Abstract
MicroRNAs (miRNAs) are small non-coding RNAs of 18–25 nucleotides that are generally believed to either block the translation or induce the degradation of target mRNA. miRNAs have been shown to play fundamental roles in diverse biological and pathological processes including cell proliferation, differentiation, apoptosis and carcinogenesis. Fibrosis results from an imbalance in the turnover of extracellular matrix molecules and is a highly debilitating process that can eventually lead to organ dysfunction. A growing body of evidence suggests that miRNAs participate in the fibrotic process in a number of organs including the heart, kidney, liver and lung. In this review, we summarize our current understanding of the role of miRNAs in the development of tissue fibrosis and their potential as novel drug targets.
Keywords: miRNA, fibrosis, extracellular matrix molecules, collagen, fibroblasts
Introduction
miRNA-mediated RNA interference has been identified as a novel mechanism that regulates gene expression at the translational level [1;2]. These short RNA sequences of 20–23 nucleotides are produced by the processing of full length mRNA-like transcripts known as primary miRNAs [3;4]. These larger primary miRNA transcripts undergo enzymatic cleavage by the RNAse III Drosha to produce ~ 70 nt precursor miRNAs. These are then transported to the cytoplasm where they are further processed by another RNAse III enzyme, DICER, to produce ~ 21–23 double stranded RNA. One strand, the mature miRNA, is then loaded into the RNA–induced silencing complex (RISC) where it is believed to either repress mRNA translation or reduce mRNA stability following imperfect binding between the miRNA and the miRNA-recognition elements (MRE) within the 3′ untranslated region (UTR) of target genes. Specificity of the miRNA is thought to be primarily mediated by the ‘seed’ region that is localised between residues 2–8 at the 5′ end [5–7].
In most circumstances, miRNAs are believed to either repress mRNA translation or reduce mRNA stability following imperfect binding between the miRNA and the miRNA-recognition elements (MRE) within the 3′ untranslated region (UTR) of target genes. Specificity of the miRNA guide strand is thought to be mediated by the ‘seed’ region localised between residues 2–8 at the 5′ end [5–7] although this also appears to be influenced by additional factors such as the presence and cooperation between multiple MREs [8;9], the spacing between MREs [9;10], proximity to the stop codon [9], position within the 3′ UTR [9], AU composition [9] and target mRNA secondary structure [11]. However, recent studies have also suggested that miRNAs might enhance mRNA translation in non-proliferating or amino-acid starved cells. Thus, serum starvation and cell-cycle arrest was shown to increase TNFα release by a mechanism that involved miRNA-369-3 binding to the AU rich elements (ARE) within the 3′ UTR of TNFα and interaction between two miRISC associated proteins, Ago 2 and fragile-X-mental-retardation-related protein 1 (FXR1) [12;13]. In similar studies, Orom et al, have demonstrated that miRNA-10a increases ribosomal protein expression following amino acid starvation and in response to cellular stress [14]. In this case, the action of miRNA-10a was shown to mediated through binding within the 5′-UTR (and not the 3′-UTR) of mRNA at regions immediately down-stream of the regulatory 5′TOP motif [14]. It therefore appears that although miRNAs predominately repress translation through binding within the 3′ UTR of target mRNA, individual miRNAs might also increase mRNA translation and bind regions with the 5′-UTR.
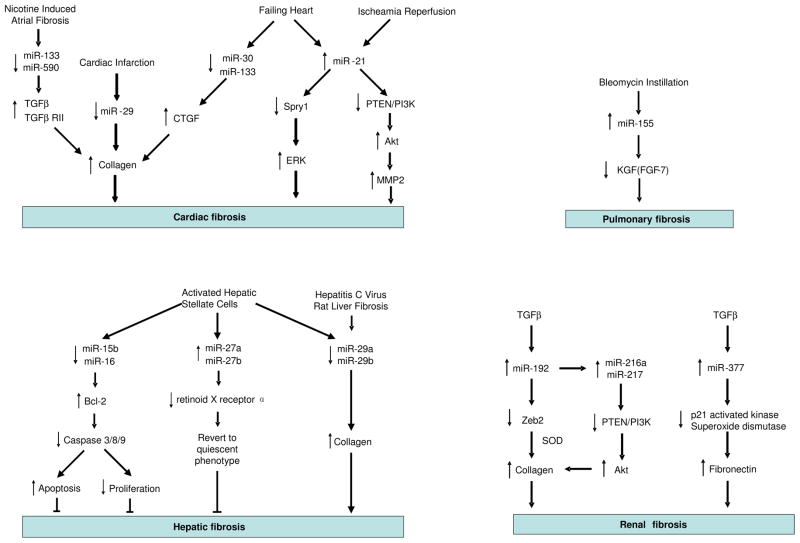
There is now overwhelming evidence that miRNAs regulate diverse biological processes including cell proliferation, differentiation and apoptosis and that aberrant miRNA expression/action can lead to the development of multiple diseases. Fibrosis is characterised by the excess deposition of extracellular matrix (ECM) components, which is the end result of an imbalance of metabolism of extracellular matrix molecule. Irregularities in multiple pathways involved in tissue repair and inflammation can lead to the development of fibrosis. Collagens are the predominant ECM proteins while other ECM proteins such as fibronectins (FNs), elastin and fibrillins also have an important role in the development of fibrosis [15]. Fibrosis is likely to result from both an increased synthesis and decreased degradation of ECM components. In particular, matrix metalloproteinases (MMPs) that degrade ECM maybe elevated while their inhibitors, tissue inhibitor of metalloproteinases (TIMPs) maybe down-regulated. Collagens are synthesised by many cell types but mainly by mesenchymal cells including fibroblasts. Following tissue injury, fibroblasts differentiate into myofibroblasts and often, persistent activation of the myofibroblast phenotype with a resistance to apoptosis ensures the development of fibrosis. A number of pro-fibrotic mediators have been identified although transforming growth factor-β (TGF-β) is proposed to play a central role in many fibrotic conditions. Extensive tissue remodelling and fibrosis can ultimately lead to failure in a number of organs. Although current treatments typically target the inflammatory response, the mechanism of fibrosis is poorly understood and there are few effective therapies. For these reasons and knowing the multitude of pathways miRNAs can affect, it is envisaged that investigating the roles of miRNAs in fibrosis could not only advance our understanding of the pathogenesis of this common condition, but might also provide new targets for therapeutic intervention. In this regard, we have reviewed the growing body of evidence that suggests miRNAs are involved in the process of fibrosis in several organs including heart, lung, kidney and liver (Figure 1).
Figure 1.
Overview of the role of miRNAs in fibrosis.
miRNA and cardiac fibrosis
Cardiac fibroblasts are the most numerous cell type in the heart and are central to the regulation of cardiac ECM metabolism [16]. Excess deposition of ECM components in the heart is associated with numerous cardiovascular diseases including hypertension, myocardial infarction and cardiomyopathy and there is now accumulating evidence that implicate miRNAs in the modulation of these conditions [17–21].
The overall importance of miRNAs was shown by Da Costa Martins et al. (2008) who reported that conditional deletion of Dicer, an enzyme that is central to miRNA metabolism, in the mouse myocardium resulted in cardiomyocytes hypertrophy and remarkable ventricular fibrosis [22]. Subsequent articles have helped to clarify the roles of individual miRNAs in cardiac fibrosis. Using in situ hybridization, Thum et al. (2008) found that miR-21 was selectively expressed in cardiac fibroblasts and that miR-21 expression was greatly enhanced in the failing heart in human, mice and rats [23]. Subsequent mechanistic studies rat cardiac fibroblasts showed that increased miR-21 enhanced extracellular regulated kinase (ERK) signaling by targeting the down-regulation of the ERK inhibitor, sprouty homologue 1 (Spry1). In turn, this promoted fibroblast survival and fibroblast growth factor (FGF) secretion leading to fibroblast proliferation and ECM/collagen deposition in vitro [23]. Significantly, in vivo silencing of miR-21 using an ‘antagomir’ was shown to reduce cardiac ERK kinase activity and inhibit interstitial fibrosis and cardiac dysfunction in a mouse mode of cardiac hypertrophy induced by overloaded pressure [23]. Increased expression of miR-21 has also been demonstrated in the infarct zone of hearts subjected to ischaemia-reperfusion (IR), especially in cardiac fibroblasts [24]. Under these circumstances increased miR-21 expression was shown to target the down-regulation of phosphatase and tension homologue (PTEN) which negatively regulates the phosphoinositol 3-kinase (PI3K)-Akt signalling pathways [24]. The subsequent activation of the PI3K-Akt pathway increased the expression of matrix metalloproteinase (MMP)-2, which is known to degrade ECM and permit the infiltration of fibroblasts [24].
The miR-29 family has also been implicated in cardiac fibrosis following a report showing down-regulation of miR-29 family, miR-29a, miR-29b and miR-29c, in the border zone of murine and human hearts during myocardial infarction [25]. This study also showed down-regulation of miR-149 and increased expression of miR-21, miR-214 and miR-223 although the functional consequences of these changes are unknown. Multiple target genes of the miR-29 family were identified including ECM proteins such as collagens, fibrillins, and elastin and it was speculated that transforming growth factor (TGF)-β-mediated down-regulation of miR-29 would enhance fibrosis. This was confirmed by demonstrating decreased collagen expression in cultured mouse cardiac fibroblasts transfected with miR-29b mimics and increased collagen expression in mouse liver, kidney and heart following the administration of cholesterol-modified inhibitor by tail vein injection [25].
Connective tissue growth factor (CTGF) is known to be a potent inducer of tissue fibrosis in multiple tissues including the heart. Interestingly, Duisters et al. have shown that miR-133 and miR-30 target the down-regulation in CTGF expression in cultured rat cardiomyocytes and fibroblasts [26]. Investigations using rodent models of cardiac hypertrophy and samples from patients with left ventricular hypertrophy showed a reduction in miR-30 and miR-133 expression that was inversely correlated with CTGF, collagen and fibrosis levels [26]. The potential importance of miR-133 has been underlined by reports that miR-133-a1 and miR-133-a2 knockout mice develop severe fibrosis and heart failure [27] and by studies showing that miR-133 and miR-590 are down-regulated in a canine model of nicotine induced atrial interstitial fibrosis [28;29]. Interestingly, mechanistic studies in the canine models and in cultured atrial fibroblasts have shown that the protective actions of miR-133 and miR-590 are mediated through targeting the down-regulation of TGF-β1 and TGF- β receptor type II, respectively [29].
Finally, van Rooij et al have demonstrated that mice containing a miR-208 deletion, unlike wild type mice, did not exhibit cardiomyocytes hypertrophy or fibrosis in response to aortic banding and transgenic expression of activated calcineurin [30]. Although deletion of miR-208 was shown to attenuate expression of β-myosin heavy chain (β-MHC) in heart, the mechanism by which miR-208 impacts on the fibrotic process is unknown.
miRNAs and pulmonary fibrosis
Pulmonary fibrosis is characterized by excessive deposition of collagen and other ECM proteins within the pulmonary interstitium and is commonly associated with the up-regulation of TGF-β [31]. Little is known regarding the role of miRNAs in lung fibrosis although a recent report by Pottier et al. using human lung fibroblasts has shown that miR-155 expression was increased following treatment with TNFα and IL-1β and reduced by TGF-β [32]. Mechanistic studies indicated that increased miR-155 down-regulates keratinocyte growth factor (KGF, FGF-7) expression and increased fibroblast migration by inducing caspase-3 expression. Studies using a bleomycin-induced mouse model of lung fibrosis confirmed that up-regulation of miR-155 was correlated with the degree of lung fibrosis in C57BL/6 and BALB/C mice [32].
miRNAs and hepatic fibrosis
Fibrosis is a common outcome of chronic hepatic diseases including viral hepatitis, alcohol abuse and metabolic diseases and can ultimately lead to liver cirrhosis and hepatic failure. Hepatic stellate cells (HSC) are believed to be the main matrix-producing cells in the liver. Following multiple injurious agents and/or exposure to inflammatory cytokines, activated HSC lose their lipid droplets, migrate to injured sites and are transformed into myofibroblast-like cells which secrete large amounts of ECM leading [33].
A number of reports have demonstrated an important role of miRNAs during HSC activation. Thus, measurement of the changes in miRNAs expression in activated rat HSC, identified 12 up-regulated miRNAs (miR-874, 29c*, -501, -349, -325-5p, -328, -138, -143, -207, -872, -140, 193) and 9 down-regulated miRNAs (miR-341, -20b-3p, -15b, -16, -375, -122, -146a, -92b, -126) [34]. Interestingly, over-expression of miR-16 and miR-15b was shown to inhibit HSC proliferation and induce apoptosis through down-regulation of the mitochondrial associated anti-apoptotic protein Bcl-2, leading to activation of caspases 3, 8 and 9 [35;36]. In contrast to these studies, a second miRNA expression profile performed in activated rat HSC showed increased miR-27a and miR-27b expression [37]. In this case, inhibition of miR-27a and 27b reverted activated HSC back to a quiescent state, a process that was mediated by preventing the increase in expression of the miR-27a/b target, retinoid X receptor α (RXRα)[37]. These findings show that miRNAs play a significant role in the progression of liver fibrogenesis via HSC activation.
Steatohepatitis resulting from chronic alcoholic abuse or metabolic syndrome is a common liver disease that usually precedes liver fibrosis [38]. Using two mice models of alcoholic and non-alcoholic steatohepatitis, Dolganiuc et al. has shown changed expression of 5 miRNAs: miR-705 and miR-1224 were increased in both groups whilst miR-182, miR-183 and miR-199a-3p were down-regulated in alcoholic group but up-regulated in the non-alcoholic group [38]. At the present time, the role of these miRNAs in the development of steatohepatitis is unknown.
As with cardiac fibrosis, reduced expression of miR-29a and miR-29b has been demonstrated during activation of primary rat HSC, in patient biopsies with hepatitis C virus infection and in a rat model of fibrosis induced by bile-duct ligation. This indicates that the miR-29 family might also have an anti-fibrogenic function in liver which is support by functional studies showing decreased collagen synthesis in HSC following over-expression of miR-29 [39].
miRNAs and renal fibrosis
Renal fibrosis involving excessive deposition of extracellular matrix in kidney, is a common response to chronic kidney diseases including many kinds of nephritis, nephropathy and other nephritic diseases, which eventually leads to irreversible renal failure [40]. Diabetic nephropathy is a common complication of diabetes and also provides a good model to study renal fibrosis, showing progressive fibrosis in the renal glomerulus and tubulo-interstitial region that correlates with the decline of renal function[41].
Kato et al. have reported that miR-192 was up-regulated in the glomeruli of type 1 and type 2 diabetic mice and cultured mesangial cells treated by TGF-β1 [42]. They found that TGF-β1-induced miR-192 expression in mesangial cells increased the expression of collagen 1α2 by down-regulating Zeb2, an E-box repressor [42]. In a subsequent report, this group showed that down-regulation of Zeb2 resulted in increased miR-216a and miR-217 expression. This also contributed to increased collagen production and the development of diabetic nephropathy through down-regulation of PTEN and the subsequent activation of Akt (Figure 1) [43]. Increased miR-377 is also thought to regulate the expression of fibronectin, that accumulates in diabetic nephropathy. Expression of miR-377 was up-regulated in mouse models of diabetic nephropathy and in cultured human and mouse mesangial cells treated with high glucose and TGF-β1 [44]. Increased expression of fibronection was shown to result from miR-377 mediated down-regulation in p21-activated kinase and superoxide dismutases levels [44].
Conclusion
As outlined in this review, there is now increasing evidence that miRNAs regulate fibrosis in multiple organs including the heart, lung, kidney and liver (Figure 1). In particular, a number of miRNA such as miR-21 and the miR-29 family are emerging as common regulator of fibrosis in multiple tissues and can be divided into two groups: those that regulate the differentiation into fibrotic cells (i.e. miR-21) and those that directly target the translation of extracellular matrix components (i.e.miR-29). Since there are few treatment options for fibrosis, modulation of miRNAs using either antisense that block their activity or over-expression from viral or plasmid vectors, has been proposed as a potentially novel therapeutic approach [45;46]. Significantly, a number of reports discussed in this review have demonstrated the feasibility of this approach in pre-clinical mouse models of fibrosis. Thus, van Rooij et al has shown that inhibiting miR-29 using cholesterol-conjugated antisense increased collagen expression in mouse liver, kidney and heart [25] whilst Thum et al used a similar approach to demonstrate that inhibition of miR-21 prevent interstitial fibrosis and cardiac hypertrophy in a mouse model of heart infarction [23]. However, future development will be crucially dependent upon understanding the function and mechanisms of action of these fibrotic miRNAs.
Acknowledgments
This work was supported by the Chinese Government Academic Exchange Programme (to X.J.), National Institute of Health Research (to E.T.) and the Wellcome Trust (076111 to M.A.L.),
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 4.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 8.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 10.Saetrom P, Heale BS, Snove O, Jr, Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 12.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 14.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Muro AF, Moretti FA, Moore BB, Yan M, Atrasz RG, Wilke CA, Flaherty KR, Martinez FJ, Tsui JL, Sheppard D, Baralle FE, Toews GB, White ES. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. Am J Respir Crit Care Med. 2008:638–45. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souders CA, Bowers SL, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res. 2009;105:1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van RE, Olson EN. Searching for miR-acles in cardiac fibrosis. Circ Res. 2009;104:138–140. doi: 10.1161/CIRCRESAHA.108.192492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diez J. Do microRNAs regulate myocardial fibrosis? Nat Clin Pract Cardiovasc Med. 2009;6:88–89. doi: 10.1038/ncpcardio1415. [DOI] [PubMed] [Google Scholar]
- 19.Haghikia A, Hilfiker-Kleiner D. MiRNA-21: a key to controlling the cardiac fibroblast compartment? Cardiovasc Res. 2009;82:1–3. doi: 10.1093/cvr/cvp058. [DOI] [PubMed] [Google Scholar]
- 20.Mishra PK, Tyagi N, Kumar M, Tyagi SC. MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med. 2009;13:778–789. doi: 10.1111/j.1582-4934.2009.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 22.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 23.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 24.Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res. 2009;82:21–29. doi: 10.1093/cvr/cvp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van dM I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–8. 6. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 27.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goette A. Nicotine, atrial fibrosis, and atrial fibrillation: do microRNAs help to clear the smoke? Cardiovasc Res. 2009;83:421–422. doi: 10.1093/cvr/cvp188. [DOI] [PubMed] [Google Scholar]
- 29.Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai B, Wang N, Li X, Feng T, Hong Y, Yang B. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- 30.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 31.du Bois RM. Strategies for treating idiopathic pulmonary fibrosis. Nat Rev Drug Discov. 2010 doi: 10.1038/nrd2958. [DOI] [PubMed] [Google Scholar]
- 32.Pottier N, Maurin T, Chevalier B, Puissegur MP, Lebrigand K, Robbe-Sermesant K, Bertero T, Lino Cardenas CL, Courcot E, Rios G, Fourre S, Lo-Guidice JM, Marcet B, Cardinaud B, Barbry P, Mari B. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS ONE. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson NC, Iredale JP. Liver fibrosis: cellular mechanisms of progression and resolution. Clin Sci (Lond) 2007;112:265–280. doi: 10.1042/CS20060242. [DOI] [PubMed] [Google Scholar]
- 34.Guo CJ, Pan Q, Cheng T, Jiang B, Chen GY, Li DG. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009;276:5163–5176. doi: 10.1111/j.1742-4658.2009.07213.x. [DOI] [PubMed] [Google Scholar]
- 35.Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Guo CJ, Pan Q, Jiang B, Chen GY, Li DG. Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis. 2009 doi: 10.1007/s10495-009-0401-3. [DOI] [PubMed] [Google Scholar]
- 37.Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759–766. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 38.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwiecinski M, Strack I, Noetel A, Schivenbusch S, Elfimova N, Dienes HP, Odenthal M. MicroRNA: antifibrogenic mediator in liver fibrogenesis. J Hepatol. 2009;50:S110. [Google Scholar]
- 40.Schnaper HW, Kopp JB. Renal fibrosis. Front Biosci. 2003;8:e68–86. e68–e86. doi: 10.2741/925. [DOI] [PubMed] [Google Scholar]
- 41.Brosius FC., III New insights into the mechanisms of fibrosis and sclerosis in diabetic nephropathy. Rev Endocr Metab Disord. 2008;9:245–254. doi: 10.1007/s11154-008-9100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 46.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]