Abstract
OBJECTIVE
Type 2 diabetes and obesity are increasingly affecting human populations around the world. Our goal was to identify early molecular signatures predicting genetic risk to these metabolic diseases using two strains of mice that differ greatly in disease susceptibility.
RESEARCH DESIGN AND METHODS
We integrated metabolic characterization, gene expression, protein-protein interaction networks, RT-PCR, and flow cytometry analyses of adipose, skeletal muscle, and liver tissue of diabetes-prone C57BL/6NTac (B6) mice and diabetes-resistant 129S6/SvEvTac (129) mice at 6 weeks and 6 months of age.
RESULTS
At 6 weeks of age, B6 mice were metabolically indistinguishable from 129 mice, however, adipose tissue showed a consistent gene expression signature that differentiated between the strains. In particular, immune system gene networks and inflammatory biomarkers were upregulated in adipose tissue of B6 mice, despite a low normal fat mass. This was accompanied by increased T-cell and macrophage infiltration. The expression of the same networks and biomarkers, particularly those related to T-cells, further increased in adipose tissue of B6 mice, but only minimally in 129 mice, in response to weight gain promoted by age or high-fat diet, further exacerbating the differences between strains.
CONCLUSIONS
Insulin resistance in mice with differential susceptibility to diabetes and metabolic syndrome is preceded by differences in the inflammatory response of adipose tissue. This phenomenon may serve as an early indicator of disease and contribute to disease susceptibility and progression.
Type 2 diabetes and obesity are major causes of mortality and morbidity worldwide (1). According to World Health Organization estimates, more than 1 billion adults are overweight and more than 200 million individuals have type 2 diabetes. The etiologies of obesity and diabetes are complex and created by interactions between environmental factors (i.e., high caloric intake and reduced energy expenditure) (1,2) and genetic background (3–5).
Inbred mouse strains are useful models for studying the role of the environment and genes in differential susceptibility to diabetes and obesity (6–14). In response to high-fat diet, aging, or genetic challenge, C57BL/6NTac (B6) mice become severely insulin resistant, hyperinsulinemic, and diabetic, whereas 129S6/SvEvTac (129) mice are resistant to these challenges (7,8). Using intercross and F2 mice, our group and others have previously shown that the difference in disease susceptibility between the strains is inherited in a dominant fashion and linked to quantitative traits on at least three different chromosomes (8,15).
In the present study, we compared gene expression profiles of B6 and 129 mice in different tissues, ages, and diets. For each comparison, we identified the differentially expressed network of genes between the strains using a novel variant of gene network enrichment analysis (GNEA) (16), an algorithm that integrates gene expression data together with protein-protein interaction networks. We subsequently intersected the results of the comparisons between B6 and 129 mice to identify genes and pathways that were significant to each or all conditions, i.e., different tissues, ages, and diets. Of the numerous, differentially expressed subnetworks in the adipose tissue of B6 versus 129 mice; the most significant related to the immune system. Importantly, these differences were identified even at 6 weeks, an age when the mouse strains were indistinguishable by fat mass or metabolic phenotyping. RT-PCR and flow cytometry confirmed higher expression of major inflammatory markers and higher infiltration of macrophages and T-cells in adipose tissue of 6-week-old B6 versus 129 mice. Weight gain associated with aging or high-fat diet further increased inflammation in adipose tissue of B6 but not in 129, mice. Taken together, these results demonstrate that measurable differences in the inflammatory milieu of the adipose tissue precede measurable differences in insulin sensitivity in response to weight gain and contribute to the differences in diabetes risk between B6 and 129 mice.
RESEARCH DESIGN AND METHODS
C57BL/6NTac and 129S6/SvEvTac male mice were obtained from Taconic (Germantown, NY). Mice were maintained on a 12-h light-dark cycle with ad libitum access to tap water (reverse osmosis purified) and a chow diet containing 21% calories from fat, 23% from protein, and 55% from carbohydrates (Mouse Diet 9F; PharmaServ, Framingham, MA). For the feeding studies, 6-week-old mice were submitted to low-fat diet (Rodent NIH-31M Auto: 14% calories from fat, 25% from protein, and 61% from carbohydrates [Taconic]) or high-fat diet (TD.93075: 55% calories from fat, 21% calories from protein, and 24% calories from carbohydrates [Harlan Teklad, Madison, WI]) for 18 weeks before they were killed. In addition to fat content, these diets differ in other components that could also influence the results of our analysis. For example, our chow diet contained fat from animal sources, whereas both low-fat and high-fat diets contained vegetable fat. These differences may have contributed to the fact that a few metabolic parameters displayed in Table 1, such as insulin levels, did not necessarily track with the percentage of fat in the diet. However, in this study and in others, these diets have been primarily used as independent paradigms to investigate the impact of the environment on weight gain of animal models, showing metabolic effects that closely resembled different characteristics of human obesity (8). Caloric-restricted animals were obtained from the National Institute on Aging. Caloric restriction was initiated at 14 weeks of age with a 10% decrease in calories, increased to 25% restriction at 15 weeks, and to 40% restriction at 16 weeks, which was maintained until the age of 6 months when the mice were killed. Protocols for animal use were reviewed and approved by the Animal Care Committee of the Joslin Diabetes Center and were in accordance with the National Institutes of Health guidelines.
TABLE 1.
Metabolic parameters in B6 vs. 129 mice at different ages and dietary conditions
| Chow diet (21% fat) |
Low-fat diet (14% fat) |
High-fat diet (55% fat) |
||||||
|---|---|---|---|---|---|---|---|---|
| 6 weeks old |
6 months old |
|||||||
| B6 | 129 | B6 | 129 | B6 | 129 | B6 | 129 | |
| Body weight (g) | 21.9 ± 0.6 | 23.5 ± 0.5 | 45.5 ± 1.0 | 30.4 ± 1.2 | 41.5 ± 1.4 | 30.9 ± 1.2 | 46.4 ± 1.1 | 39.2 ± 1.5 |
| Epididymal fat mass (g) | 0.09 ± 0.01 | 0.21 ± 0.02 | 1.67 ± 0.14 | 2.14 ± 0.30 | 1.62 ± 0.17 | 0.58 ± 0.11 | 3.39 ± 0.33 | 3.92 ± 0.14 |
| Blood glucose– random fed (mg/dl) | 155.5 ± 3.1 | 129.3 ± 2.9 | 168.2 ± 12.3 | 106.6 ± 3.5 | 148.5 ± 7.4 | 102.5 ± 3.4 | 153.3 ± 6.7 | 105.5 ± 3.7 |
| Insulin (ng/ml) | 2.26 ± 0.48 | 3.12 ± 0.58 | 92.14 ± 25.71 | 3.96 ± 2.04 | 24.78 ± 12.51 | 1.29 ± 0.32 | 32.73 ± 12.46 | 3.58 ± 0.41 |
| Leptin (ng/ml) | 7.14 ± 0.70 | 9.00 ± 0.90 | 76.18 ± 3.82 | 16.67 ± 4.5 | 72.50 ± 8.06 | 4.97 ± 0.77 | 82.38 ± 7.41 | 46.81 ± 6.02 |
Values are expressed as mean ± SE of 8 animals per group.
Gene expression analysis.
RNA pooled from tissues of two or three mice was used for microarray analysis. Two to five Affymetrix murine chips U74Av.2 (Santa Clara, CA) were used per mouse strain, for each comparison of B6 versus 129 mice. Quantitative RT-PCR was performed using a SYBR Green-based ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). For details, refer to the supplementary methods in the online appendix available at http://diabetes.diabetesjournals.org/cgi/content/full/db10-0367/DC1.
Gene network enrichment analysis.
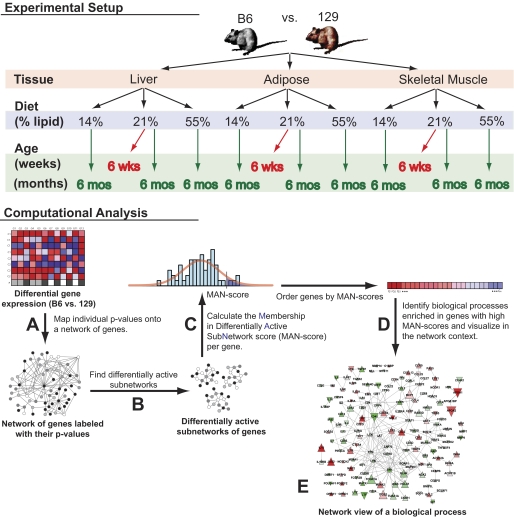
A novel version of Gene Network Enrichment Analysis (16) was applied to each comparison of B6 versus 129 mice to identify gene networks (subnetworks of genes with edges connecting physically interacting protein products) with differential activation between these mice, and to test such networks for statistically significant, over-represented pathways and processes. Briefly, the algorithm was composed of the following steps (see supplementary methods for details).
First, for each gene, a differential expression significance value was calculated in B6 versus 129 mice, converted to z scores, and used to annotate the corresponding proteins in a protein-protein interaction network curated from literature (17) (Fig. 1A).
FIG. 1.
Overview of gene network enrichment analysis. Individual comparisons of B6 and 129 mice were performed in different tissue types (adipose, skeletal muscle, and liver), ages (6 weeks and 6 months), and diets (high fat, low fat, and standard chow). A: For each comparison, a differential expression significance value was determined for each gene, converted to a z score, and mapped onto a network of protein-protein interactions. The colors in the network represent the different ranges of z scores. B: A stochastic search algorithm (18) was then applied to search for subnetworks with high mean z score (high significance). Each application of the algorithm identifies one subnetwork; hence, to detect a multitude of subnetworks, the algorithm was run many times. The consensus among these subnetworks represented approximations of the true, differentially active network in B6 vs. 129 mice. C: A membership in differentially active subnetwork (MAN) score was determined for each gene. MAN scores are P values and represent the probability of genes to belong to the true, differentially active network. This score was calculated as the random chance probability that the gene is detected in the observed number of subnetworks computed over permutations of the data. D: Biological processes and pathways were tested for enrichment in genes with high MAN scores. Statistically significant pathways and processes were predicted to be differentially active in B6 vs. 129 mice. E: For each significant process and pathway, a network visualization was generated to aid interpretation.
Subsequently, a computationally intensive, stochastic optimization algorithm (18) was employed to identify a subnetwork of genes with a high mean z score (reflecting high significance). This process was repeated many times to yield a multitude of subnetworks (Fig. 1B). The hypothesis was that a consensus among the stochastically detected subnetworks represented a prediction of the true, differentially active gene network in B6 versus 129 mice. Thus, each gene was associated with a P value based on the observed number of subnetworks in which it appeared, reflecting its chance probability of belonging to the true network (Fig. 1C).
In the next step of the algorithm, each biological process and pathway from the Gene Ontology and Molecular Signatures Database were tested for over-representation of genes with low P values, using standard statistical methods (Fig. 1D).
Finally, for each statistically significant pathway or biological process, the algorithm produced a gene network consisting of pathway member genes and all additional genes with significant, adjusted P values (false discovery rate ≤0.25) that shared an edge with a pathway member gene in the protein-protein interaction network (Fig. 1E), providing an intuitively appealing visualization aid that helped in interpreting the enrichment results.
Flow cytometry.
Erythrocyte-free stromavascular cells were isolated from epididymal fat pads after collagenase digestion as previously described (19). These cells were incubated with a mix of antibodies against different surface markers and analyzed using the LSRII flow cytometer (BD Biosciences, San Jose, CA). See supplementary METHODS for detailed protocol.
Statistics.
Statistical analyses for GNEA are described in supplementary methods. All other experiments were compared using ANOVA. Results are expressed as mean ± SEM. P values ≤ 0.05 were considered significant.
RESULTS
Gene network signatures associated with the predisposition to metabolic diseases in B6 versus 129 mice.
At 6 months of age, on a chow diet, B6 mice were overweight, hyperglycemic, and exhibited higher plasma leptin and insulin levels than 129 mice of the same age (Table 1, supplementary Fig. S1) (8). B6 mice showed clear signs of insulin resistance when compared with 129 mice, even on low-fat diet (LFD), and this difference was further exacerbated when the mice were placed on a high-fat diet (Table 1) (8). At 6 weeks of age, however, no phenotypic differences or serum markers indicative of diabetes and obesity could be detected between the two strains (Table 1) (8). Indeed, 6-week-old 129 mice have the same weight as B6 mice and actually have significantly more fat mass, as indicated by higher fat pad weight (Table 1). By studying B6 versus 129 mice at different ages and on different diets, one may therefore elucidate the background inherited phenomena predisposing to obesity-accompanied insulin resistance.
To identify gene networks that might provide early biomarkers of adult metabolic disease, we initially performed microarray analyses on RNA from adipose tissue, liver, and skeletal muscle of B6 and 129 mice at 6 weeks and 6 months of age on a standard chow diet. We then applied GNEA to detect networks that were differentially active in a predisease stage and to estimate how they changed over time in disease-prone versus diabetes-resistant strains (Fig. 1). Since disease susceptibility is an inherited phenomenon in these animals, we hypothesized that the predisposing hereditable biological processes would be those showing differential activity in young mice and maintained or further increased with age.
Among the three different tissues in which B6 versus 129 mice were compared, only adipose tissue exhibited significantly enriched biological processes (Q value <0.33) at 6 weeks of age by GNEA, with 12 enriched gene sets, and this increased to 25 enriched gene sets at 6 months of age. Of these, only two sets (“immune system process” and “regulation of cytokine secretion”) were found enriched at both ages (supplementary Table 1), and both of these related to inflammatory pathways. Furthermore, the “immune system process” was the most significantly enriched gene set at 6 months of age (Q value = 0.008; supplementary Table 1). In contrast, in the liver, 37 gene sets (Q value <0.33) were differentially regulated in B6 versus 129 at 6 months of age, but none was significantly different in the 6-week-old animals. In skeletal muscle, no significant difference was observed at either age (data not shown).
In addition to GNEA, the adipose tissue dataset was analyzed using gene set enrichment analysis (GSEA) (20) and the standard hypergeometric test (21). GSEA also identified differences in inflammation at 6 weeks. In particular, it found a significant difference in the “humoral immune response” biological process (supplementary Table 2). On the other hand, GSEA failed to identify any gene set that was differentially expressed at both ages (supplementary Table 3). By comparison, the standard hypergeometric test (21) failed to identify either a clear immune signature at 6 weeks or any enriched gene sets at 6 months (supplementary Tables 2 and 3).
Gene networks associated with the immune system process are differentially altered in the adipose tissue of B6 versus 129 mice.
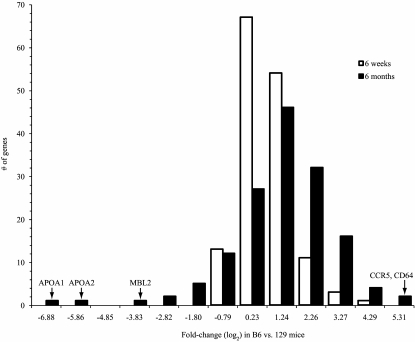
To determine how genes related to the immune response were differentially expressed in the fat tissue of young B6 and 129 mice and changed in expression as the mice aged, we identified the network associated with the “immune system process” gene set (supplementary Fig. S2) and plotted the histogram of differential expression for its genes in B6 versus 129 mice at 6 weeks and 6 months (Fig. 2). A few individual genes, such as apolipoprotein A (APOA1 and APOA2) and mannose-binding lectin (MBL2) were more strongly downregulated in B6 versus 129 mice at 6 months and less so at 6 weeks. Nonetheless, much of the immune system process network was upregulated at 6 months compared with 6 weeks (note the right-shift in the histogram for 6 months vs. 6 weeks in Fig. 2). Lymphoid and myeloid markers, such as CD45, CD3, and chemokine (C-C motif) receptor 5 (CCR5), monocyte chemotactic protein-1 (MCP1), as well as proinflammatory genes such as chemokine (C-C motif) ligand 5 (CCL5) and interleukin-6 (IL6), were more highly expressed in B6 compared with 129 mice at 6 weeks, and this increased further at 6 months of age (supplementary Fig. S2, left panel, arrows). Moreover, as shown by the histogram (Fig. 2), many of the immune system process genes already showed twofold or greater differences in expression in B6 versus 129 mice at 6 weeks, an age when no significant metabolic differences were observed between strains.
FIG. 2.
Histogram of differential expression between B6 and 129 mice at 6 weeks and 6 months of age for genes in the immune system process network. The x-axis values are the upper bounds of the histogram bins and correspond to the log2 fold-change difference in B6 vs. 129 mice. Few network genes are unchanged (absolute log2 fold change <1) in B6 vs. 129 mice at 6 months. The majority are strongly upregulated (log2 fold change ≥1), although a few are strongly downregulated (log2 fold change ≤ −1). A network view of the same biological process demonstrating the fold changes of individual genes is provided in supplementary Fig. S2.
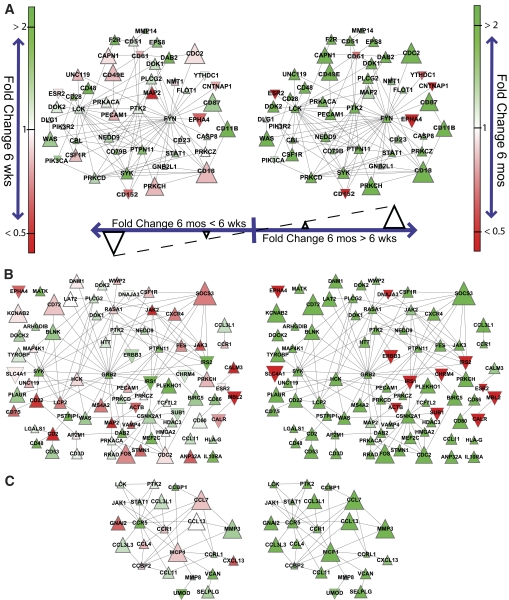
Genes that have protein products that physically interact tend to correlate in expression (22) and are likely related by function or membership in the same signaling pathways. We consequently examined the subnetworks of genes that interact at the protein level with inflammatory markers identified in the strain comparisons (Fig. 3). For the T-cell marker Thy1, minor changes in the expression of its interacting genes were observed in the 6-week-old B6 versus 129 mice comparisons (Fig. 3A, left panel). However, by 6 months of age, more than 90% of the nodes in the network were differentially active between the two strains (large green triangles in Fig. 3A, right panel). Hence, genes interacting with Thy1 were dramatically more upregulated with age-associated weight gain in adipose tissue of B6 mice than in 129 mice, suggesting that B6 mice have more T-cells recruited to adipose tissue in response to increasing adiposity than 129 mice. The same pattern was observed in subnetworks generated for CD45, a leukocyte marker (Fig. 3B); CD11c, a dendritic cell marker (supplementary Fig. S3A); CD11b, a macrophage marker (supplementary Fig. S3B); MCP1, a monocyte chemokine (Fig. 3C); and tumor necrosis factor α (TNFα), a proinflammatory cytokine (supplementary Fig. S3C). Together these data suggest that B6 mice show a proinflammatory tendency in fat at 6 weeks of age not observed in 129 mice, and that this differentially increases with weight gain promoted by aging, affecting the pool of inflammatory cells.
FIG. 3.
Gene networks associated with inflammatory markers in adipose tissue of B6 and 129 mice at 6 weeks (left) and 6 months (right) of age. Gene networks were generated by mapping genes that were significantly overrepresented (Q value < 0.25) among the gene network enrichment analysis results at 6 months intersected with those at 6 weeks onto protein-protein interaction networks involving at least one interactor to each inflammatory marker: (A) Thy1, (B) CD45, and (C) MCP1. The CD45 network was further restricted only to genes showing a greater than twofold magnitude difference in B6 vs. 129 mice between 6 weeks and 6 months. This was done to aid visualization since the unrestricted network consisted of 416 interactions and 141 genes. Colors range from bright red to green, corresponding to twofold less and twofold greater differences in expression between B6 and 129 at each age, respectively. Nodes point upward if the fold-change difference between B6 and 129 is greater at 6 months than at 6 weeks, and downward otherwise. The node size corresponds to the magnitude of that fold-change difference between the two ages. The inflammatory network around each biomarker is drawn to scale between the two ages. Networks around different biomarkers are not to scale with one another.
Changes in expression of inflammatory markers and cytokines/chemokines in adipose tissue of B6 versus 129 mice.
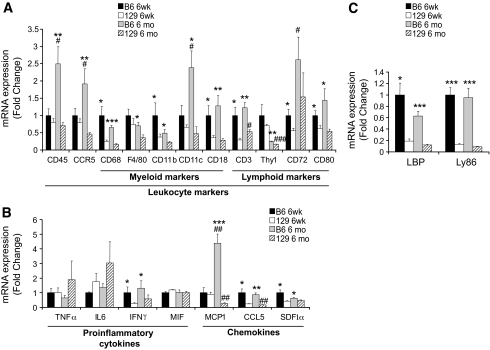
To validate the observations obtained using network analysis, we measured markers of inflammatory cells and cytokine/chemokine genes in the adipose tissue of B6 and 129 mice using quantitative RT-PCR (qPCR) (Fig. 4). In agreement with the microarray data, at 6 weeks of age, there were higher levels of macrophage/myeloid cell markers (CD68, CD11b, and CD18), lymphocyte markers (CD3, CD72, and CD80), T-cell attractants/activators CCL5/RANTES, stroma cell-derived factor-1α (SDF1α), and interferon-γ (IFNγ), among others, in B6 versus 129 mice (Fig. 4A and B). For most of these genes, the differences between strains were maintained or further exacerbated at 6 months of age. MCP1, a monocyte attractant chemokine, despite being similarly expressed in young B6 and 129 mice, exhibited a 4.4-fold increase in B6 mice between 6 weeks and 6 months of age, whereas in 129 mice, there was a 78% reduction with age (Fig. 4B). For a few cytokines, namely TNFα, IL6, and macrophage migration inhibitory factor (MIF), no differences were observed between the strains at any age (Fig. 4B).
FIG. 4.
Expression of inflammatory markers in adipose tissue of 6-week-old (6 weeks) or 6-month-old (6 months) B6 and 129 mice. A: Leukocyte markers, including selective myeloid cell markers (e.g., macrophages and dendritic cells) and lymphoid markers (e.g., lymphocytes). B: Cytokines and chemokines implicated with inflammatory responses. C: Mediators of LPS signaling. mRNA expression was assessed by qPCR. All values are normalized by TATA-binding protein (TBP) and expressed as fold change of the 6-week-old B6 average value. Results represent mean ± SEM of 5–7 animals. *P < 0.05; **P < 0.01; ***P < 0.001 vs. 129. #P < 0.05; ##P < 0.01; ###P < 0.001 vs. 6 weeks.
We also validated by RT-PCR the expression of selected genes identified as differentially present in the differentially active subnetworks identified by GNEA. Two mediators of lipopolysaccharide (LPS) signaling, i.e., the lymphocyte antigen 86 (Ly86) and LPS-binding protein (LBP), were highly ranked by GNEA and were substantially higher in B6 compared with 129 mice at both ages (Fig. 4C).
To determine if the higher expression of inflammation markers in B6 versus 129 mice was restricted to the adipose tissue or present in other tissues, we assessed the expression of two macrophage markers (CD68 and F4/80), two T-cell markers (CD3 and Thy1), and two chemokines (MCP1 and SDF1α) in the liver, skeletal muscle, and spleen of B6 and 129 mice at both ages using qPCR. The pattern of expression in liver was qualitatively similar to that in adipose tissue, but with smaller fold differences (supplementary Fig. S4A). In contrast, no differences were observed in the skeletal muscle or spleen in the expression for most of these genes (supplementary Fig. S4B and C). The two exceptions were MCP1, which was increased in all tissues in the 6-month-old B6 mice when compared with 129 mice, and CD3, which increased in both B6 and 129 mice with age in skeletal muscle only (supplementary Fig. S4).
An altered repertoire of inflammatory cells in the adipose tissue of B6 versus 129 mice.
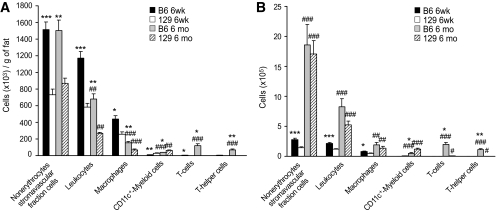
To determine if there was a cellular shift in the adipose tissue contributing to our computational and qPCR results, we analyzed the inflammatory cell repertoire in the fat tissue of B6 versus 129 mice using flow cytometry (Fig. 5). This revealed a significant twofold increase in the number of nonerythrocyte stromavascular fraction cells per gram of epididymal fat in B6 compared with 129 mice at both ages (Fig. 5A). At 6 weeks of age, 75% of this difference was explained by a higher number of leukocytes in B6 fat versus 129 fat. Although the number of leukocytes per gram of adipose tissue decreased in both strains with age as adipose mass increased, at both time points the number of leukocytes was about 2.5-fold higher in the stromavascular fraction from B6 fat than in 129 fat (Fig. 5A). Likewise, the number of macrophages per gram of fat decreased with age because of growing fat mass, but was higher in the B6 versus 129 mice at both ages (Fig. 5A). These data correlated with histologic evidence of crown-like structures, i.e., collections of macrophages surrounding dead or dying adipocytes, in the adipose tissue of 6-month-old B6 mice (supplementary Fig. S1).
FIG. 5.
Immune cell repertoire in adipose tissue of 6-week-old (6 weeks) or 6-month-old (6 months) B6 and 129 mice. Cells in the stromavascular fraction were labeled with fluorescence-conjugated antibodies against different myeloid cell and lymphoid markers and counted using flow cytometry. Nonerythrocytes stromavascular cells, PI-/Ter-119-; leukocytes, PI-/Ter-119-/CD45+; macrophages, PI-/Ter-119-/CD45+/CD11b+/F4/80+/CD11c-; CD11c+-Myeloid cells, PI-/Ter-119-/CD45+/CD11b+/F4/80+/CD11c+; T-cells, PI-/Ter-119-/CD45+/CD3+; T-helper cells, PI-/Ter-119-/CD45+/CD3+/CD4+. A: Represents number of cells per gram of fat tissue and (B) the total number of cells in the epididymal fat depot. All values are mean ± SEM of 4–8 animals. *P < 0.05; **P < 0.01; ***P < 0.001 vs. 129 mice. #P < 0.05; ##P < 0.01; ###P < 0.001 vs. 6 weeks.
Somewhat surprisingly, the density of CD11c+-myeloid cells, which have been previously associated with insulin resistance states in mice (23), was decreased in B6 versus 129 mice at 6 weeks and 6 months of age by 65 and 37%, respectively (Fig. 5A). In contrast, the T-cell population was dramatically increased by 7.1-fold in B6 mice at 6 weeks of age (Fig. 5A). Furthermore, the density of T-cells increased by 25-fold in B6 mice with age-associated weight gain, but showed only a threefold increase in 129 mice (Fig. 5A). Thus, at 6 months of age, B6 mice had >55-fold more total T-cells and >60-fold more CD4+ T-helper cells in the adipose tissue compared with the 129 mice (Fig. 5A). B-cells were also present in the adipose tissue of these mice; however, they were not considered in the analysis because of their modest numbers in comparison with other leukocytes (<1%).
A similar pattern of lymphocyte/monocyte infiltration was observed in the 6-week-old mice when the total number of cells in the epididymal fat pads was calculated, i.e., cells per gram × total grams of fat (Fig. 5B). When expressed as total cells, however, it was apparent that the number of each of the myeloid and lymphoid cell population increased with age in both strains. Importantly, the differences between strains were maintained in both younger and older mice (Fig. 5B). These results confirm the gene network and expression analysis and demonstrate that most of these differences are consistent with differences in the lymphoid/myeloid population in the adipose tissue of B6 versus 129 mice.
Effects of high-fat diet on the inflammatory status of the adipose tissue of B6 versus 129 mice.
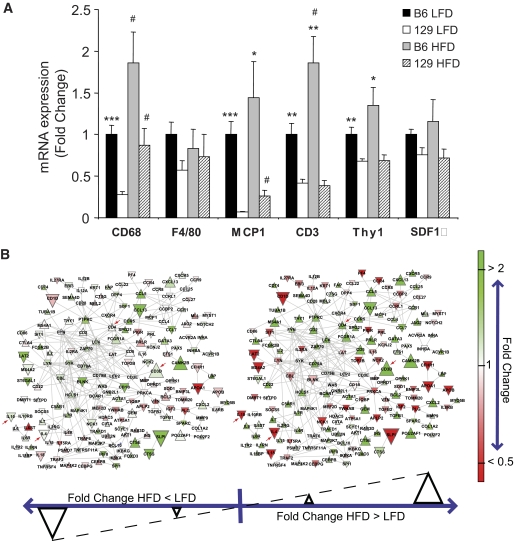
To investigate how differences in the genetic background could impact the effects of diet-induced obesity on the inflammatory cell repertoire of the adipose tissue, cohorts of B6 and 129 mice were placed on a high-fat diet (HFD) for 18 weeks and compared with mice on LFD. On both LFD and HFD, the mRNA expression of the macrophage marker CD68 was higher by twofold or greater in B6 compared with 129 mice (Fig. 6A). Similarly, MCP1 expression was higher in B6 than in 129 on LFD, and this was maintained with HFD (Fig. 6A). The macrophage marker F4/80 and SDF1α both trended higher in B6 than in 129 mice on LFD, but neither reached statistical significance (Fig. 6A). Interestingly, the HFD-induced increases in the expression of the T-cell markers CD3 and Thy1 were completely abrogated in the 129 mice (Fig. 6A). Caloric restriction, an intervention that drastically reduces fat mass, helps prevents age-associated diseases, and prolongs life span (24), decreased the expression of CD3 and Thy1 in the adipose tissue of B6 mice by 41 and 51%, respectively, without affecting the expression of macrophage markers (supplementary Fig. S5). These results demonstrate a positive correlation between adiposity and the expression of T- cells in adipose tissue of B6 mice, a phenomenon that is significantly impaired in 129 mice.
FIG. 6.
Expression of inflammatory markers in adipose tissue of 6-month-old B6 and 129 mice fed with LFD or HFD. A: Expression of mRNA was assessed by qPCR. All values are normalized by TBP and expressed as fold change of the average value of the LFD B6 mice. Results represent mean ± SEM of 4–6 animals. *P < 0.05; **P < 0.01; ***P < 0.001 vs. 129 mice. #P < 0.05 vs. LFD. B: The network view of the immune system process differences between B6 and 129 mouse strains on a LFD (left) and HFD (right). The gene network was generated by mapping genes that were significantly overrepresented in B6 vs. 129 mice (Q value <0.25) among the GNEA results on HFD intersected with those on LFD onto protein-protein interaction networks involving genes annotated with the immune system process gene set. Genes in red are more than twofold higher in 129 mice compared with B6 mice; genes in green are more than twofold higher in B6 mice compared with 129 mice. Genes are denoted by a downward triangle if the fold change of the HFD animals is less than those on the LFD, and by an upward triangle for the converse; the size of the triangle denotes the magnitude of the difference. Arrows represent genes with specific interest commented on in the main text.
Accordingly, analysis of B6 versus 129 mice under LFD and HFD using network analysis revealed a substantial number of genes in the “immune system process” gene set with higher expression in B6 compared with 129 mice, as evidenced by the large, bright green nodes in Fig. 6B. The majority of these genes had greater differences between B6 and 129 mice under HFD than under LFD conditions, as shown by the high number of upward triangles in Fig. 6. In particular, T-cell marker CD3 and the subnetwork around T-helper marker CD4 were among the most upregulated genes with HFD in B6 versus 129 mice (arrows in Fig. 6B). Conversely, anti-inflammatory cytokines, such as interleukin-4 (IL-4) and interleukin-10 (IL-10) were reduced after HFD in the B6 mice, but not in 129 mice (arrows in Fig. 6B).
DISCUSSION
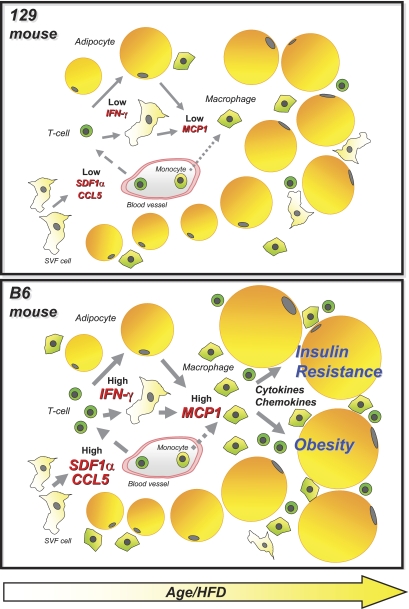
Although a relationship between obesity and inflammation in fat has been previously observed, in most cases these inflammatory changes have been viewed as being secondary to obesity. Our data, comparing diabetes-prone B6 mouse and diabetes-resistant 129 mouse at 6 weeks of age, show alterations in the inflammatory process in adipose tissue even before differences in metabolic parameters can be detected. Thus, B6 mice exhibit increased expression of the T-cell chemokines SDF1α and CCL5/RANTES and an increased number of T-cells in the fat tissue. These differences are associated with higher IFNγ and CD80 levels in the B6 mice—both molecules are known to participate in T-cell function and activation (25) (summarized in the model shown in Fig. 7).
FIG. 7.
Schematic model illustrating the potential causes and consequences related to the different repertoire of immune cells in adipose tissue of B6 and 129 mice. Solid arrows, secretion; dashed arrows, migration; dotted arrows, migration/differentiation.
Recent work has shown that T-cells infiltrate into the visceral adipose tissue of obese animals and humans with type 2 diabetes, and this is followed by recruitment of macrophages and development of insulin resistance (26–29). Based on these findings, a model for the role of T-cells in the pathogenesis of obesity and insulin resistance has been proposed in which increases in SDF1α and CCL5/RANTES levels in adipose tissue occur in response to an obesogenic environment and promote infiltration with T lymphocytes (29). IFNγ derived from these T-cells then promotes MCP1 secretion by preadipocytes (and possibly other cell types), resulting in recruitment of macrophages that further contribute to insulin resistance by production of proinflammatory cytokines (29).
Our study provides evidence to support a major impact of the genetic background of different mouse strains in the migration of T-cells to the adipose tissue, both in the basal state and in response to weight gain. In B6 mice, the number of T-cells in the adipose tissue correlates positively with the increase in adipose mass as a consequence of aging or HFD. By contrast, this response is practically absent in 129 mice despite a significant increase in adiposity in response to age or HFD. Thus, similar to mice with ablation of T-cells (27,28), 129 mice develop only mild insulin resistance in response to obesity. To what extent aging and the composition of the diet impact the infiltration of T-cells into adipose tissue, in addition to the effects of weight gain, remain to be determined. It is clear though that insulin resistance in response to increased adiposity differ substantially between mouse strains, and this phenomenon is correlated with the migration of T-cell to the adipose tissue. Thus, inflammation in the adipose tissue does not always correlate with weight gain and is strongly dependent on the genetic background of the host. Similar differences dependent on genetic background are likely to occur in humans and contribute to differences in obesity-induced diabetes risk in different ethnic groups or even different individuals, allowing for some of the “fat-fit” phenotype.
Among other implications, these observations clearly impact the choice of models for metabolic studies, in particular when knockout mice are used. Knockout mice are often derived from the embryonic stem cells of 129 mice, and therefore, in many cases, studies are performed on mice on a pure 129 background or mixed 129/B6 background. In these cases, backcrossing to B6 mice for several generations usually potentiates the inflammatory and metabolic response of the model to environmental factors. However, it is not clear which of these backgrounds (B6, 129, or mixed) most closely mimic the human condition.
Other recent reports support our findings that metabolic disease traits can be associated with alterations of inflammatory gene expression networks in the adipose tissue and liver (30,31). However, to our knowledge, no study has previously demonstrated a measurable difference in inflammation preceding any measurable phenotypic difference associated with metabolic diseases. The novelty of our study is that, by comparing young B6 and 129 animals using a sensitive computational approach, we can focus on factors that can potentially predispose to disease in a prospective manner and avoid findings that are mainly secondary to obesity or metabolic differences. Our investigation also examined differences between B6 and 129 mice in multiple organs, allowing us to conclude that inflammation in adipose tissue, and to a lesser extent in liver, but not in skeletal muscle or spleen, is associated with the predisposition to insulin resistance. In addition, our computational analysis was able to set forth hypotheses that led to subsequent biologic validation experiments which provided further insight into the components of the immune system that may contribute to metabolic diseases (i.e., T-cell recruitment).
In addition to inflammation, other phenomena have been shown to participate in the multifactorial pathophysiology of insulin resistance. Alterations in insulin receptor levels and insulin signaling through IRS-proteins (32), induction of the unfolded protein response (33) and oxidative stress pathways (34), and changes in lipid (35) and amino acid metabolism (36) can all promote insulin resistance and contribute to the final phenotype. Many of these pathways act by producing post-translational modifications of signaling proteins, such as phosphorylation or alterations in compartmentalization, which would not be detected as changes at the gene expression level. In this regard it is worth noting that in addition to differences in inflammation, GNEA analysis was able to identify other gene sets differentially expressed between the mouse strains in adipose tissue at 6 weeks of age (supplementary Table S2), including networks related to signal transduction, protein secretion pathways, and glucose catabolism. Many of these pathways can interact with inflammatory pathways, and this crosstalk could represent an entry point to the manifestation of metabolic diseases. Thus, although changes in the immune response are definitely one of the factors that precedes and predicts the tendency of B6 mice to have greater insulin resistance than 129, it is unlikely that inflammation is the only predisposing risk factor associated with diabetes between these two mouse strains.
In summary, it has been proposed that type 2 diabetes and obesity are diseases associated with an immune system that cannot cope appropriately with environmental threats (37–39). Based on this hypothesis, anti-inflammatory drugs, such as salicylates (40,41) and interleukin-1 blockers (42), have been used to improve glycemia in individuals with type 2 diabetes. In this study, we demonstrate that pre-existing differences in the inflammatory milieu in metabolically active tissues may represent an important component of the genetic background as a risk factor to metabolic diseases. Thus, inflammation cannot be viewed as only a mechanism that links susceptibility factors such as overfeeding, underactivity, aging, and stress to metabolic diseases—it may also link these pathologies to heritability. Our study indicates that inflammation is an important early variable in the metabolic response to environmental challenges and suggests several potential targets for intervention, including LBP, Ly86, SDF1α, CCL5/RANTES, and MCP1. This provides new strategies for reducing the epidemic of type 2 diabetes and metabolic diseases in spite of increasing obesity by attacking the variable genetic risk.
Supplementary Material
ACKNOWLEDGMENTS
M.A.M., O.B., K.A., and C.R.K. were supported by grants from the National Institutes of Health: DK-082659, DK-033201, and DK-060837 (Diabetes Genome Anatomy Project), as well as the Joslin Diabetes and Endocrinology Research Center cores (DK-036836) and the Mary K. Iacocca Professorship. The work of M.L. and S.K. was supported by grants from the National Human Genome Research Institute (R01 HG003367-01A1) and the National Institutes of Health (U54 LM008748) National Center for Biomedical Computing.
No potential conflicts of interest relevant to this article were reported.
M.A.M. and M.L. researched data, contributed to discussion, and wrote the manuscript. O.B. researched data and contributed to discussion. K.A. planned the study and researched data. H.S. researched data. S.K. researched data, contributed to discussion, and reviewed/edited the manuscript. C.R.K. planned the study, researched data, contributed to discussion, and reviewed/edited the manuscript.
The authors thank J. LaVecchio, G. Buruzula, and the flow cytometry core at the Joslin Diabetes Center for help with cytometry; J. Schroeder and the genomics core at the Joslin Diabetes Center for help with microarray processing; M. Rourk, Joslin Diabetes Center, for his expertise in animal care; and S.E. Shoelson, MD, PhD, Joslin Diabetes Center, for his helpful advice and discussion.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world—a growing challenge. N Engl J Med 2007;356:213–215 [DOI] [PubMed] [Google Scholar]
- 2.Lazar MA. How obesity causes diabetes: not a tall tale. Science 2005;307:373–375 [DOI] [PubMed] [Google Scholar]
- 3.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab 2008;8:186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644 [DOI] [PubMed] [Google Scholar]
- 5.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet 2009;10:431–442 [DOI] [PubMed] [Google Scholar]
- 6.Almind K, Kulkarni RN, Lannon SM, Kahn CR. Identification of interactive loci linked to insulin and leptin in mice with genetic insulin resistance. Diabetes 2003;52:1535–1543 [DOI] [PubMed] [Google Scholar]
- 7.Kulkarni RN, Almind K, Goren HJ, Winnay JN, Ueki K, Okada T, Kahn CR. Impact of genetic background on development of hyperinsulinemia and diabetes in insulin receptor/insulin receptor substrate-1 double heterozygous mice. Diabetes 2003;52:1528–1534 [DOI] [PubMed] [Google Scholar]
- 8.Almind K, Kahn CR. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes 2004;53:3274–3285 [DOI] [PubMed] [Google Scholar]
- 9.Haluzik M, Colombo C, Gavrilova O, Chua S, Wolf N, Chen M, Stannard B, Dietz KR, Le RD, Reitman ML. Genetic background (C57BL/6J versus FVB/N) strongly influences the severity of diabetes and insulin resistance in ob/ob mice. Endocrinology 2004;145:3258–3264 [DOI] [PubMed] [Google Scholar]
- 10.Bachmanov AA, Reed DR, Tordoff MG, Price RA, Beauchamp GK. Nutrient preference and diet-induced adiposity in C57BL/6ByJ and 129P3/J mice. Physiol Behav 2001;72:603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossmeisl M, Rim JS, Koza RA, Kozak LP. Variation in type 2 diabetes-related traits in mouse strains susceptible to diet-induced obesity. Diabetes 2003;52:1958–1966 [DOI] [PubMed] [Google Scholar]
- 12.West DB, York B. Dietary fat, genetic predisposition, and obesity: lessons from animal models. Am J Clin Nutr 1998;67:505S–512S [DOI] [PubMed] [Google Scholar]
- 13.Leiter EH, Lee CH. Mouse models and the genetics of diabetes: is there evidence for genetic overlap between type 1 and type 2 diabetes? Diabetes 2005;54(Suppl. 2):S151–S158 [DOI] [PubMed] [Google Scholar]
- 14.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, Ilkayeva OR, Keller MP, Blasiole DA, Kendziorski C, Yandell BS, Newgard CB, Attie AD. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet 2008;4:e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt C, Gonzaludo NP, Strunk S, Dahm S, Schuchhardt J, Kleinjung F, Wuschke S, Joost HG, Al-Hasani H. A meta-analysis of QTL for diabetes-related traits in rodents. Physiol Genomics 2008;34:42–53 [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Liberzon A, Kong SW, Lai WR, Park PJ, Kohane IS, Kasif S. Network-based analysis of affected biological processes in type 2 diabetes models. PLoS Genet 2007;3:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database–2009 update. Nucleic Acid Res 2009;37:D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics 2002;18(Suppl. 1):S233–S240 [DOI] [PubMed] [Google Scholar]
- 19.Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A 2006;103:6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher RA. On the interpretation of χ-square from contingency tables, and the calculation of P. J Royal Statistical Society 1922;85:87–94 [Google Scholar]
- 22.Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet 2001;29:482–486 [DOI] [PubMed] [Google Scholar]
- 23.Nguyen MT, Favelyukis S, Nguyen AK, Reichart D, Scott PA, Jenn A, Liu-Bryan R, Glass CK, Neels JG, Olefsky JM. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem 2007;282:35279–35292 [DOI] [PubMed] [Google Scholar]
- 24.Fontana L, Partridge L, Longo VD. Extending healthy life span–from yeast to humans. Science 2010;328:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedman AS, Freeman GJ, Rhynhart K, Nadler LM. Selective induction of B7/BB-1 on interferon-γ stimulated monocytes: a potential mechanism for amplification of T cell activation through the CD28 pathway. Cell Immunol 1991;137:429–437 [DOI] [PubMed] [Google Scholar]
- 26.Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, Mathis D. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med 2009;15:930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, Dorfman R, Wang Y, Zielenski J, Mastronardi F, Maezawa Y, Drucker DJ, Engleman E, Winer D, Dosch HM. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med 2009;15:921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, Otsu M, Hara K, Ueki K, Sugiura S, Yoshimura K, Kadowaki T, Nagai R. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med 2009;15:914–920 [DOI] [PubMed] [Google Scholar]
- 29.Kintscher U, Hartge M, Hess K, Foryst-Ludwig A, Clemenz M, Wabitsch M, Fischer-Posovszky P, Barth TF, Dragun D, Skurk T, Hauner H, Bluher M, Unger T, Wolf AM, Knippschild U, Hombach V, Marx N. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol 2008;28:1304–1310 [DOI] [PubMed] [Google Scholar]
- 30.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature 2008;452:423–428 [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, Macneil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature 2008;452:429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 1997;88:561–572 [DOI] [PubMed] [Google Scholar]
- 33.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 2004;306:457–461 [DOI] [PubMed] [Google Scholar]
- 34.Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 2006;440:944–948 [DOI] [PubMed] [Google Scholar]
- 35.Boden G. Role of fatty acids in the pathogenesis of insulin resistance and NIDDM. Diabetes 1997;46:3–10 [PubMed] [Google Scholar]
- 36.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Real JM, Pickup JC. Innate immunity, insulin resistance and type 2 diabetes. Trends Endocrinol Metab 2008;19:10–16 [DOI] [PubMed] [Google Scholar]
- 38.Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol 2008;8:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest 2008;118:2992–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldfine AB, Silver R, Aldhahi W, Cai D, Tatro E, Lee J, Shoelson SE. Use of salsalate to target inflammation in the treatment of insulin resistance and type 2 diabetes. Clin Transl Sci 2008;1:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hundal RS, Petersen KF, Mayerson AB, Randhawa PS, Inzucchi S, Shoelson SE, Shulman GI. Mechanism by which high-dose aspirin improves glucose metabolism in type 2 diabetes. J Clin Invest 2002;109:1321–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, Mandrup-Poulsen T, Donath MY. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med 2007;356:1517–1526 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.