Abstract
Experimentally, paramyxoviruses are conventionally considered good inducers of type I interferons (IFN-α/β), and have been used as agents in the commercial production of human IFN-α. However, in the last few years it has become clear that viruses in general mount a major challenge to the IFN system, and paramyxoviruses are no exception. Indeed, most paramyxoviruses encode mechanisms to inhibit both the production of, and response to, type I IFN. Here we review our knowledge of the type I IFN-inducing signals (by so-called pathogen-associated molecular patterns, or PAMPs) produced during paramyxovirus infections, and discuss how paramyxoviruses limit the production of PAMPs and inhibit the cellular responses to PAMPs by interfering with the activities of the pattern recognition receptors (PRRs), mda-5, and RIG-I, as well as downstream components in the type I IFN induction cascades.
Introduction
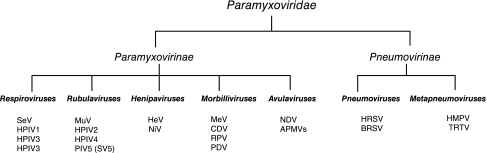
Paramyxoviruses are small enveloped viruses with a single-stranded negative sense genome of 15–19 kb (reviewed by Lamb and Parks (2006)). The Paramyxoviridae family (see Fig. 1) includes a number of important disease-causing viruses, including measles virus (MeV), mumps virus (MuV), the human parainfluenza viruses (HPIV), and human respiratory syncytial virus (HRSV) of man, as well as Newcastle disease virus (NDV), bovine respiratory syncytial virus (BRSV), rinderpest virus (RPV), turkey rhinotracheitis virus (TRTV), and Sendai virus (SeV) of mammals and birds. Certain paramyxoviruses also have zoonotic potential (Wang and others 2001) observed during outbreaks of the newly emergent Hendra virus (HeV) and Nipah virus (NiV), which appear to have natural reservoirs in fruit bats but have also infected farm animals, domestic animals, and humans.
FIG. 1.
Classification of paramyxoviruses. The Paramyxoviridae family is divided into Paramyxovirinae and the Pneumovirinae subfamilies. The Paramyxovirinae subfamily contains five genera: respiroviruses, rubulaviruses, henipaviruses, morbilliviruses, and avulaviruses. The Pneumovirinae subfamily contains 2 genera: pneumoviruses and metapneumoviruses. This classification is based predominantly on sequence homology and genome organization. Abbreviations: SeV, Sendai virus; HPIV, human parainfluenza viruses; BPIV3, bovine parainfluenza virus 3; MuV, mumps virus; SV5, simian virus 5; HeV, Hendra virus; NiV, Nipah virus; MeV, measles virus; CDV, canine distemper virus; RPV, rinderpest virus; PDV, phocine distemper virus; NDV, Newcastle disease virus; APMVs, avian paramyxoviruses; HRSV, human respiratory syncytial virus; BRSV, bovine respiratory syncytial virus; HMPV, human metapneumovirus; TRTV, turkey rhinotrachetis virus.
The life cycle of paramyxoviruses (reviewed in Lamb and Parks (2006)) takes place in the cytoplasm of infected cells after the viral nucleoprotein/genome complex has been introduced by a process of fusion between the cell membrane and viral envelope. The 3′ terminus of genomic RNA has promoters for initiation of both virus transcription and replication whereas the 3′ terminus of antigenomic RNA only functions in replication. Viral transcription occurs when the viral polymerase binds to the 3′ promoter and processes along the template, producing first a 5′-triphosphorylated leader sequence and then initiating stable transcription at gene-specific sequences that flank each viral open-reading frame (ORF) to produce 5′-capped RNAs that are also polyadenylated by the polymerase (reviewed in Whelan and others (2004)). As expected from the small size of the viral genome, the paramyxoviruses have a limited genetic repertoire. All paramyxoviruses have genes encoding nucleoprotein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), attachment protein (HN for respiroviruses, rubulaviruses, and avulaviruses, H for morbilliviruses, and G for henipaviruses and members of the Pneumovirinae subfamily), and RNA-dependent RNA polymerase (L). Some of the rubulaviruses and avulaviruses, and all of the Pneumovirinae also encode a small protein called SH. Pneumoviruses and metapneumoviruses also contain an M2 gene, and in addition pneumoviruses have 2 genes that encode the nonstructural NS1 and NS2 proteins, whose function will be discussed later.
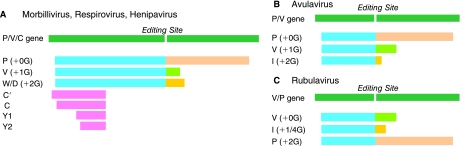
The number of proteins encoded by viruses within the subfamily Paramyxovirinae is larger than the number of genes contained in their genome, as their P genes have overlapping ORFs that give rise to multiple, distinct gene products (see Fig. 2). The morbilliviruses, respiroviruses, henipaviruses, and avulaviruses generate a primary transcript whose mRNA is translated authentically to generate the P protein; however, a unique process of “RNA editing” (also known as pseudo-template addition) can result in the insertion of G residues at an “editing site” in the middle of the transcript to generate mRNAs where there is a frameshift in the coding sequence downstream from this site. The insertion of a single G generates an mRNA that translates into a protein called the “V protein,” which has a common N-terminus to the P protein, but a unique C-terminus. The insertion of a second G residue creates an mRNA that encodes a protein with a different C-terminus called, variously, “W”, “D,” or “I.” Interestingly, in the rubulaviruses it is the V protein that is genomically templated, and production of the P protein requires an insertion of 2Gs into the mRNA by RNA editing; the addition of 1 or 4 Gs creates an mRNA for the I protein. In addition to these products, morbilliviruses, respiroviruses, and henipaviruses translate the P/V/W/D mRNAs using an alternative reading frame(s) to generate poorly conserved “C” protein(s); although only a single C protein is normally made, SeV can utilize 4 distinct start codons to generate C′, C, Y1, and Y2 proteins that have a different N-terminus but a common C-terminus. The P genes of viruses within the Pneumovirinae subfamily do not encode more than 1 protein, but rather some of these viruses, including HRSV and BRSV, have 2 extra genes, NS1 and NS2 that, like the V and C proteins, act as interferon (IFN) antagonists (see below).
FIG. 2.
Organization of the P/V/C genes of Paramyxovirinae and their relationship to their accessory proteins. (A) The P proteins of morbilliviruses, respiroviruses, and henipaviruses are translated from mRNAs produced as faithful copies of their P/V/C genes (green bar). Insertion of a single G at the RNA-editing site generates a transcript that encodes the V protein, and insertion of 2 G residues generates a transcript that encodes the W proteins and D proteins (HPIV3). The P, V, W, and D proteins share a common N-terminus (blue bar), but distinct C-termini (P = pink bar, V = light green bar, W and D = yellow bar). The C protein(s) (lilac bars) are generated by translation of the P/V/W/D mRNAs using alternative initiation codons. Translation of the prototypical C protein begins at an AUG that resides downstream of P protein initiation codon, and the Y1 and Y2 proteins of Sendai virus are translated from AUGs that reside even further downstream. The C′ protein made by Sendai virus is translated from an ACG codon that resides 5′ to the AUG of the P protein. (B) The P proteins of avulaviruses are also translated from mRNAs produced as faithful copies of their P/V genes (green bar). Insertion of a single G at the RNA-editing site generates a transcript that encodes the V protein, and insertion of 2 G residues generates a transcript that encodes the I protein. (C) In contrast, the V proteins of rubulaviruses are translated from mRNAs produced as faithful copies of their P/V genes (green bar). Insertion of 1 or 4 G residues at the RNA-editing site generates a transcript that encodes the I protein, and insertion of 2 G residues generates a transcript that encodes the P protein.
The alternative products of the P gene are rarely essential for viral replication, leading to them being described as “luxury functions” or “accessory proteins,” but their deletion frequently leads to severe attenuation. Although this attenuation is primarily associated with a failure to control the host IFN response, these proteins have additional functions, as recombinant viruses that cannot make these accessory proteins often replicate poorly in “IFN-compromised” cells. As discussed elsewhere in this issue (Ramachandran and Horvath 2009), many V and C proteins and the Pneumovirus NS1 and NS2 proteins have the capacity to interfere with IFN signaling. However, blocking IFN signaling alone is not sufficient to allow these viruses to fully circumvent the IFN response because if a virus only blocked IFN signaling, infected cells may still respond to infection by releasing large amounts of type I IFN. This would induce an antiviral state in surrounding uninfected cells, making it difficult for the virus to spread from the initial foci of infection (Andrejeva and others 2002; Carlos and others 2005; Precious and others 2007). Thus, paramyxoviruses also attempt to limit type I IFN induction. Here we review this active area of research.
Paramyxoviruses and Type I IFN Induction
As discussed elsewhere in this volume (Gale and Sen 2009), the induction of type I IFN is brought about by the detection of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) in infected cells. Viral PAMPs are usually nucleic acids with structures not found in uninfected cells, such as dsRNA or uncapped ssRNA with a 5′ triphosphate, although it has been reported that HRSV can induce type I IFN through TLR4 via recognition of its F protein (Kurt-Jones and others 2000; Haynes and others 2001). Viral nucleic acids can become associated with PRRs present either in endosomes as a result of endocytosis of virus particles/extracellular debris or autophagy of cytoplasmic material (dsRNA, ssRNA, or CpG DNA, recognized by TLR3, TLR7, and TLR9, respectively), or in the cytoplasm where they are recognized by the related RNA helicases, RIG-I and mda-5. Our knowledge of the nature of the ligands for RIG-I and mda-5 is incomplete, although the current consensus is that RIG-I responds to ssRNAs containing uncapped 5′ triphosphorylated ends (Hornung and others 2006; Pichlmair and others 2006; Cui and others 2008; Takahasi and others 2008) and short dsRNA molecules (Kato and others 2008), whereas mda-5 responds to longer dsRNA molecules (Kato and others 2008). Once activated, RIG-I and mda-5 interact with a common adaptor VISA (also known as Cardif/MAVS/IPS-1) through an N-terminal CARD domain. Recruitment of VISA initiates the activation of the transcription factors IRF-3 and NF-κB and ultimately results in the transcription of proinflammatory cytokine genes such as IFN-χ (reviewed in Randall and Goodbourn (2008), Gale and Sen (2009)).
A considerable number of studies on the regulation of type I IFN induction have used laboratory preparations of paramyxoviruses as potent inducers, especially SeV and NDV. Despite this and despite the fact that the replicative strategies of the Paramyxoviridae are understood in detail (reviewed in Lamb and Parks (2006), Whelan and others (2004)), we know relatively little about the nature of the IFN-inducing paramyxoviral PAMPs. Although paramyxoviruses efficiently encapsidate both their genomic and antigenomic RNAs by linking the process of replication to encapsida-tion, thereby “hiding” the free 5′ triphosphate on these molecules, they have nevertheless been shown to activate RIG-I (Kato and others 2006; Plumet and others 2007; Strahle and others 2007; Habjan and others 2008). Paramyxoviruses appear to generate little free dsRNA during the course of an infection (Weber and others 2006) and thus should also be poor activators of mda-5. Nevertheless, there are a number of reports that demonstrate that paramyxoviruses activate mda-5 (Andrejeva and others 2004; Yoneyama and others 2004; Melchjorsen and others 2005; Yoneyama and others 2005; Berghall and others 2006; Yount and others 2008) and, as discussed later, many paramyxoviruses encode products that interfere directly with mda-5.
One striking feature to emerge from studies on SeV is that plaque-purified virus is a poor inducer of type I IFN, and that a good induction profile only emerges when significant levels of defective interfering (DI) particles accumulate within the virus stock preparations (Johnston 1981; Poole and others 2002); we have made similar observations for PIV5 and MuV (unpublished observations), and it has also been reported that vaccine strains of MeV are associated with the induction of type I IFN that correlates with DI particle production (Shingai and others 2007). DI particles generated by SeV are the best understood, and are of 2 types, internal deletion or copyback. The latter are generated by promoter exchange during replication and thus contain ssRNA with a terminal pan-handle of dsRNA. Internal deletion genomes lack segments of the viral genome but are otherwise normal. In a molecular analysis of type I IFN induction by SeV, Kolakfsky and colleagues have shown that induction is strongly correlated to the levels of copyback genomes in their laboratory stocks and commercial preparations (Strahle and others 2006); these particles have been shown to activate RIG-I (Strahle and others 2007). However, equivalent preparations of purified DI particles of SeV are also strong activators of an mda-5-dependent antiviral innate immunity program in macrophages (Yount and others 2008). These studies raise an interesting question of what the physiological inducers of type I IFN really are during infections in vivo—are they PAMPs generated by the virus as a result of its normal repli-cative strategy (we propose to call these “essential PAMPs”), or are they PAMPs generated as a result of aberrant transcription or replication (“corrupted PAMPs”)? This question is also relevant when we consider virally encoded type I IFN antagonists—have these evolved to limit the action of “essential PAMPs,” or limit the activities of potentially more potent “corrupted PAMPs”? The issue about the potency of “essential” versus “corrupted PAMPs” also suggests that the strategies by which viruses control the fidelity of replication and transcription will have an important bearing on the control of type I IFN induction and viral fitness.
Although the requirement for accessory proteins in generating fully virulent paramyxoviruses has long been appreciated, a link to the IFN system was not established until relatively recently. Initially using PIV5 (formerly known as SV5), it was first shown that PIV5 specifically blocks IFN signaling by targeting STAT1 for proteasome-mediated degradation (Didcock and others 1999a, 1999b) and, later, that it also specifically blocks type I IFN production (He and others 2002; Poole and others 2002; Wansley and Parks 2002). Subsequent studies have shown that these observations hold true for most other paramyxoviruses (effects of paramyxoviruses on IFN signaling are reviewed in Ramachandran and Horvath (2009)). The ability to regulate the production of type I IFN also appears to be a common feature of paramyxoviruses, and a number of studies have demonstrated that viruses bearing deletions of the V gene and/or the C gene have the phenotype of enhanced type I IFN production (see below). These studies were often unable to distinguish between viral functions that controlled the generation of PAMPs from those that encoded specific type I IFN antagonists. However, analyses of the properties of individually expressed genes have demonstrated that a number of them possess the ability to directly interfere with type I IFN induction. This will be discussed later and is summarized in Figure 3. Additionally, it is becoming increasingly clear that viral products actively limit the production of PAMPs by regulating viral transcription and replication.
FIG. 3.
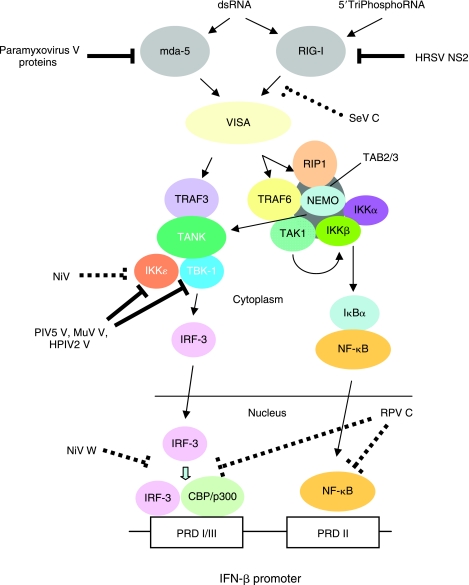
Paramyxovirus accessory proteins target the intracellular viral pathogen-associated molecular patterns (PAMP) signaling pathways. The signaling pathways leading from the RNA helicases mda-5 and RIG-I to IFN-β induction are shown (reviewed in Randall and Goodbourn (2008), Gale and Sen (2009)). As discussed in the text, paramyxovirus V proteins interact with mda-5 and prevent its activation. Sendai virus (SeV) C protein targets RIG-I, although a specific molecular interaction has yet to be shown. The NS2 protein of human respiratory syncytial virus (HRSV) directly binds to RIG-I and inhibits its activity. The V proteins of human parainfluenza virus 2 (HPIV2), simian virus 5 (PIV5, formerly SV5), and mumps virus (MuV) interact with and inhibit TBK1 and IKK-ɛ, and NiV V inhibits IKK-ɛ (although not TBK1). The C protein of rinderpest virus (RPV) and the W protein of Nipah virus (NiV) have uncharacterized nuclear targets that act downstream of transcription factors.
V Proteins
The role of mda-5 as a PRR was first unveiled by studies on the V proteins of paramyxoviruses; the V proteins of most paramyxoviruses are able to antagonize both the NF-κB and IRF-3 arms of the dsRNA signaling responses (Poole and others 2002; Childs and others 2007), and mda-5 was identified as an interacting partner for the V proteins of, initially, PIV5 (Andrejeva and others 2004), but subsequently HPIV2, MuV, SeV, MeV, HeV, NiV, Menangle virus, Mapuera virus, Salem virus, Porcine rubulavirus, Tioman virus, and NDV (Andrejeva and others 2004; Childs and others 2007). The interaction with mda-5 was shown to require the cysteine-rich C-terminus of the V protein, which is the only region of the V protein that shows significant conservation, and for each of the V proteins tested, the interaction inhibits activation of mda-5 (Childs and others 2007)—see Figure 3. In contrast, RIG-I does not appear to be an interaction partner for the V proteins examined (Childs and others 2007). By analogy to RIG-I (Cui and others 2008; Takahasi and others 2008), the activation of mda-5 requires the binding of dsRNA to 2 separate sites (one at the C-terminus and one in the helicase domain), which triggers multimerization (Childs and others 2009) and a conformational change leading to the formation of an mda-5 structure that enables the N-terminal CARD domains to align in such a way as to permit recruitment of the VISA adaptor molecule. The PIV5 V protein binds to several sites on mda-5 within the C-terminus and helicase domain, thus preventing the RNA from binding and the activated multimer from forming (Childs and others 2009). Whilst most paramyxovirus V proteins bind mda-5, it is not a universal feature, since the V protein of RPV does not interact with mda-5 (Boxer and others 2009). Furthermore, HPIV3 does not encode a V protein as the RNA-editing event within the Respirovirus HPIV3 P gene generates a protein that is truncated before the cysteine-rich C-terminus.
As discussed earlier, there is a significant body of evidence that indicates that paramyxoviruses can induce type I IFN through the activation of RIG-I as well mda-5, and therefore it seems reasonable that they encode products to block RIG-I activation of the type I IFN induction cascade. In this regard it is of note that it has recently been reported that the V proteins of the rubulaviruses, PIV5, HPIV2, and MuV inhibit TBK1 and IKK-ɛ (Lu and others 2008), kinases which reside downstream of both mda-5 and RIG-I in the type I IFN induction cascade (see Fig. 3). The interaction between the V proteins and TBK1/IKK-ɛ appears to be direct and under these circumstances the V proteins become a decoy substrate to prevent the phosphorylation of IRF-3. However, these interactions may be a cell type-specific effect since V does not inhibit TLR3, TRIF, TBK1, or RIG-I signaling in some cell lines tested (Hilton and others 2006; Childs and others 2007). The V protein of NiV has also been shown to be able to inhibit transactivation by IKK-ɛ but not TBK1 (Shaw and others 2005), although no biochemical characterization of this phenomenon is available (see Fig. 3).
A recent observation suggests that the V proteins of some paramyxoviruses may have further functions in blocking type I IFN induction. MeV is highly immunotropic and possesses the ability to infect both plasmacytoid and conventional dendritic cells (pDCs and cDCs). The former are a major source of systemic type I IFN during many viral infections and MeV has been shown to be able to profoundly down-regulate type I IFN production by TLR7 and TLR9 agonists in these cells (Schlender and others 2005). pDCs appear to be unusual in that they constitutively express high levels of IRF-7, and in contrast to the signaling pathway described in Figure 3, pDCs activate type I IFN transcription through a distinct pathway that leads to the phosphorylation of IRF-7. The signaling pathway requires that ligand-activated TLR7 or TLR9 recruits MyD88, which in turn recruits a complex containing TRAF3, TRAF6, IRAK1, and IRAK4 and leads to the activation of IKK-α that ultimately phosphorylates IRF-7 (see Fig. 3 in Randall and Goodbourn 2008). Pfaller and Conzelmann have recently shown that the V protein of MeV can block the IKK-α-mediated activation of IRF-7 by directly binding to IKK-α and acting as a decoy substrate (Pfaller and Conzelmann 2008). Additionally, MeV V can bind directly to IRF-7 and inhibit its transactivation potential (Pfaller and Conzelmann 2008).
Clearly, it is important that paramyxoviruses tightly control the type, quantity, and temporal production of PAMPs during their replication cycle. For example, it would be advantageous to limit the production of PAMPs prior to the synthesis of specific antagonists. The control of virus transcription and replication is complicated (reviewed in Whelan and others (2004), Lamb and Parks (2006)) with many proteins, including NP, P, and L, being involved, and hence the function of any of these proteins may potentially influence the generation of PAMPs. The V proteins of paramyxovi-ruses may also play an indirect role in controlling type I IFN induction. It is notable that a number of paramyxovirus V proteins have been shown to inhibit viral RNA synthesis in minireplicon systems (Curran and others 1991; Curran and others 1995; Horikami and others 1997; Lin and others 2005; Parks and others 2006; Witko and others 2006; Nishio and others 2008), and recombinant viruses with mutations in their V proteins often show increased viral RNA synthesis (Delenda and others 1997; Kato and others 1997; Schneider and others 1997; Tober and others 1998; Durbin and others 1999; Baron and Barrett 2000; Kawano and others 2001; Gainey and others 2008). The V proteins may help limit the production of PAMPs by binding soluble NP (Precious and others 1995; Curran and others 1995; Horikami and others 1997; Tober and others 1998), thereby influencing the control of virus transcription and replication, and thus helping to limit the amount of protein-free viral RNA in the cytoplasm. Additionally, PIV5 V negatively regulates the P subunit of the viral RNA polymerase by directly interacting with the kinase Akt1 and inhibiting its ability to phosphorylate the P protein (Sun and others 2008). The ability of the V protein to negatively regulate the activity of the P protein appears of paramount importance in controlling PIV5 viral RNA synthesis (Timani and others 2008). Paramyxovirus V proteins may play other roles in regulating viral replication; HPIV2 V binds to the large RNA polymerase (L) protein (ie, the major RNA polymerase subunit) and inhibits replication (Nishio and others 2008).
C Proteins
The phenotypes of respiroviruses, morbilliviruses, and henipaviruses lacking the C ORF have shown that the C protein(s) play direct and indirect (by controlling virus transcription and replication—see below) roles in limiting type I IFN induction. SeV mutants unable to express either the V gene or the C gene show elevated levels of host type I IFN production (Strahle and others 2003; Komatsu and others 2004; Strahle and others 2006). The product of the SeV C open-reading frame can directly limit type I IFN induction (Komatsu and others 2004) and it has been recently proposed that the C proteins of SeV can act to inhibit RIG-I (Strahle and others 2007), although it is not known if this inhibition is direct. The SeV C protein has also been shown to be a negative regulator of viral RNA polymerase (Curran and others 1991; Curran and others 1992; Cadd and others 1996; Horikami and others 1997), as has the C protein of the related HPIV3 (Malur and others 2004), and again thus may influence the generation of PAMPs.
MeV that cannot express the C protein also induces more type I IFN than the wild-type virus (Nakatsu and others 2006). Furthermore, C knockout MeV and RPV show reduced growth on cells that produce and respond to IFN, and in this regard are more impaired than their equivalent V-knockout mutants (Escoffier and others 1999; Baron and Barrett 2000; Devaux and Cattaneo 2004; Takeuchi and others 2005). Like the SeV C protein, the MeV C protein has also been shown to negatively regulate viral RNA synthesis in minireplicon systems (Reutter and others 2001; Bankamp and others 2005). Significantly, it has been recently shown that the MeV C protein lacks any detectable ability to directly block type I IFN induction, but rather acts entirely by blocking the production of PAMPs (Nakatsu and others 2008). Importantly, limiting the production of type I IFN by MeV has been shown to require both the mda-5 antagonistic function of the V protein and the RNA polymerase inhibitory functions of the C protein (Nakatsu and others 2008). Surprisingly, it has been recently shown that the V protein of RPV has no ability to inhibit mda-5 and is dispensable for the control of type I IFN production, with the C protein being entirely responsible for this activity (Boxer and others 2009). The RPV C protein appears to act downstream of IRF-3 and NF-κB (Boxer and others 2009)—see Figure 3. The RPV C protein is localized in the nucleus, and may act like the W protein of NiV, which also blocks the activity of IRF-3 in an uncharacterized manner that depends upon its localization to the nucleus (Shaw and others 2005)—see Figure 3. These data suggest that the RPV C protein and NiV W protein are targeting a currently poorly characterized step in IFN-β transcription such as the recruitment of transcriptional components such as RNA polymerase II. The C proteins of HPIV1 also inhibit the activation of IRF-3 and the production of IFN-α/β (Van Cleve and others 2006), as does the C protein of BPIV3, although in these cases their mechanisms of action are unclear (Komatsu and others 2007). Finally, it has been shown that the C, V, and W proteins of NiV inhibit minigenome replication, suggesting that these proteins all play a role in controlling PAMP generation (Sleeman and others 2008).
NS Protein and Attachment Proteins of RSV
The pneumoviruses are unique within the Paramyxoviridae in not producing alternative products from their P gene. Since these viruses are not noted for their ability to induce type I IFN, it can be argued that they must use other gene products for this function. Recombinant HRSVs lacking either the NS1 or NS2 genes induce more type I IFN than the wild-type virus, and a mutant virus lacking both NS1 and NS2 genes is an even better inducer of type I IFN (Spann and others 2004). These data, and similar data on BRSV (Bossert and others 2003), suggest that NS1 and NS2 independently antagonize type I IFN production. When expressed on their own NS1 and NS2 from BRSV (Bossert and others 2003), and NS2 from HRSV (Ling and others 2009), can indeed block type I IFN induction by heterologous inducers. The HRSV NS2 protein can block both the RIG-I and TLR3 pathways, and has been shown to bind directly to the N-terminal CARD domain of RIG-I (Ling and others 2009), preventing it from interacting with the downstream adaptor VISA. The mechanism of inhibition of TLR3, and the mechanism of inhibition by NS1 remain unknown.
The attachment protein (G) of RSV is known to potentiate disease and respiratory symptoms. As well as forming part of the virus envelope, it is also rapidly secreted (sG) in large amounts from infected cells (Hendricks and others 1988). As well as its other functions (reviewed in Collins and Graham (2008)), the G protein also inhibits innate responses, including inflammatory cytokines and IFN (Polack and others 2005; Shingai and others 2008) by inhibiting TLR3/4-mediated activation. Whilst sG does not effect signaling through the RIG-I/mda-5 pathway, it does inhibit the TLR adaptor TRIF/TICAM-1 pathway. The need for RSV to inhibit these pathways may be related to fact that, in contrast to sG, the RSV F protein activates TLR4 signaling (Kurt-Jones and others 2000; Haynes and others 2001).
Conclusions
Despite their limited coding capacity, paramyxoviruses have had to evolve mechanisms to circumvent the IFN system. Interestingly, the specific mechanisms by which even very closely related viruses achieve this can differ subtly and clearly these differences must influence the types of diseases these viruses cause. Nevertheless, the way in which paramyxoviruses try to circumvent the IFN response can basically be considered as (i) limiting the production of PAMPs, (ii) inhibiting specific cellular components in the type I IFN induction cascade (with mda-5 being a near universal target for all members of the Paramyxovirinae subfamily), and (iii) blocking IFN signaling (Ramachandran and Horvath 2009). To date, other than by limiting the amount of PAMPs (such as dsRNA) that can activate enzymes such as PKR with antiviral activity, there is no evidence of paramyxovirus products that directly inhibit IFN-induced proteins with antiviral activity. However, despite these evasion mechanisms, it is clear that paramyxoviruses cannot completely circumvent the IFN response, and the IFN system remains critical in controlling paramyxovirus infections, buying time for the generation of an adaptive immune response.
Although much has been learnt in recent years about how paramyxoviruses limit the production of type I IFN, there are still significant gaps in our knowledge. For example, we have a poor understanding of the molecular structures of the PAMPs that stimulate type I IFN induction in natural infections, and whether they are what we term “essential PAMPs” or “corrupted PAMPs.” It is interesting to speculate that where paramyxoviruses have been used to induce type I IFN in experimental animals; the observed induction may be due to the occurrence of “corrupted PAMPs,” such as those produced by DI particles. Furthermore, it remains unclear what the main sources of IFN are within an infected animal, and which cells are infected by any specific para-myxovirus. Hence, the knowledge gained from studies on the induction of type I IFN from continuous cell lines may be only an approximation of the situation in response to paramyxovirus infections in vivo.
Acknowledgment
S.G. and R.E.R. gratefully acknowledge the Wellcome Trust for supporting their studies on paramyxoviruses and the interferon response.
References
- Andrejeva J. Childs KS. Young DF. Carlos TS. Stock N. Goodbourn S. Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva J. Young DF. Goodbourn S. Randall RE. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J Virol. 2002;76:2159–2167. doi: 10.1128/jvi.76.5.2159-2167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankamp B. Wilson J. Bellini WJ. Rota PA. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology. 2005;336:120–129. doi: 10.1016/j.virol.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Baron MD. Barrett T. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J Virol. 2000;74:2603–2611. doi: 10.1128/jvi.74.6.2603-2611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghall H. Siren J. Sarkar D. Julkunen I. Fisher PB. Vainionpaa R. Matikainen S. The interferon-inducible RNA helicase, mda-5, is involved in measles virus-induced expression of antiviral cytokines. Microbes Infect. 2006;8:2138–2144. doi: 10.1016/j.micinf.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Bossert B. Marozin S. Conzelmann KK. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J Virol. 2003;77:8661–8668. doi: 10.1128/JVI.77.16.8661-8668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer EL. Nanda SK. Baron MD. The rinderpest virus non-structural C protein blocks the induction of type 1 interferon. Virology. 2009;385:134–142. doi: 10.1016/j.virol.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Cadd T. Garcin D. Tapparel C. Itoh M. Homma M. Roux L. Curran J. Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos TS. Fearns R. Randall RE. Interferon-induced alterations in the pattern of parainfluenza virus 5 transcription and protein synthesis and the induction of virus inclusion bodies. J Virol. 2005;79:14112–14121. doi: 10.1128/JVI.79.22.14112-14121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KS. Andrejeva J. Randall RE. Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J Virol. 2009;83:1465–1473. doi: 10.1128/JVI.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K. Stock N. Ross C. Andrejeva J. Hilton L. Skinner M. Randall R. Goodbourn S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Collins PL. Graham BS. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol. 2008;82:2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S. Eisenacher K. Kirchhofer A. Brzozka K. Lammens A. Lammens K. Fujita T. Conzelmann KK. Krug A. Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Curran J. Boeck R. Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. Marq JB. Kolakofsky D. The Sendai virus nonstruc-tural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- Curran J. Marq JB. Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol. 1995;69:849–855. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delenda C. Hausmann S. Garcin D. Kolakofsky D. Normal cellular replication of Sendai virus without the trans-frame, non-structural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- Devaux P. Cattaneo R. Measles virus phosphoprotein gene products: conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. J Virol. 2004;78:11632–11640. doi: 10.1128/JVI.78.21.11632-11640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didcock L. Young DF. Goodbourn S. Randall RE. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999a;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didcock LJ. Young DF. Goodbourn S. Randall RE. Sendai Virus and Simian Virus 5 block activation of interferon-responsive genes; importance for virus pathogenesis. J Virol. 1999b;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP. McAuliffe JM. Collins PL. Murphy BR. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology. 1999;261:319–330. doi: 10.1006/viro.1999.9878. [DOI] [PubMed] [Google Scholar]
- Escoffier C. Manie S. Vincent S. Muller CP. Billeter M. Gerlier D. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J Virol. 1999;73:1695–1698. doi: 10.1128/jvi.73.2.1695-1698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainey MD. Dillon PJ. Clark KM. Manuse MJ. Parks GD. Paramyxovirus-induced shutoff of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J Virol. 2008;82:828–839. doi: 10.1128/JVI.02023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M., Jr Sen GC. Viral evasion of the interferon system. J Interferon Cytokine Res. 2009;29(9):475–476. doi: 10.1089/jir.2009.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M. Andersson I. Klingstrom J. Schumann M. Martin A. Zimmermann P. Wagner V. Pichlmair A. Schneider U. Muhlberger E. Mirazimi A. Weber F. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes LM. Moore DD. Kurt-Jones EA. Finberg RW. Anderson LJ. Tripp RA. Involvement of toll-like receptor 4 in innate immunity to respiratory syncytial virus. J Virol. 2001;75:10730–10737. doi: 10.1128/JVI.75.22.10730-10737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. Paterson RG. Stock N. Durbin JE. Durbin RK. Goodbourn S. Randall RE. Lamb RA. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology. 2002;303:15–32. doi: 10.1006/viro.2002.1738. [DOI] [PubMed] [Google Scholar]
- Hendricks DA. McIntosh K. Patterson JL. Further characterization of the soluble form of the G glycoprotein of respiratory syncytial virus. J Virol. 1988;62:2228–2233. doi: 10.1128/jvi.62.7.2228-2233.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton L. Moganeradj K. Zhang G. Chen YH. Randall RE. McCauley JW. Goodbourn S. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol. 2006;80:11723–11732. doi: 10.1128/JVI.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikami SM. Hector RE. Smallwood S. Moyer SA. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- Hornung V. Ellegast J. Kim S. Brzozka K. Jung A. Kato H. Poeck H. Akira S. Conzelmann KK. Schlee M. Endres S. Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Johnston MD. The characteristics required for a Sendai virus preparation to induce high levels of interferon in human lymphoblastoid cells. J Gen Virol. 1981;56:175–184. doi: 10.1099/0022-1317-56-1-175. [DOI] [PubMed] [Google Scholar]
- Kato A. Kiyotani K. Sakai Y. Yoshida T. Shioda T. Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Mikamo-Satoh E. Hirai R. Kawai T. Matsushita K. Hiiragi A. Dermody TS. Fujita T. Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Sato S. Yoneyama M. Yamamoto M. Matsui K. Uematsu S. Jung A. Kawai T. Ishii KJ. Yamaguchi O. Otsu K. Tsujimura T. Koh CS. Reis e Sousa C. Matsuura Y. Fujita T. Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawano M. Kaito M. Kozuka Y. Komada H. Noda N. Nanba K. Tsurudome M. Ito M. Nishio M. Ito Y. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology. 2001;284:99–112. doi: 10.1006/viro.2001.0864. [DOI] [PubMed] [Google Scholar]
- Komatsu T. Takeuchi K. Gotoh B. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes Infect. 2007;9:954–962. doi: 10.1016/j.micinf.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Komatsu T. Takeuchi K. Yokoo J. Gotoh B. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology. 2004;325:137–148. doi: 10.1016/j.virol.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones EA. Popova L. Kwinn L. Haynes LM. Jones LP. Tripp RA. Walsh EE. Freeman MW. Golenbock DT. Anderson LJ. Finberg RW. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- Lamb RA. Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, editor; Howley PM, editor; Griffin DE, editor; Lamb RA, editor; Martin MA, editor; Roizman B, editor; Straus SE, editor. Fields' Virology. 5th. Philadelphia: Lippincott Williams and Wilkins; 2006. pp. 1449–1496. [Google Scholar]
- Lin Y. Horvath F. Aligo JA. Wilson R. He B. The role of simian virus 5 V protein on viral RNA synthesis. Virology. 2005;338:270–280. doi: 10.1016/j.virol.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Ling Z. Tran KC. Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol. 2009;83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LL. Puri M. Horvath CM. Sen GC. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKɛ)/TBK1. J Biol Chem. 2008;283:14269–14276. doi: 10.1074/jbc.M710089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malur AG. Hoffman MA. Banerjee AK. The human parainfluenza virus type 3 (HPIV3) C protein inhibits viral transcription. Virus Res. 2004;99:199–204. doi: 10.1016/j.virusres.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Melchjorsen J. Jensen SB. Malmgaard L. Rasmussen SB. Weber F. Bowie AG. Matikainen S. Paludan SR. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol. 2005;79:12944–12951. doi: 10.1128/JVI.79.20.12944-12951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y. Takeda M. Ohno S. Koga R. Yanagi Y. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J Virol. 2006;80:11861–11867. doi: 10.1128/JVI.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y. Takeda M. Ohno S. Shirogane Y. Iwasaki M. Yanagi Y. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J Virol. 2008;82:8296–8306. doi: 10.1128/JVI.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M. Ohtsuka J. Tsurudome M. Nosaka T. Kolakofsky D. Human parainfluenza virus type 2 V protein inhibits genome replication by binding to the L protein: possible role in promoting viral fitness. J Virol. 2008;82:6130–6138. doi: 10.1128/JVI.02635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks CL. Witko SE. Kotash C. Lin SL. Sidhu MS. Udem SA. Role of V protein RNA binding in inhibition of measles virus minigenome replication. Virology. 2006;348:96–106. doi: 10.1016/j.virol.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Pfaller CK. Conzelmann KK. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J Virol. 2008;82:12365–12373. doi: 10.1128/JVI.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A. Schulz O. Tan CP. Naslund TI. Liljestrom P. Weber F. Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Plumet S. Herschke F. Bourhis JM. Valentin H. Longhi S. Gerlier D. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE. 2007;2:e279. doi: 10.1371/journal.pone.0000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP. Irusta PM. Hoffman SJ. Schiatti MP. Melendi GA. Delgado MF. Laham FR. Thumar B. Hendry RM. Melero JA. Karron RA. Collins PL. Kleeberger SR. The cysteine-rich region of respiratory syncytial virus attachment protein inhibits innate immunity elicited by the virus and endotoxin. Proc Natl Acad Sci USA. 2005;102:8996–9001. doi: 10.1073/pnas.0409478102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E. He B. Lamb RA. Randall RE. Goodbourn S. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology. 2002;303:33–46. doi: 10.1006/viro.2002.1737. [DOI] [PubMed] [Google Scholar]
- Precious BL. Carlos TS. Goodbourn S. Randall RE. Catalytic turnover of STAT1 allows PIV5 to dismantle the interferon-induced anti-viral state of cells. Virology. 2007;368:114–121. doi: 10.1016/j.virol.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Precious B. Young DF. Bermingham A. Fearns R. Ryan M. Randall RE. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J Virol. 1995;69:8001–8010. doi: 10.1128/jvi.69.12.8001-8010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A. Horvath CM. Paramyxovirus disruption of interferon signal transduction: STATus Report. J Interferon Cytokine Res. 2009;29(9):531–538. doi: 10.1089/jir.2009.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE. Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Reutter GL. Cortese-Grogan C. Wilson J. Moyer SA. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology. 2001;285:100–109. doi: 10.1006/viro.2001.0962. [DOI] [PubMed] [Google Scholar]
- Schlender J. Hornung V. Finke S. Gunthner-Biller M. Marozin S. Brzozka K. Moghim S. Endres S. Hartmann G. Conzelmann KK. Inhibition of toll-like receptor 7- and 9-mediated alpha/ beta interferon production in human plasmacytoid dendritic cells by respiratory syncytial virus and measles virus. J Virol. 2005;79:5507–5515. doi: 10.1128/JVI.79.9.5507-5515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H. Kaelin K. Billeter MA. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- Shaw ML. Cardenas WB. Zamarin D. Palese P. Basler CF. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and toll-like receptor 3-triggered signaling pathways. J Virol. 2005;79:6078–6088. doi: 10.1128/JVI.79.10.6078-6088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M. Azuma M. Ebihara T. Sasai M. Funami K. Ayata M. Ogura H. Tsutsumi H. Matsumoto M. Seya T. Soluble G protein of respiratory syncytial virus inhibits Toll-like receptor 3/4-mediated IFN-beta induction. Int Immunol. 2008;20:1169–1180. doi: 10.1093/intimm/dxn074. [DOI] [PubMed] [Google Scholar]
- Shingai M. Ebihara T. Begum NA. Kato A. Honma T. Matsumoto K. Saito H. Ogura H. Matsumoto M. Seya T. Differential type I IFN-inducing abilities of wild-type versus vaccine strains of measles virus. J Immunol. 2007;179:6123–6133. doi: 10.4049/jimmunol.179.9.6123. [DOI] [PubMed] [Google Scholar]
- Sleeman K. Bankamp B. Hummel KB. Lo MK. Bellini WJ. Rota PA. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89:1300–1308. doi: 10.1099/vir.0.83582-0. [DOI] [PubMed] [Google Scholar]
- Spann KM. Tran KC. Chi B. Rabin RL. Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected] J Virol. 2004;78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle L. Garcin D. Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;20:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Strahle L. Garcin D. Le Mercier P. Schlaak JF. Kolakofsky D. Sendai virus targets inflammatory responses, as well as the interferon-induced antiviral state, in a multifaceted manner. J Virol. 2003;77:7903–7913. doi: 10.1128/JVI.77.14.7903-7913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle L. Marq JB. Brini A. Hausmann S. Kolakofsky D. Garcin D. Activation of the beta interferon promoter by unnatural Sendai virus infection requires RIG-I and is inhibited by viral C proteins. J Virol. 2007;81:12227–12237. doi: 10.1128/JVI.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M. Fuentes SM. Timani K. Sun D. Murphy C. Lin Y. August A. Teng MN. He B. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J Virol. 2008;82:105–114. doi: 10.1128/JVI.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K. Yoneyama M. Nishihori T. Hirai R. Kumeta H. Narita R. Gale M., Jr Inagaki F. Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Takeuchi K. Takeda M. Miyajima N. Ami Y. Nagata N. Suzaki Y. Shahnewaz J. Kadota S. Nagata K. Stringent requirement for the C protein of wild-type measles virus for growth both in vitro and in macaques. J Virol. 2005;79:7838–7844. doi: 10.1128/JVI.79.12.7838-7844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timani KA. Sun D. Sun M. Keim C. Lin Y. Schmitt PT. Schmitt AP. He B. A single amino acid residue change in the P protein of parainfluenza virus 5 elevates viral gene expression. J Virol. 2008;82:9123–9133. doi: 10.1128/JVI.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tober C. Seufert M. Schneider H. Billeter MA. Johnston IC. Niewiesk S. ter Meulen V. Schneider-Schaulies S. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J Virol. 1998;72:8124–8132. doi: 10.1128/jvi.72.10.8124-8132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cleve W. Amaro-Carambot E. Surman SR. Bekisz J. Collins PL. Zoon KC. Murphy BR. Skiadopoulos MH. Bartlett EJ. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology. 2006;352:61–73. doi: 10.1016/j.virol.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Wang L. Harcourt BH. Yu M. Tamin A. Rota PA. Bellini WJ. Eaton BT. Molecular biology of Hendra, Nipah viruses. Microbes Infect. 2001;3:279–287. doi: 10.1016/s1286-4579(01)01381-8. [DOI] [PubMed] [Google Scholar]
- Wansley EK. Parks GD. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J Virol. 2002;76:10109–10121. doi: 10.1128/JVI.76.20.10109-10121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F. Wagner V. Rasmussen SB. Hartmann R. Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP. Barr JN. Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- Witko SE. Kotash C. Sidhu MS. Udem SA. Parks CL. Inhibition of measles virus minireplicon-encoded reporter gene expression by V protein. Virology. 2006;348:107–119. doi: 10.1016/j.virol.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Yoneyama M. Kikuchi M. Matsumoto K. Imaizumi T. Miyagishi M. Taira K. Foy E. Loo YM. Gale M., Jr Akira S. Yonehara S. Kato A. Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M. Kikuchi M. Natsukawa T. Shinobu N. Imaizumi T. Miyagishi M. Taira K. Akira S. Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yount JS. Gitlin L. Moran TM. Lopez CB. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai Virus defective interfering particles. J Immunol. 2008;180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]