Abstract
Transforming growth factor β2 (TGFβ2) is highly expressed in a variety of different cancer cell lines. Using Z12 cells, a mutant of 293 cells with overexpression of TGFβ2, we found that the cyclic adenosine monophosphate (cAMP)-responsive element (CRE) sequence in the promoter of the TGFβ2 gene is crucial for its increased expression. Further, constitutive phosphorylation of CRE-binding protein (CREB) is increased in these cells. Treating Z12 cells with either the PI3 kinase inhibitor LY294002 or the p38 MAPK inhibitor SB203580 significantly inhibited both the phosphorylation of CREB and expression of TGFβ2. In addition, treating Z12 or cancer cell lines with either of these 2 inhibitors significantly decreased their secretion of TGFβ2. These data suggest that activated PI3 kinase and p38 MAPK play important roles in high expression of TGFβ2 in cancer cells by stimulating the phosphorylation of CREB, which activates the CRE in the promoter of the TGFβ2 gene. We have identified an important link between PI3 kinase, p38 MAPK, and TGFβ2, providing an additional rationale for using inhibitors of these kinases as therapeutic drugs in cancer.
Introduction
The role of transforming growth factor β (TGFβ) in tumorigenesis is complex. Depending on cell type and stage, TGFβ can have either positive or negative effects (Wakefield and Roberts 2002). To date, 3 TGFβ genes have been identified in mammals, and they are structurally and functionally similar (O'Reilly and others 1992). Secretion of high levels of TGFβ2 is seen in many tumor cell lines (Lu and others 2004a). In addition to other effects, high levels of TGFβ2 promote the survival of tumor cells by activating nuclear factor-κB (NF-κB) (Lu and others 2004a, 2004b, 2007). Therefore, a better understanding of the mechanism of TGFβ2 overexpression in cancer is of great importance for TGFβ2-related therapy. Previously, a cyclic adenosine monophosphate (cAMP)-responsive-element (CRE) site in the promoter of the TGFβ2 gene was shown to be essential for its transcription (O'Reilly and others 1992). Here we analyze the mechanism of increased TGFβ2 expression in Z12 cells, a 293-derived cell line with excessive production of TGFβ2, but not TGFβ1 or 3 (Lu and others 2004a).
Materials and Methods
Cell culture and reagents
293C6 (293IL1R cells transfected with E-selectin-driven zeocin resistance and thymidine kinase genes) and the derived mutant cell line Z12 were established in our laboratory (Sathe and others 2004). Human glioma U87, T98G, and D54; human prostate cancer LNCap, Du145, and PC3; human breast cancer HS578T; human lung cancer HOP62; and human kidney cancer CAKI cell lines were purchased from the American Type Culture Collection. Human lung cancer NCI-H522 and NCI-H460, and human kidney cancer ACHN, UO31, and 7860 cell lines were kind gifts of Dr. Dennis Stacey, Cleveland Clinic. Human breast cancer MCF7, T47D, and BT-549 cell lines were kind gifts of Dr. Zhenghe Wang, Case Western Reserve University. The human kidney cancer cell line RCC54 was a gift of Dr. Andrei Gudkov, Roswell Park Cancer Institute.
C6, Z12, U87, T98G, and D54 cells were cultured in Dulbecco's modified Eagle's medium. All other cell lines were cultured in RPMI-1640 medium. All media were supplemented with penicillin 100 U/mL, streptomycin 100 mg/mL, and 10% fetal calf serum. Antibody to Ser-133 phosphorylated CRE-binding protein (CREB) was from Cell Signaling (Cat. No. 9198). LY294002 (LY2), and SB203580 were from Calbiochem.
Plasmids and transfections
A 1,729-bp fragment of the TGFβ2 promoter was cloned from a human cDNA library by using the forward primer 5′ CCATTCCCGGGTACCACAGTGATAGCTAATTCACG 3′ and the reverse primer 5′ GCGATCTCGAGTCTGTCTTTCTCTTGTCAGG 3′. This fragment was cut out by endonuclease KpnI and XhoI and cloned into luciferase reporter plasmid pGL3 (Promega). Forward primers to clone shorter TGFβ2 promoter fragments were as follows: −781 construct, 5′ CCATTCCCGGGTACCAATGTCAAGTTATGAGTAGTGTAG 3′; −191 construct, 5′ AATCCCGGGTACCGCGCACATTCCACCTC 3′; −80 construct, 5′ CCATTCCCGGGTACCATCCTAGCACGTCACTT 3′; and the same reverse primer as for the −1729 construct. The forward primer for the CRE mutant construct was 5′ CTTCTGACTGTAATCCTGGCATGGCACTTTGTTGAAGGCAGAC 3′ and the reverse primer was 5′ GTCTGCCTTCAACAAAGTGCCATGCCTAGGATTACAGTCAGAAG 3′. All transfections were carried out with the Lipofectamine Plus reagent (Promega). Efficiencies of transfection were normalized to β-galactosidase activity from a cotransfected pCMVLacZ β-galactosidase reporter plasmid.
Western analyses
Cells were washed with 1× phosphate-buffered saline and pelleted at 3,000 g at 4°C for 4 min. Cell pellets were lysed with immunoprecipitation assay buffer (Sathe and others 2004). Cellular debris was removed by centrifugation at 16,000 g for 10 min. The amount of protein in the supernatant solution was determined, and samples were heat-treated in 2× sodium dodecyl sulfate (SDS) sample loading buffer (Sathe and others 2004) at 100°C for 5 min. Equal amounts of samples were fractionated by SDS/polyacrylamide gel electrophoresis. Western analysis was performed with primary antibodies, which were observed with horseradish-peroxidase-coupled secondary antibodies, using the ECL Western detection system (PerkinElmer).
Northern analyses and real-time polymerase chain reaction
Northern analyses were performed as described by Lu and others (2004b). Real-time polymerase chain reaction (PCR) was performed as described by Wan and others (2009), and the results were normalized to human glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The specific primer sequences used for GAPDH were 5′ GAGCTGAACGGGAAGCTCAC 3′ and 5′ TGTCATACCAGGAAATGAGC 3′; those for TGFβ2 were 5′ CGACGAAGAGTACTACGCCA 3′ and 5′ CGGGCAGAGCTAAACCTCAG 3′.
Enzyme-linked immunosorbent assay for TGFβ2
Conditioned media were collected as described by Lu and others (2004a). Enzyme-linked immunosorbent assay (Quantikine-Human TGFβ2 Immunoassay) was carried out following by the protocol from R&D Systems. The amount of TGFβ2 was normalized to cell numbers.
Results
TGFβ2 is highly expressed in Z12 and a variety of cancer cell lines
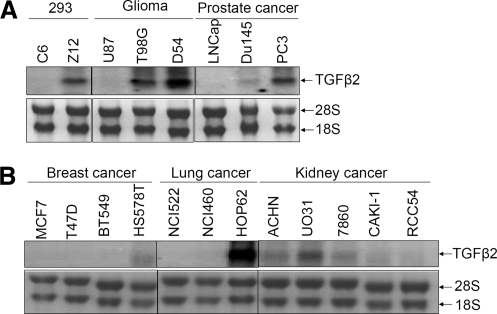
The Z12 mutant cell line was established in our laboratory following chemical mutagenesis of 293C6 cells (Sathe and others 2004). Previously, we found that Z12 cells secrete a high level of TGFβ2 (Lu and others 2004a). Northern analysis shows that the level of TGFβ2 mRNA in Z12 cells is much higher than that in C6 cells (Fig. 1A), consistent with our previous result (Lu and others 2004a, 2004b). We also assayed expression of TGFβ2 in several groups of human cancer cell lines, including lines derived from gliomas and cancers of the prostate, breast, lung, and kidney (Fig. 1A, B). Many of these cells have high TGFβ2 expression, eg, glioma T98G and D54 cells, prostate cancer PC3 and DU145 cells, breast cancer HS578T cells, lung cancer HOP62 cells, and kidney cancer UO31 cells, showing that overexpression of TGFβ2 is common in many different cancer cell lines.
FIG. 1.
TGFβ2 mRNA is highly expressed in many different cancer cell lines. Total RNAs (10 μg) were analyzed by the Northern method, using a TGFβ2 probe. (A) Analysis of TGFβ2 in 293C6 and Z12 cells, and in glioma and prostate cancer cells. (B) Analysis of TGFβ2 in breast, lung, and kidney cancer cells. TGFβ2, transforming growth factor β2.
Elevation of TGFβ2 mRNA in Z12 cells is not caused by altered mRNA turnover
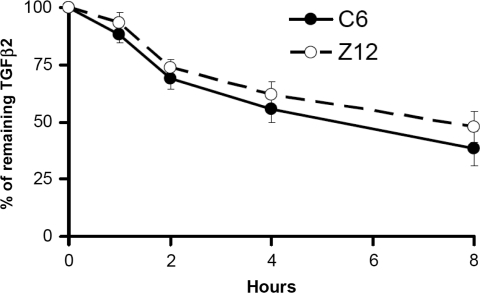
Since elevated mRNA levels can be caused by either increased transcription or decreased decay, we determined the decay rate of TGFβ2 mRNA. To have an appropriate control, we compared Z12 cells with their parental 293C6 cells. The cells were treated with actinomycin D to block mRNA synthesis, and TGFβ2 mRNAs were measured using real-time PCR. The decay rates of TGFβ2 are very similar in C6 and Z12 cells (Fig. 2).
FIG. 2.
TGFβ2 mRNA decays at similar rates in C6 and Z12 cells. The cells were treated with actinomycin D (5 μg/mL) for the indicated times. TGFβ2 mRNA was analyzed by real-time polymerase chain reaction, normalized to glyceraldehyde 3-phosphate dehydrogenase. The results shown are the means and standard deviations of triplicate experiments. TGFβ2, transforming growth factor β2.
The CRE is critical for increased TGFβ2 transcription in Z12 cells
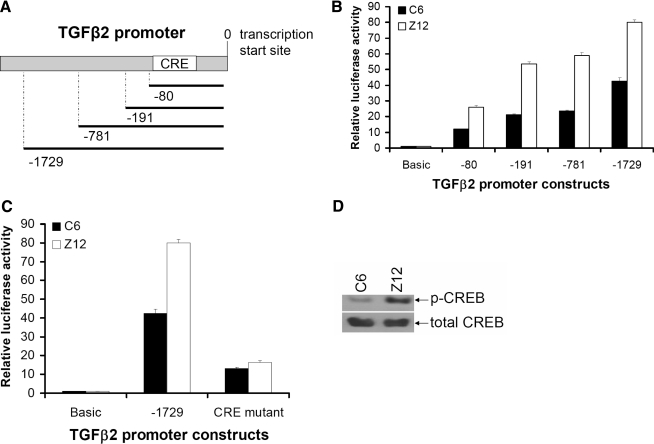
To study the mechanism of increased TGFβ2 transcription in Z12 cells, we used promoter deletion to identify the crucial sequence in the TGFβ2 promoter. Different lengths of promoter sequence were cloned into a promoterless luciferase reporter vector (Fig. 3A), and these constructs were transfected into C6 and Z12 cells. The luciferase assay shows that even the shortest fragment, which contains only 80 nucleotides upstream of the TGFβ2 transcription start site, can drive luciferase transcription (Fig. 3B). It is striking that even the short 80 bp promoter shows different transcription efficiencies in C6 and Z12 cells (Fig. 3B), indicating that this sequence is critical for increased TGFβ2 transcription in Z12 cells. O'Reilly and others (1992) have shown that a functional CRE within this region is critical for TGFβ2 transcription. To test whether the CRE is crucial for overexpression of TGFβ2 in Z12 cells, we mutated it and expressed the mutant construct into 293C6 and Z12 cells. Not only does the mutant construct have much reduced luciferase activity (Fig. 3C), but also the mutation eliminated the difference between C6 and Z12 cells (Fig. 3C), strongly suggesting that the CRE site is not only responsible for basal TGFβ2 production in both C6 and Z12 cells, but also involved in overexpression of TGFβ2 in Z12 cells. We analyzed the activation status of CREB in both C6 and Z12 cells by the Western method (Fig. 3D). CREB was more highly activated by phosphorylation in Z12 cells than in C6 cells, suggesting that pathways leading to the activation of CREB are the major cause of the increased transcription of TGFβ2 in Z12 cells.
FIG. 3.
Promoter assays show that the CRE site is crucial for increased TGFβ2 transcription in Z12 cells. (A) Schematic diagram of TGFβ2 promoter. (B) TGFβ2 promoter assay. Luciferase constructs with different lengths of the promoter were transfected into C6 or Z12 cells. The luciferase readings were normalized to β-galactosidase. The results shown are the means and standard deviations of triplicate experiments. (C) The CRE site was mutated in the TGFβ2 promoter construct, followed by transfection into C6 or Z12 cells. The luciferase readings were normalized to β-galactosidase. The results shown are the means and standard deviations of triplicate experiments. (D) C6 and Z12 cells were analyzed by the Western method with an antibody to phosphorylated CREB. CRE, cAMP-responsive element; TGFβ2, transforming growth factor β2; CREB, cyclic adenosine monophosphate (cAMP)-responsive element binding protein.
Suppression of TGFβ2 expression by PI3 kinase and p38 MAPK inhibitors in Z12 and cancer cell lines
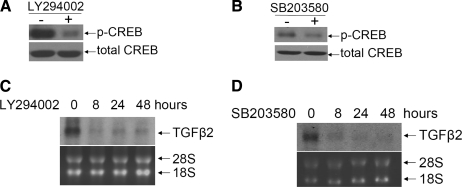
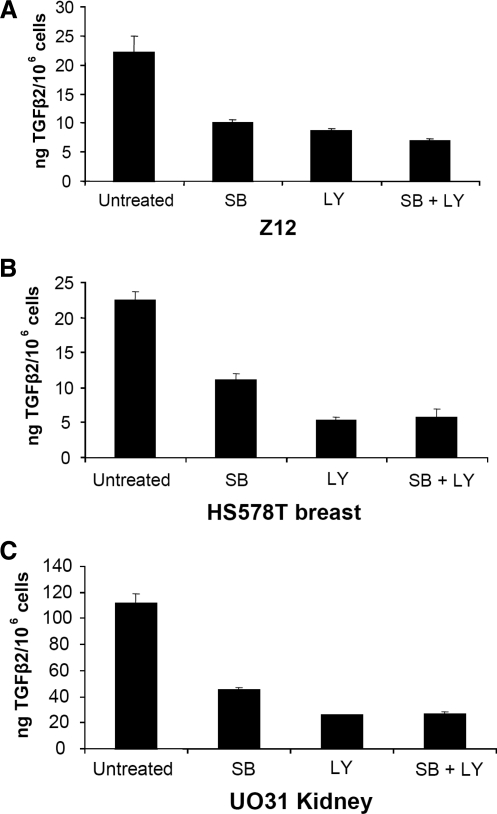
The CREB transcription factors are regulated by multiple kinases, including PI3 kinase and p38 MAPK (Mayr and Montminy 2001). To identify the kinases responsible for increased TGFβ2 expression in Z12 cells, we used the chemical inhibitors LY294002 for PI3 kinase and SB203580 for p38 MAPK. Both the phosphorylation of CREB (Fig. 4A, B) and the levels of TGFβ2 mRNA (Fig. 4C, D) were greatly decreased upon treatment with either of these two inhibitors. We then checked TGFβ2 production from Z12 and cancer cells (Fig. 5) treated with LY294002 and SB203580, separately or together. TGFβ2 secretion was substantially reduced following all three treatments, indicating that both PI3 kinase and p38 MAPK help to drive overexpression of TGFβ2 in Z12 and cancer cells.
FIG. 4.
Treatment with the PI3 kinase inhibitor LY294002 or the p38 MAPK inhibitor SB203580 decreased both the phosphorylation of CREB and TGFβ2 expression in Z12 cells. (A, B) Western analyses, showing that phosphorylated CREB decreased following treatment with LY294002 (40 μM) or SB203580 (20 μM). Z12 cells were treated for 24 h and analyzed by the Western method with an antibody to phosphorylated CREB. (C, D) Northern analyses, showing that the level of TGFβ2 mRNA in Z12 cells was decreased following treatment with LY294002 (40 μM) or SB203580 (20 μM). Total RNAs (10 μg) were analyzed by the Northern method, with a TGFβ2 probe. CREB, cyclic adenosine monophosphate (cAMP)-responsive element binding protein; TGFβ2, transforming growth factor β2.
FIG. 5.
Treatment with inhibitors of PI3 kinase or p38 MAPK decreased TGFβ2 production in cancer cells. (A) Z12 cells, (B) breast cancer HS578T cells, or (C) kidney cancer UO31 cells were treated with LY294002 (40 μM), SB203580 (20 μM), or combined LY294002 (40 μM) and SB203580 (20 μM) in 500 μL of a fresh medium in 12-well plates. Twenty-four hours later, the conditioned media were analyzed by enzyme-linked immunosorbent assay for secretion of TGFβ2. TGFβ2, transforming growth factor β2.
Discussion
Members of the TGFβ family have both positive and negative effects in tumorigenesis. While the inhibitory effects dominate at early stages, oncogenic effects are observed late in tumorigenesis (Wakefield and Roberts 2002), where altered expression of the TGFβ receptor and downstream effector molecules such as Smad can lead to loss of tumor suppressive effects (Xu and others 2000). It is thought that TGFβ benefits late-stage tumorigenesis by promoting invasiveness and metastasis (Oft and others 1998).
We found that TGFβ2 is overexpressed in cancer cells derived from several different tumors (Fig. 1), suggesting that the positive effects of TGFβ2 on tumorigenesis might be shared by many different cancers. Our previous work showed that high levels of TGFβ2 activate NF-κB, inhibiting apoptosis in PC3 prostate cancer cells (Lu and others 2004a). Suppression of TGFβ2 expression in PC3 cells by an anti-TGFβ2 antibody or siRNA promotes cell death (Lu and others 2004a). For the future, it is important to determine whether the activation of NF-κB is observed in all tumor cell lines with high levels of TGFβ2 expression, and whether blocking TGFβ2 expression causes the death of all of these cancer cells.
It is virtually impossible to find an appropriate normal control for analysis of the mechanism of TGFβ2 overexpression in cancer cells. To overcome this problem, we compared TGFβ2 overexpression in the Z12 mutant cell line with its normal expression in C6 parental cells (Lu and others 2004a, 2004b; Sathe and others 2004). We now find that the CRE site of the TGFβ2 promoter is essential for its increased activity in Z12 cells, and increased TGFβ2 expression in Z12 cells is caused by the activation of CREB, through phosphorylation. Akt mediates the activation of CREB in response to serum stimulation, and treatment with LY294002, a kinase inhibitor of PI3 kinase, an upstream activator of Akt, abolishes the activation of CREB (Du and Montminy 1998). In addition, p38 MAPK is required for the activation of CREB in response to insulin-like growth factor-1, and treatment of cells with the p38 MAPK inhibitor SB203580 abolishes CREB-mediated gene expression (Pugazhenthi and others 1999). In our study, treatment with LY294002 or SB203580 decreased CREB phosphorylation in Z12 cells (Fig. 4A, B). Further, treatment with these two inhibitors, separately or together, had a major effect in decreasing the TGFβ2 production in Z12 cells, breast cancer HS578T cells, and kidney cancer UO31 cells (Fig. 5), suggesting that signaling pathways mediated by PI3 kinase and p38 MAPK are involved in TGFβ2 overexpression in these cell lines. Importantly, both PI3 kinase and p38 MAPK are often found to be activated in cancer, and chemical inhibitors of both pathways are considered to have great potential for cancer therapy (Cuenda and Rousseau 2007; Garcia-Echeverria and Sellers 2008). Our current findings provide further insight into the complex interactions among several major signaling pathways in cancer and also provide an additional rationale for using inhibitors of PI3 kinase and p38 MAPK as potential anticancer drugs.
Acknowledgment
This work was supported by National Institutes of Health Grant P01 62220.
Author Disclosure Statement
No competing financial interests exist.
References
- Cuenda A. Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases [Review] Biochim Biophys Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Du K. Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- Garcia-Echeverria C. Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer [Review] Oncogene. 2008;27:5511–5526. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- Lu T. Burdelya LG. Swiatkowski SM. Boiko AD. Howe PH. Stark GR. Secreted transforming growth factor-β2 activates NF-κB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci U S A. 2004a;101:7112–7117. doi: 10.1073/pnas.0402048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T. Sathe SS. Swiatkowski SM. Hampole CV. Stark GR. Secretion of cytokine and growth factors as a general cause of constitutive NF-κB activation in cancer. Oncogene. 2004b;23:2138–2145. doi: 10.1038/sj.onc.1207332. [DOI] [PubMed] [Google Scholar]
- Lu T. Tian L. Han Y. Vogelbaum M. Stark GR. Dose-dependent cross-talk between the transforming growth factor-β and interleukin-1 signaling pathways. Proc Natl Acad Sci U S A. 2007;104:4365–4370. doi: 10.1073/pnas.0700118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B. Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Oft M. Heider KH. Beug H. TGF-β signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8:1243–1252. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- O'Reilly MA. Geiser AG. Kim SJ. Bruggeman LA. Luu AX. Roberts AB. Sporn MB. Identification of an activating transcription factor (ATF) binding site in the human transforming growth factor-β2 promoter. J Biol Chem. 1992;267:19938–19943. [PubMed] [Google Scholar]
- Pugazhenthi S. Boras T. O'Cnnor D. Meintzer MK. Heidenriech KA. Reusch JE. Insulin-like growth factor 1-mediated activation of the transcription factor cAMP response element-binding protein in PC12 cells. J Biol Chem. 1999;274:2829–2837. doi: 10.1074/jbc.274.5.2829. [DOI] [PubMed] [Google Scholar]
- Sathe SS. Sizemore N. Li X. Vithalani K. Commane M. Swiatkowski SM. Stark GR. Mutant human cells with constitutive activation of NF-κB. Proc Natl Acad Sci U S A. 2004;101:192–197. doi: 10.1073/pnas.0306812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield LM. Roberts AB. TGF-β signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- Wan Y. Xiao H. Affolter J. Kim TW. Bulek K. Chaudhuri S. Carlson D. Hamilton T. Mazumder B. Stark GR. Thomas J. Li X. Interleukin-1 receptor-associated kinase 2 is critical for lipopolysaccharide-mediated post-transcriptional control. J Biol Chem. 2009;284:10367–10375. doi: 10.1074/jbc.M807822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. Brodie SG. Yang X. Im Y. Parks WT. Chen L. Zhou YX. Weinstein M. Kim ST. Deng C. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]