Abstract
OBJECTIVES
Several transcription factors determine the cell fate decision between granulocytes and monocytes, but the upstream signal transduction pathways that govern myelopoiesis are largely unknown. Based on our observation of aberrant myeloid cell representation in hematopoietic tissues of 12/15-lipoxygenase (12/15-LOX)-deficient (Alox15) mice, we tested the hypothesis that polyunsaturated fatty acid metabolism regulates myelopoiesis.
METHODS
Multi-color flow cytometric analysis and methylcellulose assays were used to compare myelopoiesis and the differentiative capacity of progenitors from Alox15 and wild-type mice. Furthermore, we elucidated the mechanism by which 12/15-LOX is involved in regulation of myelopoiesis.
RESULTS
Granulopoiesis in Alox15 mice is increased while monopoiesis is reduced. Moreover, there is an accumulation of granulocyte-macrophage progenitors that exhibit defective differentiation. Mechanistically, we demonstrate that transcriptional activity of Irf8, which regulates myelopoiesis, is impaired in Alox15 progenitors and bone marrow-derived macrophages due to loss of 12/15-LOX-mediated redox regulation of Irf8 nuclear accumulation. Restoration of redox signaling in Alox15 bone marrow cells and GMP reversed the defect in myeloid differentiation.
CONCLUSIONS
These data establish 12/15-LOX-mediated redox signaling as a novel regulator of myelopoiesis and Irf8.
Introduction
Myelopoiesis is the process whereby myeloid cells, granulocytes and monocytes, are generated from hematopoietic stem cells (HSCs). Progression along this differentiation pathway gives rise to cells that are increasingly restricted in their potential fate as they commit to various lineages. Although phenotypic profiles can be used to distinguish hematopoietic progenitors at various stages of differentiation, hematopoiesis occurs along a continuum and phenotypically defined progenitors exhibit heterogeneity in their ability to differentiate to alternative lineages.
Differentiation of granulocyte-macrophage progenitors (GMPs) into granulocytes or monocytes(1) is a mutually exclusive cell fate decision. Coordinated actions of cytokine signaling and transcription factors promote one cell fate while inhibiting the alternative cell fate(2). The transcription factor Interferon Regulatory Factor-8 (Irf8), also known as Interferon Consensus Sequence Binding Protein (ICSBP), is required for monocytic differentiation. Irf8 in conjunction with PU.1 activates gene transcription that promotes monocytic differentiation while inhibiting gene transcription that promotes granulocytic differentiation(3). Loss of Irf8 transcriptional activity results in accumulation of GMP(4), defective monocytic development and enhanced granulocytic differentiation(5, 6). We previously demonstrated that Irf8 nuclear accumulation requires 12/15-lipoxygenase (12/15-LOX)-mediated signaling in macrophages and splenocytes(7,8). However, the role of 12/15-LOX in myelopoiesis and the mechanism whereby 12/15-LOX promotes nuclear accumulation of Irf8 has not been elucidated.
The signaling pathways that regulate myeloid cell specification remain under active investigation. In this study, we demonstrate that 12/15-LOX-dependent signaling regulates myelopoiesis. 12/15-LOX is an oxidative enzyme that mediates polyunsaturated fatty acid metabolism of substrates such as arachidonic (AA) and lineolic acids (LA). 12/15-LOX-mediated reactions generate bioactive lipid metabolites, including 12(S)-hydroxyeicosatetraenoic acid (12(S)-HETE), 15(S)-HETE, 13(S)-HODE, lipoxins, and hepoxilins(9). ROS generated as byproducts of 12/15-LOX can function as second messengers by modifying thiol groups in cysteine residues. Oxidative modifications regulate the activity of proteins including phosphatases, transcription factors and signal transducers(10). However, little is known of the consequences of 12/15-LOX signaling during myelopoiesis.
In view of our observations of reduced numbers of monocytes(11) and increased percentages of granulocytes(7) in 12/15-LOX-deficient (Alox15) mice, we hypothesized that 12/15-LOX regulates myeloid cell fate decisions. Herein, we establish that lack of 12/15-LOX in Alox15 mice results in accumulation of myeloid progenitors due to defective differentiation which is skewed towards the granulocyte lineage. Loss of 12/15-LOX-dependent redox signaling in part mediates nuclear accumulation of Irf8 to drive monopoiesis and inhibit granulopoiesis.
Materials and Methods
Animals
C57BL/6 (B6) and Alox15 8–10 week mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA), and bred in the Wistar Institute. All procedures were approved by the Institutional Animal Care and Use Committee of the Wistar Institute.
Flow Cytometry
Single cell suspensions were prepared and RBC lysed using ammonium chloride. Immunocytochemistry reagents included: cKit (Invitrogen, Carlsbad, CA, USA), streptavidin-conjugates (BD Bioscience, San Jose, CA, USA), all others (eBioscience, San Diego, CA, USA). Lineage markers included antibodies against: Gr-1, B220, CD3, IL-7Rα, Ter119, NK1.1, Mac1 and CD11c. Flow cytometric analysis was performed on FACSCalibur or LSR II and analyzed using Flowjo software (Treestar, Ashland, OR, USA). For cell sorting, BM cells from 2–4 mice were pooled, stained and sorted on an Aria.
BMM- 1×106 BM cells/ml was cultured in 10%FCS, 10%L929 supernatant in RPMI on non-tissue culture-treated plates. After 5 days, adherent cells were isolated. At least 90% of the cells were F4/80+ by flow cytometric analysis
Immunoblot and Quantitative PCR (Q-PCR)
Lin−cKit+ cells were obtained using MACS kit according to manufacturer’s instructions (Miltenyi Biotech, Auburn, CA, USA). BMM were collected, counted and re-plated in the presence or absence of Tiron (Mallinckrodt Baker, Inc., Phillipsburg, NJ, USA), or BSO (Sigma-Aldrich, St. Louis, MO, USA). All experiments were performed under steady state conditions. Nuclear extracts were prepared using Nucbuster kit (EMD Biosciences, San Diego, CA, USA). Lysates were normalized using Bradford assay (Pierce, Rockford, IL, USA), resolved by 7.5% SDS-PAGE, transferred to PVDF membranes, and immunoblotted with antibodies specific for Irf8, actin or Rb (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and secondary antibodies (Jackson Immunoresearch, West Grove, PA, USA). RNA was isolated from BMM using Trizol (Invitrogen, Carlsbad, CA, USA) and treated with turbo DNAse (Ambion, Austin, TX, USA). RNA was isolated from enriched progenitors using RNeasy microRNA kit (Qiagen, Valcenia, CA, USA). cDNA was synthesized using reverse transcriptase, and Q-PCR was performed by normalizing to GAPDH using syber green master mix on ABI7000 cycler (Applied Biosystems, Foster City, CA, USA). For primer sequences see supplemental methods.
ROS and Lipid Analysis
ROS levels were determined by loading with 10 μM CM-H2DCFDA (Invitrogen, Carlsbad, CA, USA) for 30min at 37°C and analyzed by flow cytometry. For lipid analysis, BMM were loaded for 30min with 50 μM AA (Cayman Chemical, Ann Arbor, MI, USA). Supernatants were extracted and analyzed by stable isotope dilution normal phase chiral liquid chromatography coupled with electron capture atmospheric pressure chemical ionization/mass spectrometry.
Methylcellulose Assays
1.5×105 splenocytes, 1.5×104 bone marrow cells, 5×102 LSK, 5×102 CMP and 103 GMP were plated in methylcellulose containing IL-3, IL-6, SCF and EPO (M3434, Stem Cell Technologies). 5×105 BM cells or 103 GMP were also plated in methylcellulose (M3234, Stem Cell Technologies, Vancouver, BC, Canada) with 10 ng/ml M-CSF, G-CSF or GM-CSF (Peprotech, Rocky Hill, NJ, USA). After 10 days, colonies were enumerated using light microscopy in a blinded fashion.
Statistical Analysis
Student’s t-tests were applied using Excel.
Results
Alox15 progenitors exhibit excess granulopoiesis and reduced monopoiesis
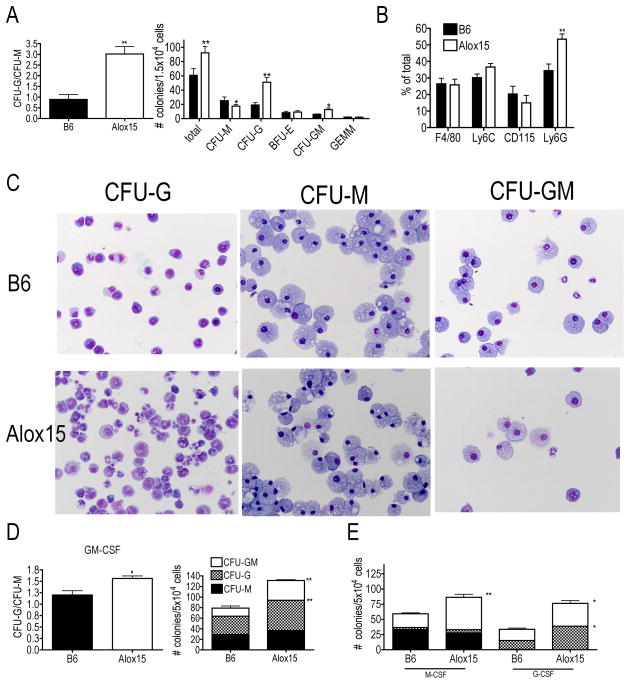
We confirmed the decrease in Alox15 blood monocytes that we previously reported(11). While the numbers of granulocytes were similar between B6 and Alox15 mice, there was a decrease in WBC number resulting in an increased percentage of granulocytes (Supplemental Figure 1A, B) (7). Moreover, we also confirmed the increased percentage of granulocytes in the Alox15 splenocytes (Supplemental Figure 1C) (7). 12/15-LOX was expressed in myeloid progenitors (Supplemental Figure 1D), therefore we hypothesized that the decrease in monocytes and increased percentage of granulocytes in Alox15 mice is due to defective myelopoiesis. To test this, we plated cells from B6 and Alox15 mice in methylcellulose assays containing IL-6, IL-3, stem cell factor (SCF) and erythropoietin (Epo) and compared the frequency of colony-forming units-granulocyte (CFU-G) to that of CFU-macrophage (CFU-M). We calculated the ratio of CFU-G/CFU-M to assess skewing towards differentiation independently of total colony number. BM cells from Alox15 mice generated an increased ratio of CFU-G/CFU-M compared to B6 (Figure 1A). Alox15 BM gave rise to an increased number of total colonies, due to an increased number of CFU-G and CFU-granulocyte-macrophage (CFU-GM), but a decreased number of CFU-M compared to B6 (Figure 1A). We also determined the percentage of cells recovered from whole methylcellulose plates that express monocyte-specific markers (F4/80, Ly6C, CD115) and granulocyte-specific marker Ly6G. The percentage of cells that expressed monocyte markers was similar between B6 and Alox15 methylcellulose plates (Figure 1B) likely because of the increase in Alox15 CFU-GM. However, the percentage of cells that expressed the granulocyte-specific marker Ly6G was increased in Alox15 methylcellulose plates, confirming the increase in granulocyte differentiation (Figure 1B). Methylcellulose assays with Alox15 splenocytes gave similar results (Supplemental Figure 1E). Moreover, we confirmed colony identifications by Wright-Giemsa stain of cytospins of representative colonies (Figure 1C). Interestingly, the CFU-G colonies from Alox15 mice contained immature granulocytic cells while the CFU-M colonies appeared similar to B6. These data demonstrate that absence of 12/15-LOX results in skewing of myelopoiesis towards aberrant granulocytic development.
Figure 1.
Enhanced granulopoiesis at the expense of monopoiesis of Alox15 progenitors in methylcellulose assays. (A) B6 and Alox15 BM cells were plated in methylcellulose in the presence of IL-3, IL-6, SCF and Epo. After 10 days, colonies were enumerated using light microscopy. Shown are the ratio of CFU-G/CFU-M and the numbers of types of identifiable colonies. (B) Cells from methylcellulose plates were recovered and analyzed for monocytic markers, Ly6C, F4/80 and CD115, and the granulocytic marker Ly6G by flow cytometry. (C) Wright-Giemsa stain from cytospin of representative B6 and Alox15 methylcellulose colonies. 20X magnification. (D) B6 and Alox15 BM cells were cultured in methylcellulose in the presence of GM-CSF. Shown are a summary of three independent experiments as a ratio of CFU-G/CFU-M and numbers of colonies CFU-M (black), CFU-G (checkered) and CFU-GM (white) (E) B6 and Alox15 BM cells were cultured in methylcellulose in the presence of M-CSF or G-CSF as indicated on the x-axis. Shown are numbers and phenotypes of colonies. n=6 in 3 independent experiments for all data shown. *p<.05, **p<.01. Error bars represent ± Standard Error of the Mean (SEM).
Similarly, Alox15 BM cells grown in the presence of GM-CSF were also skewed towards granulocyte development as indicated by an increased CFU-G/CFU-M ratio (Figure 1D). The skewing towards granulocyte development in the presence of GM-CSF was a result of increased numbers of Alox15 CFU-G colonies. Moreover, Alox15 CFU-GM was increased in the presence of GM-CSF (Figure 1D). These data provide additional evidence for enhanced granulopoiesis in Alox15 mice.
To determine the effect of instructive cytokines, we plated B6 and Alox15 BM cells in methylcellulose in the presence of M-CSF or G-CSF. Interestingly, cells from Alox15 BM gave rise to an increased number of total colonies in the presence of both M-CSF and G-CSF compared to cells from B6 BM. However, in the presence of M-CSF, Alox15 BM cells generated more CFU-GM, but not CFU-M. In the presence of G-CSF, Alox15 BM cells gave rise to an increased number of both CFU-GM and CFU-G compared to B6 BM cells (Figure 1E). Taken together, these data demonstrate that Alox15 BM cells exhibit an increased frequency of CFU-GM and enhanced granulocytic differentiation.
The increased number of total myeloid colonies that developed from Alox15 cells suggested that there are increased numbers of myeloid progenitors present. Specifically, the increase in CFU-GM in the presence of all cytokines, suggested that the GMP population is increased in Alox15 mice.
Accumulation of defective GMP in Alox15 mice
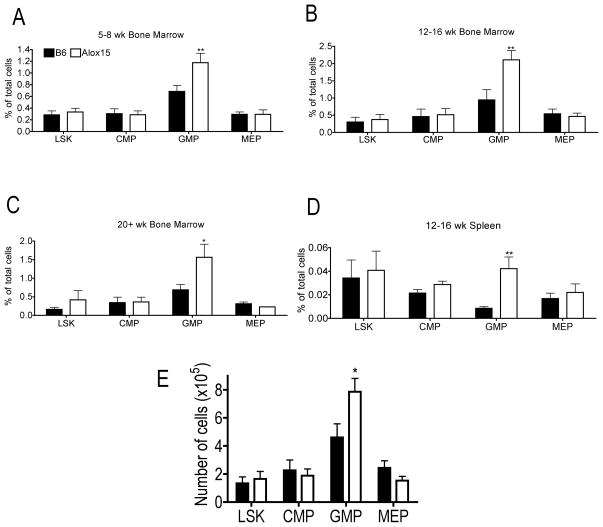
We employed multicolor flow cytometric analysis to determine the numbers of phenotypic progenitor populations. At all ages tested, Alox15 BM had similar percentages of Lin-Sca1+cKit+ cells (LSK), Common Myeloid Progenitors (CMP) and Megakaryocyte Erythroid Progenitors (MEP) but an increased percentage of GMP compared to B6 (Figure 2A–C). The increased percentage of GMP was also evident in Alox15 spleen (Figure 2D). In addition to an increase in relative percentage, the absolute frequency of Alox15 GMP was increased (Figure 2E).
Figure 2.
Accumulation of GMP in Alox15 mice. BM cells from 5–8 week (A), 12–16 week (B, E), 20+ week (C) and spleens from 12–16 week (D) old B6 and Alox15 mice were isolated and subjected to multicolor flow cytometric analysis to determine the relative frequency (A–D) and absolute number (E) of myeloid progenitors in tibias and fibulas of each mouse: LSK (Lin−Sca1+cKit+), CMP (Lin−Sca1−cKit+CD34+CD16/32lo), GMP (Lin−Sca1−cKit+CD34+CD16/32hi), and MEP (Lin−Sca1−cKit+CD34−CD16/32−) n=3 independent experiments for all data shown. *p<.05, **p<.01. Error bars represent ± SEM.
There are several potential explanations for the increased number of GMP progenitors in Alox15 mice. First, CMP may produce more GMP at the expense of MEP. However, this is not likely as there were similar numbers of erythroid progenitors (BFU-E) produced from Alox15 BM (Figure 1A), and similar numbers of MEP in B6 and Alox15 mice (Figure 2). Second, Alox15 GMP may expand by proliferating and/or through enhanced survival. Alternatively, Alox15 GMP differentiation may be decreased resulting in accumulation of GMP as they fail to progress from this differentiative stage.
To determine whether Alox15 GMP exhibit enhanced proliferation, we performed cell cycle analysis ex vivo which was comparable between B6 and Alox15 GMP (Supplemental Figure 2A). Moreover, cell survival was similar between B6 and Alox15 GMP as measured by the percentage of cells binding Annexin V ex vivo and after stimulation with SCF in vitro (Supplemental Figure 2B–C). Therefore, Alox15 GMP are not increased due to enhanced proliferation or cell survival.
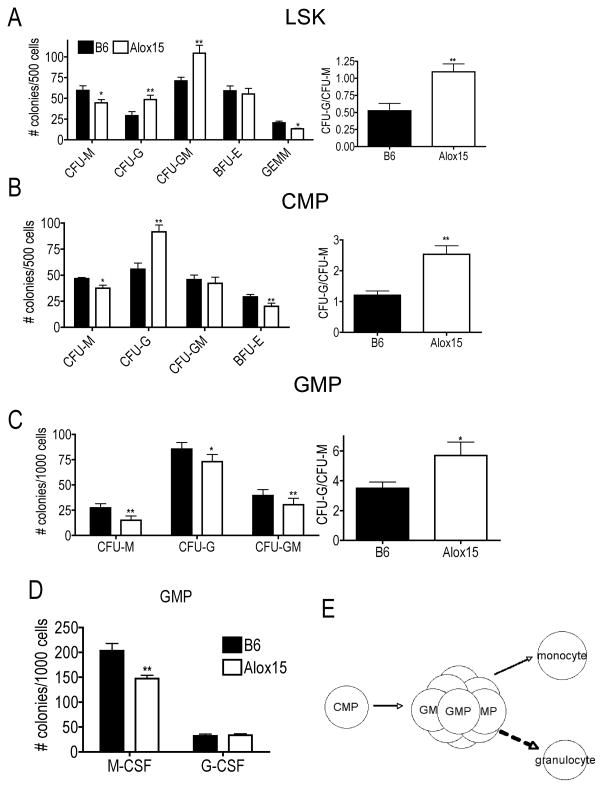
To test whether GMP are accumulated in Alox15 mice due to a decreased differentiation, we isolated enriched populations of LSK, CMP and GMP from B6 and Alox15 BM by cell sorting prior to methylcellulose assays. Similar to whole BM cells, Alox15 LSK and CMP generated increased numbers of CFU-G but decreased numbers of CFU-M compared to B6 resulting in increased ratios of CFU-G/CFU-M compared to B6 (Figure 3A–B).
Figure 3.
Alox15 GMP exhibit defective differentiation. B6 and Alox15 BM were isolated and sorted for myeloid progenitors. LSK (A), CMP (B) and GMP (C) were plated in methylcellulose in the presence of IL-3, IL-6, SCF and Epo. Shown are a summary of three experiments as colonies enumerated and the CFU-G/CFU-M ratio. (D) B6 and Alox15 GMP were plated in the presence of M-CSF or G-CSF in methylcellulose assays. n=6 in three independent experiments for all data shown*p<.05, **p<.01 Error bars represent ± SEM. (E) Diagram of GMP differentiation in Alox15 mice. There is an accumulation of Alox15 GMP which exhibit reduced monocyte differentiation and enhanced granulopoiesis.
On the other hand, GMP from Alox15 mice exhibit defective differentiation by generating fewer total colonies, significantly fewer CFU-M and subtle decreases in CFU-G and CFU-GM. There were no differences in the percentages of myeloid markers (Ly6C, Ly6G, CD115, F4/80) by flow cytometry likely due to the decrease in all colony types (data not shown). In spite of the subtle decrease in CFU-G, the ratio of CFU-G/CFU-M that arose from Alox15 GMP was increased compared to B6 (Figure 3C). To further analyze the defective differentiation, we directed differentiation of B6 and Alox15 GMP with the instructive cytokines M-CSF or G-CSF. B6 and Alox15 GMP produced similar numbers of granulocytic colonies in response to G-CSF demonstrating that Alox15 GMP did not exhibit defective differentiation or cytokine hypersensitivity to G-CSF per se. However, Alox15 GMP gave rise to fewer monocytic colonies in response to M-CSF than GMP from B6 mice (Figure 3D). Therefore, these data illustrate that Alox15 GMP exhibit reduced monocytic differentiation when directed by instructive cytokines that may in part underlie Alox15 GMP accumulation. In addition, the presence of immature granulocytes in CFU-G from Alox15 BM and the reduction of all colonies from GMP further support reduced differentiation of Alox15 GMP. Therefore, lack of 12/15-LOX results in an accumulation of GMP, reduced numbers of monocytes and increased numbers of mature and immature granulocytes (Figure 3E). 12/15-LOX regulates myelopoeisis by promoting GMP differentiation to monocytes and preventing the aberrant granulopoiesis that occurs in its absence.
Redox regulation of IRF-8 nuclear accumulation
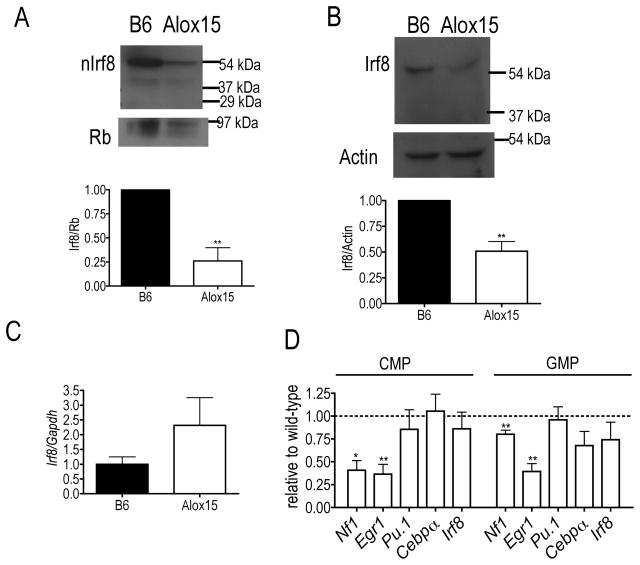
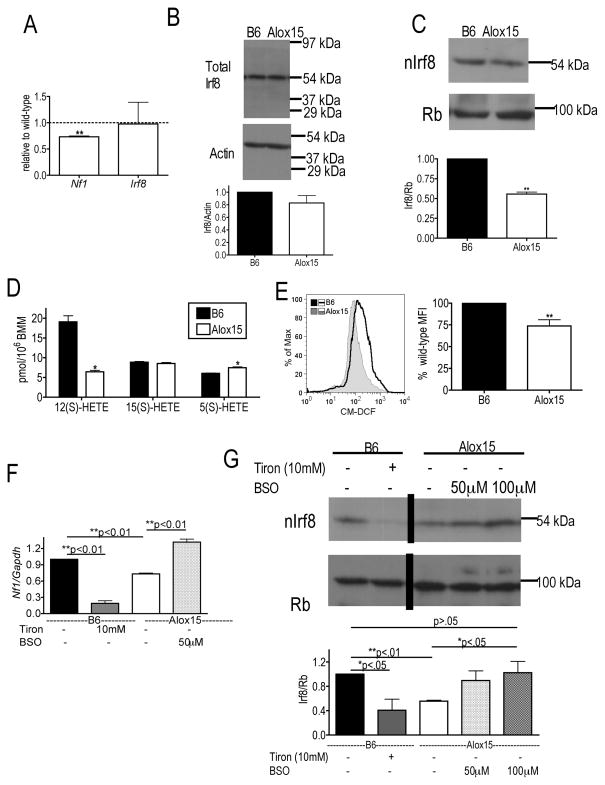
Because 12/15-LOX regulates Irf8 nuclear accumulation in mature myeloid cells(7,8) and since Irf8 critically promotes monocyte differentiation and inhibits granulocyte development(3,5,6), we hypothesized that defective 12/15-LOX regulation of Irf8 in myeloid progenitor cells may underlie the skewed granulopoiesis and reduced monopoiesis in Alox15 mice. To test this, we determined Irf8 expression in Alox15 Lin−cKit+ myeloid progenitors. Alox15 myeloid progenitors exhibited defective nuclear accumulation and total protein of Irf8 compared to B6 although the defect in Alox15 nuclear Irf8 was greater than the defect in total Irf8 (Figure 4A–B). Meanwhile, Irf8 mRNA had a trend towards increased expression in the Alox15 progenitors compared to B6 (Figure 4C). These data suggest that 12/15-LOX regulates Irf8 in a post-transcriptional manner.
Figure 4.
Decreased Irf8 nuclear accumulation and transcriptional activity in Alox15 myeloid progenitors. (A) Decreased accumulation of Irf8 in nuclear lysates of Alox15 Lin−cKit+ myeloid progenitors determined by immunoblot. Shown are a representative experiment and a summary of 3 experiments as Irf8/Retinoblastoma protein (Rb) relative to wild-type (B) Immunoblot of total lysates from B6 and Alox15 Lin−cKit+ cells demonstrating decreased levels of Irf8. Shown are a representative blot and a summary of 3 experiments as Irf8/Actin relative to wild-type. (C) Real time analysis of Irf8 mRNA in B6 and Alox15 Lin−cKit+ cells (n=3 independent experiments). (D) Real time analysis of enriched B6 and Alox15 CMP and GMP demonstrate that Alox15 myeloid progenitors exhibit decreased expression of the Irf8-mediated transcripts Nf1 and Egr1 and similar levels of Pu.1, C/ebpα and Irf8. Gene expression was normalized by Gapdh. (n=3 independent experiments for all experiments shown). *p<.05, **p<.01. Error bars represent ± SEM.
To determine the impact of decreased Irf8 nuclear accumulation on its transcriptional activity, we analyzed gene expression in myeloid progenitors. CMP and GMP from Alox15 mice exhibited decreased expression of Irf8-regulated genes Nf1(12) and Egr1(3) compared to B6 (Figure 4D) but similar expression of Pu.1 and C/ebpα(3) which are important in myeloid differentiation but not regulated by Irf8 (Figure 4D), indicating a selective defect in Irf8-mediated gene transcription. The defect in Irf8-mediated gene transcription was not due to decreased transcription of Irf8 itself, which was similar between B6 and Alox15 CMP and GMP (Figure 4D)
To determine the mechanism whereby 12/15-LOX regulates Irf8 transcriptional activity in developing monocytes, we utilized B6 and Alox15 BMM, which recapitulated the defect in Nf1 gene expression found in Alox15 CMP and GMP and expressed Irf8 mRNA similarly (Figure 5A). Although B6 and Alox15 BMM exhibit comparable expression of total Irf8 protein (Figure 5B), there is a decrease in the nuclear accumulation of Irf8 in Alox15 BMM (Figure 5C). To delineate the mechanism that may regulate Irf8-mediated gene transcription and nuclear accumulation, we compared 12/15-LOX-products. Alox15 BMM were defective in 12(S)-HETE and in basal levels of ROS compared to B6 (Figure 5D-E). Alox15 BMM also produced more of the 5-LOX product 5(S)-HETE (Figure 5D) suggesting substrate re-diversion into the 5-LOX pathway.
Figure 5.
ROS-signaling regulates Irf8 nuclear accumulation in Alox15 cells. (A) Alox15 BMM have decreased expression of Nf1 and similar levels of Irf8 compared to B6 by real time analysis normalized to Gapdh (n=3 independent experiments). (B–C) Total and nuclear lysates were isolated from B6 and Alox15 BMM and immunoblotted for Irf8 demonstrating (B) Alox15 exhibit comparable expression of total Irf8 protein but (C) decreased expression of nuclear Irf8 protein (n=3 independent experiments.) (D) Lipid product formation in B6 and Alox15 BMM after stimulation with 50 μM AA demonstrating decreased 12(S)HETE and increased 5(S)HETE in Alox15 BMM compared to B6. (n=4 in two independent experiments). (E) Flow cytometric analysis of B6 and Alox15 BMM loaded with the ROS sensitive dye H2DCFDA demonstrate decreased levels of ROS in Alox15 BMM. Shown are a representative experiment and a summary of 5 experiments. (F) Addition of 10 mM Tiron, a ROS scavenger, to B6 BMM overnight decreases Nf1 transcription while addition of 50 μM BSO, which increases ROS, to Alox15 BMM increases Nf1 transcription measured by real time analysis. n=3 independent experiments. (G) Modulation of ROS regulates Irf8 nuclear accumulation. Nuclear lysates were isolated from B6 BMM in the presence and absence of 10 mM Tiron and Alox15 BMM cultured alone or in the presence of 50 μM and 100 μM BSO overnight. Protein expression of IRF-8 was determined by immunoblot. Shown is an immunoblot from a representative experiment in which an irrelevant lane was removed and a summary of three experiments. *p<.05, **p<.01. Error bars represent ± SEM.
To determine the potential impact of the reduced levels of ROS on Irf8 mediated gene transcription, we treated B6 BMM overnight with a ROS scavenger, Tiron, and measured transcription of the Irf8 target gene Nf1. Conversely, we treated Alox15 BMM with BSO, which increases ROS by inhibiting synthesis of the cellular anti-oxidant glutathione. These treatments caused changes in ROS levels but did not cause apoptosis (Supplemental Figure 3A–B). Interestingly, Nf1 gene expression was regulated in part by ROS levels. The ROS scavenger Tiron decreased Nf1 expression in B6 BMM while the ROS inducer, BSO, restored Nf1 transcription in Alox15 BMM (Figure 5F). We determined whether ROS-signaling alters Nf1 transcription by regulating Irf8 nuclear accumulation. Both B6 BMM treated with Tiron and Alox15 BMM had decreased Irf8 nuclear accumulation compared to untreated B6 BMM, indicating that low ROS levels decrease nuclear accumulation of Irf8 (Figure 5G). Conversely, elevation of ROS levels in Alox15 BMM by the addition of BSO increased Irf8 nuclear accumulation (Figure 5G). This regulation was not due to alterations in Irf8 gene expression (data not shown) or total protein expression (Supplemental Figure 3C). The addition of BSO in Alox15 BMM restored nuclear IRF-8 protein expression to untreated B6 BMM, suggesting that redox signaling is the primary mediator of 12/15-LOX-dependent activation of Irf8. These data indicate that 12/15-LOX regulates Irf8 nuclear accumulation and transcriptional activity through ROS-mediated signaling.
Redox regulation of monocyte development in Alox15 progenitors
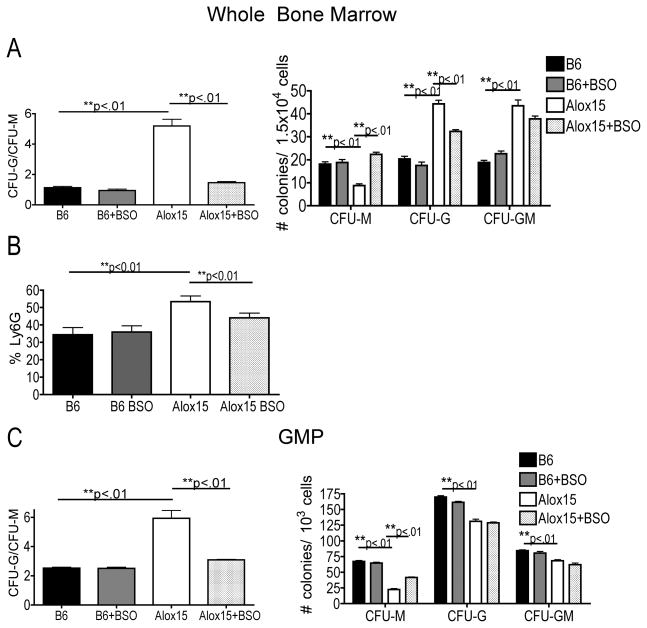
If 12/15-LOX regulates nuclear Irf8 accumulation through ROS-mediated signaling, then increasing ROS-signaling would restore Irf8 nuclear accumulation, thereby enhancing monocyte development and inhibiting aberrant granulocyte development in Alox15 progenitors. To determine the impact of redox signaling on Alox15 myeloid development, we plated BM cells from B6 and Alox15 mice in methylcellulose in the presence or absence of BSO. Interestingly, the presence of BSO increased Alox15 monocyte development (CFU-M) and decreased granulocyte development (CFU-G) resulting in a decreased ratio of CFU-G/CFU-M (Figure 6A). On the other hand, the presence of BSO had no effect on B6 myeloid development (Figure 6A), suggesting that addition of BSO selectively rescues Alox15 myeloid cell development. Treatment of BSO also decreased the percentage of granulocytic Ly6G+ cells in Alox15 plates confirming the shift from granulocyte development (Figure 6B). Addition of the 12/15-LOX product 12(S)-HETE or the 5-LOX inhibitor MK-886 had no effect on B6 and Alox15 myeloid cell differentiation (Supplemental Figure 4). GMP from Alox15 mice plated in the presence of BSO also generated more CFU-M colonies resulting in a decreased ratio of CFU-G/CFU-M (Figure 6C). Thus, 12/15-LOX regulates myelopoiesis at least in part through redox signaling and restoration of redox signaling in Alox15 progenitors restored monocyte development.
Figure 6.
ROS-signaling rescues myeloid differentiation in Alox15 BM and GMP. B6 and Alox15 BM (A–B) and sorted GMP (C) were plated in methylcellulose in the presence and absence of 0.025 μM BSO. (A, C) Shown are colony number and ratio CFU-G/CFU-M. (B) The percentage of Ly6G+ cells were determined by flow cytometry. n=6 in three independent experiments for all data shown *p<.05, **p<.01. Error bars represent ± SEM.
Discussion
This study demonstrates that 12/15-LOX promotes myelopoiesis through redox regulation of IRF-8. Lack of 12/15-LOX results in accumulation of GMP, increased numbers of mature and immature granulocytes and reduced numbers of monocytes in Alox15 mice. The accumulation of functional (CFU-GM) and phenotypic (GMP) myeloid progenitors in Alox15 mice is accompanied by defective differentiation, which is likely a result of decreased Irf8-mediated gene transcription. We show that 12/15-LOX-mediated redox signaling regulates Irf8 nuclear accumulation which subsequently promotes monocyte differentiation and inhibits granulocyte differentiation.
Similar to Alox15 BM cells, Alox15 LSK and CMP exhibited decreased numbers of CFU-M and increased numbers of CFU-G compared to B6. However, Alox15 CMP exhibited similar numbers of CFU-GM as B6. Alox15 GMP had a decreased capacity to differentiate. Interestingly, we previously demonstrated that Alox15 HSC have a reduced capacity to self-renew and an increased propensity to differentiate(11). The increased ability of Alox15 HSC to differentiate and 12/15-LOX regulation of Irf8 in CMP (Figure 4D) may also contribute to the accumulation of Alox15 GMP.
12/15-LOX supports myeloid differentiation towards monocyte development but is not absolutely required for monocyte differentiation. Rather, 12/15-LOX generates signaling intermediates that regulate Irf8, which in turn govern myeloid cell development. In the absence of 12/15-LOX, Irf8 still functions, albeit at reduced levels (Figures 4, 5). Importantly, there is a slight reduction of CFU-M and monocytes in Alox15 mice, which appear phenotypically normal (Figure 1C). The normal number of Alox15 splenic macrophages may be due to homeostatic regulation.
A redox-dependent mechanism for Irf8 nuclear accumulation suggests that other mediators of redox signaling may also regulate myelopoiesis. Though increased levels of ROS promote monocyte to macrophage transition(13), our studies implicate an additional role for ROS-signaling at an earlier stage of myeloid differentiation. Our finding that increasing ROS levels by the addition of BSO in Alox15 BM decreased granulocyte development is in agreement with a previous study that demonstrated ROS signaling, generated by dominant negative NF-κB, inhibits granulocyte differentiation(14). However, that study did not consider potential effects on monocyte development and the ROS generated mediated apoptosis. On the other hand, ROS generated in our studies did not cause apoptosis (Supplemental Figure 3). ROS-signaling is known to promote cytokine receptor signal transduction, including downstream of the cytokines Epo and GM-CSF(15, 16). Therefore, 12/15-LOX redox signaling may also regulate other mediators in addition to Irf8 to regulate myeloid differentiation.
Because leukemia can result from aberrant myeloid cell development, elucidation of the processes that govern myelopoiesis may provide insight into leukemogenesis. Although excluded from these studies, a small percentage of Alox15 mice develop a myeloproliferative disease over the course of a year(7). Irf8-deficient mice, which exhibited reduced monocyte development and enhanced granulocyte development, developed a similar myeloproliferative disease(17). Reduced monocyte development coupled with a skewing towards granulocyte development in Alox15 and Irf8-deficient mice may contribute to or provide for a favorable environment for granulocytic leukemogenesis.
In summary, we demonstrate that ROS-signaling regulates Irf8 nuclear accumulation and subsequent myeloid cell differentiation. This results in an accumulation of defective GMP and a skewing towards granulopoiesis at the expense of monopoiesis in Alox15 mice. Restoration of ROS rescues the aberrant Alox15 myeloid differentiation in vitro. These data establish a novel role for redox-signaling in the regulation of nuclear accumulation and gene transcription by Irf8 and in myeloid cell differentiation. These findings have implications for leukemogenesis.
Supplementary Material
Acknowledgments
We thank Drs. Warren Pear and Melissa Middleton for scientific discussions. We acknowledge the technical support of Wistar Institute flow cytometry, microscopy and animal facilities, and the University of Pennsylvania flow cytometry facility. These studies were funded in part by National Institutes of Health training grants T32GM07229 and T32CA09171 (M.K.), RO1CA091016 and P30ES013508 (I.A.B.), Ludwig Institute for Cancer Research (E.P.) and P30CA10815 (Wistar Institute).
Footnotes
Conflict-of-interest disclosure
authors have no conflicting financial interests.
References
- 1.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 2.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 3.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 4.Koenigsmann J, Rudolph C, Sander S, Kershaw O, Gruber AD, Bullinger L, Schlegelberger B, Carstanjen D. Nf1 haploinsufficiency and Icsbp deficiency synergize in the development of leukemias. Blood. 2009;113:4690–4701. doi: 10.1182/blood-2008-05-158485. [DOI] [PubMed] [Google Scholar]
- 5.Scheller M, Foerster J, Heyworth CM, Waring JF, Lohler J, Gilmore GL, Shadduck RK, Dexter TM, Horak I. Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood. 1999;94:3764–3771. [PubMed] [Google Scholar]
- 6.Tsujimura H, Nagamura-Inoue T, Tamura T, Ozato K. IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J Immunol. 2002;169:1261–1269. doi: 10.4049/jimmunol.169.3.1261. [DOI] [PubMed] [Google Scholar]
- 7.Middleton MK, Zukas AM, Rubinstein T, Jacob M, Zhu P, Zhao L, Blair I, Pure E. Identification of 12/15-lipoxygenase as a suppressor of myeloproliferative disease. J Exp Med. 2006;203:2529–2540. doi: 10.1084/jem.20061444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Middleton MK, Rubinstein T, Pure E. Cellular and molecular mechanisms of the selective regulation of IL-12 production by 12/15-lipoxygenase. J Immunol. 2006;176:265–274. doi: 10.4049/jimmunol.176.1.265. [DOI] [PubMed] [Google Scholar]
- 9.Conrad DJ. The arachidonate 12/15 lipoxygenases. A review of tissue expression and biologic function. Clin Rev Allergy Immunol. 1999;17:71–89. doi: 10.1007/BF02737598. [DOI] [PubMed] [Google Scholar]
- 10.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinder M, Wei C, Shelat SG, Kundu M, Zhao L, Blair IA, Pure E. Hematopoietic stem cell function requires 12/15-lipoxygenase-dependent fatty acid metabolism. Blood. doi: 10.1182/blood-2009-09-243139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu C, Saberwal G, Lu Y, Platanias LC, Eklund EA. The interferon consensus sequence-binding protein activates transcription of the gene encoding neurofibromin 1. J Biol Chem. 2004;279:50874–50885. doi: 10.1074/jbc.M405736200. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto T, Sakaguchi N, Hachiya M, Nakayama F, Yamakawa M, Akashi M. Role of catalase in monocytic differentiation of U937 cells by TPA: hydrogen peroxide as a second messenger. Leukemia. 2009;23:761–769. doi: 10.1038/leu.2008.353. [DOI] [PubMed] [Google Scholar]
- 14.Nakata S, Matsumura I, Tanaka H, Ezoe S, Satoh Y, Ishikawa J, Era T, Kanakura Y. NF-kappaB family proteins participate in multiple steps of hematopoiesis through elimination of reactive oxygen species. J Biol Chem. 2004;279:55578–55586. doi: 10.1074/jbc.M408238200. [DOI] [PubMed] [Google Scholar]
- 15.Sattler M, Winkler T, Verma S, Byrne CH, Shrikhande G, Salgia R, Griffin JD. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood. 1999;93:2928–2935. [PubMed] [Google Scholar]
- 16.Iiyama M, Kakihana K, Kurosu T, Miura O. Reactive oxygen species generated by hematopoietic cytokines play roles in activation of receptor-mediated signaling and in cell cycle progression. Cell Signal. 2006;18:174–182. doi: 10.1016/j.cellsig.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, Bachmann MF, Zinkernagel RM, Morse HC, 3rd, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.