Abstract
Background
Interleukin-15 (IL-15) plays important roles in the immune system and in the development of hematopoietic cells. Previous studies revealed that five SNPs in IL-15, rs10519612, rs10519613, rs35964658, rs17007695 and rs17015014, were significantly associated with childhood Acute Lymphoblastic Leukemia (ALL) treatment response. In adult ALL, the expression of IL-15 was also correlated with the immunophenotypes of ALL. Therefore, we hypothesize that SNPs of IL-15 might also be associated with adult ALL.
Methods and Findings
We genotyped the above five SNPs of IL-15 gene by PCR-RFLP assays in adult ALL case-control studies. The current study included 121 adult ALL patients and 263 healthy controls. IL-15 genotypes and haplotypes were determined and the associations with the risk of ALL were analyzed by logistic regression. SNPs rs10519612 and rs17007695 were significantly associated with ALL (P = 0.013 and P = 0.001). We observed a 2-fold and 2.4-fold excess risk of developing ALL for the rs10519612 CC and rs17007695 TC genotype carriers compared with non-carriers, respectively. Haplotype analysis revealed that haplotypes ACAC, CAGT and CCAT were significantly associated with adult B-ALL, while haplotype CCAT conferred susceptibility to T-ALL.
Conclusion
These findings suggest that IL-15 gene polymorphisms are significantly associated with ALL in adult Chinese population.
Introduction
The characteristic of acute lymphoblastic leukemia (ALL) is the clonal proliferation and accumulation of immature lymphoid cells [1]. Although 80% of children with ALL are cured with contemporary treatments, many patients still develop serious acute and late complications owing to the side effects of the treatments [2], [3]. Furthermore, the survival rate for adults with ALL remains below 40% [4]–[9]. The relatively poor outcome in adult ALL has been explained by an increased frequency of high-risk molecular subtypes with more aggressive clinical behavior and greater drug resistance, poorer tolerance and compliance with treatment, and less effective treatment regimens compared with childhood ALL [6], [8], [9]. Now many of techniques are used in diagnosing the biological and clinical differences of leukemic cells, including the acquisition of new chromosomal abnormalities, gene mutations, and reduced responsiveness to chemotherapeutic agents [10]–[13]. DNA copy number abnormalities (CNAs) and loss of heterozygosity (LOH) in cancer can be genome-wide analyzed by Single Nucleotide Polymorphism (SNP) arrays, which has provided important insights into the pathogenesis of newly diagnosed ALL [14].
Interleukin-15 (IL-15) was found to share many biological activities with IL-2, which is a pleiotropic cytokine of the 4-α-helix bundle cytokine family [15], [16]. IL-15 has many functions in immune system, including the activation of the proliferation and differentiation of natural killer (NK), T, and B cells, as well as the maintenance of memory T cells [15]–[20]. IL-15 appears to play a major role in the hematopoietic development controlling the survival, proliferation, and differentiation of both normal and leukemic progenitors, and can promote pathogenesis of leukemic cells [21]–[25]. Some studies have demonstrated that IL-15 can promote tumor cell growth, expansion, and organ infiltration of malignancies [23]–[27]. With respect to the potential clinical and therapeutic relevance of IL-15 in hematopoietic malignancies, high levels of IL-15 affect the initial response to therapy and have a relationship with malignant transformation, including organ infiltration and disease progression [26]–[30]. Five SNPs in IL-15 gene identified by genome-wide scan significantly associated with childhood ALL treatment response [31]. So, IL-15 may play an important role in the development and progression of ALL.
Because of the possible roles of IL-15 in ALL development, we performed case–control studies to investigate the association between the five SNPs in human IL-15 gene and adult ALL susceptibility in a Chinese population.
Materials and Methods
Study Population
The study group consisted of 121 Han Chinese patients with ALL, whose age ranged from 20 to 75. The group included 41 T-ALL patients and 80 B-ALL patients. These patients were recruited from March 2003 to May 2008 at the First Affiliated Hospital of Soochow University (Suzhou, China). Diagnosing ALL began with a medical history and physical examination, complete blood count and blood smears. Pathological examination, cytogenetics and immunophenotyping established whether the blast cells began from the B or T lymphocytes. All eligible patients diagnosed at the hospital during the study period were recruited, with a response rate of 94%. Population controls were cancer-free people living in the same region; they were selected from a nutritional survey conducted in the same period as the cases were collected. All control subjects had no history of cancer, and their age and gender frequency were matched to cases. The clinical characteristics of patients with acute lymphoblastic leukemia are shown in Table 1 [32].
Table 1. Clinical characteristics of patients with acute lymphoblastic leukemia.
| No.(121) | % | ||
| Gender | |||
| Male | 66 | 54.5 | |
| Female | 55 | 45.5 | |
| Age at diagnosis, y | |||
| 20–40 | 69 | 57 | |
| 41–60 | 44 | 36.4 | |
| ≥61 | 8 | 6.6 | |
| White blood cell count at diagnosis, /µl | |||
| <50000 | 58 | 47.9 | |
| ≥50000 | 63 | 52.1 | |
| Lineage | |||
| T-ALL | 41 | 33.9 | |
| B-ALL | 80 | 66.1 | |
| Karyotype | |||
| Aberrant | 67 | 55.4 | |
| Normal | 54 | 44.6 | |
| Molecular subtype | |||
| BCR-ABL | 22 | 18.2 | |
| E2A-PBX1 | 2 | 1.7 | |
| MLL rearrangement | 10 | 8.3 | |
| No common translocations | 87 | 71.9 | |
| Prognosis Risk Rank* | |||
| Low risk | 8 | 6.6 | |
| Intermediate risk | 57 | 47.1 | |
| High risk | 56 | 46.3 | |
*According to the risk stratification of adult ALL proposed by Hoelzer et al [32].
Ethics Statement
The bone marrow samples of patients and blood samples of healthy individuals used in this study were part of the samples taken for clinical diagnostic tests. Since no extra amount of samples were collected from the study subjects, verbal consent was received from all patients and healthy control individuals. The study procedure was approved by the ethics committee of Soochow University.
DNA extraction and genotyping analysis
Genomic DNA was extracted from 2 ml frozen whole blood or bone marrow using the RelaxGene Blood DNA system according to the manufacturer's protocol (Tiangen, China). A polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay was used to detect polymorphisms. The sequences of the primers (Shenggong, China) for the PCR and annealing temperatures for the PCRs are listed in Table 2.
Table 2. Primers and Restriction Endonucleases used for genotyping IL-15 gene.
| SNP ID | Primer sequences | Product | Annealing | Restriction |
| Length (bp) | temperature(°C) | Endonucleases | ||
| rs10519612 | F: 5′-CTCAATGTCCTTAACCCATTATTCGA-3′ | 137 | 56.1 | BspT104 I |
| R: 5′-CGTTTGAACCATATGGTGAGGTCT-3′ | ||||
| rs10519613 | F: 5′-TAGACATAACAAAACACTCGGCATTT-3′ | 122 | 59.3 | Dra I |
| R: 5′-CTCAATGACATTTTTCTGCCTTCA-3′ | ||||
| rs35964658 | F: 5′-GTTCTGAAACTTTTCATCAAATGAGC-3′ | 149 | 62.4 | Hha I |
| R: 5′-CTAGCCTTACTGGGGAAAATGG-3′ | ||||
| rs17007695 | F: 5′-GTCTTTCTCATGGTCCTCATTGA-3′ | 126 | 56.1 | BspT104 I |
| R: 5′-AAAATTTACAAATGTGTGATTTTCGA-3′ | ||||
| rs17015014 | F: 5′-CGGACTGCTGGGTCTAAGAAGCTA-3′ | 119 | 62.4 | Xsp I |
| R: 5′-GTCTGACTCATCAGCCAACACCC-3′ |
F = Forward Primer; R = Reverse Primer.
The polymorphic region was amplified by PCR in a 25 µl reaction solution containing 50 to 100 ng genomic DNA, 1×Taq PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 5 U Taq DNA polymerase (Fermentas, EU) and 0.2 uM of each primer. PCR products were digested 3 hrs with restriction enzymes (Takara, Japan) according to the manufacturer's protocol and analyzed by 3% agarose gel electrophoresis. The length of PCR products and restriction enzymes are shown in Table 2.
Statistical analysis
SNP allele frequencies were tested against departure from Hardy-Weinberg equilibrium (HWE) before analysis. χ2 tests and independent-samples t test were used to compare the difference in gender and age between cases and controls. Comparisons of the distributions of the allele, genotype and haplotype frequencies were performed using the χ2 test. The relative risk associated with rare alleles was estimated as an odds ratio (OR) with a 95% confidence interval (CI). The P values of OR and 95% CI indicated in the text are used to estimate the significance of the contribution of corresponding genotype to disease risk. The P values of genotypes indicated in the tables are used to estimate the significance of the distribution of genotype between cases and controls. A P value less than 0.05 was considered statistically significant. The statistical software package SPSS v. 17.0 was used for analyses. Haplotype frequencies for pairs of alleles as well as Linkage Disequilibrium (LD) coefficients D' = D/D max and r 2 values for pairs of the most common alleles at each locus were estimated using the SHEsis program (http://analysis.bio-x.cn/myAnalysis.php)[33], [34].
Results
rs10519612 and rs17007695 of IL-15 gene were significantly associated with adult ALL
There was no statistical difference in the age and gender distribution between ALL patients and controls (P = 0.381, P = 0.111). The allele and genotype distributions for five SNPs of IL-15 among cases and controls were summarized in Table 3. The genotype frequencies of these polymorphisms were in Hardy-Weinberg equilibrium (P>0.05) in controls in this study. When the genotype frequencies were compared between cases and controls, rs10519612 and rs17007695 of IL-15 gene showed statistically significant association with the disease. The distribution of rs10519612 and rs17007695 genotypes were significantly different between cases and controls (P = 0.013; P = 0.001) and the rs10519612 CC genotype and the rs17007695 TC genotype were more prevalent among the patients. Increased risks for ALL were found for the homozygous variant CC (OR 1.96, 95%CI 1.06–3.61) and TC (OR 2.38, 95%CI 1.43–3.97). The rs10519612 C allele and rs17007695 C allele carrier frequencies were higher in patients than in controls (P = 0.042, OR 1.37, 95%CI 1.01–1.87; P = 0.041, OR 1.38, 95%CI 1.01–1.88).
Table 3. Genotype and allele frequencies of IL-15 SNPs among ALL cases and controls and associations with risk of ALL.
| SNP | ALL Subjects (Cases/controls) | T-ALL Subjects (Cases/controls) | B-ALL Subjects (Cases/controls) | |||||||
| n (%) | OR (95% CI)* | P | n (%) | OR (95% CI)* | P | n (%) | OR (95% CI)* | P | ||
| rs10519612 | ||||||||||
| Genotype | AA | 38/91 (31.4/34.6) | 1.00 | 10/91 (24.4/34.6) | 1.00 | 28/91 (35.0/34.6) | 1.00 | |||
| AC | 51/135 (42.1/51.3) | 0.88 (0.53–1.45) | 19/135 (46.3/51.3) | 1.25 (0.55–2.84) | 32/135 (40.0/51.3) | 0.74 (0.42–1.32) | ||||
| CC | 32/37 (26.4/14.1) | 1.96 (1.06–3.61)& | 0.013 | 12/37 (29.3/14.1) | 2.68 (1.05–6.83)& | 0.041 | 20/37 (25.0/14.1) | 1.62 (0.81–3.26) | 0.049 | |
| Allele | A | 127/317 (52.5/60.3) | 1.00 | 39/317 (47.6/60.3) | 1.00 | 88/317 (55.0/60.3) | 1.00 | |||
| C | 115/209 (47.5/39.7) | 1.37 (1.01–1.87) | 0.042 | 43/209 (52.4/39.7) | 1.67 (1.05–2.67) | 0.030 | 72/209 (45.0/39.7) | 1.24 (0.87–1.77) | 0.236 | |
| rs10519613 | ||||||||||
| Genotype | CC | 40/103 (33.1/39.2) | 1.00 | 12/103 (29.3/39.2) | 1.00 | 28/103 (35.0/39.2) | 1.00 | |||
| CA | 62/131 (51.2/49.8) | 1.17 (0.73–1.89) | 23/131 (56.1/49.8) | 1.46 (0.69–3.10) | 39/131 (48.7/49.8) | 1.05 (0.60–1.83) | ||||
| AA | 19/29 (15.7/11.0) | 1.56 (0.78–3.11) | 0.315 | 6/29 (14.6/11.0) | 1.48 (0.50–4.40) | 0.450 | 13/29 (16.3/11.0) | 1.49 (0.68–3.28) | 0.436 | |
| Allele | C | 142/337 (58.7/64.1) | 1.00 | 47/337 (57.3/64.1) | 1.00 | 95/337 (59.4/64.1) | 1.00 | |||
| A | 100/189 (41.3/35.9) | 1.26 (0.92–1.72) | 0.152 | 35/189 (42.7/35.9) | 1.33 (0.83–2.13) | 0.238 | 65/189 (40.6/35.9) | 1.22 (0.85–1.75) | 0.282 | |
| rs35964658 | ||||||||||
| Genotype | AA | 31/85 (26.1/32.3) | 1.00 | 10/85 (24.4/32.3) | 1.00 | 21/85 (26.9/32.3) | 1.00 | |||
| AG | 67/142 (56.3/54.0) | 1.25 (0.76–2.08) | 26/142 (63.4/54.0) | 1.48 (0.68–3.25) | 41/142 (52.6/54.0) | 1.12 (0.62–2.02) | ||||
| GG | 21/36 (17.6/13.7) | 1.52 (0.77–3.00) | 0.368 | 5/36 (12.2/13.7) | 1.09 (0.34–3.44) | 0.511 | 16/36 (20.5/13.7) | 1.69 (0.79–3.64) | 0.298 | |
| Allele | A | 129/312 (54.2/59.3) | 1.00 | 46/312 (56.1/59.3) | 1.00 | 83/312 (53.2/59.3) | 1.00 | |||
| G | 109/214 (45.8/40.7) | 1.23 (0.91–1.68) | 0.185 | 36/214 (43.9/40.7) | 1.14 (0.71–1.83) | 0.582 | 73/214 (46.8/40.7) | 1.28 (0.90–1.84) | 0.175 | |
| rs17007695 | ||||||||||
| Genotype | TT | 27/104 (22.3/39.5) | 1.00 | 9/104 (22.0/39.5) | 1.00 | 18/104 (22.5/39.5) | 1.00 | |||
| TC | 80/124 (66.1/47.1) | 2.38 (1.43–3.97)& | 25/124 (61.0/47.1) | 2.28 (1.01–5.17)& | 55/124 (68.8/47.1) | 2.40 (1.32–4.36)& | ||||
| CC | 14/35 (11.6/13.3) | 1.40 (0.65–3.01) | 0.001 | 7/35 (17.1/13.3) | 2.19 (0.74–6.47) | 0.095 | 7/35 (8.7/13.3) | 1.02 (0.39–2.69) | 0.003 | |
| Allele | T | 134/332 (55.4/63.1) | 1.00 | 43/332 (52.4/63.1) | 1.00 | 91/332 (56.9/63.1) | 1.00 | |||
| C | 108/194 (44.6/36.9) | 1.38 (1.01–1.88) | 0.041 | 39/194 (47.6/36.9) | 1.55 (0.97–2.48) | 0.064 | 69/194 (43.1/36.9) | 1.30 (0.91–1.86) | 0.155 | |
| rs17015014 | ||||||||||
| Genotype | GG | 30/75 (24.8/28.5) | 1.00 | 9/75 (22.0/28.5) | 1.00 | 21/75 (26.3/28.5) | 1.00 | |||
| GC | 67/133 (55.4/50.6) | 1.22 (0.73–2.05) | 24/133 (58.5/50.6) | 1.43 (0.63–3.27) | 43/133 (53.7/50.6) | 1.12 (0.62–2.03) | ||||
| CC | 24/55 (19.8/20.9) | 1.07 (0.56–2.03) | 0.659 | 8/55 (19.5/20.9) | 1.20 (0.43–3.36) | 0.600 | 16/55 (20.0/20.9) | 1.03 (0.49–2.17) | 0.879 | |
| Allele | G | 127/283 (52.5/53.8) | 1.00 | 42/283 (51.2/53.8) | 1.00 | 85/283 (53.1/53.8) | 1.00 | |||
| C | 115/243 (47.5/46.2) | 1.06 (0.78–1.43) | 0.733 | 40/243 (48.8/46.2) | 1.11 (0.70–1.77) | 0.663 | 75/243 (46.9/46.2) | 1.03 (0.72–1.47) | 0.880 | |
*Adjusted for age and gender status.
P<0.05.
P for Chi-square analysis or Fisher's exact test.
We further classified the study subjects into T-ALL and B-ALL (Table 3). The rs10519612 C allele carrier frequency was higher in the T-ALL patients than in controls (P = 0.030, OR 1.67, 95%CI 1.05–2.67). The rs10519612 CC genotype and the rs17007695 TC genotype frequencies were higher in patients with T-ALL than in controls (P = 0.039, OR 2.68, 95%CI 1.05–6.83; P = 0.049, OR 2.28, 95%CI 1.01–5.17). The distribution of rs10519612 and rs17007695 genotypes were significantly different between B-ALL patients and controls (P = 0.049; P = 0.003), while only the distribution of rs10519612, but not rs17007695 genotypes were different between T-ALL patients and controls (P = 0.041; P = 0.095). An increased risk for B-ALL was found for the rs17007695 TC genotype (P = 0.004, OR 2.40, 95%CI 1.32–4.36).
We also studied whether the IL-15 SNPs were different in BCR-ABL B-ALL patients and other B-ALL patients (Table S1). Only the distribution of rs17007695 genotypes in BCR-ABL B-ALL patients (P = 0.004) and rs10519612 genotypes in other B-ALL patients (P = 0.041) was significantly different from the controls. Increased risks were found for the rs17007695 TC genotype both in BCR-ABL patients (OR 4.35, 95%CI 1.22–15.55) and other B-ALL patients (OR 1.99, 95%CI 1.03–3.85).
Haplotypes ACAC, CAGT and CCAT were significantly associated with adult ALL (especially B-ALL), while haplotype CCAT conferred susceptibility to T-ALL
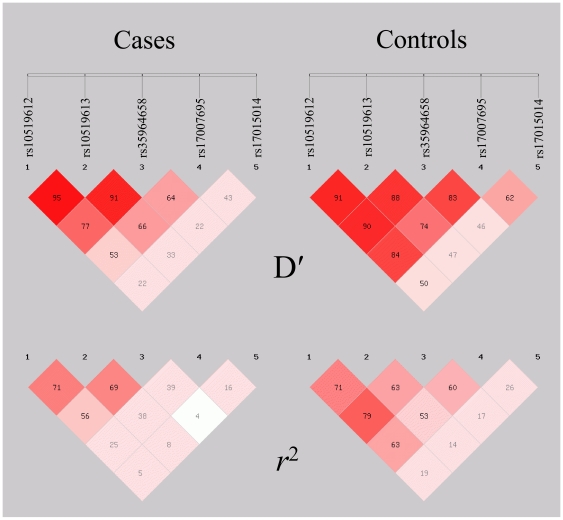
D' and r2 for these five polymorphisms were calculated according to the genotyping data reported in Table 3 and Table 4. Their LD maps measured by D' and r2 in cases and controls were shown in Figure 1. 5-SNP haplotypes were reconstructed according to the genotyping data in cases and controls. The frequencies of haplotypes ACACC, CAGCG and CCATG were higher in ALL patients (OR 2.93, 95%CI 1.26–6.82, P = 0.010; OR 2.17, 95%CI 1.15–4.11, P = 0.016; OR 5.27, 95%CI 1.71–16.26, P = 0.001). When the patients were further classified into T-ALL and B-ALL, the frequencies of haplotypes ACACC and CAGCG were also higher in B-ALL patients (OR 2.89, 95%CI 1.12–7.46, P = 0.023; OR 2.09, 95%CI 1.00–4.34, P = 0.046) and the frequency of haplotype CCATG was significantly higher in T-ALL patients compared with controls (OR 8.67, 95%CI 2.34–32.11, P<0.001) (Table S2).
Table 4. Haplotype frequencies of IL-15 in ALL cases and controls*.
| Haplotype† | All Subjects | T-ALL Subjects | B-ALL Subjects | ||||||
| Frequencies§ | OR(95%CI) | P | Frequencies§ | OR(95%CI) | P | Frequencies§ | OR(95%CI) | P | |
| A C A T | 40.2/53.2 | 1.00 | 39.3/53.2 | 1.00 | 40.9/53.2 | 1.00 | |||
| A C A C | 6.7/2.7 | 3.33 (1.57–7.08) | 0.001 | 5.4/2.7 | 2.50 (0.78–8.05) | 0.113 | 7.5/2.7 | 3.75 (1.66–8.49) | 0.001 |
| C A G C | 31.2/29.2 | 1.40 (0.98–2.01) | 0.067 | 34.4/29.2 | 1.59 (0.92–2.74) | 0.092 | 29.9/29.2 | 1.34 (0.87–2.04) | 0.181 |
| C A G T | 7.2/3.7 | 2.61 (1.30–5.23) | 0.005 | 4.4/3.7 | 1.84 (0.59–5.75) | 0.286 | 8.4/3.7 | 2.99 (1.41–6.38) | 0.003 |
| C C A T | 4.9/1.0 | 7.00 (2.40–20.38) | <0.001 | 6.2/1.0 | 8.75 (2.40–31.86) | <0.001 | 4.1/1.0 | 5.25 (1.55–17.74) | 0.003 |
*Haplotypes with frequencies of more than 5% were included.
Four SNPs alleles from left to right (rs10519612, rs10519613, rs35964658 and rs17007695) were used for reconstruction of haplotypes.
Haplotype frequencies (%) in patients and healthy controls.
Figure 1. LD maps of the five SNPs of IL-15.
LD maps of the five SNPs of IL-15 for ALL cases and controls were generated by SHEsis program. For each SNP, D' and pair r2 values were shown in diamonds in two groups, including ALL patients and healthy controls.
Four polymorphisms consisting of rs10519612, rs10519613, rs35964658 and rs17007695 were in a LD block. Accordingly, 4-SNP haplotypes (rs10519612, rs10519613, rs35964658 and rs17007695) were reconstructed according to the genotyping data in ALL patients and controls. Haplotypes with frequencies ≥5% were shown in Table 4. The four common haplotype alleles represented 89.8% of the chromosomes in the controls. The most frequent haplotypes observed in patients and controls were haplotype ACAT (40.2%, 53.2%) and CAGC (31.2%, 29.2%). The frequencies of haplotypes ACAC, CAGT and CCAT were significantly higher in ALL patients compared with healthy controls (OR 3.33, 95%CI 1.57–7.08, P = 0.001; OR 2.61, 95%CI 1.30–5.23, P = 0.005; OR 7.00, 95%CI 2.40–20.38, P<0.001). When the data were analyzed separately, the frequencies of haplotypes ACAC, CAGT and CCAT were only significantly higher in B-ALL patients compared with healthy controls (OR 3.75, 95%CI 1.66–8.49, P = 0.001; OR 2.99, 95%CI 1.41–6.38, P = 0.003; OR 5.25, 95%CI 1.55–17.74, P = 0.003). However, carriage of the CCAT haplotype conferred a 9-fold higher susceptibility to T-ALL (OR 8.75, 95%CI 2.40–31.86, P<0.001).
Discussion
IL-15 is a proinflammatory cytokine that promotes the proliferation of T, B and NK cells and induction of cytolytic effector cells [27], [35], [36]. It plays an important role not only in immune system but also in the growth and survival of immature hematopoietic cells [21]–[23], which could be the reason for its role in pathogenesis of leukemic cells [24], [25]. Some studies have already demonstrated that IL-15 can protect hematologic tumors from drug induced apoptosis in vitro [25], and high expression of IL-15 correlates with CNS disease in childhood ALL [26]. Several IL-15 SNPs which were associated with minimal residual disease (MRD) have been linked to enhanced IL-15 transcription/translation efficiency in vitro [37]. Five SNPs located in the IL-15 locus identified by genome-wide scan significantly associated with childhood ALL treatment response. Therefore, IL-15 plays a role in treatment response in childhood ALL [31]. In adult ALL, the expression of IL-15 was correlated with the immunophenotypes of ALL, patients with T-ALL and B-cell precursor (BCP)-ALL have different expression levels of IL-15 [38]. In the current study, we identified two SNPs (rs10519612, rs17007695) that were associated with the risk for adult ALL. In the haplotype analysis, we further concluded that haplotypes ACAC, CAGT and CCAT were significantly associated with adult ALL (especially B-ALL), while haplotype CCAT conferred susceptibility to T-ALL.
When we evaluated each of the selected SNPs' genotypes separately, the rs10519612 CC genotype and rs17007695 TC genotype were found to be associated with the increased risk; the other SNPs had no significant correlation with the risk for adult ALL. We demonstrated that the presence of the rs10519612 CC genotype and the rs17007695 TC genotype increased the risk of ALL about two folds. When T-ALL and B-ALL were analyzed separately, rs10519612 CC genotype and the rs17007695 TC genotype conferred risk for T-ALL (2.68 fold and 2.28 fold increase), while only the rs17007695 TC genotype was associated with B-ALL (2.4 fold increase). In B-ALL study, the distribution of rs10519612 CC and rs17007695 TC genotypes were different compared with controls, while only the distribution of rs10519612 CC genotypes were different in T-ALL.
Although five SNPs have been reported to be associated with childhood ALL treatment response [31], we only found two of them correlated with adult ALL in the present study. Out of total 121 ALL patients recruited in this study, 48 were treated relatively uniformly with induction regimen. Among those, 34 achieved CR (T-ALL 5, B-ALL 29), 4 achieved PR (T-ALL 3, B-ALL 1), and ten of them were not responding (NR, T-ALL 8, B-ALL 2). We performed association studies of the SNPs of IL-15 gene with the treatment responses, and none of them was significantly associated with the treatment responses. We have not analyzed the association of these SNPs with the disease prognosis and treatment responses, which would be extremely meaningful in future studies. The combined analysis of SNPs in IL-15 and other genes may also be needed since the polymorphisms in IL-15 might not serve as a sole risk factor for the disease prognosis. Moreover, our study may have certain limitations because of the study design. Selection bias and/or systematic error may occur because the cases were from hospital and the controls were from community. Some factors which may interact with genotype or act as potential confounders in analysis such as information of MRD is not available in our case-control studies.
In the present study, we demonstrated that the polymorphisms of IL-15 genes were associated with adult ALL. Although whether the SNPs that were significantly associated with the disease are functional needs to be further investigated, our results presented here clearly suggested that IL-15 play a role for in the tumorigenesis of adult ALL and may lead to the perspective for IL-15 as a novel therapeutic target in adult ALL.
Supporting Information
Genotype and allele frequencies of IL-15 SNPs among ALL cases and controls and associations with risk of ALL.
(0.09 MB DOC)
Haplotype frequencies of IL-15 in ALL cases and controls.
(0.04 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work has been supported by funds from the Key Laboratory of Suzhou City, startup funds and the Key Project in Science & Technology Cultivation Program from Soochow University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berman E. Recent advances in the treatment of acute leukemia: 1999. Curr Opin Hematol. 2000;7:205–211. doi: 10.1097/00062752-200007000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 4.Bassan R, Gatta G, Tondini C, Willemze R. Adult acute lymphoblastic leukaemia. Crit Rev Oncol Hematol. 2004;50:223–261. doi: 10.1016/j.critrevonc.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Hoelzer D, Gokbuget N. Recent approaches in acute lymphoblastic leukemia in adults. Crit Rev Oncol Hematol. 2000;36:49–58. doi: 10.1016/s1040-8428(00)00097-4. [DOI] [PubMed] [Google Scholar]
- 6.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 7.Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Discov. 2007;6:149–165. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 8.Goldstone AH, Rowe JM. Transplantation in adult ALL. Hematology Am Soc Hematol Educ Program. 2009:593–601. doi: 10.1182/asheducation-2009.1.593. [DOI] [PubMed] [Google Scholar]
- 9.Faderl S, O'Brien S, Pui CH, Stock W, Wetzler M, et al. Adult acute lymphoblastic leukemia: concepts and strategies. Cancer. 2010;116:1165–1176. doi: 10.1002/cncr.24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raimondi SC, Pui CH, Head DR, Rivera GK, Behm FG. Cytogenetically different leukemic clones at relapse of childhood acute lymphoblastic leukemia. Blood. 1993;82:576–580. [PubMed] [Google Scholar]
- 11.Klumper E, Pieters R, Veerman AJ, Huismans DR, Loonen AH, et al. In vitro cellular drug resistance in children with relapsed/refractory acute lymphoblastic leukemia. Blood. 1995;86:3861–3868. [PubMed] [Google Scholar]
- 12.Maloney KW, McGavran L, Odom LF, Hunger SP. Acquisition of p16(INK4A) and p15(INK4B) gene abnormalities between initial diagnosis and relapse in children with acute lymphoblastic leukemia. Blood. 1999;93:2380–2385. [PubMed] [Google Scholar]
- 13.Irving JA, Bloodworth L, Bown NP, Case MC, Hogarth LA, et al. Loss of heterozygosity in childhood acute lymphoblastic leukemia detected by genome-wide microarray single nucleotide polymorphism analysis. Cancer Res. 2005;65:3053–3058. doi: 10.1158/0008-5472.CAN-04-2604. [DOI] [PubMed] [Google Scholar]
- 14.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabstein KH, Eisenman J, Shanebeck K, Rauch C, Srinivasan S, et al. Cloning of a T cell growth factor that interacts with the beta chain of the interleukin-2 receptor. Science. 1994;264:965–968. doi: 10.1126/science.8178155. [DOI] [PubMed] [Google Scholar]
- 16.Bazan JF. Haemopoietic receptors and helical cytokines. Immunol Today. 1990;11:350–354. doi: 10.1016/0167-5699(90)90139-z. [DOI] [PubMed] [Google Scholar]
- 17.Klebanoff CA, Finkelstein SE, Surman DR, Lichtman MK, Gattinoni L, et al. IL-15 enhances the in vivo antitumor activity of tumor-reactive CD8+ T cells. Proc Natl Acad Sci U S A. 2004;101:1969–1974. doi: 10.1073/pnas.0307298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yajima T, Yoshihara K, Nakazato K, Kumabe S, Koyasu S, et al. IL-15 regulates CD8+ T cell contraction during primary infection. J Immunol. 2006;176:507–515. doi: 10.4049/jimmunol.176.1.507. [DOI] [PubMed] [Google Scholar]
- 19.Carson WE, Giri JG, Lindemann MJ, Linett ML, Ahdieh M, et al. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J Exp Med. 1994;180:1395–1403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armitage RJ, Macduff BM, Eisenman J, Paxton R, Grabstein KH. IL-15 has stimulatory activity for the induction of B cell proliferation and differentiation. J Immunol. 1995;154:483–490. [PubMed] [Google Scholar]
- 21.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 22.Lodolce JP, Burkett PR, Koka RM, Boone DL, Ma A. Regulation of lymphoid homeostasis by interleukin-15. Cytokine Growth Factor Rev. 2002;13:429–439. doi: 10.1016/s1359-6101(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 23.Giron-Michel J, Giuliani M, Fogli M, Brouty-Boye D, Ferrini S, et al. Membrane-bound and soluble IL-15/IL-15Ralpha complexes display differential signaling and functions on human hematopoietic progenitors. Blood. 2005;106:2302–2310. doi: 10.1182/blood-2005-01-0064. [DOI] [PubMed] [Google Scholar]
- 24.Trentin L, Cerutti A, Zambello R, Sancretta R, Tassinari C, et al. Interleukin-15 promotes the growth of leukemic cells of patients with B-cell chronic lymphoproliferative disorders. Blood. 1996;87:3327–3335. [PubMed] [Google Scholar]
- 25.Tinhofer I, Marschitz I, Henn T, Egle A, Greil R. Expression of functional interleukin-15 receptor and autocrine production of interleukin-15 as mechanisms of tumor propagation in multiple myeloma. Blood. 2000;95:610–618. [PubMed] [Google Scholar]
- 26.Cario G, Izraeli S, Teichert A, Rhein P, Skokowa J, et al. High interleukin-15 expression characterizes childhood acute lymphoblastic leukemia with involvement of the CNS. J Clin Oncol. 2007;25:4813–4820. doi: 10.1200/JCO.2007.11.8166. [DOI] [PubMed] [Google Scholar]
- 27.Fehniger TA, Caligiuri MA. Interleukin 15: biology and relevance to human disease. Blood. 2001;97:14–32. doi: 10.1182/blood.v97.1.14. [DOI] [PubMed] [Google Scholar]
- 28.Budagian V, Bulanova E, Paus R, Bulfone-Paus S. IL-15/IL-15 receptor biology: a guided tour through an expanding universe. Cytokine Growth Factor Rev. 2006;17:259–280. doi: 10.1016/j.cytogfr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Blaser BW, Roychowdhury S, Kim DJ, Schwind NR, Bhatt D, et al. Donor-derived IL-15 is critical for acute allogeneic graft-versus-host disease. Blood. 2005;105:894–901. doi: 10.1182/blood-2004-05-1687. [DOI] [PubMed] [Google Scholar]
- 30.VanBuskirk AM, Pidwell DJ, Adams PW, Orosz CG. Transplantation immunology. JAMA. 1997;278:1993–1999. [PubMed] [Google Scholar]
- 31.Yang JJ, Cheng C, Yang W, Pei D, Cao X, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoelzer D. Prognostic factors in acute lymphoblastic leukemia. Leukemia. 1992;6(Suppl 4):49–51. [PubMed] [Google Scholar]
- 33.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Zhang Z, He Z, Tang W, Li T, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 2009;19:519–523. doi: 10.1038/cr.2009.33. ( http://analysis.bio-x.cn) [DOI] [PubMed] [Google Scholar]
- 35.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 36.Bulfone-Paus S, Bulanova E, Budagian V, Paus R. The interleukin-15/interleukin-15 receptor system as a model for juxtacrine and reverse signaling. Bioessays. 2006;28:362–377. doi: 10.1002/bies.20380. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XJ, Yan KL, Wang ZM, Yang S, Zhang GL, et al. Polymorphisms in interleukin-15 gene on chromosome 4q31.2 are associated with psoriasis vulgaris in Chinese population. J Invest Dermatol. 2007;127:2544–2551. doi: 10.1038/sj.jid.5700896. [DOI] [PubMed] [Google Scholar]
- 38.Wu S, Fischer L, Gokbuget N, Schwartz S, Burmeister T, et al. Expression of interleukin 15 in primary adult acute lymphoblastic leukemia. Cancer. 2010;116:387–392. doi: 10.1002/cncr.24729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype and allele frequencies of IL-15 SNPs among ALL cases and controls and associations with risk of ALL.
(0.09 MB DOC)
Haplotype frequencies of IL-15 in ALL cases and controls.
(0.04 MB DOC)