Abstract
OBJECTIVE
We have previously observed that genetic profiles determined by the combination of five functionally significant single nucleotide polymorphisms (SNPs) (rs1800795, rs5498, rs5361, rs1024611, and rs679620) of genes encoding prototypical inflammatory molecules are associated with history of ischemic stroke. Here we tested the ability of this multigenic model to predict stroke risk in a large population-based prospective cohort of subjects with type 2 diabetes.
RESEARCH DESIGN AND METHODS
This study was conducted using a prospective cohort of individuals with type 2 diabetes participating in the Go-DARTS (Genetics of Diabetes Audit and Research in Tayside Scotland) study, which includes genetic and clinical information of patients with diabetes within the Tayside region of Scotland, U.K. The above-mentioned inflammatory SNPs were investigated in 2,182 Go-DARTS participants. We created an inflammatory risk score (IRS), ranging from 0 to 5, according to the number of “at-risk” genotypes concomitantly carried by a given individual. The primary outcome was the occurrence of fatal or nonfatal stroke of any kind. Mean follow-up time was 6.2 ± 1.1 years.
RESULTS
The incidence of stroke increased according to the IRS. The IRS was significantly and independently associated with increased stroke risk after adjustment for other conventional risk factors (hazard ratio 1.34 [95% CI 1.1–1.7]; P = 0.009). The highest hazard ratio for stroke was found in subjects concomitantly carrying >3 proinflammatory variations and in subjects without previous cardiovascular diseases.
CONCLUSIONS
This large prospective cohort study provides evidence that SNPs of genes encoding prototypical inflammatory molecules may be used to create multigenic models that predict stroke risk in subjects with type 2 diabetes.
Proinflammatory single nucleotide polymorphisms (SNPs) may contribute to the development and progression of pathological conditions, including cardiovascular diseases (1–7). Inflammatory gene variations may act synergistically, determining genetic profiles associated with increased risk for disease (8–13). We have previously demonstrated that genetic profiles determined by the combination of SNPs of five prototypical inflammatory genes are associated with history of ischemic stroke. In particular, we have found that the combined analysis of the rs1800795, rs5498, rs5361, rs1024611, and rs679620 of the interleukin-6 (IL-6), monocyte chemoattractant protein-1 (MCP-1), intercellular adhesion molecule-1 (ICAM-1), selectin-E (sel-E), and matrix metalloproteinase-3 (MMP-3) genes may be used to create genetic profiles that are associated with different odds of stroke in a case-control scenario (10). A similar combination of inflammatory SNPs is also associated with peripheral artery disease (PAD) and critical limb ischemia (CLI) (11).
Here, we investigated whether this multigenic model may predict stroke risk in a large population-based prospective cohort of subjects with type 2 diabetes. We studied 2,182 diabetic individuals participating in the prospective Go-DARTS (Genetics of Diabetes Audit and Research in Tayside Scotland) study and found that risk for stroke is significantly and independently influenced by an inflammatory risk score (IRS) determined by the combination of the five SNPs mentioned above.
RESEARCH DESIGN AND METHODS
We studied 2,182 individuals with type 2 diabetes in the Go-DARTS study. Additional information on Go-DARTS is available in the online appendix available at http://diabetes.diabetesjournals.org/cgi/content/full/db09-1690/DC1. The study was approved by the local research ethics committee.
Genetic analyses and creation of the multigenic model.
The following SNPs were analyzed: IL-6 −174G/C rs1800795, MCP −2518A/G rs1024611, ICAM-1 −469E/K rs5498, sel-E Ser128Arg rs5361, and MMP-3 −117155A/6A rs679620. According to our previous study (10), the following genotypes were considered “at risk”: IL-6 GG and GC, MCP-1 GG, ICAM-1 EE, sel-E RR (ArgArg), and MMP-3 5A5A. The IRS model was created as a continuous variable, ranging from 0 to 5, based on the number of “at-risk” genotypes concomitantly carried by a given individual. The population was stratified prior to evaluation of the number of disease events and execution of the statistical analyses. Additional information is available in the online appendix.
Disease events.
All individuals were followed up until their first nonfatal or fatal stroke after recruitment. Nonfatal strokes were determined from both the hospital Scottish Morbidity Record and the DARTS program of nurse-facilitated validation. Fatal events were determined from death certificates obtained from the General Register Office for Scotland, as previously described (14–16). Additional information is available in the online appendix.
Statistical analyses.
Cox regression was used to determine the association of genotype with stroke after recruitment. Survival functions were adjusted for age at recruitment and sex. For smoking, a composite variable was constructed (ever-smokers genotype+, ever-smokers genotype−, never-smokers genotype+, and never-smokers genotype−) and entered as indicator variables. Fully adjusted models, which included ratio of total-to-HDL cholesterol, mean arterial pressure, and years with diabetes, were also considered. STATA version 8 was used for all analyses. Additional information is available in the online appendix.
RESULTS
The demographic and clinical characteristics of the study population are presented in Table 1. The cohort consisted of 1,147 male and 1,035 female subjects (mean age 64.5 ± 11.7 years). Mean age at diagnosis was 56.8 ± 12.2 years. Mean duration of diabetes was 7.7 ± 6.5 years. A total of 1,109 subjects were “ever-smokers” (50.8%), 720 subjects had previous cardiovascular diseases (myocardial infarction and/or stroke) (33.0%), and 200 subjects had previous history of stroke (9.2%). During the follow-up period (6.2 ± 1.1 years), the overall incidence of new fatal and nonfatal strokes was 4.9% (108 events).
TABLE 1.
Characteristics of the study cohort
| Total sample size | 2,182 |
| Female subjects | 1,035 (47.4%) |
| Male subjects | 1,147 (52.6%) |
| Age (years) | 64.5 ± 11.7 |
| BMI (kg/m2) | 30.2 ± 5.45 |
| Duration of diabetes (years) | 7.7 ± 6.5 |
| Age at diagnosis (years) | 56.8 ± 12.2 |
| Smoker (ever) | 1,109 (50.8%) |
| Previous cardiovascular disease | 720 (33.0%) |
| Previous stroke | 200 (9.2%) |
| Follow-up (years) | 6.2 ± 11 |
| Strokes (fatal and nonfatal) after enrollment | 108 (4.9%) |
Data are means ± SD or n (%).
Genotype and allele frequencies of the individual variants in the study cohort are shown in supplemental Table 1. Allele frequencies of all variants were in Hardy-Weinberg equilibrium. The association of individual variants with prospective risk for stroke is shown in Table 2. In this analysis, we also considered whether the association between individual gene variations and stroke risk was influenced by smoking. None of the investigated gene variants increased the risk for stroke when considered alone. The ICAM-1 EE genotype demonstrated a significant association with stroke in smokers (hazard ratio [HR] 2.6 [95% CI 1.2–6.0]; P = 0.02).
TABLE 2.
Association of individual increased risk variants with prospective risk of stroke
| Variant | Model | HR (95% CI)* | P |
|---|---|---|---|
| IL-6 | GG/GC genotype | 1.2 (0.6–1.9) | 0.626 |
| Smoking (ever) | 0.9 (0.4–4.3) | 0.683 | |
| MCP-1 | GG genotype | 1.4 (0.9–2.0) | 0.102 |
| Smoking (ever) | 1.0 (0.4–2.1) | 0.857 | |
| ICAM-1 | EE genotype | 1.2 (0.8–2.0) | 0.365 |
| Smoking (ever) | 2.6 (1.2–6.0) | 0.022 | |
| sel-E | RR genotype | 2.0 (0.5–8.0) | 0.346 |
| Smoking (ever) | NA** | NA | |
| MMP3 | 5A5A genotype | 1.5 (1.0–2.2) | 0.075 |
| Smoking (ever) | 1.6 (0.7–3.7) | 0.273 |
*All models adjusted for age at study recruitment and sex.
**Zero individuals in at least one cell. NA, not available.
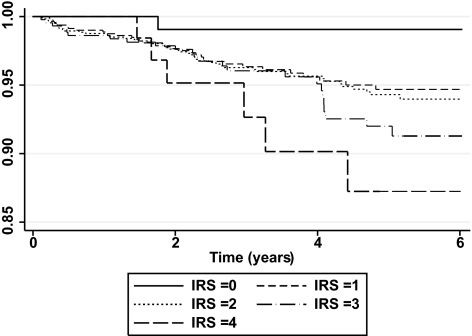
The distribution of subjects according to the IRS is shown in supplemental Fig. 1. No subjects with an IRS of 5 were identified in our cohort. The occurrence of ischemic stroke over the follow-up period increased according to the IRS, with an incidence of 1.0, 4.5, 4.7, 6.4, and 10.9% among subjects with IRS 0, 1, 2, 3, and 4, respectively (supplemental Fig. 2). Cox proportional hazards analyses were performed to assess the relative risk for stroke according to the IRS. These analyses were performed on a total of 2,123 individuals for whom full covariates were available. In this subset of subjects, the incidence of stroke (4.8%, 104 events) did not differ from that observed in the overall study cohort (n = 2,182, incidence of stroke 4.9%, 108 events). After correcting for age, smoking habit, blood pressure, cholesterol ratio, and presence of previous cardiovascular diseases, the IRS was significantly and independently associated with increased stroke risk (HR 1.34 per risk allele [95% CI 1.1–1.7]; P = 0.009) (Table 3). The effect of IRS was mainly seen in individuals free of cardiovascular diseases at entry (supplemental Table 2). The percentage of subjects that survive without stroke differs according to the IRS, with individuals who have an IRS of 4 showing the lowest stroke-free survival (Fig. 1).
TABLE 3.
IRS genotypic model and risk of stroke in the Go-DARTS study
| HR (95% CI) | P | |
|---|---|---|
| IRS | 1.34 (1.1–1.7) | 0.009 |
| Age at enrollment | 1.06 (1.0–1.1) | <0.001 |
| Smoking (ever) | 1.52 (1.0–2.3) | 0.038 |
| Previous cardiovascular disease | 2.10 (1.4–3.1) | <0.001 |
| Mean arterial pressure | 1.02 (0.8–1.1) | 0.087 |
| Cholesterol ratio | 3.93 (0.8–18.7) | 0.085 |
Analyses were performed on 2,123 genotyped individuals with full covariates available for Cox proportional hazards study. Number of incident strokes is 104.
FIG. 1.
Kaplan-Meier plot of stroke-free survival among the Go-DARTS population stratified by IRS.
DISCUSSION
Multigenic approaches have been demonstrated to be valid tools for identifying subjects at risk for complex traits, such as type 2 diabetes (17) and coronary artery disease (18). A similar strategy was used in the present study, hypothesizing that, in subjects with type 2 diabetes, an increasing number of proinflammatory alleles may confer a significant risk of developing ischemic stroke. We have previously demonstrated the utility of the Go-DARTS cohort to study the contribution of gene variations to cardiovascular risk in subjects with diabetes (14–16). To our knowledge, this is the first prospective study that attempts to look at the impact of the combined effect of multiple gene variations on incident stroke. It is also the first prospective study considering the combination of multiple inflammatory SNPs in diabetic individuals.
The decision to study the rs1800795, rs5498, rs5361, rs1024611, and rs679620 of the IL-6, MCP-1, ICAM-1, sel-E, and MMP-3 genes was based on the following criteria. First, all five SNPs are functionally important. The rs1800795 is a SNP of the IL-6 promoter and influences the transcription rate of the gene and IL-6 plasma concentration (2). Similarly, the rs1024611 is a SNP of the promoter and affects the expression rate of the MCP-1 gene (1,4). Also the rs679620 of the MMP-3 gene is a SNP promoter and regulates transcription and protein levels in an allele-specific manner (5). Regarding the rs5498 of the ICAM-1 gene and the rs5361 of the sel-E gene, these SNPs lead to changes in the amino acid sequence of the corresponding proteins that result in modifications of their biological activity. In particular, the rs5498 of the ICAM-1 gene affects the Ig-like domain 5 of the ICAM-1 protein, which is crucial for the interactions with lymphocyte function-associated antige-1 (LFA-1) and the adhesion of B-cells (6). On the other hand, the rs5361 of the sel-E gene alters the ability of the E-sel protein to regulate leukocyte-endothelial interactions (3). Previous reports have implicated these SNPs in the pathobiology of atherosclerosis and stroke (6,19). We have previously reported that these SNPs are significantly and independently associated with history of ischemic stroke in a case-control scenario (10). Finally, we have shown that a combination of SNPs similar to that investigated in this study is associated with PAD and CLI (11). It should be noted that the synergistic and interdependent effects of these SNPs are mirrored by some important physiological interactions displayed by the corresponding proteins. For instance, MCP-1 is able to stimulate IL-6 secretion, and ICAM-1 synthesis is stimulated by MCP-1 in a time- and dose-dependent manner (20,21). On the other hand, ICAM-1 induces expression of several proinflammatory cytokines, including IL-6, which in turn induces the synthesis of various acute-phase proteins, thus maintaining and promoting the inflammatory phenotype (21,22). Taken together, these data indicate that the SNPs investigated in this study create a proinflammatory state that facilitates the development of atherosclerotic and ischemic disorders, such as stroke.
In a recent prospective study, the rs1800795 of the IL-6 gene has been associated with fasting glucose levels and development of diabetes (23). This raises the possibility that at least some of the SNPs composing our IRS affect the severity of diabetes. According to this hypothesis, increased risk of stroke might depend on subjects with high IRS having more severe diabetes. However, this hypothesis cannot be tested in the Go-DARTS study, because parameters of diabetes severity are not available. The analysis of our multigenic model in prospective cohorts in which outcomes of diabetes severity are available might address this issue.
None of the SNPs evaluated in our study have been associated with stroke in recent genome-wide association studies (GWASs) (24–27). Although this might appear disappointing, it is not inconsistent with our findings, if one considers that, in our study also, none of the five investigated SNPs was significantly and independently associated with incidence of stroke when considered alone. It was the combinatorial analysis of these five SNPs that provided evidence of their association with increased stroke risk, and no GWASs have so far investigated the possibility that this or similar proinflammatory multigenic models are associated with stroke or other cardiovascular diseases. In this respect, it will be interesting to test our multigenic model in existing GWAS databases, in order to understand whether individuals carrying multiple proinflammatory SNPs are more common in the groups of subjects with stroke than in control groups. This would also indicate that GWASs should take into consideration the fact that SNPs that fall below the threshold of statistical significance when considered alone might instead be associated with a disease when evaluated in combination. This concept suggests that future studies should use genome-wide data to evaluate specific types of functional associations within biologically relevant pathways, thus combining the unbiased approach of GWASs with the hypothesis-driven approach of candidate gene studies.
Further studies are also needed to understand if the addition of multigenic models to conventional risk factors improves measures of calibration, discrimination, and risk reclassification for cardiovascular events. We have performed a receiver operating characteristic analysis to compare the predictive value of our multigenic profile with the conventional risk profile for stroke and observed only a small increase (1.1%) in predictive value (supplemental Fig. 3). However, our current model is based on a limited number of functional gene variations centered around an inflammatory hypothesis, and it is highly unlikely that we have exhausted all the possible genetic contributory factors. In its current state, the IRS model represents a work in progress that may improve as additional alleles are added, but the model is, as currently formulated, predictive of future risk of stroke in subjects with type 2 diabetes. Further validation will be necessary, and its predictive ability in the general population and for additional cardiovascular indications remains to be tested, but this study suggests a viable strategy for developing genetic profiles at risk.
This study has some potential limitations. First, our results need to be replicated in additional cohorts. Second, we did not perform an analysis of the different subtypes of ischemic stroke. Therefore, it is possible that our multigenic model predicts the risk only of some stroke subtypes. Finally, we cannot exclude that the observed associations depend on the effect of genes in linkage disequilibrium with those investigated in our study.
In conclusion, we demonstrate that variations of genes encoding prototypical inflammatory molecules synergistically interact to determine proinflammatory genetic profiles that significantly influence the risk of stroke in subjects with type 2 diabetes. This multigenic model of stroke risk merits further investigation as a new, potential, independent predictor of individual risk for cardiovascular diseases.
Supplementary Material
ACKNOWLEDGMENTS
No potential conflicts of interest relevant to this article were reported.
C.N.A.P. researched data, contributed to the discussion, and wrote/reviewed/edited the manuscript. C.H.K., A.S.P., and A.D.M. researched data. A.S.F.D. wrote/reviewed/edited the manuscript. E.G. and R.C.S. contributed to the discussion and wrote/reviewed/edited the manuscript. M.Q. reviewed/edited the manuscript. R.P. designed the study and wrote/reviewed/edited the manuscript.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
See accompanying commentary, p. 2729.
REFERENCES
- 1.Jibiki T, Terai M, Shima M, Ogawa A, Hamada H, Kanazawa M, Yamamoto S, Oana S, Kohno Y. Monocyte chemoattractant protein 1 gene regulatory region polymorphism and serum levels of monocyte chemoattractant protein 1 in Japanese patients with Kawasaki disease. Arthritis Rheum 2001;44:2211–2212 [DOI] [PubMed] [Google Scholar]
- 2.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 1998;102:1369–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida M, Takano Y, Sasaoka T, Izumi T, Kimura A. E-selectin polymorphism associated with myocardial infarction causes enhanced leukocyte-endothelial interactions under flow conditions. Arterioscler Thromb Vasc Biol 2003;23:783–788 [DOI] [PubMed] [Google Scholar]
- 4.Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem Biophys Res Commun 1999;259:344–348 [DOI] [PubMed] [Google Scholar]
- 5.Ye S, Eriksson P, Hamsten A, Kurkinen M, Humphries SE, Henney AM. Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression. J Biol Chem 1996;271:13055–13060 [DOI] [PubMed] [Google Scholar]
- 6.Staunton DE, Dustin ML, Erickson HP, Springer TA. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell 1990;61:243–254 [DOI] [PubMed] [Google Scholar]
- 7.Roy H, Bhardwaj S, Yla-Herttuala S. Molecular genetics of atherosclerosis. Hum Genet 2009;125:467–491 [DOI] [PubMed] [Google Scholar]
- 8.Eerligh P, Koeleman BP, Dudbridge F, Jan Bruining G, Roep BO, Giphart MJ. Functional genetic polymorphisms in cytokines and metabolic genes as additional genetic markers for susceptibility to develop type 1 diabetes. Genes Immun 2004;5:36–40 [DOI] [PubMed] [Google Scholar]
- 9.Machado JC, Figueiredo C, Canedo P, Pharoah P, Carvalho R, Nabais S, Castro Alves C, Campos ML, Van Doorn LJ, Caldas C, Seruca R, Carneiro F, Sobrinho-Simões M. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology 2003;125:364–371 [DOI] [PubMed] [Google Scholar]
- 10.Flex A, Gaetani E, Papaleo P, Straface G, Proia AS, Pecorini G, Tondi P, Pola P, Pola R. Proinflammatory genetic profiles in subjects with history of ischemic stroke. Stroke 2004;35:2270–2275 [DOI] [PubMed] [Google Scholar]
- 11.Flex A, Gaetani E, Angelini F, Sabusco A, Chillà C, Straface G, Biscetti F, Pola P, Castellot JJ, Jr, Pola R. Pro-inflammatory genetic profiles in subjects with peripheral arterial occlusive disease and critical limb ischemia. J Intern Med 2007;262:124–130 [DOI] [PubMed] [Google Scholar]
- 12.Caruso C, Balistreri CR, Candore G, Carruba G, Colonna-Romano G, Di Bona D, Forte GI, Lio D, Listì F, Scola L, Vasto S. Polymorphisms of pro-inflammatory genes and prostate cancer risk: a pharmacogenomic approach. Cancer Immunol Immunother 2009;58:1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krabbe KS, Mortensen EL, Avlund K, Pilegaard H, Christiansen L, Pedersen AN, Schroll M, Jørgensen T, Pedersen BK, Bruunsgaard H. Genetic priming of a proinflammatory profile predicts low IQ in octogenarians. Neurobiol Aging 2009;30:769–781 [DOI] [PubMed] [Google Scholar]
- 14.Doney AS, Fischer B, Leese G, Morris AD, Palmer CN. Cardiovascular risk in type 2 diabetes is associated with variation at the PPARG locus: a Go-DARTS study. Arterioscler Thromb Vasc Biol 2004;24:2403–2407 [DOI] [PubMed] [Google Scholar]
- 15.Doney AS, Lee S, Leese GP, Morris AD, Palmer CN. Increased cardiovascular morbidity and mortality in type 2 diabetes is associated with the glutathione S transferase theta-null genotype: a Go-DARTS study. Circulation 2005;111:2927–2934 [DOI] [PubMed] [Google Scholar]
- 16.Doney AS, Fischer B, Lee SP, Morris AD, Leese G, Palmer CN. Association of common variation in the PPARA gene with incident myocardial infarction in individuals with type 2 diabetes: a Go-DARTS study. Nucl Recept 2005;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, Zeggini E, Rayner NW, Shields B, Owen KR, Hattersley AT, Frayling TM. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med 2006;3:e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinelli N, Trabetti E, Pinotti M, Olivieri O, Sandri M, Friso S, Pizzolo F, Bozzini C, Caruso PP, Cavallari U, Cheng S, Pignatti PF, Bernardi F, Corrocher R, Girelli D. Combined effect of hemostatic gene polymorphisms and the risk of myocardial infarction in patients with advanced coronary atherosclerosis. PLoS One 2008;3:e1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada Y, Ichihara S, Nishida T. Proinflammatory gene polymorphisms and ischemic stroke. Curr Pharm Des 2008;14:3590–3600 [DOI] [PubMed] [Google Scholar]
- 20.Viedt C, Dechend R, Fei J, Hänsch GM, Kreuzer J, Orth SR. MCP-1 induces inflammatory activation of human tubular epithelial cells: involvement of the transcription factors, nuclear factor-kappaB and activating protein-1. J Am Soc Nephrol 2002;13:1534–1547 [DOI] [PubMed] [Google Scholar]
- 21.Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochem J 1990;265:621–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SJ, Drabik K, Van Wagoner NJ, Lee S, Choi C, Dong Y, Benveniste EN. ICAM-1-induced expression of proinflammatory cytokines in astrocytes: involvement of extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways. J Immunol 2000;165:4658–4666 [DOI] [PubMed] [Google Scholar]
- 23.Vaxillaire M, Veslot J, Dina C, Proença C, Cauchi S, Charpentier G, Tichet J, Fumeron F, Marre M, Meyre D, Balkau B, Froguel PDESIR Study Group Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes 2008;57:244–254 [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Fuku N, Tanaka M, Aoyagi Y, Sawabe M, Metoki N, Yoshida H, Satoh K, Kato K, Watanabe S, Nozawa Y, Hasegawa A, Kojima T. Identification of CELSR1 as a susceptibility gene for ischemic stroke in Japanese individuals by a genome-wide association study. Atherosclerosis 2009;207:144–149 [DOI] [PubMed] [Google Scholar]
- 25.Karvanen J, Silander K, Kee F, Tiret L, Salomaa V, Kuulasmaa K, Wiklund PG, Virtamo J, Saarela O, Perret C, Perola M, Peltonen L, Cambien F, Erdmann J, Samani NJ, Schunkert H, Evans AMORGAM Project The impact of newly identified loci on coronary heart disease, stroke and total mortality in the MORGAM prospective cohorts. Genet Epidemiol 2009;33:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, Debette S, Lumley T, Folsom AR, van den Herik EG, Bos MJ, Beiser A, Cushman M, Launer LJ, Shahar E, Struchalin M, Du Y, Glazer NL, Rosamond WD, Rivadeneira F, Kelly-Hayes M, Lopez OL, Coresh J, Hofman A, DeCarli C, Heckbert SR, Koudstaal PJ, Yang Q, Smith NL, Kase CS, Rice K, Haritunians T, Roks G, de Kort PL, Taylor KD, de Lau LM, Oostra BA, Uitterlinden AG, Rotter JI, Boerwinkle E, Psaty BM, Mosley TH, van Duijn CM, Breteler MM, Longstreth WT, Jr, Wolf PA. Genomewide association studies of stroke. N Engl J Med 2009;360:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gschwendtner A, Bevan S, Cole JW, Plourde A, Matarin M, Ross-Adams H, Meitinger T, Wichmann E, Mitchell BD, Furie K, Slowik A, Rich SS, Syme PD, MacLeod MJ, Meschia JF, Rosand J, Kittner SJ, Markus HS, Müller-Myhsok B, Dichgans MInternational Stroke Genetics Consortium Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann Neurol 2009;65:531–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.