Decades of research have identified numerous biomarkers for cardiovascular diseases (CVDs) and type 2 diabetes, providing molecular insights for improved treatment and prevention of the diseases (1–3). Of the biomarkers that could be objectively and systematically measured, genetic variants such as single nucleotide polymorphisms (SNPs) have some unique features in that they do not change over time, and the temporal sequence of genotype-phenotype can be clearly established for outcome prediction.
Using high-density fixed SNP arrays, recent genome-wide association studies (GWAS) have successfully identified multiple risk alleles related to CVD and type 2 diabetes. These advances in genomics present many exciting opportunities in three scientific domains: 1) integrating novel genetic variants into risk prediction models of complex diseases in humans, 2) characterizing new biological pathways involved in pathogenesis and thus improved strategies for treatment and management, and 3) enhancing inference of traditional epidemiological work relevant to public health importance. To capitalize on these opportunities, several groups have attempted to develop genetic risk scores by summing up the number of risk alleles for disease prediction. However, almost all these studies have concluded that current genetic information contributes little information in distinguishing who will or will not develop a CVD or type 2 diabetes among apparently healthy adults (4–6).
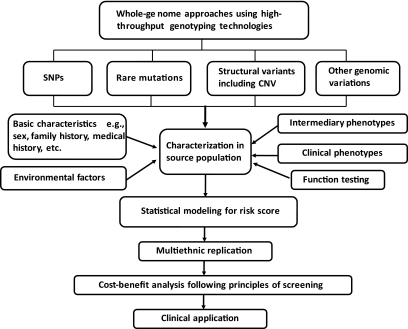
Given that most common risk variants identified so far confer relatively modest risk to these complex diseases (e.g., all risk alleles for type 2 diabetes identified by GWAS have very small relative risks [<1.50]) (7,8), the “common diseases-common variants” model has been formally challenged (9,10). In the field of complex disease genetics, it is now widely anticipated that some ongoing next-generation sequencing work covering the whole genome in diverse populations would identify rare variants of large effect sizes in the coming years (8). Yet, there still remain many questions that must be answered before genetic information can be appropriately incorporated into risk prediction models for complex diseases (Fig. 1).
FIG. 1.
Assessing and integrating reliable genomic information in the development of clinical risk prediction model. CNV, copy number variant.
In this issue of Diabetes, Palmer et al. (11) report findings of using yet another genes-based score to predict stroke risk in a cohort of 2,182 patients followed for ∼6 years. The authors selected from prior work a set of five variants involved in inflammation and developed a score by summing up “at-risk” genotypes for those variants. By assigning a score of 1 for having at least one risk allele and 0 for noncarriers, Palmer et al. implicitly assumed that these five loci follow either dominant or recessive genetic patterns. Previously, Morrison et al. (12) advocated an additive model with weighing of −1, 0, and 1, as did others (4–6).
None of these studies, however, have attempted to weigh the loci using regression coefficients from the specific proportional hazard function. Put simply in regression terms, Palmer et al. in effect converted a set of five dichotomous variables into an ordinal variable in relating genetic variants to risk of stroke in their model. Whether this is reasonable depends on the nature of the genotypes-disease relationship that is inherently defined by the specific model form. With the use of Cox proportional hazard model, an ordinal “at-risk genotype” score implies an exponential relationship in that each added “at-risk genotype” multiplies the baseline risk by a constant value corresponding to the antilogarithm of the regression coefficient (following the survival function Yi = 1− {s[t]}exp{A + B × Xi}; where Yi is predicted probability for developing stroke over time t (t was event free follow-up time for individual i); Xi represents the genotype scores [0,1,2,3,4,5]). Given that during a mean follow-up of ∼6 years none of these five variants were independently associated with stroke risk, the evidence in support of an exponential shape of relationship between these genetic variants and disease risk appeared weak. Only when converted into an ordinal variable did it become statistically significant with a hazard ratio of 1.34 for each “at-risk genotype.” This apparent gain in statistical efficiency can only be achieved with significant constraints that are model-dependent and thus has very limited implication for inference beyond the samples investigated by Palmer et al. (11).
It would be helpful to examine the distributions of traditional risk factors for specific types of stroke (e.g., family history, diet, physical activity, diabetes duration, and levels of glycemic control) by this genetic score. With ∼1% increment in the area under the receiver operating characteristic curve, this ordinal genetic score (even with strong linearity assumption in a multiplicative scale) apparently did not contribute to discrimination. Formal evaluation of prediction should also be conducted to assess improvement of fit for inclusion of each locus genotype separately and fit for the entire model by computing likelihood ratio χ2 statistics and Bayesian information criteria (fit for the entire model taking into account the number of parameters).
Aside from using genetic variants for risk prediction, recent GWAS have also started to uncover potentially new biological targets for complex diseases. Since the first GWAS for type 2 diabetes published in 2007 (13), subsequent efforts have confirmed at least 20 robust and well-replicated genetic loci associated with the disease (7). Interestingly, some identified regions have never been suspected to be involved in the pathophysiology of type 2 diabetes, including a common variant in the FTO gene (rs9939609) (14). Several studies have now confirmed the association between FTO variants and higher BMI and obesity in both children and adults (15,16). It was thus surprising that in building their risk score, Palmer et al. (11) chose to ignore recent GWAS findings for stroke (17) as well as many important candidate genes in the pathways of inflammation and endothelial dysfunction (18). It remains possible that the addition of a much larger number of common or rare risk alleles based on a better understanding of inflammatory mechanisms underlying CVD could improve risk prediction.
Meanwhile, emerging evidence indicates sex differences in genetic susceptibility to CVD among diabetic patients (19). In the U.S., CVD mortality has declined substantially in recent decades among nondiabetic individuals, but has declined only among diabetic men and increased significantly in diabetic women (20). The reason for the accelerated atherothrombotic events in diabetic women remains poorly understood. Traditional CVD risk factors such as hypertension and dyslipidemia cannot completely account for the apparent sex differences in the excess CVD risk associated with diabetes (19). Because inflammation and endothelial function are more seriously affected by diabetes in women than in men and because diabetes may cause greater shift to “android” obese pattern in women than in men (21), recent work has also intensified the search for sex-specific associations between variants of these genes and CVD risk and has developed sex-specific risk prediction models (19,22,23).
More importantly, future risk assessment for complex disease should take a much more careful consideration of gene-gene and/or gene-environmental interactions. Complex diseases such as CVD and type 2 diabetes are influenced by both genetic and environmental factors. For example, most GWAS to date have been conducted in middle-aged and older adults so that the cumulative effects of multiple environmental effects or other gene-gene or gene-environment interactions in older age may have diluted a modest but real genetic effect that may be more apparent earlier in life. Such incomplete understanding of genetic and environmental causes and their interactions appeared to have confounded those who attempted to identify a set of SNPs that could adequately explain or predict even a small fraction of complex diseases (24,25). As the field of genomics progresses, it is imperative to confirm and better characterize genetic variation (i.e., better resolution of our genomes) via fine-mapping, functional testing, integrating mechanistic analysis of intermediary phenotypes, and assessment of gene-environment interactions in multiple racial and ethnic groups. Multiethnic replications are useful in uncovering true susceptibility genes by identifying multiple significant hits within a specific region, which is particularly valuable given allelic heterogeneity of the genetic effects (different alleles may cause the disease in different populations) (26). Yet, even with these anticipated progress in genomic sciences, the preventive utility of using genetic score alone for common diseases in adults will likely be very limited, especially considering the myriad of environmental factors that also influence the development of complex diseases. With a better understanding of pathogenesis, however, integrating genetic variants with their biochemical phenotypes, as recently demonstrated in a study of sex-hormone–binding globulin and type 2 diabetes risk, should be a viable strategy to provide molecular insights and improve disease prediction (22,27). Ultimately, greater further efforts will be required to put valuable genetic information in the appropriate biological and clinical context (including cost-benefit evaluation following principles of screening) to optimize risk assessment for prevention.
ACKNOWLEDGMENTS
No potential conflicts of interest relevant to this article were reported.
Footnotes
See accompanying brief report, p. 2945.
REFERENCES
- 1.Kuller LH, Meilahn EN. Risk factors for cardiovascular disease among women. Curr Opin Lipidol 1996;7:203–208 [DOI] [PubMed] [Google Scholar]
- 2.Kullo IJ, Cooper LT. Early identification of cardiovascular risk using genomics and proteomics. Nat Rev Cardiol 2010;7:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S, Tinker L, Song Y, Rifai N, Bonds DE, Cook NR, Heiss G, Howard BV, Hotamisligil GS, Hu FB, Kuller LH, Manson JE. A prospective study of inflammatory cytokines and diabetes mellitus in a multiethnic cohort of postmenopausal women. Arch Intern Med 2007;167:1676–1685 [DOI] [PubMed] [Google Scholar]
- 4.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008;359:2220–2232 [DOI] [PubMed] [Google Scholar]
- 5.Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, Manning AK, Florez JC, Wilson PW, D'Agostino RB, Sr, Cupples LA. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 2008;359:2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paynter NP, Chasman DI, Paré G, Buring JE, Cook NR, Miletich JP, Ridker PM. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA 2010;303:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Silva NM, Frayling TM. Novel biological insights emerging from genetic studies of type 2 diabetes and related metabolic traits. Curr Opin Lipidol 2010;21:44–50 [DOI] [PubMed] [Google Scholar]
- 8.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature 2009;461:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClellan J, King MC. Genetic heterogeneity in human disease. Cell 2010;141:210–217 [DOI] [PubMed] [Google Scholar]
- 11.Palmer CNA, Kimber CH, Doney ASF, Proia AS, Morris AM, Gaetani E, Quarta M, Smith RC, Pola R. Combined effect of inflammatory gene polymorphisms and the risk of ischemic stroke in a prospective cohort of subjects with type 2 diabetes: a Go-DARTS study. Diabetes 2010;59:2945–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison AC, Bare LA, Chambless LE, Ellis SG, Malloy M, Kane JP, Pankow JS, Devlin JJ, Willerson JT, Boerwinkle E. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. Am J Epidemiol 2007;166:28–35 [DOI] [PubMed] [Google Scholar]
- 13.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 14.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007;316:889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, Chen Z, Yu L, Yang Y, Feng G, Li J, Zhang J, Liu S, Zhang Z, He L, Xu H. Meta-analysis added power to identify variants in FTO associated with type 2 diabetes and obesity in the Asian population. Obesity (Silver Spring) 2009;18:1619–1624 [DOI] [PubMed] [Google Scholar]
- 16.Song Y, You NC, Hsu YH, Howard BV, Langer RD, Manson JE, Nathan L, Niu T, F Tinker L, Liu S. FTO polymorphisms are associated with obesity but not diabetes risk in postmenopausal women. Obesity (Silver Spring) 2008;16:2472–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, Bos MJ, Beiser A, Cushman M, Launer LJ, Shahar E, Struchalin M, Du Y, Glazer NL, Rosamond WD, Rivadeneira F, Kelly-Hayes M, Lopez OL, Coresh J, Hofman A, DeCarli C, Heckbert SR, Koudstaal PJ, Yang Q, Smith NL, Kase CS, Rice K, Haritunians T, Roks G, de Kort PL, Taylor KD, de Lau LM, Oostra BA, Uitterlinden AG, Rotter JI, Boerwinkle E, Psaty BM, Mosley TH, van Duijn CM, Breteler MM, Longstreth WT, Jr, Wolf PA. Genomewide association studies of stroke. N Engl J Med 2009;360:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridker PM, Pare G, Parker A, Zee RY, Danik JS, Buring JE, Kwiatkowski D, Cook NR, Miletich JP, Chasman DI. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am J Hum Genet 2008;82:1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding EL, Song Y, Malik VS, Liu S. Sex differences of endogenous sex hormones and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 2006;295:1288–1299 [DOI] [PubMed] [Google Scholar]
- 20.Ergin A, Muntner P, Sherwin R, He J. Secular trends in cardiovascular disease mortality, incidence, and case fatality rates in adults in the United States. Am J Med 2004;117:219–227 [DOI] [PubMed] [Google Scholar]
- 21.Steinberg HO, Paradisi G, Cronin J, Crowde K, Hempfling A, Hook G, Baron AD. Type II diabetes abrogates sex differences in endothelial function in premenopausal women. Circulation 2000;101:2040–2046 [DOI] [PubMed] [Google Scholar]
- 22.Ding EL, Song Y, Manson JE, Hunter DJ, Lee CC, Rifai N, Buring JE, Gaziano JM, Liu S. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med 2009;361:1152–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding EL, Song Y, Manson JE, Rifai N, Buring JE, Liu S. Plasma sex steroid hormones and risk of developing type 2 diabetes in women: a prospective study. Diabetologia 2007;50:2076–2084 [DOI] [PubMed] [Google Scholar]
- 24.van der Net JB, Janssens AC, Sijbrands EJ, Steyerberg EW. Value of genetic profiling for the prediction of coronary heart disease. Am Heart J 2009;158:105–110 [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol 2005;34:1129–1137 [DOI] [PubMed] [Google Scholar]
- 26.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet 2009;374:340–350 [DOI] [PubMed] [Google Scholar]
- 27.Perry JR, Weedon MN, Langenberg C, Jackson AU, Lyssenko V, Sparsø T, Thorleifsson G, Grallert H, Ferrucci L, Maggio M, Paolisso G, Walker M, Palmer CN, Payne F, Young E, Herder C, Narisu N, Morken MA, Bonnycastle LL, Owen KR, Shields B, Knight B, Bennett A, Groves CJ, Ruokonen A, Jarvelin MR, Pearson E, Pascoe L, Ferrannini E, Bornstein SR, Stringham HM, Scott LJ, Kuusisto J, Nilsson P, Neptin M, Gjesing AP, Pisinger C, Lauritzen T, Sandbaek A, Sampson M, MAGIC, Zeggini E, Lindgren CM, Steinthorsdottir V, Thorsteinsdottir U, Hansen T, Schwarz P, Illig T, Laakso M, Stefansson K, Morris AD, Groop L, Pedersen O, Boehnke M, Barroso I, Wareham NJ, Hattersley AT, McCarthy MI, Frayling TM. Genetic evidence that raised sex hormone binding globulin (SHBG) levels reduce the risk of type 2 diabetes. Hum Mol Genet 2010;19:535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]