be modelled as random, fixing r at some positive integer between 1 and N (given N) results in better identified models. Thus, r = 1 implies that activation of any one of the latent factors leads to observed failure. We call this the first-activation scheme. In contrast, setting r = N implies T = max1≤k≤N Yk and delivers a different scheme where an individual is able to ‘resist’ up to N − 1 activations and fails with the last activation. We call this the last-activation scheme. In either scheme, N represents the same object (number of latent factors), but the way it brings about the observed phenomenon (e.g., relapse of cancer) is modelled differently. More generally, an exposed subject (N > 0) at any time point will not experience detectable failure if the number of latent event occurrences at that time is less than r. Henceforth, we will consider r as fixed; indeed, we will focus only on the first- and last-activation schemes.
The conditional distribution of T given N can be written down in terms of the incomplete beta function, or a beta cdf denoted by IB(S(t); N − r + 1, r), (using a standard result on order statistics using the binomial theorem, e.g., Rao21 (p. 215) as
| (1) |
where 1(·) is the indicator function and
The unconditional survival function of T, S*(t), is related to the latent distribution as
| (2) |
where the expectation is taken over the distribution of N. Here S*(t) always exists, being bounded between 0 and 1 for any valid distribution of N restricted to N ≥ r ≥ 1 as IB(S(t); N − r + 1, r) is otherwise 0. Also, as limt→∞ S(t) = 0, we have limt→∞ S*(t) = P(N = 0), showing that S*(t) is improper whenever P(N = 0) > 0. Indeed, P(N = 0) is the probability of a person being cured or immune, hence called the cure fraction, and depends only upon the distribution of N, irrespective of what r is. Despite S*(t) being improper, the hazard, say h*(t)dt ≈ P(T ∈ [t, t + dt)|T > t), is still valid so that h*(t) is evaluated as −(d/dt) log S*(t), or f*(t)/S*(t), where f*(t) is the corresponding improper density. In fact, if f (t) is the proper latent density corresponding to F(t), then
| (3) |
The variable N can never be observed and must be modelled using a probabilistic assumption. From a more theoretical perspective, one can show16 that if N ~ Po(θ) and if r or N − r is fixed at some positive integer (e.g., r = 1 or r = N), then θ is identifiable under a scale-invariant prior g(θ) ∝ 1/θ permitting regression in log(θ). These generate a well-identified class of cure models, obtained by setting r appropriately. Note that with N ~ Po(θ), we have the cure fraction exp(−θ). This theoretical identifiability permits regression (along with spatial random effects) in log(θ), amounting to a Poisson regression.
The first-activation scheme (r = 1) assumes that activation of a single latent factor will lead to observed failure. According to a biological model for patients diagnosed with cancer, N is the number of metastasis-competent clonogenic cells that are in an irreversible process towards metastasis, and Yk is the time for the kth clonogenic cell to produce ‘detectable’ tumour. Detectable metastasis occurs as soon as any one of the clonogens metastasize, so that T = min1≤k≤N Yk. This arises as a special case of equation (1) with r = 1 so that P(T ≥ t|N) = 1(N = 0) + IB(S(t); N, 1)1(N ≥ 1), which simplifies to P(T ≥ t|N) = 1(N = 0) + [S(t)]N1(N ≥ 1). The CIS models of Chen et al.13 further assume that N ~ Po(θ), from which we obtain S*(t) = EN[P(T ≥ t|N)] = P(N = 0) + EN[S(t)N1(N ≥ 1)] = exp(−θS(t)).
The physical framework of the first-activation scheme is not unique in assuming that one latent factor’s activation generates observed failure. Alternatively, N can be the number of latent factors that must all be activated for failure. For instance, biological models for certain types of cancer posit that a patient’s immune response gets activated after the initiation of N cell mutations. This immune response may be able to resist up to N − 1 promotions of mutated cells before disease manifestation or death. Here, we model Yk as the time to promotion of the kth latent factor and failure occurs after the Nth factor is activated, hence the observed failure time is T = max Yk, k = 1,…,N. Again, we have a special case of equation (1) with r = N and P(T ≥ t|N) = 1(N = 0) + IB(S(t); 1, N)1(N ≥ 1). The conditional distribution of T given N is now easily expressed in terms of the latent distribution function F(t) = 1 − S(t) as P(T ≤ t|N) = [F(t)]N1(N ≥ 1). Note that although S*(t) is different from that in the first-activation scheme, they tend to the same limit, P(N = 0), resulting in the same cure fraction. In particular, when N ~ Po(θ), we get S*(t) = 1 + exp(−θ)(1 − exp(θF(t))) which is different from that under first activation, but approaches the same cure fraction, exp(−θ).
Turning to alternative modelling of N, we note that several authors including Tucker et al.22 and Tucker and Taylor,23 have questioned the Poisson assumption for any particular cancer cure scenario. From a modelling standpoint, the number of possible latent events N can have any finite-mean integer-valued distribution (e.g., binary, geometric and so on). In traditional cure rate models,7,9 N is binary with only one latent dominant event (e.g., a dominant metastasis competent tissue-mass in breast cancer), so N ~ Ber(θ) (with θ being the probability of an activation). Here S*(t) = 1 − θ(1 − S(t)), approaches the cure fraction 1 − θ.
Another specification we consider here is N ~ Geo(θ), a geometric distribution with mean θ/(1 − θ)(P(N = n) = θn(1 − θ)). A biological motivation, different from the clonogen motivation in Chen et al. (1999),13 can be offered as follows (also see the work of Moolgavkar et al.24). Assuming a short time-interval of the mutation (initiation) period (due to exposure to genetic damage), the patient’s body produces a sequence of N mutated cells/tissues before activating the immune system. Every mutation (initiation) may give rise to an effective immune response from the body with probability 1 − θ, capable of destroying the last mutated tissue/cell and halting the mutation process. With now being the promotion times of the mutated cells, a first-activation scheme with T = mink Yk models failure with any one activation. Now we have S*(t) = (1 − θ)/(1 − θS(t)) with a cure fraction of 1 − θ. We will explore these settings in the subsequent sections, pointing out their identifiability properties in spatial regression contexts.
3 Latent spatial cure rate model
The cure rate models in Cooner et al.16 accommodate two primary regression structures, one using the mean of the latent factors, viz. the Yk’s, or using a Poisson regression in the cure fraction itself. More precisely, suppose we have I regions, with ni patients observed in the ith region. Let (tij, δij, xij) be the observations collected from the jth patient in region i (i.e., the (i, j)th patient), where tij is the time-to-event (e.g., death, relapse and so on), δij is the censoring indicator (0 if censored, 1 if dead) and xij is a set of patient-specific regressors. If we assume that Yk’s follow a two-parameter Weibull distribution Weib(ρ; η), that is S(t) = exp(−tρ eη), for each patient, then we can model patient-specific regression either in the Weibull link as , or in the respective cure fraction as log(θij) = xij. In the former, we can explore the contribution of progress rate to failure time; in the latter, we can investigate the contribution of the cure fraction.
For studying spatial variation across regions and, more specifically, smoothing across regions using adjacency information, we introduce spatial frailties4 that are spatially correlated region-specific random effects ϕi, i = 1,…, I. Interesting modelling choices arise. We could, as for the regressors, add these frailties either in η or in log(θ). If geographical variation is expected to reflect itself in lurking covariates that affect the latent factors, we opt for the former. In contrast, if we expect this variation in the cure fraction itself, we opt for the latter. However, in practice this choice will often not be clear. Therefore, more generally, one may incorporate two different sets of frailties, in the ηij’s and in the log(θij)’s. On the basis of these, we classify models as univariate or bivariate; we discuss them in detail below.
3.1 Univariate spatial cure rate model
In principle, N can be modelled using any integer-valued probability distribution. Irrespective of the distribution of N, the regression coefficients and frailties are identifiable from the data when they model the ηij, giving rise to the following models:
In contrast, covariates modelled through regression on the cure fraction will be identified only if N ~ Po(θ), thereby precluding models 1(b)–(c). The spatial cure rate models for studying the spatial associations in cure fractions are given by
Note that the interpretation of the parameters will be different for the two sets of models. In models 1(a)–(c), higher values of ηij indicate longer latent event times and hence prolonged survival, so covariates influence the latent factors that cause failure. In contrast, in model 2, covariates influence the cure fraction directly. Almost certainly, most covariates that affect the latent event times will also affect the cure. For these covariates, we have a consistency in behaviour under the two groups of models. To see this, note that the Weibull mean is a decreasing function of ηij (given by exp(−ηij/ρ)Γ(1 + 1/ρ)) and the cure fraction is a decreasing function of θij. This is sensible because covariates that adversely affect the survival time will likely affect the cure fraction adversely as well (and vice versa). Therefore, it is expected that their coefficients have the same sign under both sets of models.
Turning to the spatial modelling of the ϕi’s, we employ variants of the (univariate) conditionally autoregressive model introduced by Besag.25 For further theoretical discussions, see the works of Cressie,26 Banerjee,27 Rue and Held,19 Heikkinen and Högmander,28 Högmander and Møller29 and Hoeting et al.30 for generalizations and applications. Collecting the frailties into an I × 1 vector ϕ = (ϕ1,…, ϕI)T, a popular version of the CAR model is characterized by
| (4) |
where B is an I × I matrix with diagonal elements 0, and Dτ is a diagonal matrix with entries , i = 1,…,I. A usual assumption is Dτ = τ2D, where D is a diagonal matrix with common entries. On the basis of the formulation in equation (4), different choices of B, α ∈ (0, 1), and Dτ lead to various CAR modelling structures. Most often, we set D to be a diagonal matrix with entries mi, i = 1,…,n, where mi is the number of the spatial neighbours for region i, whence B = D−1 W is a row-normalized adjacency matrix, where W is the adjacency matrix with diagonal entries 0 and off-diagonals equalling 1, if and only if two regions are neighbours (and 0 otherwise). With α = 1, we recover the pairwise difference distribution of Besag et al.,31 also called intrinsic autoregressive model. While this model allows maximum smoothing, the spatial dispersion matrix is singular. However, it is non-singular in a subspace of dimension one less (for connected regions), and is not much cause of concern for Bayesians who can just add a linear constraint to ϕ and update it using MCMC methods. See the work of Lawson et al.32 for an excellent overview of implementing CAR models in the WinBUGS software in the disease mapping contexts.
For assessing practical benefits in modelling these effects, we also fit i.i.d models where ϕ ~ N(0, τ−2I). Two basic schemes, first and last activation, of the cure rate model class should apply in both structures. We will analyse the resulting models and investigate the performances using Deviance Information Criterion (DIC; Spiegelhalter et al.33). See section 4 for further details.
3.2 Bivariate spatial cure rate models
For investigating spatial associations in the latent link and the cure fraction jointly, the basic structure is similar to the univariate setting:
Again, both schemes proposed by Cooner et al.16 should be identifiable. When multiple parameter sets (such as frailties in cure fractions and the Weibull link) need to be modelled jointly, as opposed to independently, we resort to multivariate CAR models originally proposed by Mardia34; see also the works of Carlin and Banerjee35 and Gelfand and Vounatsou.36 Let be a vector of p variables associated with the ith region. Collecting these effects into , the joint distribution can be written down as
| (5) |
where BR is an np × np matrix with block elements (BR)ij = RiBij and 0 as diagonals, Ri and Bij are p × p matrices, and Γ is an np × np block diagonal matrix with block elements Γi, i = 1,…, n.
Mardia34 and Carlin and Banerjee35 discuss more general conditions for the positive-definiteness of Γ(I − BR), although computational feasibility often encourages simpler specifications such as Ri = αIp×p, i = 1,…, n, and Γ = D ⊗ Λ, where D is a diagonal matrix with positive entries and Λ is a p × p positive definite matrix. The dispersion matrix in equation (5) now becomes [(D(I − αB)) ⊗ Λ]−1.
Although the Kronecker structure offers computational and interpretational simplicity, there has been much research on developing more general spatial covariances, notably by Kim et al.,37 Carlin and Banerjee35 and Jin et al.20 The last work offers a Generalized Multivariate Conditionally Auto Regressive (GMCAR) model with emphasis on computational simplicity. Considering p = 2 and writing , where and , the GMCAR models the joint distribution of ϕ as p(ϕ) = p(ϕ1|ϕ2)p(ϕ2). Writing this joint distribution as
| (6) |
setting and , we have ϕ1|ϕ2 ~ N(Aϕ2, Σ11.2) and ϕ2 ~ N(0, Σ22).
Treating ϕ1|ϕ2 and ϕ2 as two univariate CARs, we assume ϕ1|ϕ2 ~ N(Aϕ2, τ1[(D − α1W)]−1), and the marginal distribution for ϕ2 is N(0, τ2[(D − α2|W)]−1). Furthermore, let D = Diag(mi), and 0 < α1, α2 < 1 to ensure that model propriety and positive spatial autocorrelation. Also define the entries of matrix A as aii = γ0, aij = γ1 if i ~ j, and 0 otherwise, hence A = γ0I + γ1W, where γ0, γ1 are called the bridging parameters that relate the associations between areal units and the associations between the variables. Jin et al.20 refer to such models as GMCAR(α1, α2, γ0, γ1, τ1, τ2) and show that many existing MCAR models are special instances of the GMCAR.
4 Bayesian estimation of spatial cure models
Our observed data comprises Dij = (tij, δij, xij), where tij is the observed failure time, δij is the failure indicator and xij is a set of covariates. We collect the model-specific parameters (and hyperparameters) into a generic set Ω = Ωij, where Ωij = (ηij(β), θij(β), ρ, ψ) denotes regression in either ηij or θij (perhaps both), and ψ as the set of other hyperparameters that may arise in specific models. Suppressing ηij and ρ, the contribution of subject (i, j) to the data likelihood (in a right-censored setting) is
where P(T ≥ t|N) is as in equation (1), and [F(t)]r−1f (t) with r = 1 or N accordingly for first or last activation. More generally, we can envision each subject-specific activation types, so that some of the r’s will be 1 and others will be N. However, it is more appropriate to think of the population (patients from a specific cancer type) as being subjected to a mechanistic generation of failure that determines the activation scheme for the first or last patient. So, we usually fix r = 1 or set it equal to N for each subject.
We seek the posterior distribution of Ω after marginalizing over the distribution of Nij’s that considerably simplifies the MCMC implementation18 by reducing the estimation space. The contribution of patient (i, j) can be written as (h*(tij))δij S*(tij), where h*(t) = f*(t)/S*(t) denotes the marginal hazard (see Section 2), yielding the marginalized data likelihood as
| (7) |
Thus, we may sample P(Ω|{Dij}) using say Metropolis–Hastings algorithms, without worrying about sampling the Nij’s.
Further computational simplifications arise as h*(t) can often be evaluated in closed forms. For instance, in the first- and last-activation schemes. Under the first-activation scheme, we obtain h*(t) = h(t)EN[N(S(t))N]/EN[(S(t))N], where h(t) = f (t)/S(t) is the latent hazard function. Furthermore, when N ~ Po(θ) with regression in log(θ) = xT β and S(t) is free of x, we obtain a proportional hazards structure for h*(t|x) = exp(xTβ)f (t). In contrast, with N ~ Geo(θ), we find that h*(t) = θf (t)/(1 − θS(t)) does not render a proportional hazards structure. Cooner et al.16 argue that a proportional hazards structure is obtained only with N ~ Po(θ). These characterization results show that the available observed data can inform about the form of S*(t|x), and thus help us deduce the distributional structures of the corresponding latent activation times and N.
For the last-activation scheme, we obtain the hazard as h*(t) = f (t)EN[N(F(t))N−1 1 (N ≥ 1)]/[1 + P(N = 0) − EN[(F(t))N]]. Here, we do not have a proportional hazards structure with Poisson or geometric. With the first, we obtain h*(t) = f (t)θe−θS(t)/[1 + e−θ (1 − eθF(t))], while for the geometric we obtain h*(t) = f (t)θ(1 − θ)/[(1 + θ(θ − 2)F(t))(1 − θF(t))]. Clearly, the hazards structure for the last-activation scheme is more complex, and perhaps less intuitive, than for the first-activation setting. Nevertheless, these provide valid regression structures and may provide better fits in situations where proportional hazards or other simpler models are inappropriate. More importantly from a computational standpoint, they are easily coded into the likelihood as a part of the MCMC implementation. These models can, in fact, be implemented in WinBUGS; www.mrc-bsu.cam.ac.uk/bugs/welcome.shtml.
Finally, we offer model comparisons using the DIC33 as a measure of model choice. The DIC has nice properties for Gaussian likelihoods (as ours) and is particularly convenient to compute from posterior samples. This criteria is the sum of the Bayesian deviance (a measure of model fit) and the (effective) number of parameters (a penalty for model complexity). It rewards better fitting models through the first term and penalizes more complex models through the second term, with lower values indicating favourable models for the data. The deviance, up to an additive quantity not depending upon Ω, is simply minus twice the log-likelihood, D(Ω) = −2 log L({Dij};Ω), where L({Dij};Ω) is the likelihood given in equation (7). The Bayesian deviance is the posterior mean, , while the effective number of parameters is given by . The DIC is then given by and is easily computed from the posterior samples.
5 Illustrations
The National Cancer Institute’s SEER database, available online at http://seer.cancer.gov, provides a national cohort of women who have been monitored for assessing breast cancer prognosis. In addition, available individual-level covariates are age at diagnosis, the number of primary cancers each woman has had diagnosed, the stage of the disease (local (200), regional (94 patients) or distant (4 patients), with local as baseline), and the county information at the time of diagnosis. We consider a sample of 298 patients from Iowa (with 99 counties), who were all diagnosed of breast cancer in January 1992 and monitored through 1998. The response here is time to death from breast cancer: only those who have been identified as having died from metastasis of nodes in the breast (there were 49 such deaths) are considered having failed, while the rest (including those who might have died from other types of cancer or other causes) are considered censored. The longest observation time of this sample is 83 months.
Under both activation schemes in our analysis of the cancer data, we assumed that the regression coefficients β had a non-informative N(0, 103) prior (flat priors are admissible here as well), while a relatively weak Gamma(2, 0.001) prior (mean 2000) was used for ρ. Specifically, for model 2, an N(0, 100) prior for η was used, a Gamma(2, 0.001) prior was used for θ in model 1(a), while U(0, 1) priors were assigned to θ in models 1(b) and (c). We also use Gamma(2, 0.001) for the precision τ in either independent case or the CAR model. These priors are vague enough to allow the data to drive the posterior inference, while still leading to acceptable MCMC convergence. We experimented with other hyperparameter values and did not observe much sensitivity to these choices in the posterior distributions.
For each analysis, we ran two initially dispersed parallel MCMC chains for 30 000 iterations each, where convergence was monitored using sample autocorrelations within the chains, cross-correlations between the parameters, and plots of the sample traces. These tools suggested discarding the first 5 000 iterations from each chain as pre-convergence burn-in. Retaining every 10th of the remaining 2 × 25 000 = 50 000 iterations yielded a final sample of size 5 000 for posterior analysis. The first- and last-activation models were implemented in WinBUGS (see www.mrc-bsu.cam.ac.uk/bugs/welcome.shtml) using R (www.r-project.org) for final posterior summarization. The relevant codes are available from www.biostat.umn.edu/~sudiptob/Software/Software.html.
5.1 Univariate analysis
We use univariate spatial models to analyse the spatial patterns in the latent link η and the cure fraction parameter θ separately. We use a single set of frailties and fit models 1(a)–(c) regressing in η and model 1(a) and model 2 with regression in log(θ). Table 1 presents the DIC scores for these models when the ϕi’s are i.i.d. Gaussian random effects. For regression in η, we see that model 1(c) has the best DIC scores in both the first- and last-activation models. Interestingly, first-activation scores show slightly greater variation between the models in their DIC scores as compared to the last-activation model. This might reflect greater sensitivity of the first activation to the distributional assumptions of N. Also, for model 1(c) the effective number of parameters, pD, seem to be prominently higher due to greater variation in the random effects. For regression in log(θ), we find that model 2 has a substantially lower DIC score compared to model 1(a) under the first-activation scheme, while the latter performs moderately better than the former for the last activation setting. A very similar story is obtained from Table 2, where the i.i.d. frailties are replaced by spatially associated CAR frailties.
Table 1.
Univariate analysis: DIC values for various models under first- and last-activation schemes and frailties (with i.i.d. normal assumption) in latent link η or cure fraction parameter θ. The model with the best DIC value in each column is highlighted
| Model | Univariate: i.i.d. assumption |
First |
Last |
|||
|---|---|---|---|---|---|---|
| i.i.d. frailty in η | pD | DIC | pD | DIC | ||
| 1(a) |
|
25.425 | 3633.430 | 27.170 | 3632.630 | |
| 1(b) |
|
24.308 | 3631.640 | 24.308 | 3631.640 | |
| 1(c) |
|
16.696 | 3624.130 | 27.262 | 3631.350 | |
| 2 |
|
5.956 | 3631.940 | 3.625 | 3634.030 | |
| 1(a) |
|
6.013 | 3632.160 | 6.594 | 3636.970 | |
| 2 |
|
8.777 | 3606.160 | 7.903 | 3640.710 | |
Table 2.
Univariate analysis: DIC values for various models under first- and last-activation schemes and spatial frailties in latent link η or cure fraction parameter θ with CAR assumption
| Model | Univariate: CAR assumption |
First |
Last |
|||
|---|---|---|---|---|---|---|
| Spatial frailty in η | pD | DIC | pD | DIC | ||
| 1(a) |
|
23.406 | 3629.240 | 23.310 | 3628.890 | |
| 1(b) |
|
20.828 | 3628.400 | 20.828 | 3628.400 | |
| 1(c) |
|
15.820 | 3621.870 | 23.454 | 3627.090 | |
| 2 |
|
5.223 | 3630.930 | 3.566 | 3634.050 | |
| 1(a) |
|
6.011 | 3632.200 | 6.343 | 3636.690 | |
| 2 |
|
14.850 | 3630.070 | 12.656 | 3640.040 | |
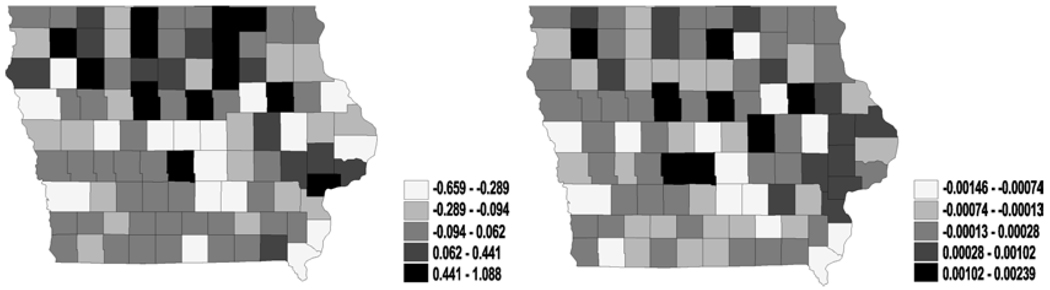
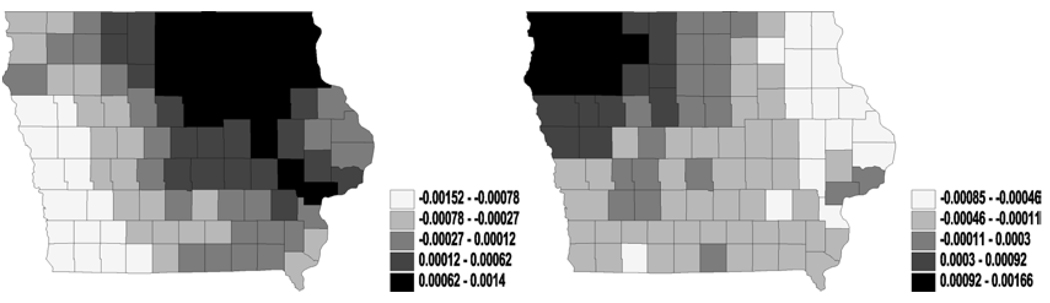
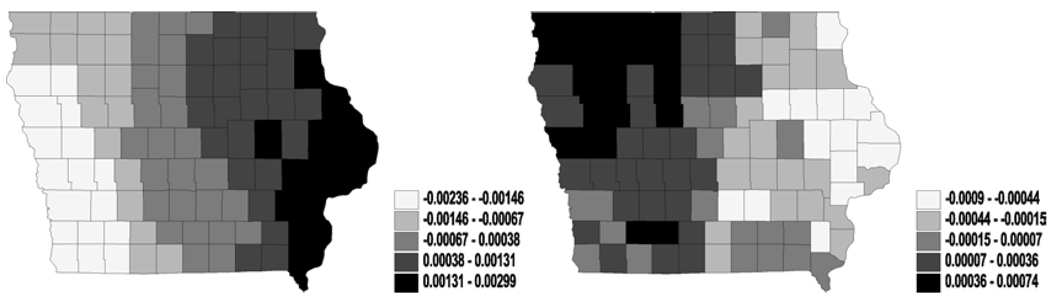
We did not observe much significance for the covariates included in our model. Indeed, the incorporation of spatial effects sometimes widen these credible intervals, as compared to models without random effects. Our preliminary results apparently indicate a rather slim impact of the activation schemes on the posterior estimation of spatial clustering. Therefore, we present the maps of the spatial posterior medians for η and θ only for model 1(a) and model 2 under first-activation schemes. Also, for η the different structures in model 1 did not cause major differences in spatial patterns, while for θ only model 1(a) and model 2 yielded stable convergence – a fact corroborated by the theoretical impropriety of the others. More specifically, the i.i.d. frailties, plotted in Figures 1 and 2, show less pronounced spatial patterns. Both figures reveal only slight differences in spatial patterns for the cure rates or the latent mean between model 1(a) and model 2. Also, we can see that the large difference of the ϕ values in model 1(a) and model 2, suggesting that the simple model (univariate and i.i.d. normal on ϕ) are not stable or reliable (perhaps due to the small sample sizes in some counties).
Figure 1.
Univariate analysis – i.i.d. normal (frailties in latent link η): maps for median frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
Figure 2.
Univariate analysis – i.i.d. normal (frailties in cure fraction parameter θ): maps for median frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
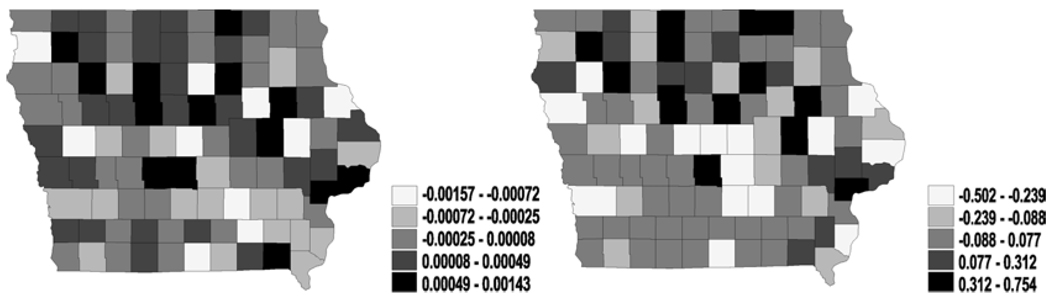
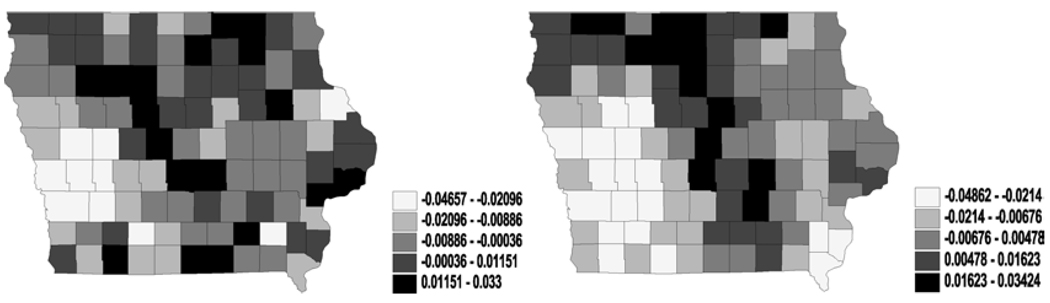
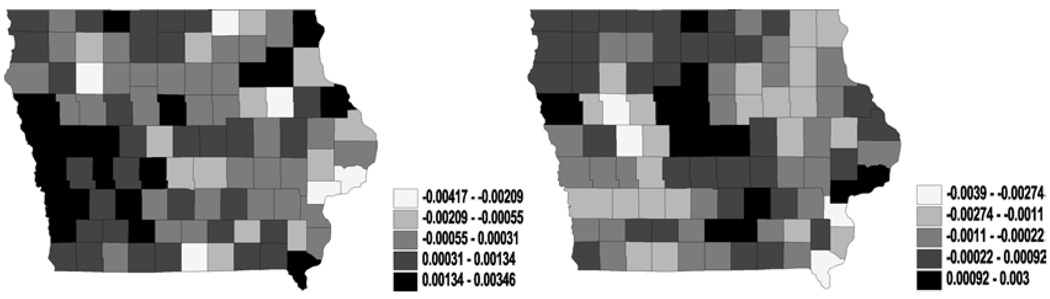
Figures 3 and 4 reveal much clearer spatial patterns from the northeast to the southwest for both η and θ. Since the latent mean survival time and the cure fractions are non-increasing functions of η and θ respectively, the maps for all the models suggest that the southwest has a better survival experience than the northeast. Although both the southwest and the northeast are relatively less populated rural regions in Iowa, these clusters might reflect unknown or missing covariates that explain differences in patient care (e.g., access to health care, quality of medical facilities and so on). Incorporating regressors containing such information might well lead to better smoothing of the maps.
Figure 3.
Univariate analysis – CAR (spatial frailties in latent link η): maps for median spatial frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
Figure 4.
Univariate analysis – CAR (spatial frailties in cure fraction parameter θ): maps for median spatial frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
5.2 Bivariate analysis
Here we consider joint spatial associations in η and θ simultaneously. In Table 3 we show the different models we consider here and their DIC scores. We classify the models as those having two sets of i.i.d. random effects, those with two independent CAR distributions and finally with jointly associated GMCAR random effects. Not much variation is seen across the DIC scores. Under the first-activation scheme we find model 2 to be slightly better for the i.i.d. effects and the two independent CAR effects, while model 1 seems to be marginally better for the GMCAR effects. The story is somewhat similar for the last-activation setting, although we did not find suitable convergence for model 2 from the data under the GMCAR setting.
Table 3.
Multivariate analysis: DIC values for various models under first- and last-activation schemes, with i.i.d. normal, CAR or GMCAR assumption on spatial frailties
| Model | Bivariate |
First |
Last |
|||
|---|---|---|---|---|---|---|
| Two independent i.i.d. normal | pD | DIC | pD | DIC | ||
| 1(a) |
|
6.022 | 3632.070 | 6.794 | 3637.110 | |
| 2 |
|
5.569 | 3631.270 | 3.867 | 3634.190 | |
| 1(a) |
|
6.311 | 3632.720 | 6.880 | 3637.370 | |
| 2 |
|
5.965 | 3632.030 | 3.954 | 3634.580 | |
| 1(a) |
|
6.352 | 3631.580 | 7.296 | 3637.050 | |
| 2 |
|
6.248 | 3631.620 | 1.159 | 3624.170 | |
In Tables 4–6, we present parameter inference results from the best model under first and last activation, with three bivariate assumptions on spatial frailties. For all six models, we find insignificant impact for age at diagnosis and the number of primary nodes, although age at diagnosis has a positive median for each of the models. As expected, the stage variable, indicating the extent of the disease, “Regional” and “Distant” lead to significant hazard increase relative to “Local”. Finally, the Weibull scale parameter ρ is robustly estimated across the models.
Table 4.
Posterior quantiles for different models (corresponding to best DIC) under first- and last-activation schemes, with i.i.d. normal assumption on spatial frailties
| Parameter | First (model 2) |
Last (model 2) |
||
|---|---|---|---|---|
| Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | |
| Intercept | 1.034 | (−3.116, 12.440) | −4.005 | (−6.645, −1.638) |
| Age | 0.008 | (−0.013, 0.029) | 0.014 | (−0.009, 0.039) |
| Primaries | −0.001 | (−0.672, 0.568) | 0.783 | (−0.458, 2.183) |
| Stage (local = 0) | ||||
| Regional | 1.281 | (0.656, 1.947) | 1.764 | (0.829, 2.937) |
| Distant | 2.980 | (2.063, 3.830) | 2.642 | (1.568, 3.621) |
| η0 (Weibull link) | −8.729 | (−19.520, −4.952) | −4.629 | (−6.214, −3.422) |
| ρ | 1.090 | (0.824, 1.524) | 1.157 | (0.827, 1.653) |
Table 6.
Posterior quantiles for different models (corresponding to best DIC) under first- and last-activation schemes, with GMCAR assumption on spatial frailties
| Parameter | First (Model 2) |
Last (Model 2) |
||
|---|---|---|---|---|
| Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | |
| Intercept | −25.230 | (−37.580, −19.390) | −4.066 | (−6.390, −1.500) |
| Age | 0.007 | (−0.012, 0.029) | 0.015 | (−0.012, 0.038) |
| Primaries | 0.018 | (−0.632, 0.584) | 0.627 | (−0.479, 1.930) |
| Stage (local = 0) | ||||
| Regional | 1.287 | (0.678, 1.941) | 1.722 | (0.826, 2.826) |
| Distant | 3.017 | (2.081, 3.856) | 2.651 | (1.629, 3.547) |
| η0 (Weibull link) | – | – | −4.723 | (−6.327, −3.475) |
| ρ | 1.054 | (0.800, 1.346) | 1.192 | (0.857, 1.684) |
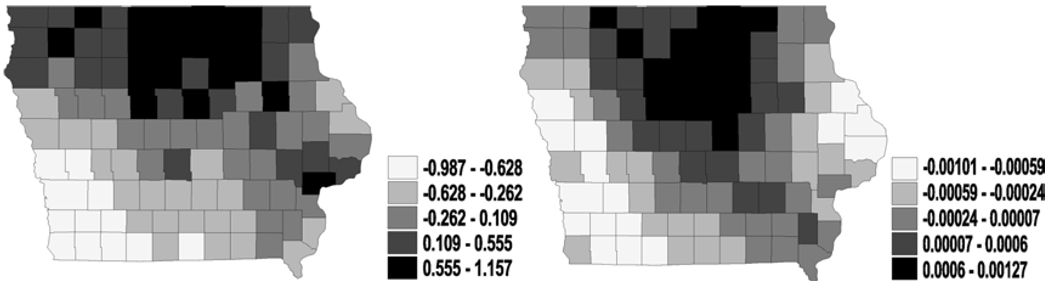
Turning to the spatial maps, two sets of i.i.d. Gaussian random effects in η and θ do not bring about much changes in the posterior behaviour for ϕ, so we do not present the maps for space. However, in Figures 5 and 6, considering the spatial frailties in the frailties together, we find that spatial clustering with frailties in η is inconsistent with that in log(θ). This happens for model 1(a) and model 2 and is in clear contrast with Figures 3 and 4 which displayed consistent behaviour. The differences between two models for both parameters are large and the spatial pattern seem reversed. It is worth mentioning that the values now are very well stabilized. Two separate i.i.d. normal or CAR frailties might still not be enough to capture the spatial patterns.
Figure 5.
Bivariate analysis – two independent CAR (spatial frailties in latent link η i.e., ϕ1): maps for median spatial frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
Figure 6.
Bivariate analysis – two independent CAR (spatial frailties in cure fraction parameter ϕ i.e., ϕ2): maps for median spatial frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
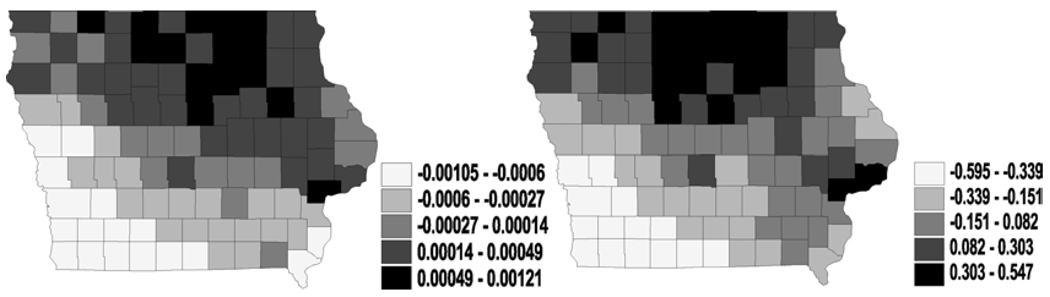
These lead us to question some of the fundamental assumptions for the independence between the spatial effects in the latent link η and the cure fraction parameter θ. Simply introducing two independent univariate spatial structures is inadequate, hence the MCAR or the GMCAR should be implemented into our models. With the GMCAR assumption, the maps (Figures 7 and 8) show somewhat similar spatial trends as the univariate cases, which may suggest a lack of significant spatial association for this set of data.
Figure 7.
Bivariate analysis – GMCAR (spatial frailties in latent link η i.e., ϕ1): maps for median spatial frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
Figure 8.
Bivariate analysis – GMCAR (spatial frailties in cure fraction parameter θ i.e., ϕ2): maps for median spatial frailties under first-activation scheme for model 1(a) (left) and model 2 (right).
6 Future work
We plan to extend our current work into several directions. First, we will substantially enhance the current illustration with the hierarchical activation scheme results. Next we propose to implement these models with more flexible MCAR specifications. Finally, we will investigate spatio-temporal effects to account for temporal associations where we may model the cure fractions θi(t) or the latent link ηi(t) as functions of time. These may include direct time-varying regression, polynomial or spline functions of time, or even completely non-parametric modelling using temporal processes.
Table 5.
Posterior quantiles for different models (corresponding to best DIC) under first- and last-activation schemes, with CAR assumption on spatial frailties
| Parameter | First (model 2) |
Last (model 2) |
||
|---|---|---|---|---|
| Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | |
| Intercept | 6.645 | (−2.279, 15.050) | −4.037 | (−6.620, −1.612) |
| Age | 0.008 | (−0.014, 0.030) | 0.014 | (−0.008, 0.038) |
| Primaries | 0.004 | (−0.650, 0.563) | 0.766 | (−0.507, 2.190) |
| Stage (local = 0) | ||||
| Regional | 1.287 | (0.668, 1.937) | 1.790 | (0.854, 2.966) |
| Distant | 2.992 | (2.067, 3.839) | 2.651 | (1.572, 3.643) |
| η0 (Weibull link) | −14.100 | (−22.730, −5.398) | −4.570 | (−6.117, −3.270) |
| ρ | 1.059 | (0.804, 1.404) | 1.144 | (0.793, 1.626) |
Footnotes
Contributor Information
Freda Cooner, Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN, USA.
Sudipto Banerjee, Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN, USA.
A Marshall McBean, Division of Health Services Research and Policy, School of Public Health, University of Minnesota, Minneapolis, MN, USA.
References
- 1.Lawson AB, Biggeri A, Boehning D, Lesaffre E, Viel J-F, Bertollini R. Disease mapping and risk assessment for public health. Wiley; 1999. [Google Scholar]
- 2.Elliot P, Wakefield JC, Best NG, Briggs DJ. Spatial epidemiology: methods and applications. Oxford University Press; 2001. [Google Scholar]
- 3.Lawson AB, Williams FLR. An introductory guide to disease mapping. Wiley Medical Sciences; 2001. [Google Scholar]
- 4.Banejee S, Wall MM, Carlin BP. Fraity modelling for spatially correlated survival data with application to infant mortality in Minnesota. Biostatistics. 2003;4:123–142. doi: 10.1093/biostatistics/4.1.123. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Ryan L. Modeling spatial survival data using semiparametric frailty models. Biometrics. 2002;58:287–297. doi: 10.1111/j.0006-341x.2002.00287.x. [DOI] [PubMed] [Google Scholar]
- 6.Cox DR, Oakes D. Analysis of survival data. Chapman & Hall; 1984. [Google Scholar]
- 7.Berkson J, Gage RP. Survival curve for cancer patients following treatment. Journal of the American Statistical Association. 1952;47:501–515. [Google Scholar]
- 8.Farewell VT. The use of mixture models for the analysis of survival data with long term survivors. Biometrics. 1982;38:1041–1046. [PubMed] [Google Scholar]
- 9.Farewell VT. Mixture models in survival analysis: are they worth the risk? Canadian Journal of Statistics. 1986;14:257–262. [Google Scholar]
- 10.Goldman AI. Survivorship analysis when cure is a possibility: a Monte Carlo study. Statistics in Medicine. 1984;3:153–163. doi: 10.1002/sim.4780030208. [DOI] [PubMed] [Google Scholar]
- 11.Ewell M, Ibrahim JG. The large sample distribution of the weighted log-rank statistic under general local alternatives. Lifetime Data Analysis. 1997;3:5–12. doi: 10.1023/a:1009690200504. [DOI] [PubMed] [Google Scholar]
- 12.Yakovlev AY, Asselain B, Bardou VJ, Fourquet A, Hoang T, Rochefordiere A, Tsodikov AD. A simple stochastic model of tumor recurrence and its application to data on premenopausal breast cancer. In: Asselain B, Boniface M, Duby C, Lopez C, Masson JP, Tranchefort J, editors. Biometrie et Analyse de Donnees Spatio-Temporelles, No. 12. Société Française de Biométrie; 1993. pp. 66–82. [Google Scholar]
- 13.Chen M-H, Ibrahim JG, Sinha D. A new Bayesian model for survival data with a surviving fraction. Journal of the American Statistical Association. 1999;94:909–919. [Google Scholar]
- 14.Ibrahim JG, Chen M-H, Sinha D. Bayesian survival analysis. Springer-Verlag; 2001. [Google Scholar]
- 15.Tsodikov A, Ibrahim J, Yakovlev A. Estimating cure rates from survival data: an alternative to two-component mixture models. Journal of the American Statistical Association. 2003;98:1063–1078. doi: 10.1198/01622145030000001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooner F, Banerjee S, Carlin BP, Sinha D. Research Report 2005–06. Division of Biostatistics, University of Minnesota; Flexible cure rate modelling under latent activation schemes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee S, Carlin BP. Parametric spatial cure rate models for interval-censored time-to-relapse data. Biometrics. 2004;60:268–275. doi: 10.1111/j.0006-341X.2004.00032.x. [DOI] [PubMed] [Google Scholar]
- 18.Carlin BP, Louis TA. Bayes and empirical Bayes methods for data analysis. second edition. Chapman & Hall/CRC Press; 2000. [Google Scholar]
- 19.Rue H, Held L. Gaussian Markov Random Fields; Theory and Application. Chapman & Hall/CRC Press; 2005. Monographs in Statistics and Applied Probability No. 104. [Google Scholar]
- 20.Jin X, Carlin BP, Banerjee S. Generalized hierarchical multivariate CAR models for areal data. Biometrics. 2006 doi: 10.1111/j.1541-0420.2005.00359.x. in press. [DOI] [PubMed] [Google Scholar]
- 21.Rao CR. Linear statistical inference and its applications. second edition. Wiley; 1973. [Google Scholar]
- 22.Tucker SL, Thames HD, Taylor JMG. How well is the probability of tumor cure after fractionated irradiation described by Poisson statistics? Radiation Research. 1990;124:273–282. [PubMed] [Google Scholar]
- 23.Tucker SL, Taylor JMG. Improved models of tumor cure. International Journal of Radiational Biology. 1996;70:539–553. doi: 10.1080/095530096144743. [DOI] [PubMed] [Google Scholar]
- 24.Moolgavkar SH, Luebeck EG, De Gunst MC. Two mutation model for carcinogenesis: relative roles of somatic mutations and cell proliferation in determining risk. In: Moolgavkar SH, editor. Scientific issues in quantitative cancer risk assessment. Birkhauser; 1990. pp. 136–152. [Google Scholar]
- 25.Besag J. Spatial interaction and the statistical analysis of lattice system (with discussion) Journal of the Royal Statistical Society, Series B. 1974;36:192–236. [Google Scholar]
- 26.Cressie NAC. Statistics for spatial data. revised edition. Wiley; 1993. [Google Scholar]
- 27.Banerjee S, Carlin BP, Gelfand AE. Hierarchical modeling and analysis for spatial data. Chapman & Hall/CRC Press; 2004. [Google Scholar]
- 28.Heikkinen J, Högmander H. Fully Bayesian approach to image restoration with an application in biogeography. Applied Statistics. 1994;43:569–582. [Google Scholar]
- 29.Högmander H, Møller J. Estimating distribution maps from atlas data using methods of statistical image analysis. Biometrics. 1995;51:393–404. [Google Scholar]
- 30.Hoeting JA, Leecaster M, Bowden D. An improved model for spatially correlated binary response. Journal of Agricultural, Biological, and Environmental Statistics. 2000;5:102–114. [Google Scholar]
- 31.Besag J, York JC, Mollié A. Bayesian image restoration, with two applications in spatial statistics (with discussion) Annals of the Institute of Statistical Mathematics. 1991;43:1–59. [Google Scholar]
- 32.Lawson AB, Browne WJ, Vidal Rodeiro CL. Disease mapping with WinBUGS and MLwiN. Wiley; 2003. [Google Scholar]
- 33.Spiegelhalter DJ, Best N, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit (with discussion) Journal of the Royal Statistical Society, Series B. 2002;64:583–639. [Google Scholar]
- 34.Mardia KV. Multi-dimensional multivariate Gaussian Markov random fields with application to image processing. Journal of Multivariate Analysis. 1998;24:265–284. [Google Scholar]
- 35.Carlin BP, Banerjee S. Hierarchical multivariate CAR models for spatio-temporally correlated survival data (with discussion) In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics. Oxford University Press; 2003. pp. 45–63. [Google Scholar]
- 36.Gelfand AE, Vounatsou P. Proper multivariate conditional autoregressive models for spatial data analysis. Biostatistics. 2003;4:11–25. doi: 10.1093/biostatistics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 37.Kim H, Sun D, Tsutakawa RK. A bivariate Bayes method for improving the estimates of mortality rates with a twofold conditional autoregressive model. Journal of the American Statistical Association. 2001;96:1506–1521. [Google Scholar]