Abstract
Context
Neuropeptide Y (NPY) is important to countering stress and is involved in neuroadaptations that drive escalated alcohol drinking following repeated alcohol exposure in rodents. In humans, haplotype-driven diminution in NPY expression is predictive of amygdala response and emotional reactivity to stress. Genetic variation that affects the NPY system could impact resilience to stress and to developing addiction with continued alcohol use.
Objective
To determine whether functional NPY variation influences CSF NPY, behavioral adaptation to stress, and alcohol consumption in a nonhuman primate model of early adversity (peer rearing).
Design
We sequenced the rhesus macaque NPY locus (rhNPY) and performed in silico analysis to identify functional variants. We performed gel shift assays for a −1002 T>G using nuclear extract from testes, brain and hypothalamus. Levels of NPY in CSF were measured by RIA, and mRNA levels were assessed in amygdala using RT-PCR. During infancy, animals were exposed to repeated social separation stress, and tested for individual differences in alcohol consumption as young adults. Animals were genotyped for −1002 T>G, and the effects of this variant on mRNA expression, CSF NPY, behavior arousal during stress, and ethanol consumption were assessed by ANOVA.
Results
The G allele altered binding of regulatory proteins in all nuclear extracts tested, and −1002 T>G resulted in lower levels of NPY expression in amygdala. Macaques exposed to adversity had lower CSF NPY and exhibited higher levels of arousal during stress, but only as a function of the G allele. We also found that stress-exposed G allele carriers consumed more alcohol and exhibited an escalation in intake over cycles of alcohol availability and deprivation.
Conclusions
Our results suggest a role for NPY promoter variation in the susceptibility to alcohol use disorders and point to NPY as a candidate for examining GxE interactions in humans.
Exposure to adversity is known to increase an individual’s risk for developing stress-related conditions, such as anxiety, depression, and addictive disorders, including alcohol dependence 1, 2. A number of studies have shown that genetic variants that increase stress reactivity interact with stressful life events to impart risk for these disorders 3, 4. Functional genetic variation that reduces stress resiliency would be equally likely to moderate risk. The neuropeptide Y (NPY) system is one whose regulation mediates stress adaptation and is, therefore, a candidate system in which functional genetic variation may impact resilience. In response to protracted or repeated periods of stress exposure, NPY is released in key regions of the brain, a mechanism proposed to be important for countering the effects of stress 5. Individuals who differ in the ability to recruit this system would be expected to differ in resilience and, therefore, vulnerability to stress-related disorders.
Studies indicate that stress exposure early in life is particularly likely to induce adult psychopathology 6. The rhesus macaque model has led the way as a controlled experimental system that permits examination of how early adversity interacts with functional genetic variants to influence stress reactivity and alcohol consumption 7. Infants that are reared with age-mates, and not by their mothers (peer reared), show evidence of harm avoidance, insecure attachment, and high levels of anxiety 8, 9. In addition to exhibiting these lifelong traits, peer-reared monkeys consume higher levels of alcohol 10, 11. Whether NPY variation influences these phenotypes in stress-exposed primates has not yet been demonstrated.
Prolonged exposure to alcohol leads to sensitization of behavioral stress responses and escalated alcohol intake. These neuroadaptations are in large part mediated through recruitment of corticotropin-releasing factor (CRF, or CRH) signaling within the amygdala complex 5. Under these conditions, rodent studies have shown that both exogenous NPY administration and local over-expression of Npy within the amygdala not only reduce stress-responses, but also suppress excessive alcohol intake 12, 13. Whether induced by genetic selection for alcohol preference 14, 15 or neuroadaptations encompassing stress circuitry 12, the emerging role of NPY is as a negative regulator of excessive alcohol consumption. It may be that NPY could also negatively regulate alcohol intake induced by other environmental stressors that recruit the CRF system. We predicted that NPY variation would modulate not only stress-reactivity but also voluntary alcohol intake, particularly as a function of prior stress or alcohol exposure.
Functional variants in the macaque are of particular interest because several key mediators of stress-responses, such as CRF, are differentially distributed between rodents and primates, and also because several rhesus variants have been identified that are functionally equivalent to those in humans 16–19. The existence of these variants and the demonstrated feasibility of modeling early life adversity in the rhesus macaque combine to provide a unique opportunity for studies of gene by environment (GxE) interactions that are likely to be relevant for humans 3, 7, 20. Here, we examined whether rhesus NPY (rhNPY) variation influenced stress resiliency and voluntary alcohol consumption. We screened the rhNPY gene and regulatory regions for variation and investigated the functionality of a single nucleotide polymorphism, or SNP (rhNPY −1002T>G), located in a region that is orthologous to one demonstrated to be important for regulation of human NPY promoter activity 21. Because of the role of the NPY system in stress and alcohol response, we examined whether −1002 T>G influenced both behavioral arousal during exposures to stress and voluntary alcohol consumption. Finally, because the NPY system becomes involved in neuroadaptations that drive escalated alcohol drinking, we also examined whether rhNPY −1002 T/G genotype differentially influenced alcohol intake over cycles of alcohol availability and deprivation.
METHODS
IDENTIFICATION OF NPY SEQUENCE VARIANTS
Genomic DNA was extracted from whole blood from rhesus macaques (Macaca mulatta) from the NIH Animal Center (NIHAC), and direct sequencing was performed using samples from 96 unrelated animals (pairwise Identity by Descent, or IBD ≤ 0.0125). We used primers designed from published human sequence and, subsequently, from rhesus sequence published on the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway) to sequence 2.5 kB of the 5′ regulatory region, exons 1–4 (Exon 1= 5′UTR, Exon 2 = NPY, Exon 3 = CPON, Exon 4 = 3′UTR), intron 1, and the exon-intron boundaries. Cycle sequencing was performed using the Big Dye Terminator Version 3.1 reaction in 96-well optical plates (Applied Biosystems, Inc., Foster City, CA). Variants were detected by visualization of electropherograms generated by ABI Sequencing Analysis software.
To identify putatively functional variants, we examined regions containing consensus sites for factors known to regulate NPY transcription 21, 22 and used web-based TFBS binding site prediction algorithms (TfSitescan, http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl 23 and TRANSFAC, http://www.cbrc.jp/research/db/TFSEARCH.html, 24. Comparative genomic analyses across anthropoid primates (H. sapiens, P. troglodytes, P. pygmaeus, M. mulatta, C. jacchus) were performed using the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
ELECTROPHORETIC MOBILITY SHIFT ASSAY
Based on the identification of a putatively functional variant (−1002 T>G) within the rhNPY regulatory region, double-stranded oligonucleotides containing the T (5′-GCA AAT TAA TGT TCA TCG TTT TTA ACA TG -3′) and G (5′-GCA AAT TAA TGT GCA TCG TTT TTA ACA TG - 5′) alleles were used to perform gel shift assays using nuclear extract from human whole brain, the osteosarcoma cell line, MG-63 (both from ActivMotif, Carlsbad, CA) and from an immortalized glucocorticoid-treated hypothalamic cell line (IVB cells treated with 100 nM Dexamethasone)25. Assays were performed using the Gel Shift Assay System (Promega, Madison, WI) per manufacturer’s instructions. After annealing complementary oligonucleotides (95°C 5 min, 25°C 30 min), double-stranded probes were [32P]-ATP labeled using T4 kinase (Promega, Madison, WI) and purified using a Bio-Spin 30 chromatography column (Bio-Rad). Incorporation of radiolabel was > 1 × 105 cpm/ng DNA. Binding assays were performed using the Gel Shift Assay System (Promega, Madison, WI) per manufacturer instructions. Nuclear extracts (5 μg/assay) were incubated for 20 min with 1 × 105 cpm of each oligonucleotide probe. Competitor oligonucleotides were added at 10× the concentration of the labeled probes. Samples were immediately separated by electrophoresis (300 V for 20 min) at 4°C on a Novex 6% DNA retardation gel along with pre-stained protein molecular weight standards (Invitrogen, Carlsbad, CA). Each gel shift assay was performed in duplicate.
Acute stress regulates NPY expression in the hypothalamus, and the temporal dynamics of this regulation are similar to those observed in other regions of the brain 26. Given that the −1002 T>G SNP disrupts a putative Glucocorticoid Response Element, or GRE, we wanted to determine whether we would observe glucocorticoid-dependent differences in the patterns of DNA-protein interactions and whether these differed according to genotype. In order to examine this, we performed gel super-shift assays using the an anti-GR antibody (Santa Cruz Biotechnology, Santa Cruz, CA) with the GR-enriched MG-63 nuclear extract. Nuclear extract (1 μl) and antibody (1 μl) were pre-incubated for 30 min at 25°C prior to performance of the assay.
NPY mRNA QUANTIFICATION BY REAL-TIME PCR
RNA was extracted from rhesus amygdala using Trizol according to manufacturer’s protocol (Invitrogen, UK). Prior to cDNA synthesis, RNA cleanup was performed using the RNeasy Mini Kit (Qiagen, USA), and RNA was treated with RQ1 RNase-free DNase (Promega, USA) following manufacturer’s instructions. Total RNA quality and integrity were verified by OD measurements (260nm/280 nm) and by measuring ribosomal 28S/18S ratios using RNA 6000 230 Nano Assay RNA chips run on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). RNA (100 ng) was then used for cDNA synthesis applying reverse transcription reagents (Applied Biosystems Inc., Foster City, CA, USA).
NPY expression in amygdala samples (n=12) was assessed by Real-Time PCR. Applied Biosystems Assay # Rh02787751_m1 was used to detect NPY mRNA levels. β-actin expression was used as an endogenous reference (ABI #Hs99999903_m1). Samples were analyzed in quadruplicate on an ABI Prism 7900HT system with Taqman universal PCR master mix. The amplification conditions were 50°C for 2 min then 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The SDS 2.0 software (Applied Biosystems Inc., Foster City, CA, USA) was used to analyze and convert the expression data into cycle threshold values (Ct-values). Data are expressed as relative NPY mRNA levels normalized to the −1002T homozygote group.
EXPERIMENTAL ANIMALS: PHYSIOLOGIC AND BEHAVIOR ASSESSMENTS
Rearing
Rhesus macaque (Macaca mulatta) infants at NIHAC were randomly selected to be reared with their mothers or in a nursery by human caregivers. 10, 27, 28. Mother-reared (MR) animals were reared in social groups composed of 8–14 females (about half of whom had same-aged infants) and two adult males. Peer-reared (PR) animals were separated from their mothers at birth and hand-reared in a neonatal nursery for the first 37 days of life. For the first 14 days, they were kept in an incubator and hand-fed. From day 15 until day 37, they were placed alone in a nursery cage and provided a blanket and a terrycloth-covered, rocking surrogate. A bottle from which the infants would feed was fixed to the surrogate. At 37 days of age, PR infants were placed in a cage with three other age-mates with whom they had continuous contact. Mother-reared infants remained in their social group. At approximately 8 months of age, both PR and MR animals were placed together into age-matched social groups and housed in large indoor-outdoor runs through late adolescence, at which point the cohorts were divided into same-sex groups. All procedures were approved by the NIAAA and NICHD ACUC.
CSF Sampling and Radioimmunoassay
Cerebrospinal fluid (CSF) levels of NPY were assessed in order to determine whether rhNPY −1002 T>G was associated with differences in central NPY release. CSF samples were obtained from the cisterna magna using a 5 ml syringe with a 22 gauge needle (Becton Dickinson, Franklin Lakes, NJ) under ketamine anesthesia (15 mg/kg, IM). All samples were collected within 30 min of investigators entrance into the animal area. CSF samples were immediately aliquoted into polypropylene tubes, frozen in liquid nitrogen, and stored at −70°C until assay using a commercially available kit (Bachem/Peninsula Laboratories, San Carlos, CA). The between and within assay coefficients of variation for all tests were less than 10%.
Social Separation Stress
When the animals reached 6 months of age, they were subjected to four sequential, 4-day-long separations 28. Subjects in the peer group were partitioned into individual sections of the home cage, which prevented the infants from seeing or touching one another. Mother-reared infants were separated from their mothers by removing the mother from the social group. Day one (Monday) of each separation week was designated as the “Acute” phase of separation. Days two through four (Tuesday through Thursday) of each separation week were designated as “Chronic” separation. Following each separation week, subjects were reunited with their attachment sources early on Friday morning and separated again at noon on Monday.
During each separation week, a total of nine behavioral observations were made, according to the following schedule (for behavior definitions, see Table S1). Three observations were made on day one- two immediately following separation and one at hour one (Acute). Two observations were made each day for days two, three, and four (Chronic). Each observation period was 300 s in duration. Behavioral data were collected by multiple observers, with inter-observer reliability of ≥ 85%.
Alcohol Consumption
Nine cohorts of young adult macaques (age 3.5–5) were allowed to freely consume an aspartame-sweetened 8.4% (v/v) alcohol solution for one hour per day, 5 days a week in the home cage. This method consisted of three phases, which have previously been reported 29: (1) Spout Training; (2) Initial Alcohol Exposure; and (3) Experimental Period. During the experimental phase, alcohol and vehicle were dispensed 5 days a week (Monday-Friday) from 1300 to 1400 while the animals were in their home-cage environment.
GENOTYPING
A portion of the rhNPY regulatory region (−1216 > −671) was amplified from 25ng of genomic DNA with flanking oligonucleotides (5-TGC TTT AAT TTC CCA ACA TGC; 5-GGA GAG TAC TTG AGG AAG GCT G) in 15-μL reactions using AmpliTaq Gold DNA Polymerase LD (Low DNA) kit from Applied Biosystems, Inc. (Foster City, CA). Amplifications were performed on a thermocycler (9700) (Applied Biosystems, Inc., Foster City, CA) with 1 cycle at 96°C for 5 minutes followed by 30 cycles of 94°C for 15 seconds, 60°C for 15 seconds, 72°C for 30 seconds, and a final 3-minute extension at 72°C. Amplicons were sequenced using the Big Dye Terminator Version 3.1 kit and the 3100 Genetic Analyzer both from Applied Biosystems, Inc. (Foster City, CA) per manufacturer’s instructions. Cycle sequencing was performed with 1 cycle at 96°C for 1 minute followed by 40 cycles of 96°C for 10 seconds, 50°C for 5 seconds, 60°C for 4 seconds, and a final 3-minute extension at 72°C. Genotypes were called by direct visualization of electropherograms using 4 Peaks (www.mekentosj.com).
STATISTICAL ANALYSES
We used archived datasets to examine the effects of NPY −1002 T>G on our phenotypes of interest. Behavioral scores during separation stress exposure were averaged for each phase (Acute and Chronic) across the four weeks of testing. As scores of behaviors relating to stress responding were inter-correlated, we performed factor analysis to reduce the dimensionality of the data. Factor analysis was then performed to yield behavioral factors relating to arousal to be used as dependent variables in ANOVA. Scores for each behavior were expressed as the mean frequency or duration of the behavior for the two testing conditions (Acute and Chronic Stress). Separate factor analyses for both phases of separation were performed using principal components extraction and varimax orthogonal rotation. Factors indicative of high levels of attachment (“Separation Anxiety”), stereotypy (“Behavioral pathology”) and arousal (“Arousal”) were identified. Although NPY has not been linked to social attachment or stereotypies, it has been repeatedly demonstrated to influence levels of arousal. 30–32 To avoid uninformative repeat testing, we, therefore, focused on effects of rhNPY −1002 T>G genotype on arousal 30–32. We performed two-way ANOVA on Acute and Chronic “Arousal”, with genotype (T/T, T/G, G/G) and rearing condition (PR vs. MR) as nominal independent variables. Two-way ANOVA was also performed to assess effects of rearing condition and genotype on CSF levels of NPY and on voluntary alcohol consumption. Under a limited access schedule, we have determined that alcohol consumption is increased following a 3-day period of deprivation (from 0.3 g/kg/h to 1 g/kg/h, Figure S1), suggesting there to be an alcohol deprivation effect. In order to examine whether genotype interacted with periods of alcohol deprivation (5 days of 1 h access with 3 days of deprivation) to influence the pattern of alcohol consumption across time, we used a mixed design repeated measures ANOVA to examine the effects of genotype and rearing on alcohol consumption using data obtained on the first day of access (Monday) over the four weeks of testing. All post-hoc comparisons were made using the Tukey-Kramer method.
The frequency of the G allele was 37%, and genotype frequencies were in Hardy-Weinberg equilibrium. Although this is an outbred colony of macaques, to verify that our effects were attributable to rhNPY variation, and not to general heritability of our traits of interest, we repeated our analyses using a set of three bi-allelic genetic markers with similar MAFs to the −1002 G allele (between 20 and 40%) 33, 34. Similar effects of the other markers tested on our phenotypes of interest were not observed, supporting the argument that our current results are attributable to effects of NPY −1002T>G. We also excluded individuals carrying alleles known to predict our phenotypes of interest (ie, rh5-HTT-LPR s allele); as results were unchanged, these individuals (N= 20) were included in the final analyses. The Kolmogorov-Smirnov Normality Test and Equality of Variances F Test were used to determine whether data deviated from normality and whether there was non-homogeneity of the data. In cases in which there was non-normality or inequality of variances, data were rank transformed and the analyses repeated. Analyses were performed using StatView 5.01 statistical software. Criterion for significance was set at P ≤ 0.05.
RESULTS
IDENTIFICATION OF A FUNCTIONAL VARIANT IN THE rhNPY PROMOTER
We sequenced the rhesus macaque NPY gene, first intron, exon-intron boundaries, and 3′ and 5′ flanking regions and identified 12 polymorphic sites (Figure 1A). Variants were assigned positions relative to the transcription start site. In silico analysis indicated that a SNP (−1002 T>G) present in a region orthologous to one shown to be important to regulation of NPY transcriptional control 21 predicted the loss of a GRE half site (Figure 1B). We found that the T>G SNP resulted in altered binding of regulatory proteins, with several bands increasing (MW of approx. 130, 210, and 260 kDa) and one of 180 kDa showing a relative decrease (Figure 1C). We also found that, in amygdala, the G allele resulted in decreased levels of NPY expression (Figure 1D, F(1,9) = 24.4, P = 0.0008). We performed gel super-shift assays using an anti-GR antibody and found that the T allele showed a relative increase in the degree of binding of the 180 kDa (and 90 kDa) bands, both of which showed decreased motility with the addition of the anti-GR antibody. The 180 kDa band (GR dimer) was preferentially bound in experiments performed with T allele oligonucleotides (Figure S2).
Figure 1. rhNPY −1002 T>G is present in a conserved portion of a NPY repressor and results in altered DNA-protein interactions and decreased amygdala NPY expression.
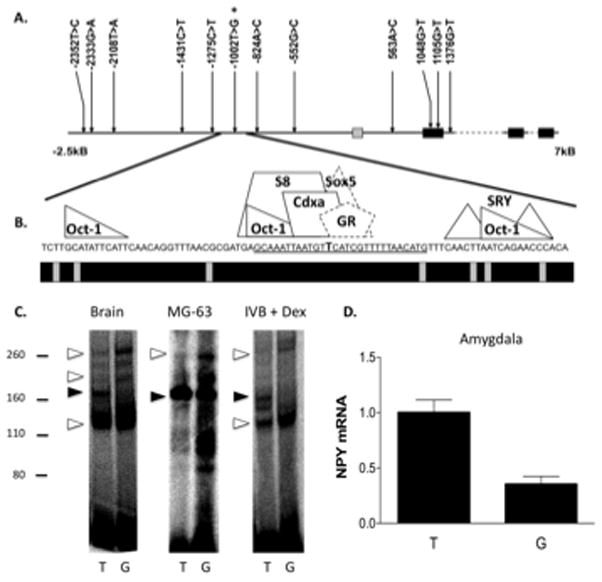
A. Schematic of the NPY gene and regulatory region and SNPs detected by sequencing of genomic DNA. B. Region 40-bp up and downstream of the −1002 T>G SNP. The precise locations of the −1002 T>G SNP (in bold) and the oligonucleotide sequence used in the gel shift assays (underlined) are indicated. Predicted sites for transcription factor binding (above) and sequence conservation among primates (below, black = conserved) are shown. Binding sites in dashed lines (Sox5 and a preferred glucocorticoid response element half site, GR) were predicted to be disrupted by the −1002 T>G SNP. C. Gel shift assay results from experiments performed using nuclear extracts from Whole Brain, osteosarcoma cells (MG-63), and glucocorticoid-treated hypothalamic cells (IVB + Dex). Relative migrations of the protein molecular weight standards are shown to the left (kDa). Open arrows indicate bands that increase with G allele probes, closed arrows indicate that which shows a relative increase with T allele probes. D. NPY mRNA expression in amygdala as a function of the −1002 T>G allele (P = 0.0008; T/T, N = 4, G carrier, N = 8). *** P < 0.001.
CSF NPY
There was a trend for an effect of rearing condition, with lower NPY levels among PR animals (F(2, 66)= 2.68, P = 0.1). There was no main effect for genotype (F(2, 66) = 0.84, P = 0.44). However, genotype interacted with rearing condition to predict CSF NPY (F(2, 66) = 4.2, P < 0.02). PR animals carrying the G allele (T/G or G/G) had lower CSF NPY levels than did PR T/T animals (Figure 2, Tukey-Kramer, P < 0.05). Among PR subjects, genotype accounted for 28 % of the variance.
Figure 2. Interaction between rhNPY genotype (T/T, T/G, G/G) and early rearing history (MR, mother- reared, vs. PR, peer-reared) on CSF levels of NPY.
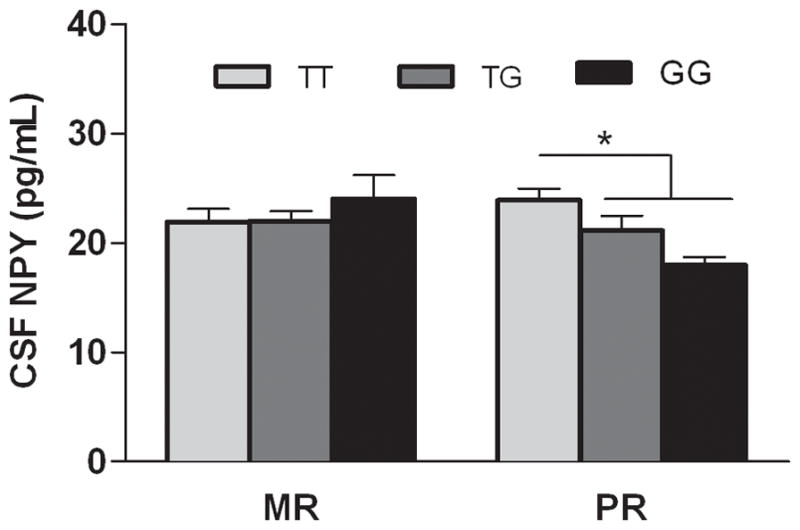
There was an interaction between genotype and rearing (F(2, 66) = 4.2, P < 0.02). The G allele dose-dependently decreased levels of NPY measured in a cisternal CSF sample among stress-exposed monkeys (Tukey-Kramer, P < 0.05) (MR T/T = 17, MR T/G = 14, MR G/G = 4; PR T/T = 16, PR T/G = 13, PR G/G = 8). Genotype accounted for 28 % of the variance in PR subjects. Values shown are mean pg/ml ± SEM. * P < 0.05.
BEHAVIORAL RESPONSES TO STRESS
Factor Analysis performed on behavioral measures taken during social separation generated three factors for each of the two phases of data collection. For the acute phase of stress, three factors were generated (“Separation Anxiety”, “Arousal”, and “Behavioral Pathology”) that accounted for 71.6% of the variance. The same three factors accounted for 77.6% of the variance for analysis performed on behaviors collected during chronic separation stress (Table S2).
During acute separation, there were main effects of rearing (F(1, 96) = 6.4, P = 0.01) and genotype (F(2, 96) = 3.2, P = 0.04) on “Arousal”, but no interaction. Post-hoc analyses demonstrated that PR infants exhibited higher levels of arousal than did MR infants and that those homozygous for the G allele had higher arousal scores than did those homozygous for the T allele (Figure 3A, Tukey-Kramer, P < 0.05). As with acute stress exposure, there was a main effect of rearing condition on “Arousal” (F(1, 96) = 25.0, P < 0.0001) during chronic separation, with PR animals exhibiting higher scores (Figure 3B, Tukey-Kramer, P < 0.05). There was no main effect for genotype. However, there was an interaction between rearing and genotype (F(2, 96) = 4.2, P = 0.02). Although PR T/T subjects responded no differently than MR animals, PR G allele carriers (T/G or G/G) exhibited higher levels of arousal (Figure 3B, Tukey-Kramer, P < 0.05). In both cases (acute and chronic stress), results remained the same following rank transformation of the data. Among PR subjects, genotype accounted for 7% and 10% of the variance during acute and chronic stress exposure, respectively.
Figure 3. Interaction between rhNPY genotype (T/T, T/G, G/G) and early rearing history (MR, mother- reared, vs. PR, peer-reared) on arousal during periods of A. Acute (1 H) and B. Chronic (96 H) separation stress.
During acute separation stress, there were main effects of both rearing (F(1, 96) = 6.4, P = 0.01) and genotype (F(2, 96) = 3.2, P = 0.04) on “Arousal”, with genotype accounting for 7% of the variance. During chronic stress, there was an interaction between rearing and genotype (F(2, 96) = 4.2, P = 0.02). Although PR T/T subjects responded no differently than did MR animals, PR G allele carriers exhibited higher levels of arousal (T/G and G/G vs. T/T, Tukey-Kramer, P < 0.05), with genotype accounting for 10% of the variance in these subjects. (MR T/T = 35, MR T/G = 27, MR G/G = 10; PR T/T = 9, PR T/G = 15, PR G/G = 6). Values shown are “Arousal” factors scores ± SEM. * P < 0.05.
ALCOHOL CONSUMPTION
There was a main effect of rearing condition on alcohol consumption in adolescent/adult subjects, with PR consuming more alcohol than MR subjects (F(1, 85) = 16.5, P = 0.0001). There was also an interaction between rearing and genotype (F(2, 85) = 3.3, P = 0.04), and this relationship remained following rank transformation of the data. Among monkeys reared in peer only groups, only those that were carriers of the G allele (T/G and G/G) consumed higher levels of alcohol than MR monkeys (Figure 4, Tukey-Kramer, P < 0.05). NPY −1002 T>G genotype accounted for 12.5% of the variance in alcohol consumption in the PR subjects.
Figure 4. Interaction between rhNPY genotype (T/T, T/G, G/G) and early rearing history (MR, mother- reared, vs. PR, peer-reared) on levels of voluntary alcohol consumption (mean ± SEM).
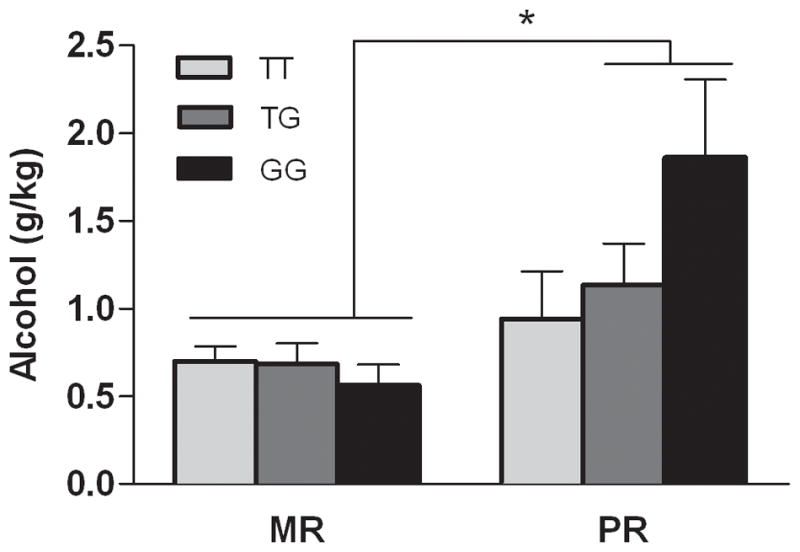
There was an interaction between rearing condition and genotype on alcohol consumption (F(2, 85) = 3.3, P = 0.04). When given simultaneous access to alcohol (8.4% v/v) and sweetened vehicle in a limited access paradigm, PR monkeys who were carriers of the G allele consumed higher levels of alcohol than did non-stress-exposed (MR) subjects genotype (Tukey-Kramer, P < 0.05). Genotype accounted for 12.5% of the variance in PR monkeys. (MR T/T = 29, MR T/G = 25, MR G/G = 8; PR T/T = 10, PR T/G = 11, PR G/G = 8). Values shown are G/KG alcohol consumed in a 1-hour session (G/KG/H) ± SEM. * P < 0.05.
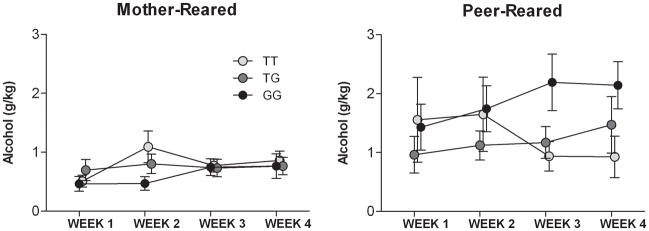
When we examined the effects of rearing and genotype on alcohol consumption following periods of deprivation across the four weeks of testing, we found a main effect of rearing (F(1, 204) = 12.5, P = 0.0007), and both genotype × time (F(6, 204) = 3.02, P < 0.008) and rearing × genotype × time (F(6, 204) = 2.2, P < 0.05) interactions. While PR monkeys that were homozygous for the T allele showed a decline in drinking over time, there was an escalation in alcohol intake among PR monkeys with the T/G and G/G genotypes (Figure 5). There was no effect of genotype in MR monkeys. When we examined the effects of genotype, rearing and time over weeks of testing during the second through fifth days for the weekly sessions, the effects of genotype (F(2,228) = 3.23, P = 0.04), rearing (F(1, 228) = 26.4, P < 0.0001), and a genotype by rearing interaction (F(2,228) = 4.9, P = 0.01) were maintained, but there were not interactions of any of these factors with week of testing (Figure S3).
Figure 5. Interaction between rhNPY genotype (T/T, T/G, G/G) and early rearing history (MR, mother- reared, vs. PR, peer-reared) on alcohol consumption over repeated weeks of alcohol deprivation (WEEKS 1, 2, 3, and 4).
There were both genotype by time (F(6, 204) = 3.02, P < 0.008) and genotype by time by rearing time (F(6, 204) = 2.2, P < 0.05) interactions. In PR monkeys, alcohol intake decreased across over time in individuals with the T/T genotype, but an escalation in consumption was observed in those carrying G allele. There was no significant effect of genotype in MR monkeys. The interaction between time and genotype accounted for 19% of the variance in PR monkeys. (MR T/T = 22, MR T/G = 19, MR G/G = 7; PR T/T = 9, PR T/G = 10, PR G/G = 7). Values shown are G/KG alcohol consumed in a 1-hour session (G/KG/H) ± SEM following a 3-day period of deprivation.
COMMENT
There is accumulating evidence that genetic and environmental factors interact to determine susceptibility to stress-related disorders later in life 3. Of particular interest for the study of GxE interactionsis variation in genes encoding stress-responsive signaling molecules that may contribute to stress vulnerability or resiliency 7. Perhaps most notable among the GxE studies are those examining interactions between life stress and the serotonin transporter-linked polymorphism (5-HTTLPR). We have previously demonstrated that a functionally similar variant in rhesus macaques, rh5-HTTLPR, interacts with early adversity to influence stress reactivity and alcohol consumption, emphasizing the utility of this model for examining gene by environment interactions that translate to the human condition.
Because the NPY system is a key modulator of behavioral adaptation to stress, we screened the rhNPY gene for variants that might impact stress resilience, with the prediction that we would observe similar interactions. We identified a SNP (−1002 T>G) in the rhNPY regulatory region that predicts loss of a GRE half site. Glucocorticoids have long been known to regulate NPY expression 35, and this regulation may be important for NPY induction during periods of stress. We performed gel shift assays with nuclear extract derived from several GR-expressing cell lines35, 36 and found that the G allele resulted in altered DNA-protein interactions with each nuclear extract tested. Among the bands that exhibited a relative decrease with the G allele (which, overall, showed increased TF binding) was one of 180 kDa, which was recognized by an anti-GR antibody. This suggests that the −1002 T>G SNP resulted in decreased preference for a functional GRE. We found that −1002 T>G predicted decreased NPY expression in amygdala, a brain region in which NPY release decreases anxious responding. Based on these functional differences, we predicted that this SNP would result in a decrease in NPY system activity and/or a failure to recruit the NPY system under stressful conditions, both of which could lead to reduced stress resiliency.
In humans, both genetic and environmental factors are suggested to influence NPY system function. Decreases in NPY levels are observed among subjects with treatment-refractory depression and PTSD 37–40. There is also evidence that a gain-of-function variant resulting in a Leu7Pro substitution of the preproNPY signal peptide 40 may protect against depression, while markers on low expressing NPY haplotypes (−399 T>C) result in decreased levels NPY levels and up-regulated stress-responses 39, 41. Here, we show that rhesus macaques exposed to adversity have lower CSF levels of NPY, but only as a function of the loss-of-function G allele. Consistent with this observation, G allele carriers are more aroused during both short-term and protracted exposures to stress. We postulate that a history of trauma and genotype may also interact to predict NPY levels in humans, and that individuals with low levels of NPY expression or who are unable to recruit the NPY system in response to stress would be less stress resilient and, therefore, more vulnerable to stress-related disorders.
There is considerable evidence suggesting that NPY regulates alcohol consumption 42–44. Npy-deficient mice consume more ethanol, while consumption is reduced in mice over-expressing Npy 44. Moreover, the Npy gene maps to a quantitative trait locus underlying alcohol consumption in genetically selected alcohol preferring rats 45. Based on these findings, a screen for functional variants was performed, identifying a marker (D4Mit7) that reduced brain expression of Npy in this line 46. In humans, linkage to the chromosomal region containing the NPY gene has been demonstrated 47, and there have been associations of NPY variation with both alcohol consumption 40 and dependence 48, 49. Other studies, however, have failed to replicate this association 50–52. Of note, our present study did not find any effects of rhNPY −1002T>G on alcohol consumption in normally reared animals, even following repeated alcohol exposure. Instead, rhNPY −1002 T>G genotype increased alcohol consumption among those exposed to both early adversity and cycles of alcohol exposure. This suggests a high degree of stress loading may be required for the G allele to produce an effect, raising the possibility that human NPY variation could potentially increase risk for alcohol dependence moreso among individuals with especially traumatic life experiences or high cumulative levels of stress exposure. In support of this argument, the only reports of a link between NPY variation and alcohol dependence have studied late-onset alcoholics 49 or samples highly represented by war veterans 48.
Dysregulation of the CRF system following repeated periods of alcohol exposure and deprivations contributes to the transition from reward- to relief- drinking 53, 5, and NPY signaling is a counter-regulatory process that buffers actions of CRF 5. When we examined patterns of alcohol intake during repeat cycles of availability and deprivation, we found an interaction between NPY genotype and alcohol exposure, such that stress-exposed carriers of the G allele exhibit an escalation in alcohol intake. This is potentially indicative of genotype-mediated inability to recruit NPY in response to induction of the CRF system among subjects consuming high levels of alcohol, suggesting that these subjects might more easily transition to the addicted state.
The NPY system is important to countering stress. We hypothesized that genetic variation that resulted in low levels of NPY expression or a failure to recruit the NPY system would render individuals less resilient to stress and to addiction with continued alcohol use. Studies in humans have demonstrated there to be haplotypes that decrease NPY expression and predict stress-induced NPY release, amygdala response and stress resiliency 41. However, whether NPY promoter variation moderates the risk for alcohol problems or interacts with life stress to moderate risk for stress-related disorders in humans has not been determined. Using an established primate model of adversity, we found that functional NPY variation influences CSF levels of NPY, behavioral arousal in response to stress, and alcohol consumption. Overall, this study suggests a role for NPY variation in the susceptibility to alcohol-related disorders and may further implicate the NPY system as a treatment target in selected individuals. Our results also suggest NPY to be a candidate for examining gene by environment interactions in humans.
Supplementary Material
Alcohol consumption on Mondays and Thursdays. During the later weeks of testing, animals exhibited increased levels of alcohol consumption following periods of deprivation (over the weekend). Values shown are G/KG alcohol consumed in a 1-hour session (G/KG/H) ± SEM. * P < 0.05.
There were effects of genotype (F(2,228) = 3.23, P = 0.04), rearing (F(1, 228) = 26.4, P < 0.0001), and a genotype by rearing interaction (F(2,228) = 4.9, P = 0.01), but no interactions of any of these factors with week of testing. (MR T/T = 22, MR T/G = 19, MR G/G = 7; PR T/T = 9, PR T/G = 10, PR G/G = 7). Values shown are G/KG alcohol consumed in a 1-hour session (G/KG/H) ± SEM averaged across Tuesday-Friday testing sessions.
Gel shift assay results from experiments performed using nuclear extract from osteosarcoma cells (MG-63) and an anti-GR antibody. Relative migrations of the protein molecular weight standards are shown to the left (in kDa). Closed arrows indicate DNA-protein complexes of approx. 90 and 180 kDa that are supershifted with the addition of the anti-GR antibody (open arrows). The 180 kDa band (GR dimer) is preferentially bound in experiments performed with T allele oligonucleotides.
Acknowledgments
We would like to thank Karen Smith for assistance in the preparation of this manuscript and the research and animal care staff at the National Institutes of Health Animal Center (NIHAC) for their assistance in data collection. This work was funded by NARSAD and the NIAAA and NICHD intramural programs.
Reference List
- 1.De Bellis MD. Developmental traumatology: a contributory mechanism for alcohol and substance use disorders. Psychoneuroendocrinology. 2002;27(1–2):155–170. doi: 10.1016/s0306-4530(01)00042-7. [DOI] [PubMed] [Google Scholar]
- 2.Reed PL, Anthony JC, Breslau N. Incidence of drug problems in young adults exposed to trauma and posttraumatic stress disorder: do early life experiences and predispositions matter? Arch Gen Psychiatry. 2007;64(12):1435–1442. doi: 10.1001/archpsyc.64.12.1435. [DOI] [PubMed] [Google Scholar]
- 3.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7(7):583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 4.Koenen KC, Nugent NR, Amstadter AB. Gene-environment interaction in posttraumatic stress disorder: review, strategy and new directions for future research. Eur Arch Psychiatry Clin Neurosci. 2008;258(2):82–96. doi: 10.1007/s00406-007-0787-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilig M, Koob GF. A key role for corticotropin-releasing factor in alcohol dependence. Trends Neurosci. 2007;30(8):399–406. doi: 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber K, Rockstroh B, Borgelt J, Awiszus B, Popov T, Hoffmann K, Schonauer K, Watzl H, Propster K. Stress load during childhood affects psychopathology in psychiatric patients. BMC Psychiatry. 2008;8:63. doi: 10.1186/1471-244X-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr CS, Goldman D. Non-human primate models of inheritance vulnerability to alcohol use disorders. Addict Biol. 2006;11(3–4):374–385. doi: 10.1111/j.1369-1600.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- 8.Suomi SJ. The development of social competence by rhesus monkeys. Ann Ist Super Sanita. 1982;18(2):193–202. [PubMed] [Google Scholar]
- 9.Ruppenthal GC, Arling GL, Harlow HF, Sackett GP, Suomi SJ. A 10-year perspective of motherless-mother monkey behavior. J Abnorm Psychol. 1976;85(4):341–349. doi: 10.1037//0021-843x.85.4.341. [DOI] [PubMed] [Google Scholar]
- 10.Higley JD, Hasert MF, Suomi SJ, Linnoila M. Nonhuman primate model of alcohol abuse: effects of early experience, personality, and stress on alcohol consumption. Proc Natl Acad Sci U S A. 1991;88(16):7261–7265. doi: 10.1073/pnas.88.16.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suomi SJ, Collins ML, Harlow HF, Ruppenthal GC. Effects of maternal and peer separations on young monkeys. J Child Psychol Psychiatry. 1976;17(2):101–112. doi: 10.1111/j.1469-7610.1976.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 12.Thorsell A, Repunte-Canonigo V, O’Dell LE, Chen SA, King AR, Lekic D, Koob GF, Sanna PP. Viral vector-induced amygdala NPY overexpression reverses increased alcohol intake caused by repeated deprivations in Wistar rats. Brain. 2007;130(Pt 5):1330–1337. doi: 10.1093/brain/awm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilpin NW, Stewart RB, Badia-Elder NE. Neuropeptide Y suppresses ethanol drinking in ethanol-abstinent, but not non--ethanol-abstinent, Wistar rats. Alcohol. 2008;42(7):541–551. doi: 10.1016/j.alcohol.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badia-Elder NE, Stewart RB, Powrozek TA, Roy KF, Murphy JM, Li TK. Effect of neuropeptide Y (NPY) on oral ethanol intake in Wistar, alcohol-preferring (P), and -nonpreferring (NP) rats. Alcohol Clin Exp Res. 2001;25(3):386–390. [PubMed] [Google Scholar]
- 15.Badia-Elder NE, Stewart RB, Powrozek TA, Murphy JM, Li TK. Effects of neuropeptide Y on sucrose and ethanol intake and on anxiety-like behavior in high alcohol drinking (HAD) and low alcohol drinking (LAD) rats. Alcohol Clin Exp Res. 2003;27(6):894–899. doi: 10.1097/01.ALC.0000071929.17974.DA. [DOI] [PubMed] [Google Scholar]
- 16.Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, Shoaf SE, Champoux M, Suomi SJ, Linnoila MV, Higley JD. Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatry. 2002;7(1):118–122. doi: 10.1038/sj.mp.4000949. [DOI] [PubMed] [Google Scholar]
- 17.Newman TK, Syagailo YV, Barr CS, Wendland JR, Champoux M, Graessle M, Suomi SJ, Higley JD, Lesch KP. Monoamine oxidase A gene promoter variation and rearing experience influences aggressive behavior in rhesus monkeys. Biol Psychiatry. 2005;57(2):167–172. doi: 10.1016/j.biopsych.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Barr CS, Dvoskin RL, Yuan Q, Lipsky RH, Gupte M, Hu X, Zhou Z, Schwandt ML, Lindell SG, McKee M, Becker ML, Kling MA, Gold PW, Higley D, Heilig M, Suomi SJ, Goldman D. CRH Haplotype as a Factor Influencing Cerebrospinal Fluid Levels of Corticotropin-Releasing Hormone, Hypothalamic-Pituitary-Adrenal Axis Activity, Temperament, and Alcohol Consumption in Rhesus Macaques. Arch Gen Psychiatry. 2008;65(8):934–944. doi: 10.1001/archpsyc.65.8.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller GM, Bendor J, Tiefenbacher S, Yang H, Novak MA, Madras BK. A mu-opioid receptor single nucleotide polymorphism in rhesus monkey: association with stress response and aggression. Mol Psychiatry. 2004;9(1):99–108. doi: 10.1038/sj.mp.4001378. [DOI] [PubMed] [Google Scholar]
- 20.Barr CS, Schwandt ML, Newman TK, Higley JD. The use of adolescent nonhuman primates to model human alcohol intake: neurobiological, genetic, and psychological variables. Ann N Y Acad Sci. 2004;1021:221–233. doi: 10.1196/annals.1308.027. [DOI] [PubMed] [Google Scholar]
- 21.Mayer CM, Cai F, Cui H, Gillespie JM, MacMillan M, Belsham DD. Analysis of a repressor region in the human neuropeptide Y gene that binds Oct-1 and Pbx-1 in GT1–7 neurons. Biochem Biophys Res Commun. 2003;307(4):847–854. doi: 10.1016/s0006-291x(03)01289-0. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi H, Nakano K, Miki N. Identification of NGF-response element in the rat neuropeptide Y gene and induction of the binding proteins. Biochem Biophys Res Commun. 1992;189(3):1553–1560. doi: 10.1016/0006-291x(92)90253-h. [DOI] [PubMed] [Google Scholar]
- 23.Kel-Margoulis OV, Kel AE, Reuter I, Deineko IV, Wingender E. TRANSCompel: a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res. 2002;30(1):332–334. doi: 10.1093/nar/30.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26(1):362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasckow J, Mulchahey JJ, Aguilera G, Pisarska M, Nikodemova M, Chen HC, Herman JP, Murphy EK, Liu Y, Rizvi TA, Dautzenberg FM, Sheriff S. Corticotropin-releasing hormone (CRH) expression and protein kinase A mediated CRH receptor signalling in an immortalized hypothalamic cell line. J Neuroendocrinol. 2003;15(5):521–529. doi: 10.1046/j.1365-2826.2003.01026.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Whang WW, Kim HT, Pyun KH, Cho SY, Hahm DH, Lee HJ, Shim I. Expression of neuropeptide Y and cholecystokinin in the rat brain by chronic mild stress. Brain Res. 2003;983(1–2):201–208. doi: 10.1016/s0006-8993(03)03087-7. [DOI] [PubMed] [Google Scholar]
- 27.Higley JD, Suomi SJ, Linnoila M. A nonhuman primate model of type II excessive alcohol consumption .1. Low cerebrospinal fluid 5-hydroxyindoleacetic acid concentrations and diminished social competence correlate with excessive alcohol consumption. Alcohol Clin Exp Res. 1996;20(4):629–642. doi: 10.1111/j.1530-0277.1996.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 28.Higley JD, Suomi SJ, Linnoila M. CSF monoamine metabolite concentrations vary according to age, rearing, and sex, and are influenced by the stressor of social separation in rhesus monkeys. Psychopharmacology (Berl) 1991;103(4):551–556. doi: 10.1007/BF02244258. [DOI] [PubMed] [Google Scholar]
- 29.Barr CS, Newman TK, Lindell S, Shannon C, Champoux M, Lesch KP, Suomi SJ, Goldman D, Higley JD. Interaction between serotonin transporter gene variation and rearing condition in alcohol preference and consumption in female primates. Arch Gen Psychiatry. 2004;61(11):1146–1152. doi: 10.1001/archpsyc.61.11.1146. [DOI] [PubMed] [Google Scholar]
- 30.Korotkova TM, Brown RE, Sergeeva OA, Ponomarenko AA, Haas HL. Effects of arousal- and feeding-related neuropeptides on dopaminergic and GABAergic neurons in the ventral tegmental area of the rat. Eur J Neurosci. 2006;23(12):3407. doi: 10.1111/j.1460-9568.2006.04792.x. [DOI] [PubMed] [Google Scholar]
- 31.Fu LY, Acuna-Goycolea C, van den Pol AN. Neuropeptide Y inhibits hypocretin/orexin neurons by multiple presynaptic and postsynaptic mechanisms: tonic depression of the hypothalamic arousal system. J Neurosci. 2004;24(40):8741–8751. doi: 10.1523/JNEUROSCI.2268-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson RM, Choe JS, Cameron HA, Thorsell A, Crawley JN, Holmes A, Heilig M. The neuropeptide Y Y1 receptor subtype is necessary for the anxiolytic-like effects of neuropeptide Y, but not the antipdepressant-lilke effects of fluoxetine in mice. Psychopharmacology (Berl) 2008;195(4):547–557. doi: 10.1007/s00213-007-0945-2. [DOI] [PubMed] [Google Scholar]
- 33.Barr CS, Newman TK, Schwandt M, Shannon C, Dvoskin RL, Lindell SG, Taubman J, Thompson B, Champoux M, Lesch KP, Goldman D, Suomi SJ, Higley JD. Sexual dichotomy of an interaction between early adversity and the serotonin transporter gene promoter variant in rhesus macaques. Proc Natl Acad Sci U S A. 2004;101(33):12358–12363. doi: 10.1073/pnas.0403763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barr CS, Schwandt ML, Lindell SG, Higley JD, Maestripieri D, Goldman D, Suomi SJ, Heilig M. Variation at the mu-opioid receptor gene (OPRM1) influences attachment behavior in infant primates. Proc Natl Acad Sci U S A. 2008;105(13):5277–5281. doi: 10.1073/pnas.0710225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higuchi H, Yang HY, Sabol SL. Rat neuropeptide Y precursor gene expression. mRNA structure, tissue distribution, and regulation by glucocorticoids, cyclic AMP, and phorbol ester. J Biol Chem. 1988;263(13):6288–6295. [PubMed] [Google Scholar]
- 36.Wenger T, Bouhdiba M, Saint PP, Ciofi P, Tramu G, Leonardelli J. Presence of neuropeptide--Y and its C-terminal flanking peptide immuno-reactivity in the seminiferous tubules of human testis. Andrologia. 1990;22(4):299–303. doi: 10.1111/j.1439-0272.1990.tb01986.x. [DOI] [PubMed] [Google Scholar]
- 37.Morgan CA3, Wang S, Southwick SM, Rasmusson A, Hazlett G, Hauger RL, Charney DS. Plasma neuropeptide-Y concentrations in humans exposed to military survival training. Biol Psychiatry. 2000;47(10):902–909. doi: 10.1016/s0006-3223(99)00239-5. [DOI] [PubMed] [Google Scholar]
- 38.Yehuda R, Brand S, Yang RK. Plasma neuropeptide Y concentrations in combat exposed veterans: relationship to trauma exposure, recovery from PTSD, and coping. Biol Psychiatry. 2006;59(7):660–663. doi: 10.1016/j.biopsych.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 39.Heilig M, Zachrisson O, Thorsell A, Ehnvall A, Mottagui-Tabar S, Sjogren M, Asberg M, Ekman R, Wahlestedt C, Agren H. Decreased cerebrospinal fluid neuropeptide Y (NPY) in patients with treatment refractory unipolar major depression: preliminary evidence for association with preproNPY gene polymorphism. J Psychiatr Res. 2004;38(2):113–121. doi: 10.1016/s0022-3956(03)00101-8. [DOI] [PubMed] [Google Scholar]
- 40.Karvonen MK, Pesonen U, Koulu M, Niskanen L, Laakso M, Rissanen A, Dekker JM, Hart LM, Valve R, Uusitupa MIJ. Association of a leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y with high serum cholesterol and LDL cholesterol levels. Nat Med. 1998;4(12):1434–1437. doi: 10.1038/4027. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, Sinha R, Virkkunen M, Mash DC, Lipsky RH, Hu XZ, Hodgkinson CA, Xu K, Buzas B, Yuan Q, Shen PH, Ferrell RE, Manuck SB, Brown SM, Hauger RL, Stohler CS, Zubieta JK, Goldman D. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452(7190):997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehlers CL, Li TK, Lumeng L, Hwang BH, Somes C, Jimenez P, Mathe AA. Neuropeptide Y levels in ethanol-naive alcohol-preferring and nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998;22(8):1778–1782. [PubMed] [Google Scholar]
- 43.Tecott LH, Heberlein U. Y do we drink? Cell. 1998;95(6):733–735. doi: 10.1016/s0092-8674(00)81695-5. [DOI] [PubMed] [Google Scholar]
- 44.Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396(6709):366–9. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- 45.Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22(4):884–887. [PubMed] [Google Scholar]
- 46.Spence JP, Liang T, Habegger K, Carr LG. Effect of polymorphism on expression of the neuropeptide Y gene in inbred alcohol-preferring and -nonpreferring rats. Neuroscience. 2005;131(4):871–876. doi: 10.1016/j.neuroscience.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz KK, Porjesz B, Li TK, Conneally PM, Nurnberger JIJ, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–215. [PubMed] [Google Scholar]
- 48.Lappalainen J, Kranzler HR, Malison R, Price LH, Van Dyck C, Rosenheck RA, Cramer J, Southwick S, Charney D, Krystal J, Gelernter J. A Functional Neuropeptide Y Leu7Pro Polymorphism Associated With Alcohol Dependence in a Large Population Sample From the United States. Arch Gen Psychiatry. 2002;59(9):825–831. doi: 10.1001/archpsyc.59.9.825. [DOI] [PubMed] [Google Scholar]
- 49.Mottagui-Tabar S, Prince JA, Wahlestedt C, Zhu G, Goldman D, Heilig M. A novel single nucleotide polymorphism of the neuropeptide Y (NPY) gene associated with alcohol dependence. Alcohol Clin Exp Res. 2005;29(5):702–707. doi: 10.1097/01.alc.0000164365.04961.b1. [DOI] [PubMed] [Google Scholar]
- 50.Zhu G, Pollak L, Mottagui-Tabar S, Wahlestedt C, Taubman J, Virkkunen M, Goldman D, Heilig M. NPY leu7pro and Alcohol Dependence in Finnish and Swedish Populations. Alcohol Clin Exp Res. 2003;27(1):19–24. doi: 10.1097/01.ALC.0000050642.62233.44. [DOI] [PubMed] [Google Scholar]
- 51.Zill P, Preuss UW, Koller G, Bondy B, Soyka M. Analysis of single nucleotide polymorphisms and haplotypes in the neuropeptide Y gene: no evidence for association with alcoholism in a German population sample. Alcohol Clin Exp Res. 2008;32(3):430–434. doi: 10.1111/j.1530-0277.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 52.Wetherill L, Schuckit MA, Hesselbrock V, Xuei X, Liang T, Dick DM, Kramer J, Nurnberger JI, Jr, Tischfield JA, Porjesz B, Edenberg HJ, Foroud T. Neuropeptide Y Receptor Genes Are Associated With Alcohol Dependence, Alcohol Withdrawal Phenotypes, and Cocaine Dependence. Alcohol Clin Exp Res. 2008 doi: 10.1111/j.1530-0277.2008.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdez GR, Koob GF. Allostasis and dysregulation of corticotropin-releasing factor and neuropeptide Y systems: implications for the development of alcoholism. Pharmacol Biochem Behav. 2004;79(4):671–689. doi: 10.1016/j.pbb.2004.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alcohol consumption on Mondays and Thursdays. During the later weeks of testing, animals exhibited increased levels of alcohol consumption following periods of deprivation (over the weekend). Values shown are G/KG alcohol consumed in a 1-hour session (G/KG/H) ± SEM. * P < 0.05.
There were effects of genotype (F(2,228) = 3.23, P = 0.04), rearing (F(1, 228) = 26.4, P < 0.0001), and a genotype by rearing interaction (F(2,228) = 4.9, P = 0.01), but no interactions of any of these factors with week of testing. (MR T/T = 22, MR T/G = 19, MR G/G = 7; PR T/T = 9, PR T/G = 10, PR G/G = 7). Values shown are G/KG alcohol consumed in a 1-hour session (G/KG/H) ± SEM averaged across Tuesday-Friday testing sessions.
Gel shift assay results from experiments performed using nuclear extract from osteosarcoma cells (MG-63) and an anti-GR antibody. Relative migrations of the protein molecular weight standards are shown to the left (in kDa). Closed arrows indicate DNA-protein complexes of approx. 90 and 180 kDa that are supershifted with the addition of the anti-GR antibody (open arrows). The 180 kDa band (GR dimer) is preferentially bound in experiments performed with T allele oligonucleotides.